Abstract
Deep learning has become an effective tool for classifying biological sex based on functional magnetic resonance imaging (fMRI). However, research on what features within the brain are most relevant to this classification is still lacking. Model interpretability has become a powerful way to understand “black box” deep-learning models, and select features within the input data that are most relevant to the correct classification. However, very little work has been done employing these methods to understand the relationship between the temporal dimension of functional imaging signals and the classification of biological sex. Consequently, less attention has been paid to rectifying problems and limitations associated with feature explanation models, e.g. underspecification and instability. In this work, we first provide a methodology to limit the impact of underspecification on the stability of the measured feature importance. Then, using intrinsic connectivity networks from fMRI data, we provide a deep exploration of sex differences among functional brain networks. We report numerous conclusions, including activity differences in the visual and cognitive domains and major connectivity differences.
Keywords: brain connectivity, deep learning, model interpretability, neuroimaging, sex differences
Introduction
Deep learning is an effective tool for both classification of biological sex and understanding the features relevant to such classification (Abrol et al. 2021, Arslan et al. 2018, Liu et al. December 2015, Long et al. 2021). However, it suffers from two critical flaws from the standpoint of model interpretability: underspecification and instability of the relevant features. Underspecified (D’Amour et al. 2020) models can have many local minima, or possible functions, which produce the same mapping between the input and the output under different parameters. This is particularly problematic for feature attribution methods such as saliency (Simonyan et al. 2013), which are very sensitive to changes in model architecture, even to initialization within a given architecture. Although saliency methods can be informative about the data, this sensitivity to small perturbations in the initial state or architecture make the models unstable. This instability is particularly pronounced for deep classification models applied to small datasets. A secondary flaw is specific to sequential/recurrent models, such as long short-term memory models (LSTMs). In this case, saliency methods become ineffective due to a phenomenon known as vanishing saliency (Ismail et al. 2019), which significantly reduces the magnitude of the salient gradients as the model backpropagates through time, providing inaccurate saliency maps.
In recent decades, functional magnetic resonance imaging (fMRI) has significantly extended our understanding of the human brain (Bandettini 2012). We have witnessed great strides in analyzing fMRI data, particularly through independent component analysis (ICA) to extract intrinsic connectivity networks (ICNs) (Calhoun and Adali 2012). These ICNs and their associated timecourses have become central to fMRI research, including research into brain activation patterns and biological sex. Although there is now a wealth of information about sex differences among brain signals, there is still a long way to go before we truly understand how brain signals relate to biological sex (Spets and Slotnick 2021, Iraji et al. 2022). One of the more promising avenues for fMRI research is the analysis of complex brain disorders such as schizophrenia, Alzheimer’s, and autism. As a great deal of research has found sex differences relating to these disorders (Häfner 2003, Kirkovski et al. 2013, Stites et al. 2021), it is imperative to better understand the relationship between sex and fMRI signals as a whole. With a deeper understanding of this relationship, researchers may better grasp how best to treat these disorders based on sex. This can include medications, dosage, and behavioral treatments.
This paper presents a methodology to mitigate instability in feature importance assessments using state-of-the-art, nonlinear models and feature attribution methods. We then apply this methodology to elucidate the relationship between biological sex and mesoscale brain dynamics. Specifically, using an LSTM model coupled with a specific saliency method known as integrated gradients (IG), we take a deep dive into understanding sex differences among functional networks estimated from fMRI data. LSTMs are important for this work because they can capture the dynamics of fMRI temporal signals. Lastly, we present evidence that deep learning can be usefully employed as a “feature explainer” or a tool that highlights aspects of the brain function most relevant to sex differences.
With the power of novel deep learning methods, we take an in-depth look at sex differences among fMRI ICN timecourses. These timecourses represent interpretable functional networks, making quantitative analysis of our results relatively easy. We use a large sample size, from the UK Biobank (UKB) repository, which aids in model stability. Our approach to investigating these networks is a novel adaptation of simple feature explanation techniques that fix several key problems, primarily the instability of the feature maps and an LSTM-specific issue, a phenomenon known as vanishing saliency. We then validate our methodology with synthetic data in which the most relevant features are known beforehand. While showing visual representations of our maps, we also quantitatively compare our proposed methodology with an existing methodology, the input-cell attention (Ismail et al. 2019). We performed these analyses to ground our work and show quantitatively that our methods can discover data-relevant signals. After this validation, we provide a broad set of post hoc analyses, showing both the validity of our model and novel results, further expanding biological sex analysis based on fMRI data. Pointedly, after comparing with static functional network connectivity, we find considerable sex-specific results within relationships between the individual ICNs, and in particular, differences within key functional domains, including the visual (VIS) and default mode network (DMN).
Data and Methods
FMRI data and preprocessing
The data, a total of 11 754 resting-state fMRI scans, was sourced from 22 sites within the United Kingdom between 2006 and 2018. Data processing and quality control were previously performed in (Hassanzadeh et al. 2022), where over 11 000 subjects were selected from the entire UKB dataset. After processing the data, this study showed that pair-wise relationships between ICNs (using Pearson correlation) could be highly predictive of individuals with a neural network. We use the same ICNs that this work computed. However, we removed subjects with inconclusive sex, meaning any subject where the documented genetic sex is not consistent with the self-reported sex at the time of the study, or subjects missing either field, which gives us the final 11 461 (5821 women and 5640 men) subjects. Of the 293 subjects removed from the final set, 157 self-reported as female but had inconclusive genetic sex, and 129 of the self-reported males had inconclusive genetic sex. 3 subjects self-reported as male with a female reported genetic sex, and 4 subjects self-reported as female with a genetic test that resulted in male. Participants were between 45 and 80 years of age (Alfaro-Almagro et al. 2018, Baecker et al. 2021) with an average age of 62.55. All participants were self-reported as being healthy. The data acquisition protocol follows: 39ms echo time, a 0.735 s repetition time (TR), 52 flip angle, and a multiband factor of 8 using 3T Siemens machines. The T2 signal was both linearly and nonlinearly warped to MNI152 space. Each volume was resampled to 3mm
flip angle, and a multiband factor of 8 using 3T Siemens machines. The T2 signal was both linearly and nonlinearly warped to MNI152 space. Each volume was resampled to 3mm for a final image size of 53
for a final image size of 53 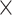 63
63 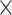 46 mm
46 mm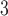 , and 160 time steps using the statistical parametric mapping (SPM12, http://www.fil.ion.ucl.ac.uk/spm/) MATLAB package.
, and 160 time steps using the statistical parametric mapping (SPM12, http://www.fil.ion.ucl.ac.uk/spm/) MATLAB package.
After the preprocessing pipeline, the ICNs were extracted using spatially constrained ICA with the GIFT package (Li and Adali 2010) via the Neuromark pipeline (Yuhui et al. 2020) for MATLAB. ICA is a robust and evidence-based method to capture regions of functional activity (Calhoun et al. 2001). Since our goal is to analyze functional brain activity, we require clean and data-driven representations of this activity. We also want to compare the functional relationships measured with our methods with other connectivity metrics. Logically, we need the network timecourses so we can compare our analyses of the relationships between networks with other robust connectivity estimations. This pipeline provides a fully automated approach to compute ICA (both spatial components and timecourses) and output labeled and ordered components. Overall, 53 networks covered 7 domains: Subcortical (SC), auditory (AUD), sensorimotor (SM), VIS, cognitive control (CC), the DMN, and the cerebellum (CB).
