Abstract
Plant organogenesis requires matching the available metabolic resources to developmental programs. In Arabidopsis, the root system is determined by primary root‐derived lateral roots (LRs), and adventitious roots (ARs) formed from non‐root organs. Lateral root formation entails the auxin‐dependent activation of transcription factors ARF7, ARF19, and LBD16. Adventitious root formation relies on LBD16 activation by auxin and WOX11. The allocation of shoot‐derived sugar to the roots influences branching, but how its availability is sensed for LRs formation remains unknown. We combine metabolic profiling with cell‐specific interference to show that LRs switch to glycolysis and consume carbohydrates. The target‐of‐rapamycin (TOR) kinase is activated in the lateral root domain. Interfering with TOR kinase blocks LR initiation while promoting AR formation. The target‐of‐rapamycin inhibition marginally affects the auxin‐induced transcriptional response of the pericycle but attenuates the translation of ARF19, ARF7, and LBD16. TOR inhibition induces WOX11 transcription in these cells, yet no root branching occurs as TOR controls LBD16 translation. TOR is a central gatekeeper for root branching that integrates local auxin‐dependent pathways with systemic metabolic signals, modulating the translation of auxin‐induced genes.
Keywords: Arabidopsis thaliana, auxin, lateral root, metabolism, TOR
Subject Categories: Plant Biology
Shoot‐derived sugar activates TOR signalling to initiate lateral root formation.

Introduction
Plants assimilate atmospheric CO2 in their leaves and convert it into simple sugars by photosynthesis. Sucrose is the predominant sugar transported from the source tissues to heterotrophic sink tissues, where it is hydrolyzed to fructose and glucose that fuels growth and development.
The root system fulfills essential functions, and its architecture is postembryonically determined by lateral roots (LRs), which originate from the growing primary root, and adventitious roots (ARs), which are formed from stems or leaf tissues in response to environmental and physiological stresses (Bellini et al, 2014). The root system is an obligate sink organ, and mounting evidence suggests that allocating sugars to roots drives primary root growth and lateral root development, two main determinants of the root system architecture. Shortly after germination, light triggers root growth via the transport of photosynthesis‐derived sugar into the root tip (Kircher & Schopfer, 2012), and increased photosynthetic rates in aboveground tissues correlate with increased LR formation (Crookshanks et al, 1998). Crosstalk between carbon metabolism and phytohormone signaling, mainly auxin signaling, has been linked to the modulation of root system architecture (Sairanen et al, 2012). Regardless of the known role sugar plays in LR development, our knowledge about how sugars and glucose modulate LR formation at the molecular level remains unknown.
Lateral root formation is an auxin‐controlled process. In Arabidopsis thaliana (hereafter Arabidopsis), it occurs through the activation of pericycle cells facing the xylem pole that become founder cells and undergo a series of cell divisions to form a primordium that emerges from the primary root (Malamy & Benfey, 1997). The earliest marker of LR initiation is their radial swelling, repolarization, and nuclei migration toward the common anticlinal wall (Schütz et al, 2021). Additional founder cells are recruited (Torres‐Martínez et al, 2020), which further proliferate and form a dome‐shaped LR primordium (LRP) (Lucas et al, 2013). Upregulation of auxin signaling (Dubrovsky et al, 2008) and GATA23 expression (De Rybel et al, 2010) are two molecular markers associated with LR founder cells and initiation. Auxin‐dependent gene regulation plays a major role in all stages of LR development. It occurs through TRANSPORT INHIBITOR RESISTANT 1/AUXIN SIGNALING F‐BOX (TIR1/AFB) induced degradation of the AUXIN/INDOLEACETIC ACID (Aux/IAA) repressors that frees the transcriptional activators AUXIN RESPONSE FACTORS (ARFs) inducing expression of downstream genes (Blázquez et al, 2020). During LR initiation, Aux/IAA 14 (IAA14, SOLITARY ROOT), ARF7, and ARF19 are necessary for cell cycle entry (Fukaki et al, 2002) and activation of LATERAL ORGAN BOUNDARY (LBD) 16, a transcription factor, is required for the asymmetric division of these cells (Okushima et al, 2007). LR and AR have similar mechanisms for primordium development but are triggered by divergent initial processes (Bellini et al, 2014). WUSCHEL‐RELATED HOMEOBOX11 (WOX11) is required to initiate ARs from hypocotyl and activates LBD16 (Sheng et al, 2017). While WOX11 is not involved in LR initiation when plants are vertically grown on medium, the primary root can produce both WOX11‐mediated roots and ARF7/19‐mediated LRs when plants are grown in soil or upon wounding (Sheng et al, 2017). This noncanonical WOX11‐mediated primary root branching (also called “adventitious lateral root,”; Ge et al, 2019) contributes to the plasticity of the Arabidopsis root system architecture. The cell and mechanistic bases of lateral root and adventitious root initiation start to be elucidated (Atkinson et al, 2014; Santos Teixeira & Ten Tusscher, 2019). However, insight into how the plant's metabolic status is integrated into regulating this developmental program is mostly unknown.
