Abstract
Extensive studies have focused on the misregulation of individual miRNAs in cancer. More recently, mutations in the miRNA biogenesis and processing machinery have been implicated in several malignancies. Such mutations can lead to global miRNA misregulation, which may promote many of the well-known hallmarks of cancer. Interestingly, recent evidence also suggests that oncogenic Kristen rat sarcoma viral oncogene homolog (KRAS) mutations act in part by modulating the activity of members of the miRNA regulatory pathway. Here, we highlight the vital role mutations in the miRNA core machinery play in promoting malignant transformation. Furthermore, we discuss how mutant KRAS can simultaneously impact multiple steps of miRNA processing and function to promote tumorigenesis. Although the ability of KRAS to hijack the miRNA regulatory pathway adds a layer of complexity to its oncogenic nature, it also provides a potential therapeutic avenue that has yet to be exploited in the clinic. Moreover, concurrent targeting of mutant KRAS and members of the miRNA core machinery represents a potential strategy for treating cancer.
Introduction
Posttranscriptional regulation through miRNAs is a highly conserved and ubiquitous process known to be misregulated across all cancers. More recently, miRNA core machinery mutations have been implicated in several malignancies (1–4). Interestingly, the oncogenic effects of global miRNA misregulation overlap with the functional consequences of mutations in the Kristen rat sarcoma viral oncogene homolog (KRAS). KRAS mutations drive tumor initiation in several types of cancer, including pancreatic ductal adenocarcinoma (PDAC), non–small cell lung cancer (NSCLC), and colorectal cancer (5–8). In fact, lung, colorectal, and pancreatic cancer now represent the top three causes of cancer-related deaths in the United States. Thus, the need for identifying novel ways to treat KRAS-driven cancers is greater than ever. KRAS is the most mutated oncogene among all cancers, and activating KRAS mutations lead to the constitutive activation of downstream signaling cascades, which induce various oncogenic characteristics such as sustained proliferation, sustained self-renewal, and increased vascularization (6, 9–12). Although much is known about how miRNAs influence KRAS expression and function, less is known about how KRAS modulates the global miRNA landscape (13). In recent years, oncogenic KRAS has been found to influence aspects of miRNA biogenesis and effector function. Similar to consequences seen with mutations in miRNA biosynthesis machinery, mutations in KRAS can analogously lead to global miRNA misregulation. Here, we review what is currently known about how mutations in the miRNA biosynthesis machinery contribute to cancer formation and relate this to how oncogenic KRAS modifies the miRNA regulatory pathway to promote tumorigenesis. Understanding this additional layer to the complex genetic misregulation induced by oncogenic KRAS may elucidate novel targetable pathways for therapeutic advancement. Moreover, with the development of enhanced small-molecule inhibitors of KRAS on the horizon, concurrent targeting of mutant KRAS and individual members of the miRNA biogenesis and processing machinery thus represents the potential for additive or synergistic therapies to treat several types of cancer (14).
Overview of miRNA Biogenesis
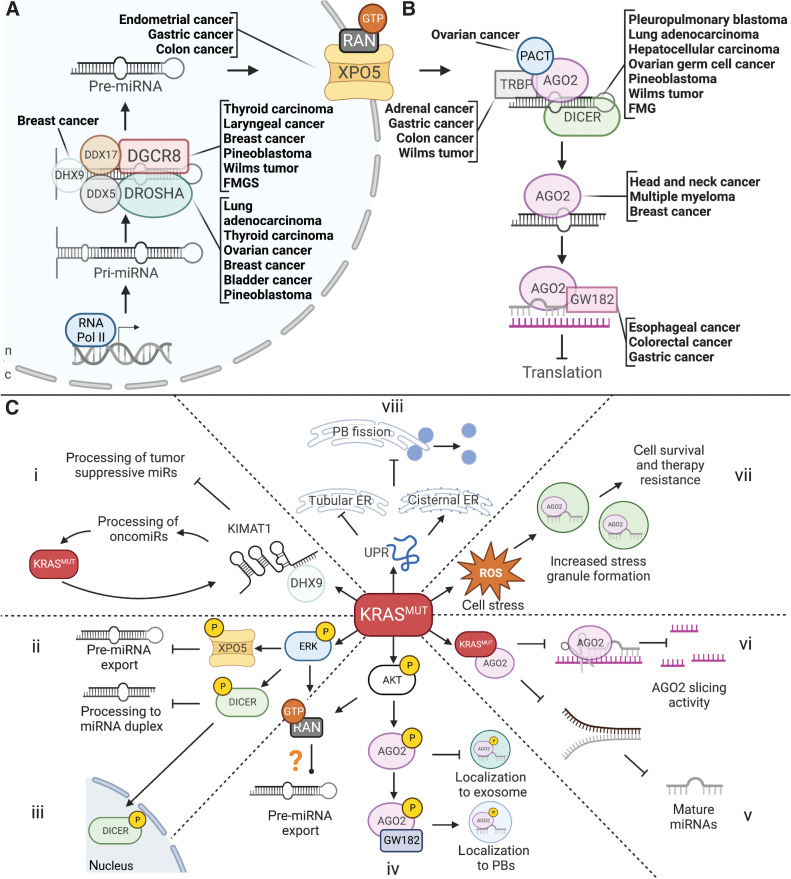
miRNAs are small, noncoding RNAs that range between 20 and 25 nucleotides in size and are involved in posttranscriptional gene regulation (15). miRNAs typically exert their function by binding to their target mRNA through the interaction of the seed region of the miRNA with the 3′ untranslated region (UTR) of the target transcript. The interaction of miRNAs with their target then regulates gene expression by promoting translational inhibition or mRNA decay. miRNA biosynthesis begins in the nucleus (Fig. 1A), where canonically, pri-miRNAs are transcribed by RNA polymerase II and processed into pre-miRNAs by the nuclear microprocessor complex, which comprises DROSHA and DGCR8. DCGR8 is an RNA-binding protein responsible for binding the pri-miRNA in complex with other members of the microprocessing machinery such as DEAD-box helicase 5 (DDX5), DexH-box helicase 9 (DHX9), and DEAD-Box helicase 17 (DDX17; refs. 15–17). DROSHA is another nuclear miRNA microprocessing complex member and a binding partner of DGCR8. After transcription by RNA pol II, DROSHA cleaves off the stem-loop of the pri-miRNA bound by DGCR8, generating a pre-miRNA.
Figure 1.
The miRNA regulatory pathway, associated cancers, and the effect of KRASMUT on miRNA processing. A, Nuclear miRNA processing and cancers associated with mutations in the microprocessor. B, Cytoplasmic miRNA processing and cancers associated with mutations in cytoplasmic factors. C, KRASMUT-induced effects on miRNA biogenesis, processing, and function. i, Increased oncomirs and decreased tumor suppressive miRNAs via KIMAT1 and DHX9. ii, Decreased miRNA export. iii, Decreased cytoplasmic DICER processing function. iv, Decreased exosomal miRNA secretion and increased PB localization of AGO2. v, Inhibition of AGO2 miRNA processing function. vi, Inhibition of AGO2 slicing activity. vii, Upregulation of SG formation and increased cell survival and therapy resistance. viii, Upregulation of UPR and shift from tubular to cisternal ER, causing decreased PB fission events and decreased PB numbers.
After pri-miRNAs are processed into pre-miRNAs, they exit the nucleus via Exportin-5 and GTP-bound Ras-related nuclear protein (RAN-GTP; ref. 18). They are then processed into mature duplex miRNAs by the endoribonuclease DICER (Fig. 1B). Next, argonaute (AGO) proteins bind to one strand of the mature miRNA duplex and GW182, a scaffolding protein that promotes the recruitment of other RNA-binding proteins. In humans, the AGO family of proteins consists of AGO1–4 and is involved in miRNA-mediated mRNA silencing (19, 20). Although all four members of AGO are equally loaded with miRNAs, AGO1, and AGO2 are generally expressed at higher levels than AGO3 or AGO4 (21–23). The AGO-bound miRNA and GW182 constitute the core miRNA–induced silencing complex (miRISC) that is guided to the target mRNA for silencing. Although miRNA-mediated regulation typically results in translational inhibition, certain miRNA families can induce AGO-slicing activity, resulting in miRNA degradation.