Static Functional Network Connectivity
Static functional network connectivity (sFNC) is the functional relationships or connections between ICNs. It contrasts with functional connectivity (FC) in that it is a connectivity map for estimated networks within the brain rather than voxels or neurons. We estimate the sFNC as the pair-wise correlation (specifically Pearson correlation) between each network timecourse.
Our Model
A key aspect of our model that mitigates vanishing saliency is an additive attention mechanism (Bahdanau et al. 2014) which creates a direct gradient flow path from the classification to the input via the attention parameters (Lewis et al. 2021). A diagram of our model can be seen in Fig. 1. A bi-directional LSTM (Schuster and Paliwal 1997) was chosen because we do not consider streaming data, the additional parameters aid training, and the extra directional flow for gradients may also improve the quality of the saliency maps.
Fig. 1.
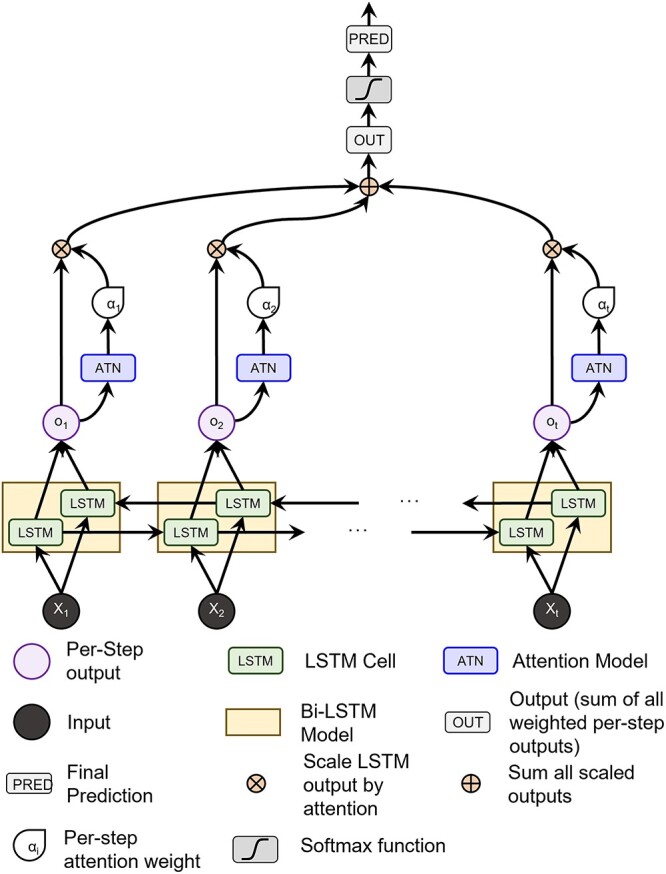
A diagram of our model. The data pass through an LSTM, from which the hidden states parameterize the attention model. The attention weights scale the hidden states, which are summed over all states (i.e. step in the LSTM), and is finally fed through a classification layer.
The attention mechanism (Bahdanau et al. 2014) is a powerful way to amalgamate temporal information and “attend” only to the most important steps in the LSTM output by assigning a weight to each step. To parameterize the attention mechanism, we pass the LSTM output at each step through an attention network of two feed-forward layers to create a single, per-step weight value. The weight values from all time steps are jointly softmaxed and used to adjust the LSTM output at the individual time steps. As the model is bi-directional, we use the output from both the forward and backward directions concatenated into a single vector as our context for the attention mechanism. In other words, h_backwardT is concatenated with h_forwardT and passed through the attention mechanism to give us the respective attention weight for that time step. Once the hidden outputs have been individually weighted by the attention scalar, they are summed along the time dimension and pushed through a linear transform for classification.
Gradient-based Feature Attribution
Gradient-based feature attribution methods, commonly known as saliency, are model interpretability methods that leverage the gradients of a trained model to better understand why the model makes its predictions. It is defined as the gradients of the prediction of the correct class w.r.t. the input, or 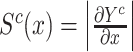 .
.
With this paradigm of using calculated gradients to interpret and understand the input data, there are numerous methods to compute the gradients as 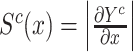 . For our purposes, we choose a method called IG (Sundararajan et al. 2017). IG is defined, for any model,
. For our purposes, we choose a method called IG (Sundararajan et al. 2017). IG is defined, for any model,  as
as 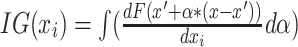 , where
, where 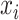 is feature
is feature 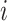 for a given input sample,
for a given input sample, 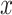 , and
, and 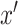 is defined as a baseline sample, which, for our purposes we use a zero-valued ”blank” sample. Associated with the baseline is an interpolation constant,
is defined as a baseline sample, which, for our purposes we use a zero-valued ”blank” sample. Associated with the baseline is an interpolation constant, 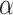 . Essentially, for each feature, we interpolate between the baseline and the given input sample with a constant interpolation factor,
. Essentially, for each feature, we interpolate between the baseline and the given input sample with a constant interpolation factor, 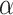 . For each interpolation step, we pass this modified input through the model,
. For each interpolation step, we pass this modified input through the model, 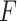 , and then compute the gradients of the correct class with respect to input feature
, and then compute the gradients of the correct class with respect to input feature 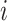 . An example of these maps can be seen in Fig. 4. Finally, we integrate over the entire set of gradient maps from these interpolated steps. One thing to note about our implementation of IGs is that we do not multiply our gradient maps with the input element-wise, unlike the original formulation. We do this to ensure no information from the input is enforced upon the gradient maps, meaning the attribution assignments from our maps are separate from the input itself. We argue that, based on (Kindermans et al. 2019) and (Adebayo et al. 2018), the element-wise multiplication can negatively impact the resulting maps. This element-wise multiplication can essentially act as an edge detector. All of our feature attribution calculations come from the Captum python library (Kokhlikyan et al. 2020).
. An example of these maps can be seen in Fig. 4. Finally, we integrate over the entire set of gradient maps from these interpolated steps. One thing to note about our implementation of IGs is that we do not multiply our gradient maps with the input element-wise, unlike the original formulation. We do this to ensure no information from the input is enforced upon the gradient maps, meaning the attribution assignments from our maps are separate from the input itself. We argue that, based on (Kindermans et al. 2019) and (Adebayo et al. 2018), the element-wise multiplication can negatively impact the resulting maps. This element-wise multiplication can essentially act as an edge detector. All of our feature attribution calculations come from the Captum python library (Kokhlikyan et al. 2020).
Fig. 4.