Energy availability perception is mediated in plants by two evolutionarily conserved and counteracting kinases (Shi et al, 2018). The SUCROSE NONFERMENTING1‐RELATED PROTEIN KINASE1 (SnRK1) promotes catabolic metabolism, contrasting the TARGET OF RAPAMYCIN (TOR) kinase that promotes anabolic, energy‐consuming processes. TOR forms a complex (TORC) consisting of TOR and the TOR‐interacting proteins RAPTOR (regulatory‐associated protein of mTOR, RAPTOR1A, and RAPTOR1B) and LST8 (small lethal with SEC13 protein 8, LST8‐1, and LST8‐2) (Menand et al, 2002; Moreau et al, 2012). While tor‐null mutants are embryonic lethal (Menand et al, 2002), the predominantly expressed regulatory proteins RAPTOR1B and LST8‐1 show viable mutant phenotypes (Salem et al, 2017). TORC is activated by nutrients (Dobrenel et al, 2016) such as glucose and branched‐chain amino acids (Cao et al, 2019) as well as phytohormones such as auxin (Schepetilnikov et al, 2017). It phosphorylates targets linked to the cell cycle, translation, lipid synthesis, N assimilation, autophagy, and ABA signaling (Shi et al, 2018). The usage of inducible TOR knockdown lines (Xiong et al, 2013) and specific chemical inhibitors such as AZD8055 (Montané & Menand, 2013) led to the discovery of a mechanistic connection between TOR and its phosphorylation substrates to a multitude of developmental processes (Van Leene et al, 2019). In particular, TOR is essential for activating the embryo‐derived root and shoot meristems during the photoautotrophic transition (Xiong et al, 2013). While TOR plays a central role in coordinating the energy status of plants with several developmental programs from embryo development to senescence (Shi et al, 2018), it remains unknown whether TOR plays a role in the postembryonic establishment of new meristems like in the case of LR formation. Two reports suggest a possible link between energy availability sensing pathways and LR formation. Using an engineered rapamycin‐sensitive version of TOR in potato, it was shown that TOR is necessary for hypocotyl‐borne (adventitious) root formation (Deng et al, 2017) and, recently, SnRK1 was shown to be required for LR formation induced by unexpected darkness (Muralidhara et al, 2021).
Here, we examine the role of TOR in LR formation and its interplay with auxin signaling. Using a combination of root metabolomics profiling, sugar metabolism manipulation, chemical/genetic and tissue‐specific inhibition of TOR‐dependent signaling, and genome‐wide profiling of TOR effects on transcriptome and translatome, we present evidence that TOR activation in the LR founder cells and subsequent LR initiation are coordinated by high glycolysis rates depending on shoot‐derived sugar. We further show that TOR inhibition marginally affects the auxin‐induced transcriptional response of the pericycle but attenuates the translation of ARF19, ARF7, and LBD16. TOR inhibition also induces the AR program by inducing WOX11 transcription, yet as it simultaneously attenuates the translation of LBD16, no primary root branching occurs. These data place TOR as a central gatekeeper for postembryonic root branching that integrates local auxin‐dependent pathways with systemic metabolic signals, modulating the translation of auxin‐induced gene expression.
