Abstract
Gastric cancer (GC) is a complex disease influenced by multiple genetic and epigenetic factors. Chronic inflammation caused by Helicobacter pylori infection and dietary risk factors can result in the accumulation of aberrant DNA methylation in gastric mucosa, which promotes GC development. Tensin 4 (TNS4), a member of the Tensin family of proteins, is localized to focal adhesion sites, which connect the extracellular matrix and cytoskeletal network. We identified upregulation of TNS4 in GC using quantitative reverse transcription PCR with 174 paired samples of GC tumors and adjacent normal tissues. Transcriptional activation of TNS4 occurred even during the early stage of tumor development. TNS4 depletion in GC cell lines that expressed high to moderate levels of TNS4, i.e., SNU-601, KATO III, and MKN74, reduced cell proliferation and migration, whereas ectopic expression of TNS4 in those lines that expressed lower levels of TNS4, i.e., SNU-638, MKN1, and MKN45 increased colony formation and cell migration. The promoter region of TNS4 was hypomethylated in GC cell lines that showed upregulation of TNS4. We also found a significant negative correlation between TNS4 expression and CpG methylation in 250 GC tumors based on The Cancer Genome Atlas (TCGA) data. This study elucidates the epigenetic mechanism of TNS4 activation and functional roles of TNS4 in GC development and progression and suggests a possible approach for future GC treatments.
Keywords: cell migration, cell proliferation, DNA methylation, gastric cancer, TNS4
INTRODUCTION
Gastric cancer (GC) is a complex heterogeneous disease that is influenced by a variety of genetic, epigenetic, and environmental factors (Lim et al., 2016). The most important risk factors for GC are Helicobacter pylori infection and certain diet-related behaviors, such as a high salt intake, both of which cause a long-term inflammatory response (Crew and Neugut, 2006). Traditionally, GC is histologically divided into two types, intestinal- and diffuse-type GC, according to Lauren’s classification (Lauren, 1965). Intestinal-type GCs originate from gastric mucosal cells that are characterized by well-differentiated glandular structures and develop through well-characterized sequential pathological stages such as chronic gastritis, atrophy, intestinal metaplasia, and dysplasia (Yuasa, 2003). Diffuse-type GCs are characterized by poorly differentiated growth without clear precancerous stages and are associated with aggressive tumor growth and poor prognosis (Chiaravalli et al., 2012). Recently, The Cancer Genome Atlas (TCGA) network investigated the exome sequence, copy number alterations, gene expression, DNA methylation, and protein activity in GC tissues. This resulted in classifying GCs according to four subtypes: Epstein–Barr virus (EBV)-positive, microsatellite instability (MSI), genomically stable (GS), and chromosomal instability (CIN) (Cancer Genome Atlas Research Network, 2014).
DNA methylation at the fifth position of cytosine (5mC) is a stable epigenetic mark that plays an important role in mammalian development, differentiation, and maintenance of cellular identity through the regulation of gene expression (Kim and Costello, 2017; Yoon et al., 2018). Changes in DNA methylation are observed in many human cancers, particularly GC (Ebrahimi et al., 2020). Hypermethylation of tumor suppressor genes is a major abnormality in EBV-positive GCs (Kaneda et al., 2012), and aberrant DNA methylation accumulates in the gastric mucosa as a result of chronic inflammation caused by H. pylori infection (Maeda et al., 2017). We previously reported the hypermethylation of tumor suppressor genes such as LIMS2 (Kim et al., 2006), PRKD1 (Kim et al., 2008a), DCBLD2 (Kim et al., 2008c), LRRC3B (Kim et al., 2008b), BVES, and POPDC3 (Kim et al., 2010) in GC.
Here we describe the hypomethylation of Tensin 4 (TNS4). The Tensin family comprises four members, TNS1, TNS2, TNS3, and TNS4, all of which are localized to focal adhesion sites and regulate cell adhesion, migration, proliferation, differentiation, and tissue development (Liao and Lo, 2021). TNS4 shares sequence homology with other tensins only in its C-terminal region, which contains the SH2 and PTB domains, but, unlike TNS1-3, it lacks an actin-binding domain at its N terminus. Thus, it is also known as C terminal tensin like (CTEN). TNS4 has oncogenic roles during cancer progression (Lu et al., 2018; Wu and Liao, 2018). We investigated the expression of TNS4 in GC cell lines and tumors derived from GC patients and adjacent normal tissues. We also examined the role of TNS4 in GC progression. Finally, we analysed the correlation between promoter methylation and expression of TNS4 in GC.
MATERIALS AND METHODS
Clinical samples
Samples from gastric tumor tissues and paired adjacent normal tissues were provided by Chungnam National University Hospital (Korea) with clinicopathological information. Authorization for the use of GC samples for research purposes and ethical approval were obtained from the Institutional Review Board of Chungnam National University Hospital (CNUH 2018-01-056).
Immunohistochemistry (IHC) of GC patient tissue samples
IHC was performed as described (Seo et al., 2020). Five cases of intestinal-type GC and one case of diffused-type GC were chosen randomly from among the samples and analysed using TNS4 monoclonal antibody (1:100; Abnova, Taiwan). All samples were independently reviewed by a pathologist (K.-S.S.) in a blinded manner.
Cell lines
HEK293T cells were cultured in DMEM (Welgene, Korea) supplemented with 1% antibiotics (Thermo Fisher Scientific, USA) and 10% fetal bovine serum (Welgene). GC cell lines, SNU-16, SNU-216, SNU-520, SNU-601, SNU-620, SNU-638, SNU-668, SNU-719, AGS, KATO III, MKN1, MKN45, and MKN74 were cultured in RPMI1640 (Welgene) supplemented with 1% antibiotics and 10% fetal bovine serum. All cell lines were purchased from the Korean Cell Line Bank and maintained at 37°C in a humidified atmosphere containing 5% CO2.
Quantitative reverse transcription PCR (qRT-PCR)
Cells were harvested by trypsinization. RNA was isolated by using the RNeasy Plus Mini Kit (Qiagen, USA). cDNA was synthesized from 1 µg of RNA for each sample by using the iScript cDNA Synthesis Kit (Bio-Rad, USA). PCR reactions were conducted in triplicate for each sample using 200 ng of cDNA (Bio-Rad). The value for TNS4 was normalized based on that of β-actin. The primers used for TNS4 qRT-PCR were 5′-AGCAGGGCATCACTCTGACT-3′ (sense) and 5′-CTGAGGCTCTGTCTGGCTCT-3′ (antisense). The primers used for β-actin qRT-PCR were 5′- CAAGAGATGGCCACGGCTGCT-3′ (sense) and 5′- TCCTTCTGCATCCTGTCGGCA-3′ (antisense).