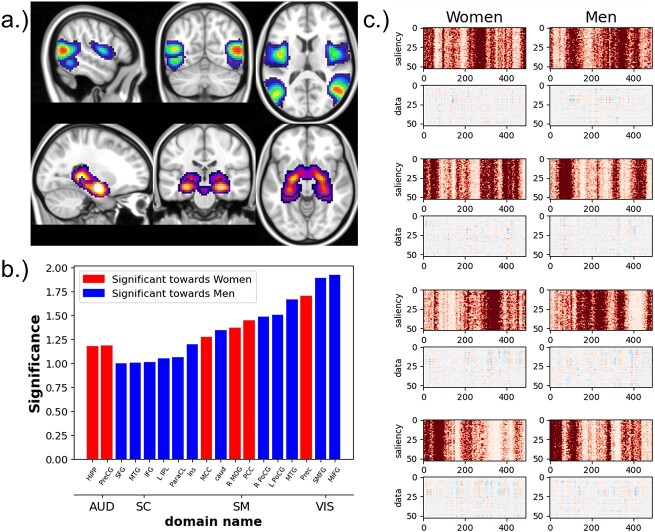
Two ICA components (a), the middle temporal gyrus (top), which is most significant for men, and the hippocampus, which is most significant for women (bottom). Bar graph (b) shows the effect size of the most significantly different components between men and women. The blue components are directed toward men, and components in red are directed toward women. And (c) saliency maps (rows labeled “saliency”) with associated ICA timecourses (rows labeled data) from four subjects (both women and men).
Our Approach
Using 10-fold cross-validation, from the trained model of each fold, we calculate the IG maps (aka saliency maps) for each test sample. This accumulates to one map for each subject when the subject was used as a test sample. This methodology ensures that none of the maps were from training subjects, which could bias the resulting maps. These saliency maps highlight the features that the prediction likelihood of the correct class is the most sensitive to. However, we observe that the maps are rarely stable and vary widely with the initial randomization. To correct for this, we train multiple models (keeping the hyperparameter settings the same) with different random initializations and calculate saliency maps from each model (for all experiments, we train 300 total models). We select the map closest to the average map from all models for each input sample using Euclidean distance (the distances are weighted by the loss for that subject/model pair to give more importance to better performing models). Then, the selected maps are normalized by the sum. A diagram of our methodological flow is in Fig. 2.
Fig. 2.
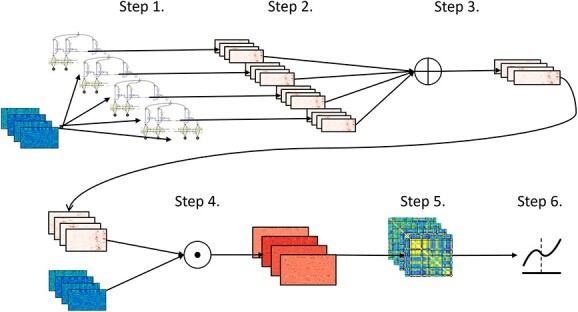
Flowchart describing our pipeline for analyzing the ICA timecourses. For all other datasets, we use only the first 4 steps to calculate the finalized maps. Step 1: we train 300 separate models (each with the same architecture) using different random initializations for each model on the same set of ICA timecourses. Step 2: we calculate the saliency maps for each sample from all 300 models. Step 3: We calculate the average saliency map for each sample over all 300 models. Step 4: We select the per-model set of maps with the lowest Euclidean distance to the average over all models, resulting in a stable saliency map for each input sample. This distance is weighted by the loss of that subject/model pair, giving more importance (shorter distances) to more accurate model/subject pairs.
Synthetic Data
We purposefully engineered the data so that the relevant information within the data was quantifiable and interpretable. In this work, we use two sets of synthetic data. These synthetic datasets consist of interpretable, ground-truth input in which the location of the relevant information was specified during its generation. In the first dataset of 30,000 samples, each sample is generated as random Gaussian noise with a sequence length of 200 and 30 channels, then randomly assigned a class label of either 0 or 1, with a 50% chance for either label to create a balanced dataset. For each sample, a window of 15 time steps is randomly chosen, within which a portion of the input data is perturbed based on the assigned label. If the label is 0, the first 15 channels in each of the 15 time steps are perturbed, and if the label is 1. The last 15 channels are perturbed. Each target feature is perturbed by adding randomly generated Gaussian noise, effectively changing the distribution in these perturbed areas from (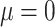 ,
, 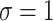 ) to (
) to (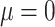 ,
, 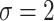 ). This creates a pattern of “boxes” for the dataset, an example of which can be seen in Fig. 3. In essence, only the channels are predictive of the class label instead of temporal patterns. This box dataset, a trivial example, is a way to show the effectiveness of our methodology in a vacuum with very few confounding variables.
). This creates a pattern of “boxes” for the dataset, an example of which can be seen in Fig. 3. In essence, only the channels are predictive of the class label instead of temporal patterns. This box dataset, a trivial example, is a way to show the effectiveness of our methodology in a vacuum with very few confounding variables.
Fig. 3.
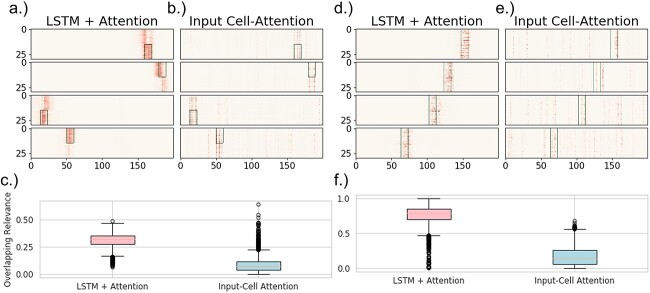
(a-c) The results from the analysis of the boxes dataset. Four examples of resulting maps from both (a) LSTM+attention and (b) input-cell attention, where the green rectangle is a mask representing the truly relevant information (i.e., box location). (c) Boxplots of the overlapping-values metric over all 3,000 samples for both models. The overlap is defined as the percentage of the total sum of the maps that are within the relevant area seen in the top figures. (d-f) Results from the analysis of the VAR dataset. The figures are organized the same as the figures for the boxes results. (d) LSTM+attention and (e) input-cell attention are four examples of the maps, where the green rectangles are the ground-truth relevancies (i.e. where the auto-regressive signal changes). Each baseline image has a certain underlying transition matrix, as computed by VAR. Each sample is interpolated with one of two different transition matrices, depending on the class label (the label along the y-axis) within the area demarcated by the green rectangles. (f) Boxplots of the overlapping-values metric over 3,000 held-out samples for both models. Both experiments represent two separate ideas or class-relevant patterns. The boxes data contains a feature specific pattern that aligns certain rows (e.g. components) with the classification label, but also disregard any temporal information. The VAR data contains connectivity patterns (estimated by the transition matrices), or how the interactions between rows/components relates to the classification label.
A second synthetic experiment shows that the saliency maps are still accurate when the relevant information is based on dynamic patterns. As with the first experiment, each sample begins as a Gaussian noise (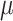 =0,
=0, 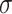 =1). However, vector autoregression (VAR) is used to control the underlying dynamics of each sample. VAR explains the evolution of a variable over time with the generalized equation:
=1). However, vector autoregression (VAR) is used to control the underlying dynamics of each sample. VAR explains the evolution of a variable over time with the generalized equation: 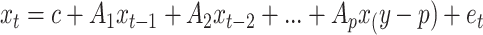 . For all samples, the VAR is computed using a positive semi-definite matrix,
. For all samples, the VAR is computed using a positive semi-definite matrix, 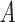 . Then, 15 successive steps are randomly chosen to be perturbed with new dynamic information. Or, two more positive semi-definite matrices,
. Then, 15 successive steps are randomly chosen to be perturbed with new dynamic information. Or, two more positive semi-definite matrices, 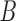 and
and 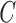 , are created, and VAR is again used to compute 15 new steps using Gaussian noise and either matrix
, are created, and VAR is again used to compute 15 new steps using Gaussian noise and either matrix 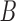 or
or 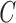 , depending on the class label of the sample. These new steps,
, depending on the class label of the sample. These new steps, 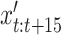 are added to the sample at a randomly selected interval (
are added to the sample at a randomly selected interval (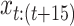 ), with an interpolation variable,
), with an interpolation variable, 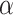 resulting in the equation:
resulting in the equation: 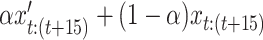 . Examples of these data are seen in Fig. 5. This VAR dataset is again built specifically to show, without a doubt, the efficacy of the methodology. However, in this case, as it is specifically engineered to highlight dynamical information, we argue that it is somewhat representative of fMRI data, where dynamical patterns are prevalent and highly influential.