Results and Discussion
Plants with impaired lateral root formation hyperaccumulate starch in foliage
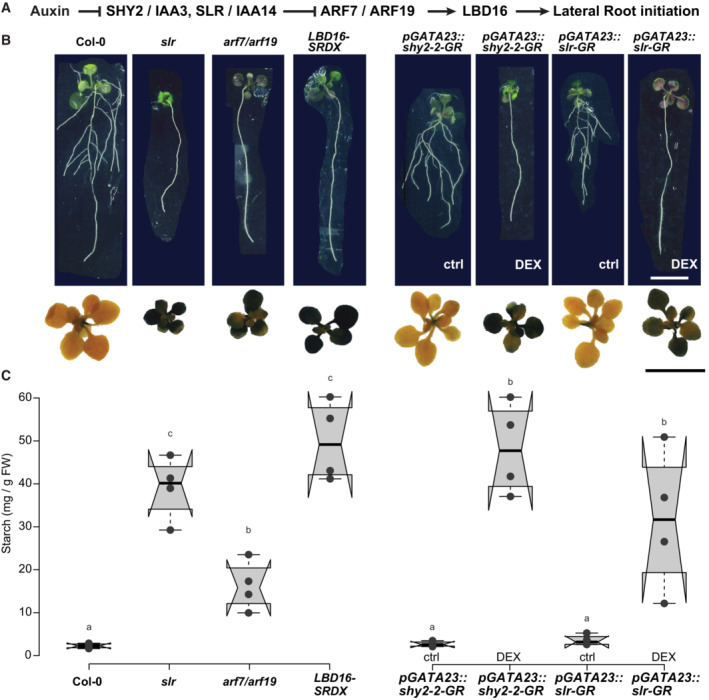
Lateral root formation is an energy‐demanding process that depends on shoot‐derived carbon. Photoassimilates not consumed immediately by the plant's foliar metabolism are stored as transitory starch granules and consumed in the dark until the onset of the light period (Graf et al, 2010). We first set out to assess how LR carbon demand impacts foliar carbon metabolism by performing starch staining in seedlings impaired in different steps of the auxin signaling cascade, specifically controlling LR formation (Fig 1A). At the end of the dark period, starch accumulation was limited in wild‐type Col‐0 seedlings, indicated by the characteristic purple stain (Fig 1B). Globally, impairing auxin signaling in LR‐less iaa14/solitary root (slr) and arf7/arf19 mutant plants resulted in dark purple coloration throughout the leaves, indicative of higher levels of starch accumulation (Fig 1B). ARF7 and ARF19 are regulators for the transcriptional activation of LBD16, and plants expressing a dominant repressor version of LBD16 (LBD16‐SRDX) do not form LR (Goh et al, 2012). Starch staining in LR‐less LBD16‐SRDX seedlings led to comparably intense dark coloration as observed for slr foliage, indicating that blocking LR formation by interfering with the auxin signaling cascade or its direct targets leads to hyperaccumulation of photoassimilates in leaves. Notably, starch accumulation in arf7/arf19 was less pronounced. To ascertain that this increased accumulation of starch was caused by the lack of LRs and not a systemic effect of interfering with auxin signaling, we performed starch staining in pGATA23::shy2‐2‐GR and pGATA23::slr1‐GR plants. These lines express dominant repressor versions of SLR and SHY2/IAA3, specifically in the pericycle upon application of dexamethasone (DEX), resulting in an inducible, pericycle‐specific inhibition of auxin signaling and LR formation (Ramakrishna et al, 2019). Whereas control‐treated lines showed faint purple coloration in leaves comparable with that in Col‐0 plants, upon DEX treatment, we observed intense starch accumulation in the leaves, indicating that, similar to the global effect observed in the slr, arf7/19 and gLBD16‐SRDX mutants, blocking LR formation specifically in the root pericycle is sufficient to induce starch hyperaccumulation in foliage (Fig 1B). This hyperaccumulation of starch in LR‐less genotypes can be observed as early as 12 DAG (Appendix Fig S1). We did not observe any differences in foliar starch accumulation between these genotypes at the end of the light period, indicating that these plants do not differ in their ability to accumulate starch but to consume starch during the dark period (Appendix Fig S2). We quantified starch abundance to assess the differences in starch accumulation in more detail (Fig 1C). The elevated starch levels in LR‐less genotypes confirmed the observations of the starch staining. Notably, starch accumulation is less pronounced in the arf7/arf19 double mutant compared with the other LR‐less mutants. This observation might be attributed to other regulatory factors of the primary metabolism in leaves that might not be affected in arf7/arf19—possibly other ARFs downstream of SLR—that may contribute to the sink strength of the root. Taken together, these observations point toward LR formation and its associated resource consumption to drive an increase in demand for shoot‐derived carbon sources, in agreement with starch being a major integrator of plant growth regulation (Sulpice et al, 2009).
Figure 1. Lateral root deficiency leads to starch hyperaccumulation in leaves.
-
ASchematic representation of key components of the auxin signaling module acting during lateral root initiation.
-
BImages of rosettes and seedlings (15 DAG) of Col‐0, arf7/arf19, slr, and gLBD16‐SRDX as well as the inducible lateral‐root‐less lines pGATA23::shy2‐2‐GR and pGATA23::slr1‐GR grown on DMSO control medium or 30 μM Dexamethasone (DEX), Scale bar 1 cm. The rosettes were stained with a Lugol's Iodine solution for starch accumulation at the end of the dark period (representative images of n = 3 biological replicates).