Western blotting
Cells were washed three times with cold phosphate-buffered saline and lysed with RIPA buffer (T&I, Korea) supplemented with protease inhibitors (Roche, Switzerland). Protein concentrations were determined with a Bradford Protein Assay Kit (Bio-Rad) from boiled cell lysates. Protein samples were loaded onto a 10% SDS-polyacrylamide gel for electrophoresis and then were transferred to a polyvinylidene fluoride membrane (Roche). After being blocked with 5% skim milk (BD, USA) with Tris-buffered saline with Tween 20 (TBS-T) (Bio-Rad) for 1 h at room temperature, membranes were incubated overnight at 4°C with specific primary antibodies. Antibodies were as follows: anti-TNS4 (1:1,000; Abnova), anti-GAPDH (1:2,000; AbFrontier, Korea), horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (1:5,000; Santa Cruz Biotechnology, USA), and HRP-conjugated goat anti-rabbit IgG (1:5,000; Santa Cruz Biotechnology). After being washed with TBS-T, membranes were incubated with appropriate secondary antibody (Invitrogen, USA). After another washing with TBS-T, immunopositive bands were detected with the ECL Kit (Advansta, USA) and visualized with Fujifilm LAS-4000 (Fujifilm, Japan).
The DepMap data analysis
To determine whether GC cell lines’ response to TNS4 depletion depends on their basal TNS4 expression levels, we investigated human cell line transcriptome data and CRISPR knockout screening data generated by the projects CCLE (Cancer Cell Line Encyclopedia) and Achilles. Both data sets were obtained from the DepMap portal (https://depmap.org/portal/; CCLE_expression.csv; CRISPR_gene_effect.csv). A total of 41 GC cell lines with CCLE transcriptome data were divided into TNS4-high (n = 21) and TNS4-low (n = 20) groups based on the median TNS4 expression as a cutoff. Among the cell lines, 30 cell lines that have TNS4 gene effect scores (TNS4-high, n = 13; TNS4-low, n = 17) were used for further analysis. The significance of the difference between the mean TNS4 gene effect scores of the two groups was assessed by a one-tailed t-test. A negative gene effect score indicates cell death following TNS4 knockout.
Short hairpin RNA and generation of stable cell lines by lentiviral transduction
Short hairpin RNAs (shRNAs) against TNS4 were purchased from Sigma-Aldrich (USA). pLKO.1-puro empty vector (Sigma-Aldrich) was used as the negative control vector. To construct lentiviruses for shRNA production, 293T cells were cotransfected with MISSION Lentiviral Packaging Mix (Sigma-Aldrich) and the negative control or TNS4 shRNA vector. SNU-601, KATO III, and MKN4 cells were infected and selected with puromycin (1 µg/ml; Sigma-Aldrich) for 2 weeks. TNS4 mRNA reduction was assessed by qRT-PCR, and TNS4 protein reduction was assessed by western blotting.
TNS4 cDNA expression plasmids
TNS4 cDNA expression plasmids were prepared by PCR amplification using pGEM-TNS4 (Sino Biological Inc., China). The primers used for TNS4 amplification were 5′-ATGCGCTAGCGCCACCATGTCCCAGGTGATGTCCAG-3′ (sense) and 5′- GCGGCCGCCTACATCCTTTCTGCGTCCTG-3′ (antisense). The TNS4 expression vector and negative control vector were constructed using pCDH-CMV-MCS-EF1α-Puro (SBI, USA). The expression vector was digested with restriction enzymes NheI and NotI (NEB, USA). All DNA expression constructs were confirmed by DNA sequencing.
Colony formation assay
Cells were seeded onto 6-well plates (1 × 103 cells/well) and incubated at 37°C in 5% CO2 for 2 weeks. The resulting colonies were fixed and stained with 0.5% crystal violet (Sigma-Aldrich), 3.7% formaldehyde (Sigma-Aldrich), and 30% ethanol (Merck Millipore, USA). This analysis was performed in triplicate and repeated in at least two independent experiments.
Cell migration assay
Cell migration was measured by using Transwell chambers (Corning, USA). The lower surface of each chamber was prepared by coating the filter with 0.25 mg/ml fibronectin (Sigma-Aldrich). The filter was air dried for >30 min. Cells that had been starved for 24 h in serum-free RPMI 1640 were then seeded into the upper chamber (5 × 104 cells/chamber) in serum-free medium. The cells were cultured at 37°C in 5% CO2 for 12 h and were then fixed and stained for >2 h with a solution of 0.5% crystal violet, 3.7% formaldehyde, and 30% ethanol.
Bisulfite sequencing
Genomic DNA (1 µg) was modified by sodium bisulfite by using the EZ DNA Methylation-Gold kit (Zymo Research, USA). Bisulfite-modified DNA was amplified using PCR and purified. Products were cloned with pGEM-T Easy Vector (Promega, USA). Multiple plasmid DNA samples were isolated from randomly picked clones using the HTS Plasmid kit (Core Bio System, Korea). The primers used for targeting the CpG sites of TNS4 were 5′-AAAGGGGGATTGTAGAGGGTTTTTT-3′ (sense) and 5′-CCAAACCTACCTTAAAAACAACTTACCC-3′ (antisense). The plasmid DNA samples were sequenced inserts using the ABI 3700 automated capillary gel electrophoresis system (Applied Biosystems, USA). The raw data were deposited in the Korea Bio Data Station (K-BDS; https://kbdsc.kisti.re.kr/) under accession number PRJKA465538.
5-aza-2′-deoxycytidine and trichostatin A (TSA) treatment
GC cells (SNU-216 and MKN1) were treated with 1 µM of 5-aza-2′-deoxycytidine (5-aza-dC; Sigma-Aldrich) and 0.5 µM of TSA (Sigma-Aldrich) every 24 h for 3 days and then harvested. Another set of cells was treated with 0.5 M TSA (Sigma-Aldrich) for 1 day. To characterize the combined effect of 5-aza-dC and TSA, cells were treated with 1 µM 5-aza-dC every 24 h for 3 days and then with 0.5 µM TSA for 1 day. Then, cells were harvested, and qRT-PCR was performed to detect TNS4 mRNA expression as described above.
Statistical analysis
The number of biological replicates (n) is indicated in the figure legends. Data are presented as the mean ± SD. A paired t-test or a Mann–Whitney U-test was used to detect statistical significance between two groups. A P value of <0.05 was considered significant. The chi-squared test was performed to assess statistical significance in the medical records of the 174 patients. The characteristics assessed included gender, Lauren’s classification, tumor progression, tumor stage, and H. pylori infection status. Statistical analysis of correlations between expression and methylation was performed using Pearson’s correlation coefficient (Prism 5.0; GraphPad Software, USA).