. Examples of these data are seen in Fig. 5. This VAR dataset is again built specifically to show, without a doubt, the efficacy of the methodology. However, in this case, as it is specifically engineered to highlight dynamical information, we argue that it is somewhat representative of fMRI data, where dynamical patterns are prevalent and highly influential.
Fig. 5.
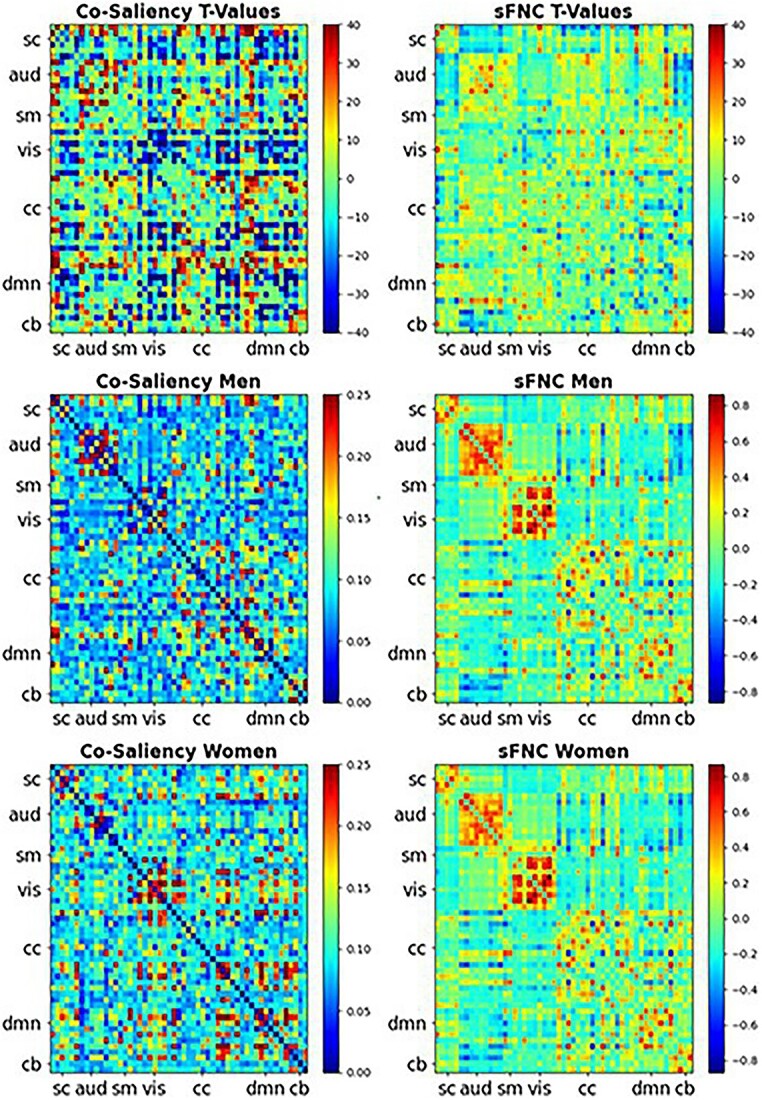
Heatmaps for the co-saliency (left): T-values masked by the FDR corrected significance, where 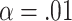 (top), the average co-saliency for men (middle), and the average co-saliency for women (bottom). The corresponding sFNCs are on the right side. We compared the co-saliency and the sFNC with show the relationships captured within the relevant information compared with relationships in the data (sFNC). The similarities between the two are striking, suggesting that the model does rely on some of these relationships. We also see that the model relies on relationships in different ways for both men and women. For example, we see that the model relies on connections within the AUD domain for men and relationships within the VIS domain for women. All of these relationships are seen in the raw data.
(top), the average co-saliency for men (middle), and the average co-saliency for women (bottom). The corresponding sFNCs are on the right side. We compared the co-saliency and the sFNC with show the relationships captured within the relevant information compared with relationships in the data (sFNC). The similarities between the two are striking, suggesting that the model does rely on some of these relationships. We also see that the model relies on relationships in different ways for both men and women. For example, we see that the model relies on connections within the AUD domain for men and relationships within the VIS domain for women. All of these relationships are seen in the raw data.
Saliency Quality Metrics
Since the relevant information of the synthetic datasets is easily quantifiable, we can use basic similarity scores between the saliency maps and proper representations of the input to understand the quality of the maps. We compare our map quality on holdout samples with those of an input-cell attention model. Firstly, as the input data are noisy, we need a reasonable representation of each sample. For both experiments, we represent each sample as a binary matrix in which only the elements within the perturbed regions are ones, and all other elements are zero. In the first experiment, the randomly selected window of 15x15 elements is set to one, and in the second experiment, the 15x30 window perturbed with added dynamics is our non-null region. Additionally, for the saliency maps from both our method and input-cell attention, we pass each sample through an absolute function (Bruce et al. 2015). To conduct a fair comparison with (Ismail et al. 2019), we use both of the similarity metrics therein: Euclidean distance and weighted Jaccard similarity. We also use a third metric, referred to as “overlapping values”, which is the sum of all salient values within the window over the sum of the entire saliency map. These overlapping values show the percentage of the total saliency map within the relevant areas.
To ensure an unbiased sampling of the timecourses with our model, we separate the data into 27,000 training samples and 3,000 test samples. We train 300 models on the non-holdout set and generate the maps for every sample. Then, we select the saliency maps using the selection criteria described in our approach section and generate the saliency maps for the holdout set. We chose 300 due to computational restrictions, as each model can take some time to train. These maps are then fed through either a rectified linear unit (ReLU) function or an absolute function (depending on the experiment) to avoid relying on both positive and negative derivatives to find the relevant information. Recent research has shown that removing negative values entirely from saliency can be beneficial (Selvaraju et al. 2019).
Salient Networks
With the selected saliency maps, we sum along the temporal axis for each subject, resulting in a vector of size 53 for each subject. We then compute the group-wise sex differences for each component using Cohen’s D (Cohen 2013). We select Cohen’s D because, due to the large sample size of our data, we prefer an effect size measure agnostic to sample size. We chose a cutoff threshold of one. This analysis highlights which networks (and brain regions) are significantly more important for correctly classifying men vs. women.
Co-Saliency
To better understand the sex differences within the ICA TCs, we computed the pairwise correlation of the processed saliency maps, which we call ”co-saliency”, using Pearson correlation. These correlation matrices describe the relationships between the relevancy of time-varying values of the ICA components. Notably, they capture relationships similar to those found in fMRI connectivity, as shown in Fig. 5.