Oncogenic mutations in the nuclear microprocessing complex and export machinery
Mutations in the factors involved in miRNA biosynthesis and processing have been implicated in many cancer types (Table 1; Fig. 1A and B). Mutations in DGCR8 have been correlated with increased cancer incidence and have been shown to act as driver mutations in Wilms tumors, certain thyroid cancers, pineoblastoma, and familial tumor susceptibility syndromes (24–29). One study identified DGCR8 missense, frameshift, and nonsense mutations in a significant percentage of Wilms tumors, all leading to nonfunctional DGCR8 peptides (24). One of the mutational hotspots identified in this study (E518K, which resides within the RNA-binding domain) has similarly been identified in follicular thyroid tumors and cases of familial multinodal goiter with schwannomatosis (26, 28–30). These studies suggest that a decrease in DGCR8 function leads to reduced miRNA biosynthesis, thus promoting the misregulation of their target transcripts.
Table 1.
miRNA machinery mutations and the effects of oncogenic KRAS on miRNA regulation in cancer.
| Gene | Function | Mutation | Cancer type | Ref. | |
|---|---|---|---|---|---|
| DGCR8 | Nuclear microprocessing complex member. Binds pri-miRNAs to facilitate processing by DROSHA | E518K | Wilms tumor thyroid carcinoma, FMGS | 30 | |
| rs417309 G/A | Laryngeal cancer, breast cancer | 26, 29 | |||
| p.R32fs | 25 | ||||
| Copy-number loss, p.S92fsa | Pineoblastoma | 27 | |||
| RNASEN/DROSHA | Nuclear microprocessing complex member. Ribonuclease that cleaves pir-miRNA to pre-miRNAs | rs640831 C/A | Lung adenocarcinoma | 31 | |
| rs1110386 G/Aa | Ovarian cancer | 32, 33 | |||
| rs486732 C/Aa | Bladder cancer | 34 | |||
| p.E500* | Breast cancer | 35 | |||
| p.R277C | Thyroid carcinoma | 37 | |||
| p.Q136*a | Pineaoblastoma | 27 | |||
| DHX9 | Nuclear microprocessing complex member. Helicase that modulates nuclear miRNA processing | p.S625C, p.P89A | Breast cancer | 42 | |
| XPO5 | Facilitates pre-miRNA export from the nucleus to the cytoplasm | p.T1182del | Colon cancer, gastric cancer, endometrial cancer | 46 | |
| rs2257082 A/G | Gastric cancer | 48 | |||
| DICER1 | Member of the miRISC. Endoribonuclease that cleaves the stem-loop from pre-miRNAs, leading to the formation of mature miRNAs | Germline: | |||
| p.Y1180* | Fetal lung adenocarcinoma, Sertoli-Leydig ovarian tumor, FMGS | 54 | |||
| p.Y819H | Hepatocellular carcinoma | 67 | |||
| p.D1713Aa | Wilms tumor | 55 | |||
| p.R187*a | Pleuropulmonary blastoma | 52 | |||
| p.G803Ra | Wilms tumor | 56 | |||
| p.E503Xa | Embryonal ovarian cancer | 51 | |||
| Somatic: | |||||
| p.A872Ta | Wilms tumor | 55 | |||
| p.E428Ka | Hepatocellular carcinoma | 67 | |||
| p.E813Qa | Sertoli-Leydig cell tumor | 58 | |||
| p.D1709Na | Yolk-sac tumor | 57 | |||
| p.D1709G | Juvenile granulosa-cell tumor | ||||
| p.D1810Y | Teratoma | ||||
| p.E1813Qa | Sertoli-Leydig cell tumor | 57 | |||
| p.Y1701*a | Pineoblastoma | 27 | |||
| TARBP1 | Member of the miRISC. Binding partner of DICER that facilitates RISC assembly | p.P144fs | Colon cancer | 68 | |
| p.R353fs | Wilms tumor | 41 | |||
| p.M145fs | Gastric cancer | 71 | |||
| Copy-number gain | Adrenal cancer | 70 | |||
| PRKRA | Member of the miRISC. Binding partner of DICER that facilitates RISC assembly | p.P127L | Ovarian cancer | 71 | |
| AGO2 | Member of the miRISC. Endonuclease that has mRNA cleavage function | Copy-number gain | Head and neck cancer | 74 | |
| Multiple myeloma | 76 | ||||
| Breast cancer | 75 | ||||
| TNRC6A | Member of the miRISC. Binding partner of AGO2 required for mRNA silencing | p.P115-Q118del | Esophageal cancer | 78 | |
| p.R1183fs, p.Trp804fs | Gastric cancer, colorectal cancer | 71, 73 | |||
| miRNA process | Influence by KRAS | Cancer type | Ref. | ||
| Nuclear miRNA processing | Increased expression of lncRNA KIMAT1 leads to increased expression of oncomiRs and decreased expression of tumor-suppressive miRNAs | Lung adenocarcinoma | 83 | ||
| miRNA export | Phosphorylation of Exportin-5 by ERK inhibits miRNA export from the nucleus | HCC | 84 | ||
| miRNA processing by DICER | Phosphorylation of DICER by ERK inhibits processing function and increases nuclear localization | NSCLS, PDAC, cholangiocarcinoma, endometrioid tumors | 87–92 | ||
| miRNA processing by AGO2 | Binding of KRAS to AGO2 inhibits miRNA unwinding and slicing function | PDAC, CRC, NSCLS | 94, 99, 101 | ||
| Phosphorylation of AGO2 by AKT increases localization to PBs and decreases export via exosomes | |||||
| Upregulation of SGs | Upregulates SG formation and transcript storage for stress protection and therapy resistance | PDAC, multiple myeloma | 118, 121 | ||
| Upregulation of the UPR | Upregulates UPR to compensate for ER stress, which promotes survival | PDAC | 118, 136 | ||
| UPR upregulation also induces SG formation | |||||
aOne mutation represented from citation.
Mutations in RNASEN, the gene encoding DROSHA, have also been identified in various tumor types. SNPs in RNASEN have been identified and correlated with decreased survival in subpopulations of patients with lung, ovarian, and bladder cancer (31–34). Other studies have identified possible oncogenic mutations in RNASEN in breast and thyroid cancer (35–37). In addition, studies of copy-number variation in genes encoding the miRNA machinery have identified RNASEN as one of the most frequently amplified genes in lung and cervical cancer, correlating with decreased survival (38, 39). Finally, mutations in RNASEN have been shown to act as driver mutations in Wilms tumors, similar to DGCR8 (40). Multiple studies have also demonstrated how several RNASEN mutations lead to decreased catalytic function and reduced miRNA biogenesis, suggesting that the overall outcome of miRNA processing inhibition is the promotion of tumorigenesis regardless of the mutation that causes it (24, 41).
DHX9 is a helicase with diverse functions, including modulating miRNA processing in the nucleus. Multiple point mutations in DHX9 have been identified in breast cancer. These mutations are thought to drive tumorigenesis in a subset of BRCA1/BRCA2 wild-type (WT) tumors, as DXH9 interacts with BRCA proteins as part of the miRNA processing function of BRCA1/BRCA2 (42, 43). DDX17, another helicase involved in nuclear miRNA processing, has also been implicated in cancer. Although the expression of DDX17 is misregulated in various cancers, including colon, lung, breast, prostate, and liver cancer, whether this misregulation is driven by genetic mutations, copy-number variations, or posttranslational modification is unknown (44, 45).
The gene XPO5 encodes exportin-5. Like members of the microprocessor, XPO5 mutations have been reported across multiple types of cancer. One study reported the presence of inactivating mutations in XPO5 in colon, endometrial, and stomach cancer cell lines (46). Various SNPs in the XPO5 gene have also been correlated with increased breast, esophageal, and gastric cancer incidence and poorer gastric cancer outcomes, while others have been associated with better survival outcomes in lung and colon cancer (47, 48). The expression of the binding partner of exportin-5, RAN-GTP, is generally upregulated across most types of cancer. However, no cancer-specific mutations have been identified within RAN-GTP as of yet (49).
Oncogenic mutation in the cytoplasmic components of miRNA core machinery
More tumor-promoting mutations have been identified in the cytoplasmic components of the miRNA biosynthesis machinery compared with their nuclear counterparts. Both germline and somatic mutations in DICER1, the gene encoding the DICER protein, have been shown to promote multiple cancers. Germline loss-of-function mutations in DICER1 have been shown to cause embryonal tumors in pediatric populations, such as Wilms tumors, pleuropulmonary blastoma, cystic nephroma, embryonal rhabdomyosarcoma, ovarian Sertoli-Leydig cell tumor, ciliary body medulloepithelioma, multinodular goiter, thyroid adenomas, pituitary blastoma, pineoblastoma, and renal sarcoma (50–58). Germline DICER1 mutations can also cause familial genetic tumor predisposition syndromes, such as DICER1 syndrome, where patients may develop multiple of the above mentioned tumor types (50, 59–65). The propensity for DICER1 mutations (and certain mutations in other miRNA core machinery factors) to promote congenital and childhood cancers may be due to the vital role that miRNAs play in the spatiotemporal regulation of critical developmental processes. However, the misregulation of these processes can also promote tumor formation in adults.