RESULTS
TNS4 is upregulated in GC cell lines
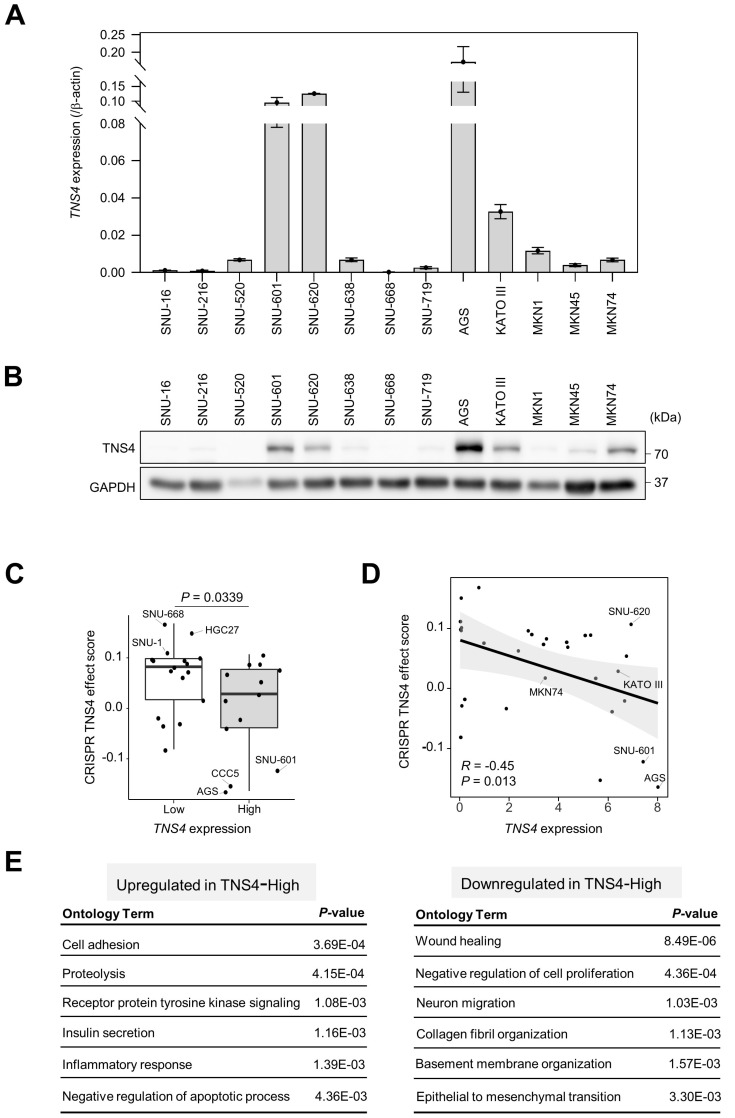
Gastric tumor cells that strongly express CD49f, a subunit of the laminin receptors, retain their sphere-forming and tumor-initiating activities (Fukamachi et al., 2013). We previously found that TNS4 is highly expressed in CD49fhigh cells (data not shown). To examine the expression patterns of TNS4 in GC cell lines, we analyzed the mRNA expression of TNS4 in 13 GC cell lines with qRT-PCR. Most GC cell lines showed TNS4 expression. In particular, SNU-601, SNU-620, AGS, and KATO III showed very high TNS4 mRNA expression levels (Fig. 1A). Western blotting also showed consistent expression of TNS4 protein in these cell lines (Fig. 1B).
Fig. 1. mRNA and protein expression of TNS4 in gastric cancer (GC) cell lines.
(A) Quantitative RT-PCR analysis for TNS4 mRNA in GC cell lines (mean ± SD; n = 3 independent samples). (B) Western blotting for TNS4 expression in GC cell lines. GAPDH was used as the loading control. (C) Distribution of TNS4 dependency of GC cell lines divided into TNS4-high (13 cell lines) and TNS4-low (17 cell lines) groups. Dots depict TNS4 effect scores. A lower effect score indicates that TNS4 was more likely to be essential for a given cell line. Data were downloaded from the DepMap web portal (https://depmap.org/portal/) based on the CRISPR Public 22Q2 dataset. (D) Pearson’s correlation analyses between TNS4 expression and TNS4 effect scores for the GC cell lines from the DepMap data. (E) Gene ontology analysis of differentially expressed genes between TNS4-high (SNU-601, SNU-620, and AGS) and -low (SNU-16, SNU-216, and SNU-668) GC cell lines using CCLE RNA-seq data.
To elucidate TNS4 dependency of the GC cell lines, we analyzed CRISPR Public 22Q2 datasets available through the DepMap web portal (https://depmap.org/portal/). In such an analysis, a lower effect score indicates a higher likelihood that the gene of interest is essential in a given cell line (Tsherniak et al., 2017). We divided the GC cell lines into two groups according to the mRNA expression level of TNS4: TNS4-high (13 cell lines) and TNS4-low (17 cell lines). TNS4-high cells such AGS, CCC5, and SNU-601 showed TNS4 dependence, whereas TNS4-low cells such as SNU-668, HGC27, and SNU-1 did not show TNS4 dependence (Fig. 1C). There was a significant negative correlation between TNS4 expression levels and CRISPR TNS4 effect scores in the 30 GC cell lines (R = –0.45, P = 0.013; Fig. 1D), indicating that TNS4 may play a pivotal role in the survival of TNS4-overexpressing GC cells.
To characterize differences between the TNS4-high and -low groups, we next analyzed differentially expressed genes using CCLE RNA-seq data from GC cell lines (Ghandi et al., 2019). TNS4-high cells showed upregulation of genes involved in cell adhesion and the inflammatory response but downregulation of genes involved in cell migration and collagen fibril organization (Fig. 1E), suggesting that TNS4 may have a key role in integrating signaling between the extracellular microenvironment and cells.
TNS4 is upregulated in primary gastric tumor tissues
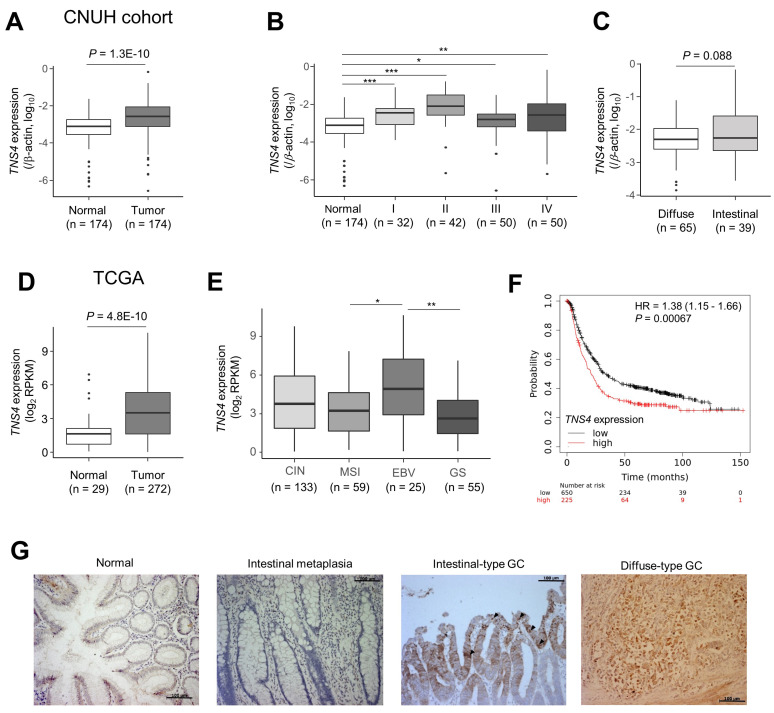
We next examined TNS4 expression in tissue samples from GC patients. We used 174 pairs of tumor and adjacent normal tissue samples provided by Chungnam National University Hospital. qRT-PCR showed that TNS4 expression was significantly upregulated in gastric tumor tissues relative to that in adjacent normal tissues (Fig. 2A). TNS4 expression was upregulated at least twofold in 64.3% (112 of 174) of the gastric tumors as compared with their corresponding normal tissues. Clinicopathological characteristics of TNS4 expression are summarized in Table 1. Increased TNS4 expression was present in early-stage as well as in advanced-stage tumors (Fig. 2B). Significant differences in TNS4 expression were not observed in tumors between intestinal and diffuse-type GC (Fig. 2C).