Results
Synthetic Verification
Results from synthetic data show that our method effectively finds genuinely relevant information. Tables 1 and 2 compare the saliency quality metric results between our method and Input-Cell Attention for the boxes and VAR datasets, respectively. The weighted Jaccard and Euclidean distance showed vast improvement for our method over a current state-of-the-art method, input-cell attention for both the boxes dataset (in Fig. 3) and the more dynamic, VAR-induced dataset (in Fig. 5). The VAR dataset is especially significant as it shows that our method can properly capture non-stationary information. It is also important to note that our method and the input-cell attention model got 99% accuracy on holdout data from the boxes dataset. However, our model achieved much higher accuracy (92%) on holdout data from the VAR dataset than input-cell attention (81%).
Table 1.
Table comparing the LSTM+attention with the input-cell attention methodology on the boxes datasets. Within each cell is the average and standard deviation of the metric over 3,000 test samples, and the 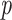 -value comparing the two methodologies using a 2-sample
-value comparing the two methodologies using a 2-sample 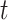 -test.
-test.
| Boxes Dataset | |||
|---|---|---|---|
| Euclidean Distance | Overlapping Values | Weighted Jaccard | |
| LSTM + Attention |
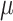 =1.24,
=1.24, 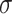 =.23, p <.0001 =.23, p <.0001 |
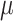 =.33,
=.33, 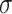 =.07, p <.0001 =.07, p <.0001 |
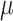 =.22,
=.22, 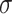 =.04, p <.0001 =.04, p <.0001 |
| Input-Cell Attention |
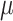 =2.35,
=2.35, 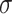 =.45, p <.0001 =.45, p <.0001 |
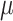 =.15,
=.15, 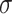 =.05, p <.0001 =.05, p <.0001 |
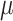 =.07,
=.07, 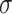 =.02, p <.0001 =.02, p <.0001 |
Table 2.
Table comparing the LSTM+attention with the input-cell attention methodology on the VAR datasets. Within each cell is the average and standard deviation of the metric over 3,000 test samples, and the 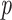 -value comparing the two methodologies using a 2-sample
-value comparing the two methodologies using a 2-sample 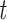 -test.
-test.
| VAR Dataset | |||
|---|---|---|---|
| Euclidean Distance | Overlapping Values | Weighted Jaccard | |
| LSTM + Attention |
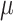 =4.57,
=4.57, 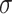 =1.16, p <.0001 =1.16, p <.0001 |
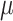 =.56,
=.56, 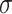 =.31, p <.0001 =.31, p <.0001 |
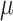 =.10,
=.10, 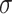 =.05, p <.0001 =.05, p <.0001 |
| Input-Cell Attention |
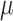 =5.34,
=5.34, 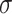 =1.42, p <.0001 =1.42, p <.0001 |
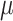 =.17,
=.17, 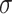 =.14, p <.0001 =.14, p <.0001 |
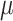 =.05,
=.05, 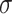 =.03, p <.0001 =.03, p <.0001 |
Performance Evaluation
To verify that our model is learning discriminatory patterns, we used stratified 10-fold cross-validation across the entire dataset and found that all models’ average overall validation accuracy was 91.3%. Using 10-fold cross-validation, we accumulated the predictions for each subject when they were used as a test sample for each model. In the end, we had 3,438,300 predictions (300 models * 11,461 subjects). 90.5% of women were correctly predicted, and 91.8% of men were correctly predicted. A confusion matrix is shown in Table 3.
Table 3.
The confusion matrix over the average of all models and folds (3,000 in total) for the UKB classification accuracies using our modality. The values are normalized to percentages.
| Predict Female | Predict Male | |
|---|---|---|
| True Female | .905 | .095 |
| True Male | .082 | .918 |
Our model performed well compared with other fMRI biological sex classification studies that we are aware of(Billmeyer and Parhi 2021, Leming and Suckling 2021, Sen and Parhi 2019), reporting between 85% and 88% accuracy, while at least one (Sen and Parhi 2021) performed better than our model with 94%. We suggest that this validates the trustworthiness of our model. The model’s performance, which is not the primary focus of this work, is relevant because if we are to trust the explanations from the model, we must trust the model’s performance.
Other studies focusing on structural MRI data have found much higher performance, between 95% and 99% accuracy (Abrol et al. 2021, Luo et al. 2019) suggesting sMRI is much more informative than fMRI. However, it should be noted that while whole-brain sMRI data can have tens of thousands of features, and the models can have millions of parameters, our model had just under 700,000 parameters, much fewer than in the sMRI studies we have found.
Sex-Relevant functional Activity
Figure 4 shows the components with the largest Cohen’s D effect size between groups based on the per-component relevancy averaged over time, where the red networks are more salient for women and the blue networks are more salient for men. We find that the AUD domain is particularly relevant for women (Jaušovec and Jaušovec 2009). The SC and VIS domains also appear to be highly relevant for male classification, with the highest biological sex differences within the VIS domain. Finally, the SM domain is highly relevant, with different networks signaling for the two sexes. Of the remaining 35 non-significant components, we found that 10 components from the CC domain had an absolute effect size between 0.5 and 1. 9 VIS components had effect sizes between 0.38 and 0.52. There were also 2 AUD components, 5 SC components, and 9 SM components with effect sizes between 0.02 and 0.5.
Co-Saliency Analysis
From the co-saliency heatmaps, Fig. 6, we find that the differences are almost “orthogonal,” showing that the saliency method starkly separates the two sexes by focusing on network-specific timepoints that are intercorrelated in functionally structured ways. The co-saliencies indicate that temporal patterning of network-specific saliencies in women is significantly more strongly correlated than in men, as can be seen in the connectivity matrices in Fig. 5. This significance is shown by the top row’s 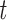 -test results. We use a
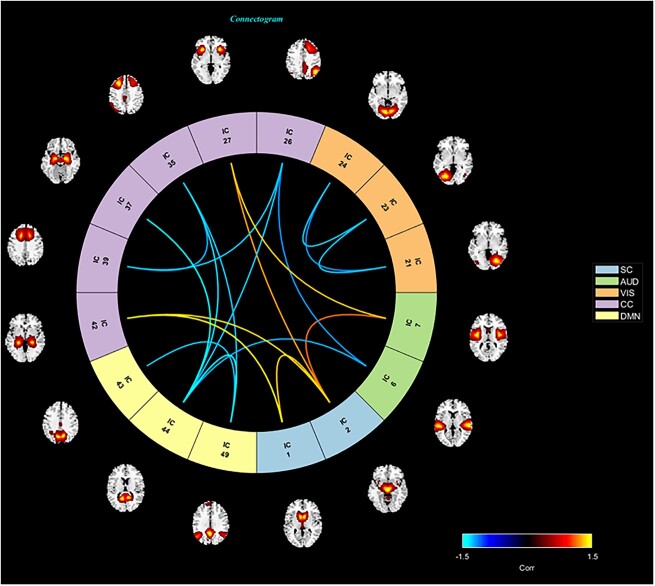
-test results. We use a  -test as we wish only to compare the means between men and women for both the co-saliency heatmaps and sFNC matrices. This strong co-saliency for women suggests that the model identifies timepoints in which networks are more tightly aligned when correctly classifying women, with networks in less tight temporal alignment at salient timepoints for the correct classification of men. Fig. 5 highlights the co-saliency differences in comparison to the differences among static connectivity. This is further elucidated in Fig. 6, highlighting our sex difference results in a connectome. Primarily, we see that both co-saliency and sFNC show sex differences in the VIS, CC, and DMN domains.