-
CBox plots of starch quantification in rosette tissues at 15 DAG (n = 4 biological replicates). Comparison between samples was performed by one‐way ANOVA and post‐hoc Tukey HSD Test (α = 0.05); different letters indicate significant differences.
Source data are available online for this figure.
Auxin‐triggered pericycle activation depends on shoot‐derived carbohydrate catabolism in the root
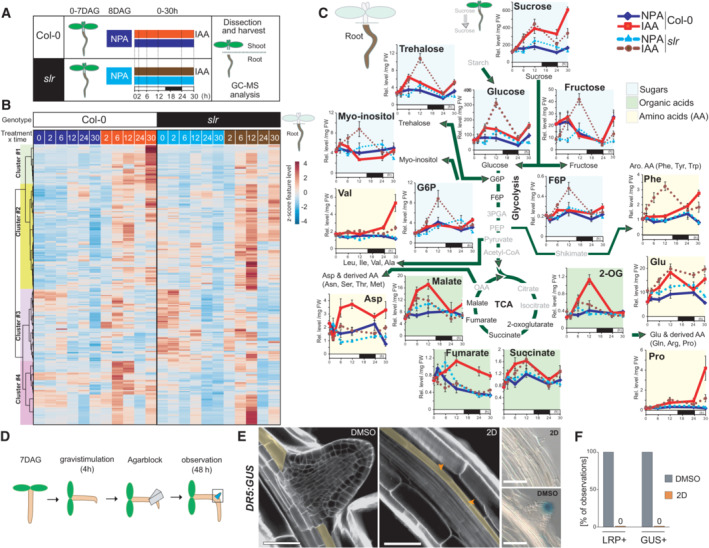
To monitor changes in central carbon pathways resulting from the metabolism of shoot‐derived carbohydrates and associated with the formation of LR, we conducted nontargeted metabolomics by gas chromatography coupled with mass spectrometry from shoot and root samples collected after synchronous activation of the pericycle (Himanen et al, 2002). Briefly, after a pretreatment with the auxin transport inhibitor NPA (N‐1‐naphthylphthalamic acid, 10 μM for 24 h) that prevents pericycle activation, 7‐day‐old seedlings were shifted to a medium containing auxin (Indole‐3‐acetic acid, IAA 10 μM) to activate the entire pericycle synchronously. This induction system allows the synchronization of the pericycle and recapitulates the onset of the lateral root initiation program (Vanneste et al, 2005; Lavenus et al, 2013; Ramakrishna et al, 2019; Vilches Barro et al, 2019; Ursache et al, 2021). By activating the entire pericycle, we circumvent that at any given time, only a few cells are activated, and the analysis of the metabolites would be dominated by the nonactivated cells. Shoot and root samples were dissected and collected at six time points (0, 2, 6, 12, 24, and 30 h) after transferring seedlings from NPA to IAA to induce LR formation or maintaining them on NPA as control (Fig 2A). This time series covers LR formation from pericycle activation to stage V. By applying this procedure to wild‐type (Col‐0) and slr mutant seedlings in which pericycle activation is prevented (Vanneste et al, 2005), we aimed to infer a metabolic signature associated specifically with the activation of the pericycle marking LR initiation. To this end, raw metabolomics data were deconvoluted to extract compound‐derived mass spectra used for annotation and statistical analysis. From a pool of more than 400 deconvoluted spectra, we conducted a hierarchical clustering analysis of the top 250 ones that exhibited nonconstant intensity levels across the genotype x treatment x time matrix (Fig 2B; Dataset EV1). This clustering analysis revealed that IAA induced several phases of reconfigurations of the root carbon metabolism in Col‐0, which were largely altered in the slr mutant. Most specifically, cluster #1 comprised IAA‐responsive compounds that were characterized in Col‐0 roots by a slow build‐up rate (reaching maximum values at 30 h post‐IAA); the latter response was mostly impaired in the slr mutant (Fig 2B). Cluster #2 and a subpart cluster #4 were characterized by IAA‐responsive compounds, which reached much greater relative levels in the slr mutant 12 h after transfer to IAA than in Col‐0 roots (Fig 2B).
Figure 2. Increased flux within sugar glycolytic catabolism and connected pathways precede and are essential for LR formation.
-
ASchematic of the experimental setup used for the GC–MS‐based metabolomics profiling.
-
BHeatmap from a hierarchical clustering analysis (HCA) with Ward's linkage showing z‐score normalized relative levels of top 250 most intense compound‐derived spectra (Dataset EV1) exhibiting non‐constant intensity (One‐way ANOVA & FDR‐adjusted P < 0.05) across experimental conditions in roots. Main HCA clusters are color labeled.