Fig. 2. TNS4 was upregulated in primary gastric cancer (GC).
(A) TNS4 mRNA expression, relative to that of β-actin, in the paired gastric normal and tumor tissues (n = 174) from the Chungnam National University Hospital (CNUH) cohort. TNS4 expression was measured by qRT-PCR. Statistical significance between normal and tumor tissues was inferred by using the paired t-test. Each box plot shows the median and 25th and 75th percentiles, and the dots represent outliers. (B and C) TNS4 expression status was stratified by (B) tumor stage (I, II, III, and IV) and (C) Lauren classification. *P < 0.05, **P < 0.01, ***P < 0.001 (Mann–Whitney U-test). (D and E) Expression of TNS4 analyzed using TCGA RNA sequencing data. Comparisons (D) between normal and GC tumor tissue samples and (E) among molecular subtypes of GC were carried out (Mann–Whitney U-test). *P < 0.05, **P < 0.01. (F) An online database (http://kmplot.com/analysis/) was used to determine the relevance of TNS4 mRNA expression in tumor tissue to the overall survival of the individual. Kaplan–Meier survival plots in which the number-at-risk is indicated below the main plot. Hazard ratio (HR) and 95% confidence intervals, as well as the log rank P were calculated and displayed on the webpage. (G) Representative immunohistochemistry for TNS4 in intestinal- and diffuse-type GC. Paraffin-embedded sections of normal mucosa, intestinal metaplasia, and gastric tumor samples from individuals with intestinal-type or diffuse-type GC were examined for TNS4 expression. Black arrowheads indicate representative TNS4 signals in well-differentiated tumor tissue. Scale bars = 100 µm (also see Table 2).
Table 1.
Clinicopathological characteristics of TNS4 expression in gastric cancer patients
| Clinicopathological parameter | Gastric tumors with increased relative TNS4 expression | P value | |
|---|---|---|---|
|
| |||
| >2-fold increase (n = 112) | ≤2-fold increase (n = 62) | ||
| Mean age (y) | 60 ± 13 | 57 ± 14 | 0.1075 |
| Sex | |||
| Male | 79 | 35 | 0.0612 |
| Female | 33 | 27 | |
| Lauren’s classification | |||
| Intestinal | 39 | 11 | 0.0238 |
| Diffuse | 65 | 44 | |
| Mixed | 8 | 6 | |
| No information | 0 | 1 | |
| Tumor progression | |||
| Early gastric cancer | 15 | 2 | 0.0305 |
| Advanced gastric cancer | 97 | 60 | |
| Stage | |||
| I | 23 | 9 | 0.0002 |
| II | 36 | 6 | |
| III | 21 | 29 | |
| IV | 32 | 18 | |
| Helicobacter pylori infection | |||
| Positive | 63 | 40 | 0.5862 |
| Negative | 29 | 15 | |
| No information | 20 | 7 | |
Values are presented as mean ± SD or number only.
According to TCGA, TNS4 expression was higher in stomach tumor tissue than in normal gastric tissue (Fig. 2D). Among the four molecular subtypes of GC, the EBV-positive subtype showed higher expression of TNS4 than MSI and GS subtypes. There was no significant difference between the EBV and CIN subtypes (Fig. 2E). According to an analysis with KM-plotter (https://kmplot.com/) of transcriptomic data from 875 GC patients (Szasz et al., 2016), the overall survival of GC patients with high TNS4 expression was significantly lower than that of patients with low TNS4 expression (Fig. 2F).
To examine whether TNS4 is already upregulated in gastric intestinal metaplasia, an intermediate lesion in the development of intestinal-type GC, we performed immunohistochemistry of paraffin-embedded sections from tumor and normal gastric tissue samples from five patients with intestinal-type GC and one with diffuse-type GC (Table 2). Despite low signals only in superficial mucosa, cytoplasmic TNS4 signals were barely detected in normal gastric tissues and intestinal metaplasia. In contrast, we noted strong staining for TNS4 in gastric tumors from both intestinal- and diffuse-type GCs (Fig. 2G, Table 2). In particular, in more-differentiated tissues, stronger signals were detected (Fig. 2G). Taken together, these data suggest that increased expression of TNS4 may have a key role in GC initiation and progression.
Table 2.
Immunohistochemistry assay for TNS4 in gastric tumors and normal tissues
| Patient No. | Lauren’s classification | Normal mucosa | Intestinal metaplasia | Gastric cancer |
|---|---|---|---|---|
| S16-7995-A5 | Intestinal type | ‐ | ‐ | ++ |
| S16-8389-A4 | Intestinal type | ‐ | ‐ | + |
| S16-10969-A7 | Intestinal type | ‐ | ‐ | + |
| S16-12683-A3 | Intestinal type | ‐ | ‐ | ++ |
| S16-12705-2 | Intestinal type | ‐ | ‐ | ++ |
| S16-11744-A2 | Diffuse type | ‐ | + |
++, score for 50%-95% of positive stained tumor cells; +, score for 10%-49% of tumor cells positive; ‐, score for less than 10% of cells or no visible staining.
Knockdown of TNS4 suppresses GC cell proliferation and migration
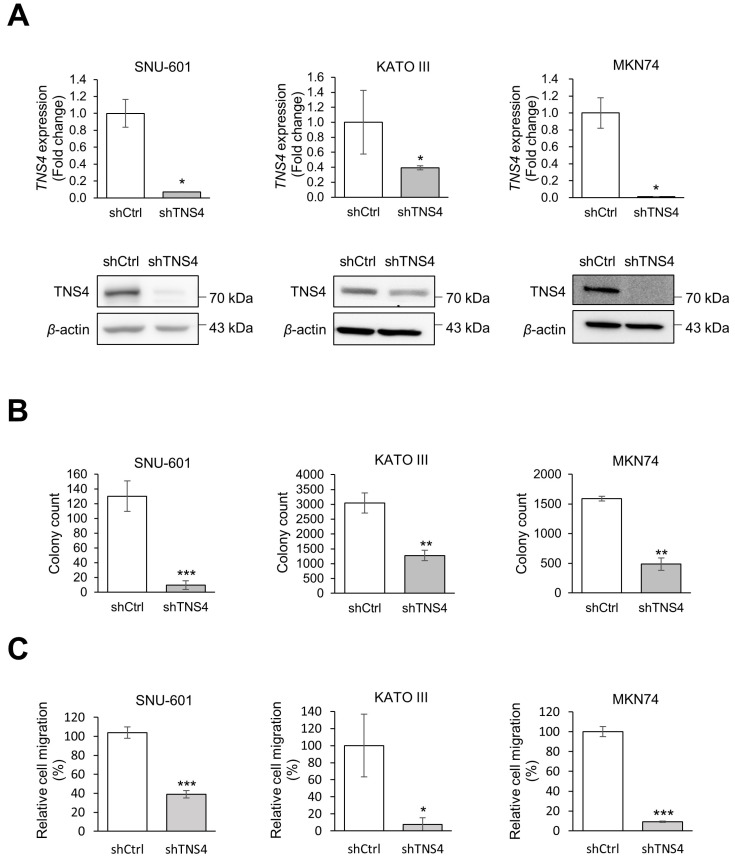
To examine the functions of TNS4 in GC progression, we depleted TNS4 in three GC cell lines with high to moderate TNS4 expression levels, SNU-601, KATO III, and MKN74 cells, using TNS4 shRNA (Fig. 3A). Although TNS4 expression was high in the SNU-620 cell line, these cells grow only in suspension culture, which is not appropriate for further study such as with colony formation assays. In addition, although MKN1 had a high level of TNS4 mRNA, it expressed TNS4 protein at a low level (Figs. 1A and 1B). As aggressive cancer requires rapid growth and metastasis (Hanahan and Weinberg, 2000), we performed cell proliferation and migration assays. TNS4 knockdown was associated with a significant decrease in colony formation (Fig. 3B) and cell migration (Fig. 3C) in all three cell lines, suggesting that TNS4 may promote cell proliferation and migration during GC progression.