-test as we wish only to compare the means between men and women for both the co-saliency heatmaps and sFNC matrices. This strong co-saliency for women suggests that the model identifies timepoints in which networks are more tightly aligned when correctly classifying women, with networks in less tight temporal alignment at salient timepoints for the correct classification of men. Fig. 5 highlights the co-saliency differences in comparison to the differences among static connectivity. This is further elucidated in Fig. 6, highlighting our sex difference results in a connectome. Primarily, we see that both co-saliency and sFNC show sex differences in the VIS, CC, and DMN domains.
Fig. 6.
A connectome highlighting the component pairs from the saliency maps that are most significantly different between men and women. The locations within the brain of the associated ICNs are also visualized. The colors of the edges define the level of significance and which group is significantly higher. The blue edges indicate pairs that are significantly higher for men, and red edges indicate pairs that are higher for women. The color of the component number indicates the domain, each of which is defined in the legend.
We capture this modularity with greedy graph modularity estimations using the Clauset–Newman–Moore greedy algorithm (CNM) (Aaron et al. 2004). Because modularity computation is scale variant, we  -score all matrices along pairs. Although there are many community detection algorithms, we chose the CNM algorithm because it is a very well-studied and widely accepted approach, which is vital for our work, as our methodology is novel and needs to be robustly validated. In Fig. 7, we can see the estimated communities of the two sexes with CNM. From these communities, we computed the overall modularity of each sex (Newman 2011), giving us a final modularity score of 0.345 for women and 0.246 for men. The modularity for the sFNCs was 0.31 for women and 0.359 for men. The sFNC and co-saliency matrices organized by community are also found in Fig. 7.
-score all matrices along pairs. Although there are many community detection algorithms, we chose the CNM algorithm because it is a very well-studied and widely accepted approach, which is vital for our work, as our methodology is novel and needs to be robustly validated. In Fig. 7, we can see the estimated communities of the two sexes with CNM. From these communities, we computed the overall modularity of each sex (Newman 2011), giving us a final modularity score of 0.345 for women and 0.246 for men. The modularity for the sFNCs was 0.31 for women and 0.359 for men. The sFNC and co-saliency matrices organized by community are also found in Fig. 7.
Fig. 7.
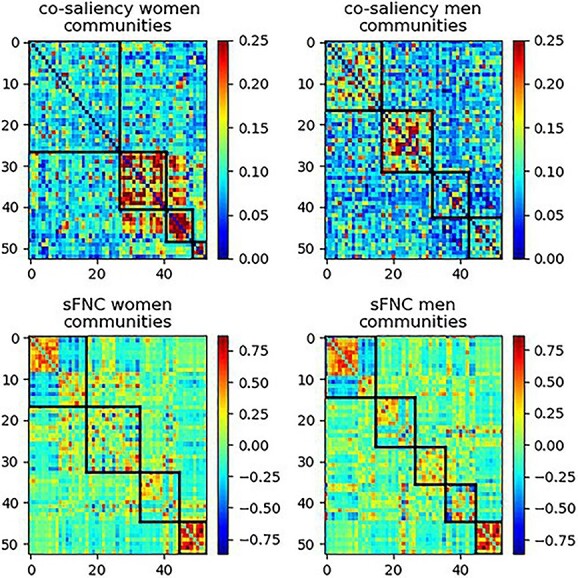
The co-saliency (top) and sFNC (bottom) matrices organized by communities (black bounding boxes) for women (left) and men (right). This illustrates how the community structure and modularity differ between the co-saliency and sFNC. As the modularity is much lower for the co-saliency, we see that the components in the co-saliency are much more uniformly connected, suggesting that the model finds relatively similar relationships over all components.
We can see two primary results. The first highlighted difference is which biological sex is most modular. In co-saliency, the women are now more modular, and their higher modularity value shows that certain groups of pairs are structurally dissimilar to the other pairs. Three of the four communities for women also have a very high correlation, meaning that these three groups of pairs are more distinctive than the communities for men, which also have a lower average correlation. We see a dissimilar result for the sFNC, where men are more modular. This suggests that the networks for women are most relevant to the LSTM when they are tightly coupled over time, even though most communities in the raw data are not modular for women.
Secondly, the absolute modularity difference between men and women is more prominent in the co-saliency. This is expected as the co-saliency is specifically wired to separate between sexes. It is, in our opinion, interesting that one of the pronounced separating signals is the communities among the most prediction-relevant components.
Discussion
After validating our methodology on synthetic data (Lewis et al. 2021), we extract classification-relevant sex differences within the ICA timecourses. Overall, the saliency maps and post hoc analyses capture several patterns found within the data. The ICN differences from the saliency maps show that at least four domains are key for sex classification, with many of these differences being backed by previous literature. The similarities between our results and past research are important, as these results are values for the ICNs averaged over time, which makes them more generalized, and presumptively more representative of global, high-level differences.
Figure 4 highlights the ICN and domain differences. The high effect size of the difference between men and women in the AUD domain shows that this domain is particularly relevant for the model’s classification of women. Our results are supported by previous research (Hofer et al. 2006), which shows major differences between the two sexes in the AUD domain. The SM domain is also highly relevant for women, supported by (Brun et al. 2009). These findings are quite interesting, as they are the only domains that are primarily more relevant for women than men. A striking aspect of our findings is that the VIS seems very relevant to how the model classifies and understands biological sex. We also see that 12 networks are significantly relevant for male classification, whereas only six are significantly relevant for female classification. This is important because, as each saliency map is normalized to be a probability map, this suggests that the relevant information for female classification is more concentrated in fewer networks than in male classification. In other words, compared with male-classified maps, a smaller number of networks from the female-classified maps have a higher percentage of the total relevancy. In addition to VIS, the SC domain is primarily relevant for the correct classification of males. Currently, there is a wealth of research connecting the SC to differences between men and women, from both MRI and functional MRI studies (Wang et al. 2018). Specifically, (Herron et al. 2015) has found sex differences in the gyrus, thus supporting our findings about differences in the SC domain. Another study by (Luders et al. 2004) showed differences in complexity, or the spatial frequency of the brain surface gyrification in the parietal lobe, a part of the SC domain, highlighting our findings that this particular domain shows sex-based differences.