Somatic mutations in DICER1 can also promote the formation of similar tumors seen in the germline syndromes, such as adult-onset pulmonary blastoma, pineoblastoma, Sertoli-Leydig cell tumors, Wilms tumors, and hepatocellular carcinoma (HCC; refs. 27, 40, 63, 64, 66, 67). In addition, SNPs in DICER1 have been significantly correlated with poorer outcomes in certain gastric tumors (48). trans-activation response element (TAR)-binding protein (TARBP1 or TRBP) and the protein activator of protein kinase R (PACT or PRKRA) are DICER-binding partners that modulate its miRNA processing ability. Whole genome sequencing of various tumor types has identified TARBP1 mutations in gastrointestinal cancer, colorectal cancers, and Wilms tumors associated with decreased expression of TARBP1 (68, 69). However, copy-number variations that lead to increased expression have also been reported in adrenal cancers (70). While less is known about PRKRA, hotspot mutations in this gene have been identified in ovarian cancer (71).
Although four AGO proteins exist in humans, only AGO2 has been implicated in cancer regarding its role in the miRNA regulatory pathway. While AGO1 does display oncogenic function in multiple cancers, it does so outside of its miRNA processing and effector function (72). Although some loss-of-function somatic AGO2 mutations have been identified in gastric and colorectal cancers, most cancers with AGO2 misregulation show genetic amplification and increased protein expression (73). Breast cancer, multiple myeloma, HCC, bladder cancer, ovarian cancer, gastric cancer, colorectal cancer, and head and neck squamous cancers are all types of cancer where AGO2 copy-number amplification or protein overexpression has been shown to drive proliferation and cancer progression (74–77). Because AGO2 is involved in both miRNA biosynthesis and effector function, the overexpression of AGO2 may affect how transcripts are regulated. AGO2 overexpression also does not necessarily lead to increased miRNA processing, as AGO2 is heavily regulated by posttranslational modification. As the various AGO family members are differentially loaded with miRNAs, AGO2 may also promote the processing of oncogenic miRNAs to a greater degree than tumor-suppressive miRNAs. Trinucleotide repeat-containing adapter 6A (TNRC6A) encodes for the AGO2-binding partner, GW182. Interestingly, a recent analysis showed that TNRC6A is the most frequently mutated miRNA biogenesis gene across 33 types of cancer. However, further studies on TNRC6A mutations in cancer are needed, as TNRC6A mutations have only been significantly correlated with poor outcomes in esophageal, gastric, and colorectal cancers (71, 73, 78).
KRAS and miRNA Regulation
Clearly, mutations in members of the miRNA processing machinery can promote cancer. Save for a few exceptions, most of these mutations have been linked to a decrease in miRNA biosynthesis, thus leading to the global misregulation of miRNAs and the mRNA transcripts they target. Interestingly, recent evidence suggests that mutant KRAS is similarly able to modulate the activity of multiple members of the core miRNA machinery as an additional mechanism to promote malignant transformation. Several studies have shown that miRNA biogenesis and global miRNA expression levels are grossly misregulated in KRAS-mutated cancers (79–82). However, over the last decade, KRAS has been shown to play a more significant role in driving miRNA misregulation by directly regulating critical factors involved in the miRNA regulatory pathway (Table 1). This additional regulatory mechanism disrupted by mutant KRAS may represent a novel vulnerability in KRAS-driven cancers that has yet to be targeted.
KRAS and nuclear miRNA biosynthesis
Recently, KIMAT1, a KRAS-dependent long noncoding RNA, was shown to regulate DHX9 (83). Increased KIMAT1 expression can stabilize DHX9 and promote an increase in the relative expression of oncogenic miRNAs while simultaneously preventing the expression of tumor-suppressive miRNAs in lung tumors (Fig. 1C, i). This suggests that oncogenic KRAS may influence the differential processing of miRNAs by modulating the microprocessing complex. Increased levels of phosphorylated ERK have also been shown to increase the phosphorylation of exportin-5 in HCC, which inhibits its ability to bind and export pre-miRNAs (Fig. 1C, ii; ref. 84). Nuclear export of pre-miRNAs also depends on RAN, a small GTPase that binds to Exportin-5. Because RAN activation occurs downstream of KRAS, mutant KRAS may increase RAN activity or expression, which is known to occur across many types of cancer (49). Thus, mutant KRAS may affect not only the nuclear processing of miRNAs but also whether those miRNAs can exit the nucleus.
KRAS and cytoplasmic miRNA processing
Once pre-miRNAs exit the nucleus, they are further processed into mature miRNAs by DICER. During C. elegans oogenesis, ERK-dependent phosphorylation of DICER at serine 1705 and 1833 (1728 and 1852 in humans) coordinates its nuclear localization and also inhibits its miRNA processing function (85, 86). As mutant KRAS increases ERK signaling, increased nuclear localization and decreased nuclease activity of DICER would be two logical consequences of KRAS mutations in cancer. The sequestration of DICER away from cytoplasmic pre-miRNAs and inhibition of its nuclease activity would presumably lead to a relative decrease in mature miRNA production (Fig. 1C, iii). Interestingly, the nuclear localization of DICER and its phosphorylation at Serine 1712 and 1836 (1728 and 1852 in humans) has indeed been shown to promote tumor formation and treatment resistance in various KRAS-driven mouse cancer models, including NSCLC and PDAC models (87–92).
In addition to DICER, oncogenic KRAS can affect other RNA-binding proteins involved in cytoplasmic miRNA processing, such as AGO2. The N-terminal domain of AGO2 can bind the switch II domain in KRAS, and this interaction inhibits AGO2-dependent miRNA maturation and stabilization (93–95). Although WT KRAS can bind AGO2, it is the interaction between AGO2 and mutant KRAS that drives cellular transformation, implying that there is some type of feed-forward mechanism between AGO2 and mutant KRAS (94). AGO2 is also required for pancreatic intraepithelial neoplasia (PanIN) progression to PDAC via EGFR–KRAS signaling (96). Indeed, increased AGO2 expression and phosphorylation have been found to promote tumorigenesis and metastasis in other cancer models, such as HCC and NSCLC (97–99). Mutant KRAS also influences the function of AGO2 by modulating AGO2-mediated miRNA secretion via exosomes. In colorectal cancer cells, mutant KRAS was found to promote the differential sorting of miRNAs into exosomes through posttranslational modifications of AGO2 mediated by increases in MAPK pathway activation (100, 101). Thus, oncogenic KRAS may exert much of its miRNA-modulating activity through direct and indirect interactions with AGO2.
KRAS and miRNA-mediated silencing and decay
Beyond its effect on miRNA biogenesis, oncogenic KRAS has been shown to modulate how miRNAs regulate their target transcript. In addition to interfering with miRNA maturation, the mutant KRAS/AGO2 interaction can also misregulate the effector function of miRNAs, as AGO2 and the miRISC are essential for miRNA localization and miRNA-mediated target silencing. AGO2 function is regulated via posttranslational modification, including phosphorylation at various residues. Cells harboring an activating KRAS mutation have higher levels of AGO2 phosphorylation at serine 387 (AGO2S387; ref. 101). As previously mentioned, AGO2-bound miRNAs can be secreted from the cell via exosomes. However, ERK and AKT-mediated phosphorylation of AGO2S387 prevents this secretion and shifts AGO2 localization to processing bodies (PB; Fig. 1C, iv; refs. 101, 102). AGO2S387 phosphorylation also increases the association of AGO2 with GW182 (102, 103). This interaction is necessary for miRNA-mediated silencing and the localization of AGO2 to PBs (103–105). Although increasing the AGO2/GW182 interaction would presumably lead to an increase in miRNA-mediated gene silencing, it is not entirely known how mutated KRAS affects this process.