Fig. 3. Knockdown of TNS4 reduced colony formation and cell migration in gastric cancer (GC) cells.
(A) Establishment of TNS4 knockdown cells. GC cell lines (SNU-601, KATO III, and MKN74) were transduced with TNS4-specific shRNA or an empty shRNA vector (shCtrl). TNS4 mRNA and protein levels were measured by qRT-PCR (top) and western blotting (bottom), respectively. β-Actin was used as the loading control. (B) Colony formation assay of control and TNS4 knockdown GC cells. (C) Cell migration assay of control and TNS4 knockdown GC cells at 24 h after seeding. Migration relative to that of the control cells is shown. In (B) and (C), n = 3 independent experiments; mean ± SD; *P < 0.05, **P < 0.01, ***P < 0.001 (Mann–Whitney U-test).
Ectopic expression of TNS4 enhances GC cell proliferation and migration
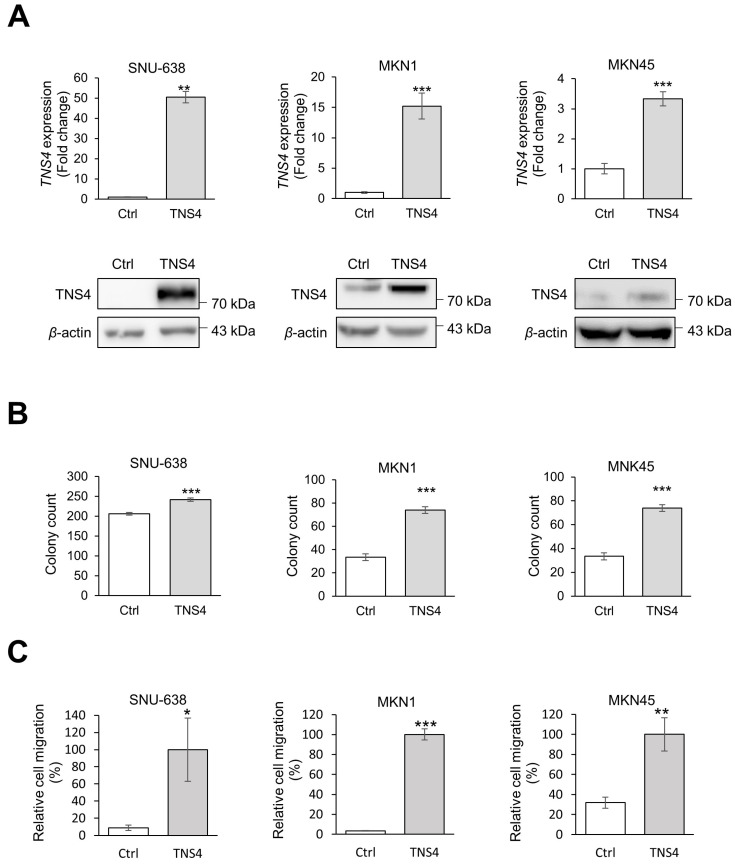
We next examined whether ectopic expression of TNS4 promotes GC cell proliferation and migration by using stable cell lines that ectopically expressed TNS4. As SNU-638, MKN1, and MKN45 cells expressed endogenous TNS4 at low levels, these cell lines were transfected with lentiviral pCDH-TNS4 vectors for stable ectopic expression of TNS4. Increased expression of TNS4 was confirmed with mRNA and protein levels (Fig. 4A). Overexpression of TNS4 significantly increased colony formation (Fig. 4B) and cell migration (Fig. 4C), suggesting that TNS4 promotes the oncogenicity of GC cells. Taken together, these results suggest that TNS4 may be involved in GC cell proliferation and metastasis.
Fig. 4. Ectopic expression TNS4 increased colony formation and cell migration in gastric cancer (GC) cells.
(A) Establishment of cells ectopically expressing TNS4. GC cells (SNU-638, MKN1, and MKN45) were transduced with lentivirus containing the TNS4 expression vector. TNS4 mRNA and protein levels were measured by qRT-PCR (top) and western blotting (bottom). β-Actin was used as the loading control. (B) Colony formation assay for control GC cells and GC cells that expressed ectopic TNS4. (C) Migration assay for control GC cells and GC cells that expressed ectopic TNS4 at 24 h after seeding. In (B) and (C), n = 3 independent experiments; mean ± SD; *P <0.05, **P <0.01, ***P < 0.001 (Mann–Whitney U-test). Ctrl, control.
TNS4 promoter is hypomethylated in GC cell lines and tumor tissues
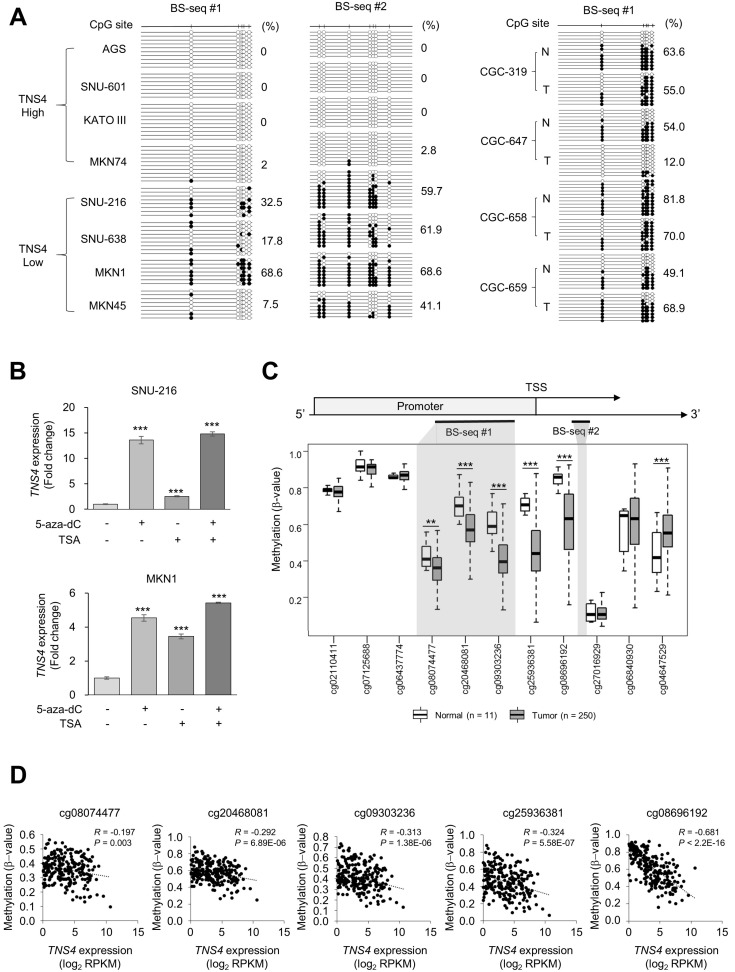
Epigenetic modification such as DNA demethylation can cause aberrant activation of oncogenes and promotes cancer development (Das and Singal, 2004). To examine TNS4 promoter methylation differences between cells that express high or low levels of TNS4, we performed bisulfite sequencing of the TNS4 promoter. Intriguingly, the promoter region in GC cells with high TNS4 expression was mostly unmethylated, whereas GC cells with low TNS4 expression showed heavy methylation in this same region (Fig. 5A). In addition, these CpG sites were hypermethylated in the normal tissue samples and moderately methylated in the tumor samples from four paired tissue samples (Fig. 5A). Consistently, treatment with 5-aza-2′-deoxycytidine (5-aza-dC), a DNA methylation inhibitor (Jones and Taylor, 1980), and/or TSA, a histone deacetylase inhibitor (Yoshida et al., 1990), enhanced TNS4 expression in cell lines that expressed low amounts of TNS4 before treatment (Fig. 5B), suggesting that TNS4 expression is regulated via epigenetic mechanisms such as DNA methylation and histone modification.