Our co-saliency analysis, elucidated in Figs. 5 and 6 provide a wealth of information; explanations of these and their relationship to current research are in the discussion section of this paper. The areas of consistency offer an indirect validation of the relatively new method presented here, which is still being developed and refined. One important finding is that the dynamics of the networks within the CC domain, in relation to networks both within and outside the CC domain, figure strongly in the model’s ability to differentiate male and female subjects. The connections between CC networks and the AUD, DMN, and SC domains significantly inform the model’s decisions. Six networks within the CC domain also play a major and statistically significant role in model classification. We see a split where some networks are highly relevant for male classification while others are highly relevant for female classification. Specifically, connections within and between the inferior and middle-frontal gyrus and the hippocampal networks are primarily relevant for men. The left inferior parietal lobule, the middle cingulate cortex, and the superior frontal gyrus have especially salient connectivity patterns for women. These two domains, CC and VIS also show stark coherency differences, which can be visible in the co-saliency maps, and quantified in the community detection and graph-modularity computation seen in Fig. 7. However, the differences weighted toward men appear de-modularized compared with the sFNCs. It is also interesting to note that the most significant VIS co-saliency pairs are entirely within the domain and female-centric. These patterns also appear in previous connectivity work (Ritchie et al. 2018, Tomasi and Volkow 2012). The co-saliency maps seem to show these patterns, but with much more contrast than in the raw data. This contrast enhancement highlights subtle differences that are only weakly evident in the raw data, differences that look negligible under a linear, univariate lens but prove highly relevant to biological sex when employed in a multivariate nonlinear classification model. Our results also show several key findings that are missed by nonlinear analysis. Specifically, we see substantial differences in how certain domains are organized. We find that VIS and DMN domains and part of the CC are prominently modular for women. The community and general graph analyses also show an overall lack of modularity and even coherence for men. Fig. 7 highlights our community/graph analysis results, which show the overall modularity differences. The findings we report are complex and do not admit straightforward interpretations framed by previous results. They suggest that our understanding of biological sex has been limited by the ubiquitous use of linear univariate models. Expanding the traditional model space could help better realize the scientific promise of noninvasive functional brain imaging.
The fundamental goal of our work is to build upon previous research and expand on the scientific community’s understanding of sex differences in the brain. As described in previous work (Häfner 2003, Kirkovski et al. 2013, Stites et al. 2021, Wang et al. 2018), these understandings can help future researchers better understand how certain disorders vary based on biological sex. In the future, this work could lead to clinical advancements such as new treatments or medication.
The field of model interpretability is still young and rapidly evolving. While this work aims to introduce approaches to stabilize and quantify deep-learning interpretability methods for analyzing brain imaging data, new and more effective methods may emerge, revealing different information. As we highlight in this paper, recurrent models are architecturally challenging to interpret using gradient-based approaches. Although we suggest that our methods mitigate the gradient bias against distal timepoints and model instability across initializations, the computed saliencies remain sensitive to changes in hyperparameters and architecture. Overall, they can still be underspecified.
Another impediment to model interpretability is the complexity of the data itself. Although we have many carefully crafted and effective methods for quantifying information from ICN timecourses, these timecourses are highly processed dimensional reductions of the original scan data. This adds another layer of complexity to understanding brain functionality through the lens of saliency maps, increasing the possibility of erroneous interpretation of the findings. These flaws can be mitigated in due time. As more researchers focus on these topics, specifically neuroimaging and general, we will find more robust and effectively interpretable maps. We also suggest that new methods to analyze the maps (such as co-saliency) will become more prominent, opening the door for a better understanding of intricate datasets, especially neuroimaging.
Funding
FUNDING: NIH grants R01MH118695 and R01MH123610. NSF grant 2112455.
Contributor Information
Noah Lewis, Computational Science and Engineering, Georgia Institute of Technology, North Ave, 30332, GA, United States.
Robyn Miller, Tri-institutional Center for Translational Research in Neuroimaging and Data Science (TReNDS), 55 Park Pl NE, 30303, GA, United States; Georgia State University, 33 Gilmer St SE, 30303, GA, United States.
Harshvardhan Gazula, Athinoula A. Martinos Center for Biomedical Imaging, 149 13th Street, 02129, MA, United States; Harvard Medical School, 25 Shattuck St, 02115, MA, United States.
Vince Calhoun, Computational Science and Engineering, Georgia Institute of Technology, North Ave, 30332, GA, United States; Tri-institutional Center for Translational Research in Neuroimaging and Data Science (TReNDS), 55 Park Pl NE, 30303, GA, United States; Georgia State University, 33 Gilmer St SE, 30303, GA, United States.
References
- Aaron C, Newman MEJ, Moore C. Finding community structure in very large networks. Pys Rev E. 2004:70:6. [DOI] [PubMed] [Google Scholar]
- Abrol A, Zening F, Salman M, Silva R, Yuhui D, Plis S, Calhoun V. Deep learning encodes robust discriminative neuroimaging representations to outperform standard machine learning. Nat Commun. 2021:12:01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adebayo J, Gilmer J, Muelly M, Goodfellow I, Hardt M, Kim B. Sanity checks for saliency maps. Adv Neural Inf Proces Syst. 2018:31. [Google Scholar]
- Alfaro-Almagro F, Jenkinson M, Bangerter NK, Andersson JLR, Griffanti L, Douaud G, Sotiropoulos SN, Jbabdi S, Hernandez-Fernandez M, Vallee E, et al. Image processing and quality control for the first 10,000 brain imaging datasets from uk biobank. NeuroImage. 2018:166:400–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arslan S, Ktena SI, Glocker B, Rueckert D, Stoyanov D, Taylor Z, Ferrante E, Dalca AV, Martel A, et al. Graph saliency maps through spectral convolutional networks: Application to sex classification with brain connectivity. In: Arslan S, Ktena SI, Glocker B, Rueckert D, editors, Graphs in Biomedical Image Analysis and Integrating Medical Imaging and Non-Imaging Modalities. Cham: Springer International Publishing; 2018. pp. 3–13 ISBN 978-3-030-00689-1. [Google Scholar]
- Baecker L, Dafflon J, Da Costa PF, Garcia-Dias R, Vieira S, Scarpazza C, Calhoun VD, Sato JR, Mechelli A, Pinaya WHL. Brain age prediction: A comparison between machine learning models using region-and voxel-based morphometric data. Hum Brain Mapp. 2021:42(8):2332–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahdanau D, Cho K, Bengio Y. Neural machine translation by jointly learning to align and translate. arXiv preprint arXiv:1409.0473. 2014.
- Bandettini PA. Twenty years of functional mri: The science and the stories. NeuroImage. 2012:62(2):575–588 ISSN 1053-8119. 20 YEARS OF fMRI. [DOI] [PubMed] [Google Scholar]
- Billmeyer R, Parhi KK. Biological gender classification from fmri via hyperdimensional computing. In: 2021 55th Asilomar Conference on Signals, Systems, and Computers. Pacific Grove, California: IEEE; 2021. pp. 578–582 [Google Scholar]
- Bruce NDB, Wloka C, Frosst N, Rahman S, Tsotsos JK. On computational modeling of visual saliency: Examining what’s right, and what’s left. Vision Research, 116: 95–112, 2015. ISSN 0042–6989. Elsevier. [DOI] [PubMed] [Google Scholar]
- Brun CC, Lepore N, Luders E, Chou Y-Y, Madsen SK, Toga AW, Thompson PM. Sex differences in brain structure in auditory and cingulate regions. Neuroreport. 2009:20(10):930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun V, Adali T, Pearlson G. Independent component analysis applied to fmri data: a generative model for validating results. In: Neural Networks for Signal Processing XI: Proceedings of the 2001 IEEE Signal Processing Society Workshop (IEEE Cat. No.01TH8584); 2001. pp. 509–518
- Calhoun VD, Adali T. Multisubject independent component analysis of fmri: A decade of intrinsic networks, default mode, and neurodiagnostic discovery. IEEE Rev Biomed Eng. 2012:5:60–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. Statistical power analysis for the behavioral sciences. Routledge; 2013 [Google Scholar]
- D’Amour A, Heller KA, Moldovan DI, Adlam B, Alipanahi B, Beutel A, Chen C, Deaton J, Eisenstein J, Hoffman MD, et al. Underspecification presents challenges for credibility in modern machine learning. Journal of Machine Learning Research. 2020.