AGO2 is also regulated by phosphorylation at tyrosine 393 (AGO2Y393), which also increases in cells harboring KRAS mutations, such as in PDAC and NSCLC. AGO2Y393 phosphorylation inhibits the miRNA unwinding and miRISC loading function of AGO2 (Fig. 1C, v; ref. 96). During physiologic conditions, AGO2Y393 is phosphorylated by tyrosine kinases, such as EGFR and c-Src kinase (106, 107). Phosphorylation of AGO2Y393 leads to the dissociation of KRASWT and AGO2 but is insufficient to interrupt the mutant KRAS–AGO2 interaction, thus preventing proper miRNA binding by AGO2 (96). In addition, mutant KRAS has been shown to prevent AGO2Y393 dephosphorylation by protein tyrosine phosphatase 1B (PTP1B), further interfering with AGO2 function (108). Although there are multiple other sites in which AGO2 may be phosphorylated, it is not yet known how oncogenic KRAS affects these sites.
Despite the conservation of the N-terminal domain across the four AGO members, KRAS has only been shown to interact with and modulate the activity of AGO2. AGO2 can catalyze the degradation of target transcripts through its mRNA-slicing function. However, this only occurs in certain contexts with specific miRNAs that bind with perfect sequence complementarity to their target transcripts. Increased phosphorylation of AGO2S387 was also shown to prevent the slicing activity of AGO2. (Fig. 1C, vi; ref. 102). Although the consequences of mutant KRAS inhibiting AGO2 slicing are unknown, one possibility is that transcripts usually degraded by AGO2 slicing may be stabilized, such as the HOX family of genes. HOX genes are regulated by miR-196-directed cleavage by AGO2 (109). Many HOX genes have been shown to act as oncogenes in KRAS-driven cancers such as lung, colorectal, and pancreatic cancers (110). Thus, inhibition of HOX transcript cleavage by AGO2 may contribute to HOX upregulation in cancer.
KRAS and intracellular condensates
Beyond directly regulating members of the miRNA regulatory pathway, oncogenic KRAS may also coordinate the storage of miRNA-targeted transcripts in intracellular condensates. Two types of intracellular condensates involved in transcript regulation are stress granules (SG) and PBs; refs. 111–114). Components of these RNP granules can modulate the expression of transcripts that control disrupted functions in cancer, such as signal transduction, metabolism, and cellular stress. Thus, understanding their misregulation in cancer has become of great interest (115).
SGs are stress-induced and are storage sites for translationally arrested mRNAs (116). miRNAs and members of the miRNA core machinery have been shown to localize to SGs, suggesting that SGs may regulate miRNA-bound transcripts during cell stress (117). Mutated KRAS has been shown to upregulate SG formation in PDAC and multiple myeloma via the upregulation of 15-Deoxy-delta (12, 14)-prostaglandin J (15d-PGJ2; ref. 2), which subsequently targets eIF4A for inactivation, promoting SG formation (118–120). This upregulation of SGs has been shown to promote survival and enhanced stress protection, both of which drive therapy resistance (Fig. 1C, vii; refs. 118, 121). Moreover, another SG protein, DDX3, has been shown to enhance the transcription of oncogenic KRAS (122). This suggests that although mutated KRAS likely affects components of SGs, factors that localize to SGs may also affect the expression of KRAS. This is not surprising, considering that most of these factors have some function in regulating mRNA transcript levels.
PBs are composed of decapping enzymes, decapping activators, exonucleases, deadenylases, translational repressors, and miRNA machinery. The specific roles of these proteins and how PBs function under physiologic conditions have been extensively described in other reviews (113, 123–125). As stated above, oncogenic KRAS affects the posttranslational modification of AGO2, leading to its increased localization to PBs. KRAS has also been shown to interact with DDX6, one of the few components required for PB formation. This interaction was found to play a role in modulating KRAS signaling downstream of HER2 by promoting the translation of HER2 in gastric cancer (126). In addition, it has been recently shown that receptor tyrosine kinase fusion proteins have the capacity to drive oncogenic KRAS signaling at membraneless protein granules (127). Similar to SGs, this suggests that KRAS may not only affect RNP granule dynamics, but signaling from RNP granules may also regulate KRAS.
KRAS has also been identified as an upstream regulator of mRNA transcripts targeted by the CCR4-NOT deadenylase machinery in PBs (128). In addition to regulating the expression of CCR4-NOT–bound transcripts, KRAS has been shown to regulate TOB, a cofactor in the CCR4–NOT complex, which regulates cyclin D1 and prevents proliferation (129). Interestingly, TOB has been shown to act as a tumor suppressor in certain cancers (130). MAPK and c-Jun JNK, both downstream effectors of KRAS, can phosphorylate TOB, decreasing its antiproliferative function and preventing its tumor suppressive function. JNK also phosphorylates multiple PB components, such as DCP1A and 4E-T. Upon phosphorylation by JNK, DCP1a is redistributed from small, punctate PB to larger cytoplasmic inclusions (131). JNK can also phosphorylate 4E-T, increasing PB size during cell stress (132). Together, these data indicate that mutant KRAS may affect multiple components of granules involved in posttranscriptional regulation, further supporting the notion that some interplay exists between oncogenic KRAS, miRNAs, and PBs.
KRAS and the endoplasmic reticulum
Regulation of mRNA translation and silencing is also tightly connected to the endoplasmic reticulum (ER). AGO2, which oncogenic KRAS heavily influences, was recently shown to regulate mRNA degradation at the ER (133). Interestingly, oncogenic KRAS also affects ER homeostasis. Mutations in KRAS, along with other RAS family members, have been shown to lead to ER expansion via the upregulation of the unfolded protein response (UPR; Fig. 1C, viii; ref. 134). The UPR is also critically important in KRAS-mutated PDAC, as KRAS is required to activate the UPR in response to ER stress (135). Under physiologic conditions, the upregulation of the UPR can lead to cell-cycle arrest or stress-induced apoptosis. However, in KRAS-transformed cancers, chronic upregulation of the UPR can lead to tumor cell resistance to acute stress–induced apoptosis (134, 136). Thus, some of the transformative characteristics of mutant KRAS likely depend on transcript regulation by AGO2 and the ER. Activation of the UPR also induces SG formation, which KRAS-mutated cells have coopted to promote survival (118). Interestingly, the ratio of cisternal to tubular ER also plays a role in the fission of PB granules, thus affecting the size and number of PBs (137). When induced, the UPR leads to a decrease in PB numbers, presumably due to a shift to a more cisternal-dominant ER. However, how this phenomenon alters posttranscriptional control through PBs has yet to be discovered.
Conclusion and Future Perspectives
Recently, more effective inhibitors against mutant KRAS subtypes have been developed, such as MRTX849 (adagrasib), which has shown potent inhibition of KRASG12C (138). Despite its success and current status in clinical trials, a significant percentage of preclinical models still showed no effective response. In addition, most of the successes in targeting mutant KRAS subtypes have centered on KRASG12C. While KRASG12C is the predominant driver mutation in lung cancers, KRASG12D driver mutations occur far more frequently in PDAC and colorectal cancer (139). Although MRTX1133 was recently announced (January 2023) as a novel KRASG12D inhibitor cleared to enter clinical trials, it will still be years before clinical implementation if it is proven effective in human patients (140).
In addition to KRAS-specific inhibitors, other inhibitors of both upstream (i.e., EGFR) and downstream (i.e., MEK and AKT) KRAS signaling pathways have shown promise in treating specific subsets of NSCLC and other cancer types where KRAS is less frequently mutated (139). Regardless of instances where clinical success is seen, almost all treatment modalities are plagued by nonresponders or the development of resistance mechanisms. Because of these issues, identifying novel targetable pathways for therapeutic development in KRAS-driven cancers is still vitally important. As summarized in this review, the miRNA regulatory pathway represents one such pathway that has yet to be therapeutically exploited. Most intriguing is the numerous ways oncogenic KRAS can influence several steps of miRNA biogenesis and function simultaneously, thus presenting several potential avenues for treating KRAS-driven cancers that have yet to be explored.