Fig. 5. Methylation analysis of the TNS4 promoter.
(A) Bisulfite sequencing (BS-seq) of gastric cancer (GC) cell lines and paired normal and tumor tissue samples. Methylation status of the TNS4 promoter in GC cell lines with high or low expression of TNS4 (Fig. 1). BS-seq regions are also indicated in Fig. 5C. These regions include the infinium human methylation 450 K BeadChip CpG probes cg08074477, cg20468081, and cg09303236. Black circles, methylated; white circles, unmethylated. Total methylation percentages are indicated. (B) TNS4 mRNA levels in GC cells treated with 5-aza-dC and/or trichostatin A (TSA). n = 8 independent experiments; mean ± SD; ***P < 0.001 (Mann–Whitney U-test). (C) The methylation level of the TNS4 promoter in normal gastric tissue (n = 11) and GC tumor tissue (n = 250) from TCGA data using Infinum Human BeadChip 450K. BS-seq regions in Fig. 5A are indicated with gray shading. TSS, transcription start site. Each box plot shows the median and 25th and 75th percentiles. Statistical significance between normal and tumor tissues was inferred by using the Mann–Whitney U-test; **P < 0.01, ***P < 0.001. (D) Pearson’s correlation analyses between TNS4 CpG methylation and TNS4 expression levels for the indicated probes from TCGA data.
To investigate whether DNA methylation affects TNS4 expression in GC patients, we analysed DNA methylation data from TCGA. Most CpG probes within the promoter region of TNS4 were hypomethylated in GC tumor samples (n = 250) as compared with normal tissue samples (n = 11) (Fig. 5C). Moreover, we found a significant correlation between CpG methylation and TNS4 expression (Fig. 5D). These data suggest that increased expression of TNS4 in GC may be at least partially due to DNA demethylation.
DISCUSSION
Upregulation of TNS4 expression has been reported in many cancers such as colon (Raposo et al., 2020b), lung (Misono et al., 2019), breast (Albasri et al., 2011), skin (Sjoestroem et al., 2013), and pancreatic (Al-Ghamdi et al., 2013) cancer and GC (Sakashita et al., 2008), but transcriptional activation mechanisms of TNS4 during tumorigenesis are not well understood. Here we describe the hypomethylation of the TNS4 promoter in GC as a possible mechanism for its increased expression.
TNS4 is located in focal adhesions, which regulate cell architecture during cell movement and are involved in cell signaling such as epidermal growth factor receptor (EGFR) signaling. EGF downregulates TNS3 expression, whereas it upregulates TNS4 expression. TNS4 displaces TNS3 from the cytoplasmic tail of integrin β1, triggering actin fibre disassembly and cell migration (Katz et al., 2007). TNS4 expression is also correlated with high EGFR and HER2 levels as well as with metastasis to lymph nodes in invasive breast cancer (Katz et al., 2007). TNS4 reduces ligand-induced EGFR degradation by binding to the E3 ubiquitin ligase c-Cbl and decreasing the ubiquitination of EGFR (Hong et al., 2013). TNS4 also increases MET protein stability through a direct interaction with phosphorylated MET via the SH2 domain and positively regulates cell survival, proliferation, and migration (Muharram et al., 2014).
TNS4 has been suggested to play a pivotal role in proliferation and metastasis of GC. TNS4 promotes the epithelial-mesenchymal transition via AKT/GSK-3β signaling in GC cells (Qi et al., 2020). Clinical analysis showed that GC patients with high TNS4 expression exhibited significantly reduced 5-year overall survival relative to those individuals with lower expression (Sawazaki et al., 2017). Consistent with previous studies (Sakashita et al., 2008), we found that TNS4 was upregulated in GC tissue samples.
Epigenetic modification comprising DNA methylation, histone modifications, and noncoding RNAs plays a critical role in cancer development (Kim et al., 2020; Sharma et al., 2010). CpG island hypomethylation promotes tumorigenesis via aberrantly activating oncogenes (Das and Singal, 2004). Using TCGA data, we found that the promoter region of TNS4 was hypomethylated in GC tumor tissues, and there was a significant negative correlation between TNS4 expression and DNA methylation in these tissues. We also found that treatment with 5-aza-dC enhanced TNS4 expression in GC cell lines. However, 5-aza-dC is incorporated into DNA, inhibits DNMT1 activity, and causes proteolysis of DNMT1 that results in genome-wide DNA demethylation. A recent CRISPR-based epigenome editing technology may overcome this technical limitation and could demonstrate a direct effect of DNA methylation on TNS4 expression (Nunez et al., 2021).
TNS4 is upregulated by Wnt signaling during intestinal tumorigenesis (Raposo et al., 2020a). SWI/SNF related matrix-associated actin-dependent regulator of chromatin subfamily A member 4 (SMARCA4) acts as a positive regulator of Wnt signaling in the small intestinal epithelium (Holik et al., 2014). A recent study showed that PRMT1-mediated modification of histone H4 on arginine 3 (H4R3me2a) recruits SMARCA4 to activate TNS4 and EGFR expression in colorectal cancer (Yao et al., 2021). Intriguingly, activation of Wnt signaling is involved in the development and progression of GC (Chiurillo, 2015). Wnt signaling is associated with the development and progression of gastric adenoma, which is considered a premalignant lesion of gastric adenocarcinoma, and this process appears to be accelerated by H. pylori infection (Wang et al., 2012). It would be interesting to elucidate the relationship between Wnt signaling and epigenetic activation of TNS4 during GC development and progression.