- Yuhui D, Zening F, Sui J, Gao S, Xing Y, Lin D, Salman M, Anees Abrol M, Rahaman A, Jiayu Chen L, et al. Neuromark: An automated and adaptive ica based pipeline to identify reproducible fmri markers of brain disorders. NeuroImage: Clinical. 2020:28:102375 ISSN 2213-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanzadeh R, Silva RF, Abrol A, Salman M, Bonkhoff A, Yuhui D, Zening F, DeRamus T, Damaraju E, Baker B, et al. Individualized spatial network predictions using siamese convolutional neural networks: A resting-state fmri study of over 11,000 unaffected individuals. PLoS One. 2022:17:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron TJ, Kang X, Woods DL. Sex differences in cortical and subcortical human brain anatomy. F1000Research. 2015:4(88):88. [Google Scholar]
- Häfner H. Gender differences in schizophrenia. Psychoneuroendocrinology. 2003:28:17–54 ISSN 0306-4530. [DOI] [PubMed] [Google Scholar]
- Hofer A, Siedentopf CM, Ischebeck A, Rettenbacher MA, Verius M, Felber S, Wolfgang Fleischhacker W. Gender differences in regional cerebral activity during the perception of emotion: A functional mri study. NeuroImage. 2006:32(2):854–862 ISSN 1053-8119. [DOI] [PubMed] [Google Scholar]
- Iraji A, Faghiri A, Fu Z, Rachakonda S, Kochunov P, Belger A, Calhoun VD. Multi-spatial-scale dynamic interactions between functional sources reveal sex-specific changes in schizophrenia. Network Neuroscience. 2022:6(2):357–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail AA, Gunady MK, Pessoa L, Bravo HC, Feizi S. Input-cell attention reduces vanishing saliency of recurrent neural networks. In: Advances in Neural Information Processing Systems 32: Annual Conference on Neural Information Processing Systems. Vol. 32, 2019. pp. 10813–10823 [Google Scholar]
- Jaušovec N, Jaušovec K. Gender related differences in visual and auditory processing of verbal and figural tasks. Brain Res. 2009:1300:135–145 ISSN 0006-8993. [DOI] [PubMed] [Google Scholar]
- Kindermans P-J, Hooker S, Adebayo J, Alber M, Schütt KT, Dähne S, Erhan D, Kim B. The (un) reliability of saliency methods. In: Explainable AI: Interpreting, Explaining and Visualizing Deep Learning. Cham: Springer International Publishing; 2019. pp. 267–280 [Google Scholar]
- Kirkovski M, Enticott P, Fitzgerald P. A review of the role of female gender in autism spectrum disorders. J Autism Dev Disord. 2013:43(11):2584–2603 ISSN 01623257. [DOI] [PubMed] [Google Scholar]
- Kokhlikyan N, Miglani V, Martin M, Wang E, Alsallakh B, Reynolds J, Melnikov A, Kliushkina N, Araya C, Yan S, et al. Captum: A unified and generic model interpretability library for pytorch. arXiv preprint arXiv:2009.07896. 2020.
- Leming M, Suckling J. Deep learning for sex classification in resting-state and task functional brain networks from the uk biobank. NeuroImage. 2021:241:118409 ISSN 1053-8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis N, Miller R, Gazula H, Rahman MM, Plis S, Vince C. Can recurrent models know more than we do? In: IEEE International Conference on Healthcare Informatics (ICHI). IEEE; 2021. pp. 243–247.
- Li X-L, Adali T. Blind spatiotemporal separation of second and/or higher-order correlated sources by entropy rate minimization. In: 2010 IEEE International Conference on Acoustics, Speech and Signal Processing. IEEE; 2010. pp. 1934–1937 [Google Scholar]
- Liu Z, Luo P, Wang X, Tang X. Deep learning face attributes in the wild. Proceedings of the IEEE International Conference on Computer Vision (ICCV). 2015:3730–3738. [Google Scholar]
- Long H, Fan M, Yang X, Guan Q, Huang Y, Xu X, Xiao J, Jiang T. Sex-related difference in mental rotation performance is mediated by the special functional connectivity between the default mode and salience networks. Neuroscience. 2021:478:65–74 ISSN 0306-4522. [DOI] [PubMed] [Google Scholar]
- Luders E, Narr KL, Thompson PM, Rex DE, Jancke L, Steinmetz H, Toga AW. Gender differences in cortical complexity. Nat Neurosci. 2004:7(8):799–800. [DOI] [PubMed] [Google Scholar]
- Luo Z, Hou C, Wang L, Dewen H. Gender identification of human cortical 3-d morphology using hierarchical sparsity. Front Hum Neurosci. 2019:13:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MEJ. Networks: An Introduction. Oxford, England: Oxford University Pres; 2011. [Google Scholar]
- Ritchie SJ, Cox SR, Shen X, Lombardo MV, Reus LM, Alloza C, Harris MA, Alderson HL, Hunter S, Neilson E, et al. Sex Differences in the Adult Human Brain: Evidence from 5216 UK Biobank Participants. Cereb Cortex. 2018:28(8):2959–2975 ISSN 1047-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M, Paliwal K. Bidirectional recurrent neural networks. IEEE Transactions on Sig Proc. 1997:45:2673–2681. [Google Scholar]
- Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-cam: Visual explanations from deep networks via gradient-based localization. Int J Comput Vis. 2019:128(2):336–359. [Google Scholar]
- Sen B, Parhi KK. Predicting male vs. female from task-fmri brain connectivity. In: 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). IEEE; 2019. pp. 4089–4092. [DOI] [PubMed] [Google Scholar]
- Sen B, Parhi KK. Predicting biological gender and intelligence from fmri via dynamic functional connectivity. IEEE Trans Biomed Eng. 2021:68(3):815–825. [DOI] [PubMed] [Google Scholar]
- Simonyan K, Vedaldi A, Zisserman A. Deep inside convolutional networks: Visualising image classification models and saliency maps. arXiv preprint arXiv:1312.6034. 2013.
- Spets DS, Slotnick SD. Are there sex differences in brain activity during long-term memory? a systematic review and fmri activation likelihood estimation meta-analysis. Cogn Neurosci. 2021:12(3–4):163–173. [DOI] [PubMed] [Google Scholar]
- Stites SD, Cao H, Harkins K, Flatt JD. Measuring Sex and Gender in Aging and Alzheimer’s Research: Results of a National Survey. The Journals of Gerontology: Series B. 2022:77(6):1005–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararajan M, Taly A, Yan Q. Axiomatic attribution for deep networks. In Precup D, Teh, YW, editors, Proceedings of the 34th International Conference on Machine Learning, volume 70 of Proceedings of Machine Learning Research, pages 3319–3328. PMLR, 2017. [Google Scholar]
- Tomasi D, Volkow ND. Gender differences in brain functional connectivity density. Hum Brain Mapp. 2012:33(4):849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xu Q, Li S, Li G, Zuo C, Liao S, Yang L, Li S, Joshi RM. Gender differences in anomalous subcortical morphology for children with adhd. Neurosci Lett. 2018:665:176–181 ISSN 0304-3940. [DOI] [PubMed] [Google Scholar]