Although intriguing, targeting the miRNA regulatory pathway in KRAS-driven cancers would not come without challenges. Because individual miRNAs can target multiple transcripts, inhibiting, or activating members of the miRNA core machinery could induce off-target responses. Despite this concern, there may be ways to mitigate potential adverse effects. Many of the ways oncogenic KRAS modulates members of the miRNA regulatory pathway involve posttranslational modifications, such as the phosphorylation of DICER and AGO2 on various residues. Kinase inhibitors are one class of drugs that may modulate the activity of KRAS-induced phosphorylation of DICER and AGO2. However, kinase inhibitors have classically failed to treat KRAS-driven cancers, often due to a lack of specificity or resistance mechanisms. Another appealing option for targeting posttranslationally modified proteins involves using an in silico approach to identifying potential interactions between small-molecule inhibitors and site-specific phosphorylation events in proteins. With the advancement of various cheminformatics platforms, high-throughput screening of small molecules has been used to identify inhibitors of distinct phosphorylation events on specific proteins (141). A similar approach may be able to identify site-specific small-molecule inhibitors of DICER, AGO2, and other posttranslationally modified proteins involved in the miRNA-driven oncogenicity of KRAS. In addition, next-generation sequencing technology has advanced significantly over the last ten years, thus allowing for fast and accurate sequencing of the small RNA transcriptome. Because of this, it is possible to identify potential off-target responses that occur if members of the miRNA regulatory pathway are inhibited/ activated (i.e., the undesired upregulation or downregulation of specific miRNAs). Off-target changes in miRNA expression can be controlled using miRNA mimics or antagomirs (142).
Another possible strategy to inhibit miRNA-dependent oncogenesis of mutant KRAS could involve targeting how miRNAs or transcripts are spatially misregulated by oncogenic KRAS. Recently, the subcellular localization of specific transcripts was shown to be critical for driving tumor invasion (143). Similarly, specific miRNAs or miRNA-targeted transcripts can be differentially enriched in subcellular compartments such as the surface of the ER or PBs, which could also be targeted in KRAS-mutant cancer cells. Although studies are beginning to identify how mutant KRAS alters some of these regulatory pathways, many questions remain to be answered. What are the key downstream effectors driving miRNA misregulation in KRAS-dependent cancers? Can the miRNA regulatory pathway be targeted therapeutically without significant off-target effects? What other role do SGs, PBs, and the ER play in oncogenesis? Are there targetable interactions between oncogenic KRAS, its downstream effectors, and proteins involved in miRNA regulation? Does oncogenic KRAS lead to differential regulation of transcripts in SGs and PBs? Answering these questions and understanding precisely how mutant KRAS alters posttranscriptional regulation through miRNAs seem essential to identifying novel treatment modalities for KRAS- and other oncogene-driven cancers.
Acknowledgments
The authors would like to thank Joel Neilson and Bruno Di Stefano for their valuable feedback and support. The Parchem lab is supported by the NICHD (R01HD099252 and R01HD098131), CPRIT Scholar in Cancer Research (RR150106), Andrew McDonough B+ Foundation, V Scholar in Cancer Research (V2017–017), and the NHLBI (T32 HL092332 to A.S. Bortoletto).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authors' Disclosures
No disclosures were reported.
References
- 1. Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med 2014;20:460–9. [DOI] [PubMed] [Google Scholar]
- 2. Rachagani S, Macha MA, Heimann N, Seshacharyulu P, Haridas D, Chugh S, et al. Clinical implications of miRNAs in the pathogenesis, diagnosis and therapy of pancreatic cancer. Adv Drug Deliv Rev 2015;81:16–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Peng Y, Croce CM. The role of microRNAs in human cancer. Signal Transduct Target Ther 2016;1:15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ali Syeda Z, Langden SSS, Munkhzul C, Lee M, Song SJ. Regulatory mechanism of microrna expression in cancer. Int J Mol Sci 2020;21:1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Le Rolle A-F, Chiu TK, Zeng Z, Shia J, Weiser MR, Paty PB, et al. Oncogenic KRAS activates an embryonic stem cell-like program in human colon cancer initiation. Oncotarget 2016;7:2159–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Waters AM, Der CJ. KRAS: the critical driver and therapeutic target for pancreatic cancer. Cold Spring Harb Perspect Med 2018;8:a031435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shi L, Middleton J, Jeon Y-J, Magee P, Veneziano D, Laganà A, et al. KRAS induces lung tumorigenesis through microRNAs modulation. Cell Death Dis 2018;9:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buscail L, Bournet B, Cordelier P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat Rev Gastroenterol Hepatol 2020;17:153–68. [DOI] [PubMed] [Google Scholar]
- 9. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 10. Haigis KM. KRAS alleles: the devil is in the detail. Trends Cancer 2017;3:686–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang M-T, Fer N, Galeas J, Collisson EA, Kim SE, Sharib J, et al. Blockade of leukemia inhibitory factor as a therapeutic approach to KRAS driven pancreatic cancer. Nat Commun 2019;10:3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Drosten M, Barbacid M. Targeting the MAPK Pathway in KRAS-Driven Tumors. Cancer Cell 2020;37:543–50. [DOI] [PubMed] [Google Scholar]
- 13. Shui B, La Rocca G, Ventura A, Haigis KM. Interplay between K-RAS and miRNAs. Trends Cancer 2022;8:384–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang L, Guo Z, Wang F, Fu L. KRAS mutation: from undruggable to druggable in cancer. Signal Transduct Target Ther 2021;6:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. O'Brien J, Hayder H, Zayed Y, Peng C. Overview of microrna biogenesis, mechanisms of actions, and circulation. Front Endocrinol 2018;9:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kawai S, Amano A. BRCA1 regulates microRNA biogenesis via the DROSHA microprocessor complex. J Cell Biol 2012;197:201–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nguyen TA, Jo MH, Choi Y-G, Park J, Kwon SC, Hohng S, et al. Functional anatomy of the human microprocessor. Cell 2015;161:1374–87. [DOI] [PubMed] [Google Scholar]
- 18. Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 2003;17:3011–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet 2013;14:447–59. [DOI] [PubMed] [Google Scholar]
- 20. Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song J-J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science 2004;305:1437–41. [DOI] [PubMed] [Google Scholar]
- 21. O'Carroll D, Mecklenbrauker I, Das PP, Santana A, Koenig U, Enright AJ, et al. A slicer-independent role for argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev 2007;21:1999–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 2007;131:1097–108. [DOI] [PubMed] [Google Scholar]
- 23. Cheloufi S, Dos Santos CO, Chong MMW, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 2010;465:584–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wegert J, Ishaque N, Vardapour R, Geörg C, Gu Z, Bieg M, et al. Mutations in the SIX1/2 pathway and the DROSHA/DGCR8 miRNA microprocessor complex underlie high-risk blastemal type wilms tumors. Cancer Cell 2015;27:298–311. [DOI] [PubMed] [Google Scholar]
- 25. Wen J, Lv Z, Ding H, Fang X, Sun M. Association of miRNA biosynthesis genes DROSHA and DGCR8 polymorphisms with cancer susceptibility: a systematic review and meta-analysis. Biosci Rep 2018;38:BSR20180072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rivera B, Nadaf J, Fahiminiya S, Apellaniz-Ruiz M, Saskin A, Chong A-S, et al. DGCR8 microprocessor defect characterizes familial multinodular goiter with schwannomatosis. J Clin Invest 2020;130:1479–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li BK, Vasiljevic A, Dufour C, Yao F, Ho BLB, Lu M, et al. Pineoblastoma segregates into molecular sub-groups with distinct uppre-pathologic features: a Rare Brain Tumor Consortium registry study. Acta Neuropathol 2020;139:223–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Paulsson JO, Rafati N, DiLorenzo S, Chen Y, Haglund F, Zedenius J, et al. Whole-genome sequencing of follicular thyroid carcinomas reveal recurrent mutations in MicroRNA processing subunit DGCR8. J Clin Endocrinol Metab 2021;106:3265–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rodrigues L, Canberk S, Macedo S, Soares P, Vinagre J. DGCR8 microprocessor subunit mutation and expression deregulation in thyroid lesions. Int J Mol Sci 2022;23:14812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vardapour R, Kehl T, Kneitz S, Ludwig N, Meese E, Lenhof H-P, et al. The DGCR8 E518K mutation found in Wilms tumors leads to a partial miRNA processing defect that alters gene expression patterns and biological processes. Carcinogenesis 2022;43:82–93. [DOI] [PubMed] [Google Scholar]