In summary, TNS4 was upregulated during GC progression. Knockdown of TNS4 suppressed GC cell proliferation and migration, whereas overexpression of TNS4 enhanced GC cell proliferation and migration. TNS4 expression was correlated with promoter hypomethylation in GC cell lines and tissues. Within this molecular mechanism, it is not clear how GC risk factors such as H. pylori infection may lead to epigenetic changes and activation of TNS4 transcription. In addition, the feasibility of activation of TNS4 by EGFR or Wnt signaling as a technique for controlling GC development and progression will be further validated in the future.
ACKNOWLEDGMENTS
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (2019R1A2C1087104), the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare, Republic of Korea (HI21C0538), National Research Council of Science & Technology (NST) Aging Convergence Research Center (CRC22011-400), and the Korea Research Institute of Bioscience & Biotechnology (KRIBB) Research Initiative Program (KGM5192221).
Footnotes
AUTHOR CONTRIBUTIONS
H.H. and M.K. generated the hypothesis and designed the study. H.H., H.-J.K., H.A.S., Y.-J.S., K.H., and H.-J.J. performed the experiments. S.-I.L. and K.-S.S. performed the immunohistochemistry and contributed to its analysis. H.G. contributed to qRT-PCR data analyses. H.H., J.-H.K., M.-J.K., H.L., E.-S.K., S.-Y.K., Y.S.K., and M.K. contributed to the data analyses. H.H. and M.K. wrote the manuscript with contributions from all authors. M.K. supervised the research. M.K. is the guarantor of this work. All authors have read and approved the final manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Al-Ghamdi S., Cachat J., Albasri A., Ahmed M., Jackson D., Zaitoun A., Guppy N., Otto W.R., Alison M.R., Kindle K.B., et al. C-terminal tensin-like gene functions as an oncogene and promotes cell motility in pancreatic cancer. Pancreas. 2013;42:135–140. doi: 10.1097/MPA.0b013e3182557ceb. [DOI] [PubMed] [Google Scholar]
- Albasri A., Aleskandarany M., Benhasouna A., Powe D.G., Ellis I.O., Ilyas M., Green A.R. CTEN (C-terminal tensin-like), a novel oncogene overexpressed in invasive breast carcinoma of poor prognosis. Breast Cancer Res. Treat. 2011;126:47–54. doi: 10.1007/s10549-010-0890-3. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaravalli A.M., Klersy C., Vanoli A., Ferretti A., Capella C., Solcia E. Histotype-based prognostic classification of gastric cancer. World J. Gastroenterol. 2012;18:896–904. doi: 10.3748/wjg.v18.i9.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiurillo M.A. Role of the Wnt/β-catenin pathway in gastric cancer: an in-depth literature review. World J. Exp. Med. 2015;5:84–102. doi: 10.5493/wjem.v5.i2.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crew K.D., Neugut A.I. Epidemiology of gastric cancer. World J. Gastroenterol. 2006;12:354–362. doi: 10.3748/wjg.v12.i3.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das P.M., Singal R. DNA methylation and cancer. J. Clin. Oncol. 2004;22:4632–4642. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- Ebrahimi V., Soleimanian A., Ebrahimi T., Azargun R., Yazdani P., Eyvazi S., Tarhriz V. Epigenetic modifications in gastric cancer: focus on DNA methylation. Gene. 2020;742:144577. doi: 10.1016/j.gene.2020.144577. [DOI] [PubMed] [Google Scholar]
- Fukamachi H., Seol H.S., Shimada S., Funasaka C., Baba K., Kim J.H., Park Y.S., Kim M.J., Kato K., Inokuchi M., et al. CD49f (high) cells retain sphere-forming and tumor-initiating activities in human gastric tumors. PLoS One. 2013;8:e72438. doi: 10.1371/journal.pone.0072438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R., 3rd, Barretina J., 3rd, Gelfand E.T., 3rd, Bielski C.M., 3rd, Li H., 3rd, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019;569:503–508. doi: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Holik A.Z., Young M., Krzystyniak J., Williams G.T., Metzger D., Shorning B.Y., Clarke A.R. Brg1 loss attenuates aberrant wnt-signalling and prevents wnt-dependent tumourigenesis in the murine small intestine. PLoS Genet. 2014;10:e1004453. doi: 10.1371/journal.pgen.1004453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.Y., Shih Y.P., Li T., Carraway K.L., 3rd, Lo S.H., 3rd CTEN prolongs signaling by EGFR through reducing its ligand-induced degradation. Cancer Res. 2013;73:5266–5276. doi: 10.1158/0008-5472.CAN-12-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P.A., Taylor S.M. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- Kaneda A., Matsusaka K., Aburatani H., Fukayama M. Epstein-Barr virus infection as an epigenetic driver of tumorigenesis. Cancer Res. 2012;72:3445–3450. doi: 10.1158/0008-5472.CAN-11-3919. [DOI] [PubMed] [Google Scholar]
- Katz M., Amit I., Citri A., Shay T., Carvalho S., Lavi S., Milanezi F., Lyass L., Amariglio N., Jacob-Hirsch J., et al. A reciprocal tensin-3-cten switch mediates EGF-driven mammary cell migration. Nat. Cell Biol. 2007;9:961–969. doi: 10.1038/ncb1622. [DOI] [PubMed] [Google Scholar]
- Kim D., Lee Y.S., Kim D.H., Bae S.C. Lung cancer staging and associated genetic and epigenetic events. Mol. Cells. 2020;43:1–9. doi: 10.14348/molcells.2020.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Costello J. DNA methylation: an epigenetic mark of cellular memory. Exp. Mol. Med. 2017;49:e322. doi: 10.1038/emm.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Jang H.R., Haam K., Kang T.W., Kim J.H., Kim S.Y., Noh S.M., Song K.S., Cho J.S., Jeong H.Y., et al. Frequent silencing of popeye domain-containing genes, BVES and POPDC3, is associated with promoter hypermethylation in gastric cancer. Carcinogenesis. 2010;31:1685–1693. doi: 10.1093/carcin/bgq144. [DOI] [PubMed] [Google Scholar]
- Kim M., Jang H.R., Kim J.H., Noh S.M., Song K.S., Cho J.S., Jeong H.Y., Norman J.C., Caswell P.T., Kang G.H., et al. Epigenetic inactivation of protein kinase D1 in gastric cancer and its role in gastric cancer cell migration and invasion. Carcinogenesis. 2008a;29:629–637. doi: 10.1093/carcin/bgm291. [DOI] [PubMed] [Google Scholar]
- Kim M., Kim J.H., Jang H.R., Kim H.M., Lee C.W., Noh S.M., Song K.S., Cho J.S., Jeong H.Y., Hahn Y., et al. LRRC3B, encoding a leucine-rich repeat-containing protein, is a putative tumor suppressor gene in gastric cancer. Cancer Res. 2008b;68:7147–7155. doi: 10.1158/0008-5472.CAN-08-0667. [DOI] [PubMed] [Google Scholar]