- 31. Rotunno M, Zhao Y, Bergen AW, Koshiol J, Burdette L, Rubagotti M, et al. Inherited polymorphisms in the RNA-mediated interference machinery affect microRNA expression and lung cancer survival. Br J Cancer 2010;103:1870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Permuth-Wey J, Kim D, Tsai Y-Y, Lin H-Y, Chen YA, Barnholtz-Sloan J, et al. LIN28B polymorphisms influence susceptibility to epithelial ovarian cancer. Cancer Res 2011;71:3896–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liang D, Meyer L, Chang DW, Lin J, Pu X, Ye Y, et al. Genetic variants in MicroRNA biosynthesis pathways and binding sites modify ovarian cancer risk, survival, and treatment response. Cancer Res 2010;70:9765–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yuan L, Chu H, Wang M, Gu X, Shi D, Ma L, et al. Genetic variation in DROSHA 3’UTR regulated by I-miR-27b is associated with bladder cancer risk. PLoS One 2013;8:e81524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Felicio PS, Grasel RS, Campacci N, de Paula AE, Galvão HCR, Torrezan GT, et al. Whole-exome sequencing of non-BRCA1/BRCA2 mutation carrier cases at high-risk for hereditary breast/ovarian cancer. Hum Mutat 2021;42:290–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Paulsson JO, Backman S, Wang N, Stenman A, Crona J, Thutkawkorapin J, et al. Whole-genome sequencing of synchronous thyroid carcinomas identifies aberrant DNA repair in thyroid cancer dedifferentiation. J Pathol 2020;250:183–94. [DOI] [PubMed] [Google Scholar]
- 37. Mohammadpour-Gharehbagh A, Heidari Z, Eskandari M, Aryan A, Salimi S. Association between genetic polymorphisms in microRNA machinery genes and risk of papillary thyroid carcinoma. Pathol Oncol Res 2020;26:1235–41. [DOI] [PubMed] [Google Scholar]
- 38. Czubak K, Lewandowska MA, Klonowska K, Roszkowski K, Kowalewski J, Figlerowicz M, et al. High copy number variation of cancer-related microRNA genes and frequent amplification of DICER1 and DROSHA in lung cancer. Oncotarget 2015;6:23399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Harden ME, Munger K. Perturbation of DROSHA and DICER expression by human papillomavirus 16 oncoproteins. Virology 2017;507:192–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rakheja D, Chen KS, Liu Y, Shukla AA, Schmid V, Chang T-C, et al. Somatic mutations in DROSHA and DICER1 impair microRNA biogenesis through distinct mechanisms in Wilms tumours. Nat Commun 2014;2:4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Torrezan GT, Ferreira EN, Nakahata AM, Barros BDF, Castro MTM, Correa BR, et al. Recurrent somatic mutation in DROSHA induces microRNA profile changes in Wilms tumour. Nat Commun 2014;5:4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guénard F, Labrie Y, Ouellette G, Beauparlant CJ, Durocher F, INHERIT BRCAs. Genetic sequence variations of BRCA1-interacting genes AURKA, BAP1, BARD1 and DHX9 in French Canadian families with high risk of breast cancer. J Hum Genet 2009;54:152–61. [DOI] [PubMed] [Google Scholar]
- 43. Lee T, Pelletier J. The biology of DHX9 and its potential as a therapeutic target. Oncotarget 2016;7:42716–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xue Y, Jia X, Li C, Zhang K, Li L, Wu J, et al. DDX17 promotes hepatocellular carcinoma progression via inhibiting Klf4 transcriptional activity. Cell Death Dis 2019;10:814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wu K-J. The role of miRNA biogenesis and DDX17 in tumorigenesis and cancer stemness. Biomed J 2020;43:107–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, Spizzo R, et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell 2010;18:303–15. [DOI] [PubMed] [Google Scholar]
- 47. Wu K, He J, Pu W, Peng Y. The role of Exportin-5 in MicroRNA biogenesis and cancer. Genomics Proteomics Bioinformatics 2018;16:120–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liao Y, Liao Y, Li J, Liu L, Li J, Wan Y, et al. Genetic variants in miRNA machinery genes associated with clinicopathological characteristics and outcomes of gastric cancer patients. Int J Biol Markers 2018;33:301–7. [DOI] [PubMed] [Google Scholar]
- 49. Boudhraa Z, Carmona E, Provencher D, Mes-Masson A-M. Ran gtpase: A key player in tumor progression and metastasis. Front Cell Dev Biol 2020;8:345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Moke DJ, Thomas SM, Hiemenz MC, Nael A, Wang K, Shillingford N, et al. Three synchronous malignancies in a patient with DICER1 syndrome. Eur J Cancer 2018;93:140–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schultz KAP, Pacheco MC, Yang J, Williams GM, Messinger Y, Hill DA, et al. Ovarian sex cord-stromal tumors, pleuropulmonary blastoma and DICER1 mutations: a report from the international pleuropulmonary blastoma registry. Gynecol Oncol 2011;122:246–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Brenneman M, Field A, Yang J, Williams G, Doros L, Rossi C, et al. Temporal order of RNase IIIb and loss-of-function mutations during development determines phenotype in pleuropulmonary blastoma/DICER1 syndrome: a unique variant of the two-hit tumor suppression model. F1000Res 2015;4:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Luke AM, Moroney JW, Snitchler A, Whiteway SL. Ovariuppresoli-leydig cell tumor with elevated inhibin B as a cause of secondary amenorrhea in an adolescent with germ line DICER1 mutation. J Pediatr Adolesc Gynecol 2017;30:598–600. [DOI] [PubMed] [Google Scholar]
- 54. Wu Y, Chen D, Li Y, Bian L, Ma T, Xie M. DICER1 mutations in a patient with an ovarian Sertoli-Leydig tumor, well-differentiated fetal adenocarcinoma of the lung, and familial multinodular goiter. Eur J Med Genet 2014;57:621–5. [DOI] [PubMed] [Google Scholar]
- 55. Wu MK, Sabbaghian N, Xu B, Addidou-Kalucki S, Bernard C, Zou D, et al. Biallelic DICER1 mutations occur in Wilms tumours. J Pathol 2013;230:154–64. [DOI] [PubMed] [Google Scholar]
- 56. Palculict TB, Ruteshouser EC, Fan Y, Wang W, Strong L, Huff V. Identification of germline DICER1 mutations and loss of heterozygosity in familial Wilms tumour. J Med Genet 2016;53:385–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Witkowski L, Mattina J, Schönberger S, Murray MJ, Choong CS, Huntsman DG, et al. DICER1 hotspot mutations in non-epithelial gonadal tumours. Br J Cancer 2013;109:2744–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Heravi-Moussavi A, Anglesio MS, Cheng S-WG, Senz J, Yang W, Prentice L, et al. Recurrent somatic DICER1 mutations in nonepithelial ovarian cancers. N Engl J Med 2012;366:234–42. [DOI] [PubMed] [Google Scholar]
- 59. Schultz KA, Yang J, Doros L, Williams GM, Harris A, Stewart DR, et al. DICER1-pleuropulmonary blastoma familial tumor predisposition syndrome: a unique constellation of neoplastic conditions. Pathol Case Rev 2014;19:90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Durieux E, Descotes F, Mauduit C, Decaussin M, Guyetant S, Devouassoux-Shisheboran M. The co-occurrence of an ovarian Sertoli-Leydig cell tumor with a thyroid carcinoma is highly suggestive of a DICER1 syndrome. Virchows Arch 2016;468:631–6. [DOI] [PubMed] [Google Scholar]
- 61. Dehner LP, Messinger YH, Schultz KAP, Williams GM, Wikenheiser-Brokamp K, Hill DA. Pleuropulmonary blastoma: evolution of an entity as an entry into a familial tumor predisposition syndrome. Pediatr Dev Pathol 2015;18:504–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hill DA, Ivanovich J, Priest JR, Gurnett CA, Dehner LP, Desruisseau D, et al. DICER1 mutations in familial pleuropulmonary blastoma. Science 2009;325:965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Terry W, Carlisle EM, Mallinger P, Nelson AT, Gordon D, Messinger YH, et al. Thoracic Sertoli-Leydig cell tumor: An alternative type of pleuropulmonary blastoma associated with DICER1 variation. Pediatr Blood Cancer 2021;68:e29284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Verrier F, Dubois d'Enghien C, Gauthier-Villars M, Bonadona V, Faure-Conter C, Dijoud F, et al. Mutiple DICER1-related lesions associated with a germline deep intronic mutation. Pediatr Blood Cancer 2018;65:e27005. [DOI] [PubMed] [Google Scholar]
- 65. Klein SD, Martinez-Agosto JA. Hotspot mutations in DICER1 causing GLOW syndrome-associated macrocephaly via modulation of specific microRNA populations result in the activation of PI3K/ATK/mTOR signaling. Microrna 2020;9:70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. de Kock L, Bah I, Brunet J, Druker H, Astigarraga I, Bosch-Barrera J, et al. Somatic DICER1 mutations in adult-onset pulmonary blastoma. Eur Respir J 2016;47:1879–82. [DOI] [PubMed] [Google Scholar]
- 67. Caruso S, Calderaro J, Letouzé E, Nault J-C, Couchy G, Boulai A, et al. Germline and somatic DICER1 mutations in familial and sporadic liver tumors. J Hepatol 2017;66:734–42. [DOI] [PubMed] [Google Scholar]
- 68. Garre P, Pérez-Segura P, Díaz-Rubio E, Caldés T, de la Hoya M. Reassessing the TARBP2 mutation rate in hereditary nonpolyposis colorectal cancer. Nat Genet 2010;42:817–8. [DOI] [PubMed] [Google Scholar]
- 69. Yu X, Li Z. The role of TARBP2 in the development and progression of cancers. Tumour Biol 2016;37:57–60. [DOI] [PubMed] [Google Scholar]
- 70. Caramuta S, Lee L, Ozata DM, Akçakaya P, Xie H, Höög A, et al. Clinical and functional impact of TARBP2 over-expression in adrenocortical carcinoma. Endocr Relat Cancer 2013;20:551–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Galka-Marciniak P, Urbanek-Trzeciak MO, Nawrocka PM, Kozlowski P. A pan-cancer atlas of somatic mutations in miRNA biogenesis genes. Nucleic Acids Res 2021;49:601–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nowak I, Sarshad AA. Argonaute proteins take center stage in cancers. Cancers (Basel) 2021;13:788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kim MS, Oh JE, Kim YR, Park SW, Kang MR, Kim SS, et al. Somatic mutations and losses of expression of microRNA regulation-related genes AGO2 and TNRC6A in gastric and colorectal cancers. J Pathol 2010;221:139–46. [DOI] [PubMed] [Google Scholar]
- 74. Chang SS, Smith I, Glazer C, Hennessey P, Califano JA. EIF2C is overexpressed and amplified in head and neck squamous cell carcinoma. ORL J Otorhinolaryngol Relat Spec 2010;72:337–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Casey MC, Prakash A, Holian E, McGuire A, Kalinina O, Shalaby A, et al. Quantifying argonaute 2 (Ago2) expression to stratify breast cancer. BMC Cancer 2019;19:712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhou Y, Chen L, Barlogie B, Stephens O, Wu X, Williams DR, et al. High-risk myeloma is associated with global elevation of miRNAs and overexpression of EIF2C2/AGO2. Proc Natl Acad Sci USA. 2010;107:7904–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhang K, Pomyen Y, Barry AE, Martin SP, Khatib S, Knight L, et al. AGO2 mediates MYC mRNA stability in hepatocellular carcinoma. Mol Cancer Res 2020;18:612–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Guo J, Huang J, Zhou Y, Zhou Y, Yu L, Li H, et al. Germline and somatic variations influence the somatic mutational signatures of esophageal squamous cell carcinomas in a Chinese population. Bmc Genomics 2018;19:538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Szafranska AE, Davison TS, John J, Cannon T, Sipos B, Maghnouj A, et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene 2007;26:4442–52. [DOI] [PubMed] [Google Scholar]
- 80. Jiao LR, Frampton AE, Jacob J, Pellegrino L, Krell J, Giamas G, et al. MicroRNAs targeting oncogenes are down-regulated in pancreatic malignant transformation from benign tumors. PLoS One 2012;7:e32068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Schetter AJ, Okayama H, Harris CC. The role of microRNAs in colorectal cancer. Cancer J 2012;18:244–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wu K-L, Tsai Y-M, Lien C-T, Kuo P-L, Hung AJ-Y. The roles of microrna in lung cancer. Int J Mol Sci 2019;20:1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Shi L, Magee P, Fassan M, Sahoo S, Leong HS, Lee D, et al. A KRAS-responsive long non-coding RNA controls microRNA processing. Nat Commun 2021;12:2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sun H-L, Cui R, Zhou J, Teng K-Y, Hsiao Y-H, Nakanishi K, et al. ERK activation globally downregulates miRNAs through phosphorylating exportin-5. Cancer Cell 2016;30:723–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Burger K, Schlackow M, Potts M, Hester S, Mohammed S, Gullerova M. Nuclear phosphorylated dicer processes double-stranded RNA in response to DNA damage. J Cell Biol 2017;216:2373–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Drake M, Furuta T, Suen KM, Gonzalez G, Liu B, Kalia A, et al. A requirement for ERK-dependent dicer phosphorylation in coordinating oocyte-to-embryo transition in C. elegans. Dev Cell 2014;31:614–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Aryal NK, Pant V, Wasylishen AR, Rimel BJ, Baseler L, El-Naggar AK, et al. Dicer1 phosphomimetic promotes tumor progression and dissemination. Cancer Res 2019;79:2662–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Cheng W, Qi Y, Tian L, Wang B, Huang W, Chen Y. Dicer promotes tumorigenesis by translocating to nucleus to promote SFRP1 promoter methylation in cholangiocarcinoma cells. Cell Death Dis 2017;8:e2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Su Y-H, Hsu T-W, Chen H-A, Su C-M, Huang M-T, Chuang T-H, et al. ERK-mediated transcriptional activation of Dicer is involved in gemcitabine resistance of pancreatic cancer. J Cell Physiol 2021;236:4420–34. [DOI] [PubMed] [Google Scholar]
- 90. Li C, Chen L, Song W, Peng B, Zhu J, Fang L. DICER activates autophagy and promotes cisplatin resistance in non-small cell lung cancer by binding with let-7i-5p. Acta Histochem 2021;123:151788. [DOI] [PubMed] [Google Scholar]
- 91. Morris JP, Greer R, Russ HA, von Figura G, Kim GE, Busch A, et al. Dicer regulates differentiation and viability during mouse pancreatic cancer initiation. PLoS One 2014;9:e95486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wang X, Chen H, Wen Y, Yang X, Han Q, Jiang P, et al. Dicer affects cisplatinmediated apoptosis in epithelial ovarian cancer cells. Mol Med Report 2018;18:4381–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Winter J, Diederichs S. Argonaute proteins regulate microRNA stability: increased microRNA abundance by argonaute proteins is due to microRNA stabilization. RNA Biol 2011;8:1149–57. [DOI] [PubMed] [Google Scholar]
- 94. Shankar S, Pitchiaya S, Malik R, Kothari V, Hosono Y, Yocum AK, et al. KRAS engages AGO2 to enhance cellular transformation. Cell Rep 2016;14:1448–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Waninger JJ, Beyett TS, Gadkari VV, Siebenaler RF, Kenum C, Shankar S, et al. Biochemical characterization of the interaction between KRAS and argonaute 2. Biochem Biophys Rep 2022;29:101191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Shankar S, Tien JC-Y, Siebenaler RF, Chugh S, Dommeti VL, Zelenka-Wang S, et al. An essential role for argonaute 2 in EGFR-KRAS signaling in pancreatic cancer development. Nat Commun 2020;11:2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Cheng N, Li Y, Han Z-G. Argonaute2 promotes tumor metastasis by way of up-regulating focal adhesion kinase expression in hepatocellular carcinoma. Hepatology 2013;57:1906–18. [DOI] [PubMed] [Google Scholar]
- 98. Turhal NS, Savaş B, Çoşkun Ö, Baş E, Karabulut B, Nart D, et al. Prevalence of K-Ras mutations in hepatocellular carcinoma: a Turkish oncology group pilot study. Mol Clin Oncol 2015;3:1275–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Tien JC-Y, Chugh S, Goodrum AE, Cheng Y, Mannan R, Zhang Y, et al. AGO2 promotes tumor progression in KRAS-driven mouse models of non-small cell lung cancer. Proc Natl Acad Sci USA. 2021;118:e2026104118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Cha DJ, Franklin JL, Dou Y, Liu Q, Higginbotham JN, Demory Beckler M, et al. KRAS-dependent sorting of miRNA to exosomes. eLife 2015;4:e07197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. McKenzie AJ, Hoshino D, Hong NH, Cha DJ, Franklin JL, Coffey RJ, et al. KRAS-MEK signaling controls Ago2 sorting into exosomes. Cell Rep 2016;15:978–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Horman SR, Janas MM, Litterst C, Wang B, MacRae IJ, Sever MJ, et al. Akt-mediated phosphorylation of argonaute 2 downregulates cleavage and upregulates translational repression of MicroRNA targets. Mol Cell 2013;50:356–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Lian SL, Li S, Abadal GX, Pauley BA, Fritzler MJ, Chan EKL. The C-terminal half of human Ago2 binds to multiple GW-rich regions of GW182 and requires GW182 to mediate silencing. RNA 2009;15:804–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Ding L, Han M. GW182 family proteins are crucial for microRNA-mediated gene silencing. Trends Cell Biol 2007;17:411–6. [DOI] [PubMed] [Google Scholar]