- Kim M., Lee K.T., Jang H.R., Kim J.H., Noh S.M., Song K.S., Cho J.S., Jeong H.Y., Kim S.Y., Yoo H.S., et al. Epigenetic down-regulation and suppressive role of DCBLD2 in gastric cancer cell proliferation and invasion. Mol. Cancer Res. 2008c;6:222–230. doi: 10.1158/1541-7786.MCR-07-0142. [DOI] [PubMed] [Google Scholar]
- Kim S.K., Jang H.R., Kim J.H., Noh S.M., Song K.S., Kim M.R., Kim S.Y., Yeom Y.I., Kim N.S., Yoo H.S., et al. The epigenetic silencing of LIMS2 in gastric cancer and its inhibitory effect on cell migration. Biochem. Biophys. Res. Commun. 2006;349:1032–1040. doi: 10.1016/j.bbrc.2006.08.128. [DOI] [PubMed] [Google Scholar]
- Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol. Microbiol. Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- Liao Y.C., Lo S.H. Tensins - emerging insights into their domain functions, biological roles and disease relevance. J. Cell Sci. 2021;134:jcs254029. doi: 10.1242/jcs.254029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim B., Kim J.H., Kim M., Kim S.Y. Genomic and epigenomic heterogeneity in molecular subtypes of gastric cancer. World J. Gastroenterol. 2016;22:1190–1201. doi: 10.3748/wjg.v22.i3.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Gao J., Zhang Y., Zhao T., Cai H., Zhang T. CTEN induces epithelial-mesenchymal transition (EMT) and metastasis in non small cell lung cancer cells. PLoS One. 2018;13:e0198823. doi: 10.1371/journal.pone.0198823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda M., Moro H., Ushijima T. Mechanisms for the induction of gastric cancer by Helicobacter pylori infection: aberrant DNA methylation pathway. Gastric Cancer. 2017;20((Suppl 1)) doi: 10.1007/s10120-016-0650-0. [DOI] [PubMed] [Google Scholar]
- Misono S., Seki N., Mizuno K., Yamada Y., Uchida A., Sanada H., Moriya S., Kikkawa N., Kumamoto T., Suetsugu T., et al. Molecular pathogenesis of gene regulation by the miR-150 duplex: miR-150-3p regulates TNS4 in lung adenocarcinoma. Cancers (Basel) 2019;11:601. doi: 10.3390/cancers11050601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muharram G., Sahgal P., Korpela T., De Franceschi N., Kaukonen R., Clark K., Tulasne D., Carpen O., Ivaska J. Tensin-4-dependent MET stabilization is essential for survival and proliferation in carcinoma cells. Dev. Cell. 2014;29:629–630. doi: 10.1016/j.devcel.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez J.K., Chen J., Pommier G.C., Cogan J.Z., Replogle J.M., Adriaens C., Ramadoss G.N., Shi Q., Hung K.L., Samelson A.J., et al. Genome-wide programmable transcriptional memory by CRISPR-based epigenome editing. Cell. 2021;184:2503–2519.e17. doi: 10.1016/j.cell.2021.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Sun L., Wan J., Xu R., He S., Zhu X. Tensin4 promotes invasion and migration of gastric cancer cells via regulating AKT/GSK-3β/snail signaling pathway. Pathol. Res. Pract. 2020;216:153001. doi: 10.1016/j.prp.2020.153001. [DOI] [PubMed] [Google Scholar]
- Raposo T.P., Alfahed A., Nateri A.S., Ilyas M. Tensin4 (TNS4) is upregulated by Wnt signalling in adenomas in multiple intestinal neoplasia (Min) mice. Int. J. Exp. Pathol. 2020a;101:80–86. doi: 10.1111/iep.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo T.P., Susanti S., Ilyas M. Investigating TNS4 in the colorectal tumor microenvironment using 3D spheroid models of invasion. Adv. Biosyst. 2020b;4:e2000031. doi: 10.1002/adbi.202000031. [DOI] [PubMed] [Google Scholar]
- Sakashita K., Mimori K., Tanaka F., Kamohara Y., Inoue H., Sawada T., Hirakawa K., Mori M. Prognostic relevance of Tensin4 expression in human gastric cancer. Ann. Surg. Oncol. 2008;15:2606–2613. doi: 10.1245/s10434-008-9989-8. [DOI] [PubMed] [Google Scholar]
- Sawazaki S., Oshima T., Sakamaki K., Aoyama T., Sato T., Shiozawa M., Yoshikawa T., Rino Y., Imada T., Masuda M. Clinical significance of tensin 4 gene expression in patients with gastric cancer. In Vivo. 2017;31:1065–1071. doi: 10.21873/invivo.11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo E.H., Kim H.J., Kim J.H., Lim B., Park J.L., Kim S.Y., Lee S.I., Jeong H.Y., Song K.S., Kim Y.S. ONECUT2 upregulation is associated with CpG hypomethylation at promoter-proximal DNA in gastric cancer and triggers ACSL5. Int. J. Cancer. 2020;146:3354–3368. doi: 10.1002/ijc.32946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Kelly T.K., Jones P.A. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoestroem C., Khosravi S., Zhang G., Martinka M., Li G. C-terminal tensin-like protein is a novel prognostic marker for primary melanoma patients. PLoS One. 2013;8:e80492. doi: 10.1371/journal.pone.0080492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szasz A.M., Lanczky A., Nagy A., Forster S., Hark K., Green J.E., Boussioutas A., Busuttil R., Szabo A., Gyorffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsherniak A., Vazquez F., Montgomery P.G., Weir B.A., Kryukov G., Cowley G.S., Gill S., Harrington W.F., Pantel S., Krill-Burger J.M., et al. Defining a cancer dependency map. Cell. 2017;170:564–576.e16. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.K., Liu J., Liu C., Wang F.Y., Chen C.Y., Zhang X.H. Hypermethylation of adenomatous polyposis coli gene promoter is associated with novel Wnt signaling pathway in gastric adenomas. J. Gastroenterol. Hepatol. 2012;27:1629–1634. doi: 10.1111/j.1440-1746.2012.07219.x. [DOI] [PubMed] [Google Scholar]
- Wu W.M., Liao Y.C. Downregulation of C-terminal tensin-like protein (CTEN) suppresses prostate cell proliferation and contributes to acinar morphogenesis. Int. J. Mol. Sci. 2018;19:3190. doi: 10.3390/ijms19103190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao B., Gui T., Zeng X., Deng Y., Wang Z., Wang Y., Yang D., Li Q., Xu P., Hu R., et al. PRMT1-mediated H4R3me2a recruits SMARCA4 to promote colorectal cancer progression by enhancing EGFR signaling. Genome Med. 2021;13:58. doi: 10.1186/s13073-021-00871-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon B.H., Kim M., Kim M.H., Kim H.J., Kim J.H., Kim J.H., Kim J., Kim Y.S., Lee D., Kang S.J., et al. Dynamic transcriptome, DNA methylome, and DNA hydroxymethylome networks during T-cell lineage commitment. Mol. Cells. 2018;41:953–963. doi: 10.1158/1538-7445.AM2018-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M., Kijima M., Akita M., Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 1990;265:17174–17179. doi: 10.1016/S0021-9258(17)44885-X. [DOI] [PubMed] [Google Scholar]
- Yuasa Y. Control of gut differentiation and intestinal-type gastric carcinogenesis. Nat. Rev. Cancer. 2003;3:592–600. doi: 10.1038/nrc1141. [DOI] [PubMed] [Google Scholar]