- 105. Zeng Y, Sankala H, Zhang X, Graves PR. Phosphorylation of argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem J 2008;413:429–36. [DOI] [PubMed] [Google Scholar]
- 106. Shen J, Xia W, Khotskaya YB, Huo L, Nakanishi K, Lim S-O, et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature 2013;497:383–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Liu T, Zhang H, Fang J, Yang Z, Chen R, Wang Y, et al. AGO2 phosphorylation by c-Src kinase promotes tumorigenesis. Neoplasia 2020;22:129–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Yang M, Haase AD, Huang F-K, Coulis G, Rivera KD, Dickinson BC, et al. Dephosphorylation of tyrosine 393 in argonaute 2 by protein tyrosine phosphatase 1B regulates gene silencing in oncogenic RAS-induced senescence. Mol Cell 2014;55:782–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Yekta S, Shih I-H, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science 2004;304:594–6. [DOI] [PubMed] [Google Scholar]
- 110. Feng Y, Zhang T, Wang Y, Xie M, Ji X, Luo X, et al. Homeobox genes in cancers: from carcinogenesis to recent therapeutic intervention. Front Oncol 2021;11:770428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol 2007;27:3970–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Wang C, Schmich F, Srivatsa S, Weidner J, Beerenwinkel N, Spang A. Context-dependent deposition and regulation of mRNAs in P-bodies. eLife 2018;7:e29815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Zhang B, Herman PK. It is all about the process(ing): P-body granules and the regulation of signal transduction. Curr Genet 2020;66:73–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Luo Y, Na Z, Slavoff SA. P-bodies: composition, properties, and functions. Biochemistry 2018;57:2424–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Lavalée M, Curdy N, Laurent C, Fournié J-J, Franchini D-M. Cancer cell adaptability: turning ribonucleoprotein granules into targets. Trends Cancer 2021;7:902–15. [DOI] [PubMed] [Google Scholar]
- 116. Kedersha N, Ivanov P, Anderson P. Stress granules and cell signaling: more than just a passing phase? Trends Biochem Sci 2013;38:494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Leung AKL, Calabrese JM, Sharp PA. Quantitative analysis of argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci USA. 2006;103:18125–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Grabocka E, Bar-Sagi D. Mutant KRAS enhances tumor cell fitness by upregulating stress granules. Cell 2016;167:1803–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Wang R, Cao L, Thorne RF, Zhang XD, Li J, Shao F, et al. LncRNA GIRGL drives CAPRIN1-mediated phase separation to suppress glutaminase-1 translation under glutamine deprivation. Sci Adv 2021;7:eabe5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Hamada S, Matsumoto R, Tanaka Y, Taguchi K, Yamamoto M, Masamune A. Nrf2 activation sensitizes K-Ras mutant pancreatic cancer cells to glutaminase inhibition. Int J Mol Sci 2021;22:1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Qiang Y-W, Ye S, Chen Y, Epstein J, Davies FE, Morgan G, et al. Mutant KRAS enhances stress granules and resistance to proteasome inhibition Via 15-d-PGJ2 in multiple myeloma. Blood 2019;134:4383. [Google Scholar]
- 122. Wu D-W, Lin P-L, Cheng Y-W, Huang C-C, Wang L, Lee H. DDX3 enhances oncogenic KRASinduced tumor invasion in colorectal cancer via the βcatenin/ZEB1 axis. Oncotarget 2016;7:22687–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Standart N, Weil D. P-bodies: cytosolic droplets for coordinated mRNA storage. Trends Genet 2018;34:612–26. [DOI] [PubMed] [Google Scholar]
- 124. Xing W, Muhlrad D, Parker R, Rosen MK. A quantitative inventory of yeast P body proteins reveals principles of composition and specificity. eLife 2020;9:e56525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 2017;18:285–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Tokumaru Y, Tajirika T, Sugito N, Kuranaga Y, Shinohara H, Tsujino T, et al. Synthetic miR-143 inhibits growth of HER2-positive gastric cancer cells by suppressing KRAS networks including DDX6 RNA helicase. Int J Mol Sci 2019;20:1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Tulpule A, Guan J, Neel DS, Allegakoen HR, Lin YP, Brown D, et al. Kinase-mediated RAS signaling via membraneless cytoplasmic protein granules. Cell 2021;184:2649–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Faraji F, Hu Y, Yang HH, Lee MP, Winkler GS, Hafner M, et al. Post-transcriptional control of tumor cell autonomous metastatic potential by CCR4-NOT deadenylase CNOT7. PLoS Genet 2016;12:e1005820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Maekawa M, Nishida E, Tanoue T. Identification of the anti-proliferative protein Tob as a MAPK substrate. J Biol Chem 2002;277:37783–7. [DOI] [PubMed] [Google Scholar]
- 130. Lee HS, Kundu J, Kim RN, Shin YK. Transducer of ERBB2.1 (TOB1) as a tumor suppressor: a mechanistic perspective. Int J Mol Sci 2015;16:29815–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Rzeczkowski K, Beuerlein K, Müller H, Dittrich-Breiholz O, Schneider H, Kettner-Buhrow D, et al. c-Jun N-terminal kinase phosphorylates DCP1a to control formation of P bodies. J Cell Biol 2011;194:581–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Cargnello M, Tcherkezian J, Dorn JF, Huttlin EL, Maddox PS, Gygi SP, et al. Phosphorylation of the eukaryotic translation initiation factor 4E-transporter (4E-T) by c-Jun N-terminal kinase promotes stress-dependent P-body assembly. Mol Cell Biol 2012;32:4572–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Efstathiou S, Ottens F, Schütter L-S, Ravanelli S, Charmpilas N, Gutschmidt A, et al. ER-associated RNA silencing promotes ER quality control. Nat Cell Biol 2022;24:1714–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Walczak A, Gradzik K, Kabzinski J, Przybylowska-Sygut K, Majsterek I. The role of the ER-induced UPR pathway and the efficacy of its inhibitors and inducers in the inhibition of tumor progression. Oxid Med Cell Longev 2019;2019:5729710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Wu RF, Liao C, Hatoum H, Fu G, Ochoa CD, Terada LS. RasGRF couples Nox4-dependent endoplasmic reticulum signaling to Ras. Arterioscler Thromb Vasc Biol 2017;37:98–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Botrus G, Miller RM, Uson Junior PLS, Kannan G, Han H, Von Hoff DD. Increasing stress to induce apoptosis in pancreatic cancer via the unfolded protein response (UPR). Int J Mol Sci 2022;24:577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Lee JE, Cathey PI, Wu H, Parker R, Voeltz GK. Endoplasmic reticulum contact sites regulate the dynamics of membraneless organelles. Science 2020;367:eaay7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Jänne PA, Rybkin II, Spira AI, Riely GJ, Papadopoulos KP, Sabari JK, et al. KRYSTAL-1: activity and safety of adagrasib (MRTX849) in advanced/metastatic non–small-cell lung cancer (NSCLC) harboring KRAS G12C mutation. Eur J Cancer 2020;138:S1–2. [Google Scholar]
- 139. Zhu C, Guan X, Zhang X, Luan X, Song Z, Cheng X, et al. Targeting KRAS mutant cancers: from druggable therapy to drug resistance. Mol Cancer 2022;21:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Wang X, Allen S, Blake JF, Bowcut V, Briere DM, Calinisan A, et al. Identification of MRTX1133, a noncovalent, potent, and selective KRASG12D inhibitor. J Med Chem 2022;65:3123–33. [DOI] [PubMed] [Google Scholar]
- 141. Pandey V, Wang B, Mohan CD, Raquib AR, Rangappa S, Srinivasa V, et al. Discovery of a small-molecule inhibitor of specific serine residue BAD phosphorylation. Proc Natl Acad Sci USA 2018;115:E10505–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Simonson B, Das S. Microrna therapeutics: the next magic bullet? Mini Rev Med Chem 2015;15:467–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Chrisafis G, Wang T, Moissoglu K, Gasparski AN, Ng Y, Weigert R, et al. Collective cancer cell invasion requires RNA accumulation at the invasive front. Proc Natl Acad Sci USA. 2020;117:27423–34. [DOI] [PMC free article] [PubMed] [Google Scholar]