-
CMean relative levels (± SE, n = 5 biological replicates, normalized to the ribitol internal standard and per mg fresh weight) for representative metabolites of sugar, glycolytic, tricarboxylic acid, amino acid metabolic pathways in root tissues of Col‐0 (solid lines) and slr (dashed lines) at the indicated time after IAA application. The white and black boxes below the x‐axis indicate light and dark phases during the sampling. Statistical differences for genotype × treatment (NPA‐ vs. IAA‐treated roots) are summarized in Dataset EV1.
-
DSchematic of the experimental setup for induction of LR formation upon local 2‐deoxy‐D‐glucose (2D, 10 mM) treatment.
-
ERepresentative confocal sections of calcofluor counterstained (E, n = 5 biological replicates) and differential interference contrast (DIC) images of GUS‐stained root bends (F, n = 16 biological replicates) in DR5:GUS seedlings treated as indicated 48 h after gravistimulation. In (E), orange arrows indicate the pericycle (marked in yellow). Scale bar: 50 μm.
-
FFraction of root bends forming an LRP and showing DR5 GUS staining after treatment with either control or 2D containing agar blocks (n = 16 biological replicates, ± SE).
Source data are available online for this figure.
We next mined clusters for metabolites associated with these IAA−/slr‐dependent root metabolome responses. In line with the starch staining data, glucose and sucrose levels were slightly higher in slr shoot tissues than in Col‐0 (Appendix Fig S3). Upon shift to IAA, levels of shoot sucrose quickly increased within 2 h, and while they kept increasing for up to 12 h in slr, it plateaued in WT. This and the starch hyperaccumulation in shoots upon LR impairment indicate that LR formation relies on a sucrose transfer from the shoot. In seedlings, photosynthetically derived sucrose has been described as an interorgan signal and fuel to drive the primary root growth (Kircher & Schopfer, 2012). For up to 24 h after shifting to IAA, sucrose levels built up similarly in root tissues of both Col‐0 and slr and then became significantly higher in Col‐0 than slr (Fig 2C), in line with a weaker sink strength of slr resulting from its inability to form LRs. From the clustering, we looked at the levels of added sugars and glycolytic intermediates found to be deregulated in slr roots (Fig 2B and C). The root levels of glucose and fructose derived from sucrose cleavage increased for 6 h upon IAA in Col‐0 and then decreased until the end of the night period. Noticeably, glucose and fructose levels were strongly increased by the IAA treatment in the slr mutant and peaked at 12 h (Fig 2C). This suggests that upon pericycle activation, glucose and fructose derived from sucrose cleavage do not build up, probably due to their catabolism by glycolysis, the latter being reduced in the absence of pericycle activation (slr mutant). A similar IAA‐dependent overaccumulation was detected for several additional sugars enriched within the IAA‐regulated cluster visible at 12 h in slr (Fig 2B), such as the disaccharide trehalose, the polyol myo‐inositol as well as glucose‐6‐P and fructose‐6‐P. Notably, glucose‐6‐P is produced by the hexokinase1 (HXK1), which, when mutated, was reported to reduce LR formation (Gupta et al, 2015) supporting that Glucose‐6‐P levels are instrumental for LR formation, a notion further supported in a recent study that showed that WOX7, a WUSCHEL‐related transcription factor, acts downstream of HXK1 to regulate LR formation (Li et al, 2020). The accumulation of glucose‐6‐P and fructose‐6‐P, two glycolysis intermediates, was in contrast with their normally low steady‐state abundance (e.g., in Col‐0 NPA/IAA conditions) characteristic of their rapid consumption by the glycolytic flux (Arrivault et al, 2009). Interestingly, soluble carbohydrates have been demonstrated to promote auxin biosynthesis (Sairanen et al, 2012). The accumulation of carbohydrates in roots when LR formation is compromised could thus explain the previously reported elevated levels of auxin observed in slr mutants (Vanneste et al, 2005). Downstream in the carbohydrate catabolic pathway, the prolonged increases in the levels of several intermediates of the tricarboxylic acid (TCA), observed in Col‐0 but not in slr, indicate that auxin‐induced LR formation increases the catabolic flux. Levels of several amino acids whose biosynthetic pathways connect to the TCA cycle further indicated that an upregulation of energy‐releasing and amino acid production pathways, previously reported at the transcriptomic level (Dembinsky et al, 2007; Appendix Fig S4) and here backed up by metabolite data, underpins early stages of LR formation and is impaired in slr. Together these data indicate that the auxin‐induced pericycle activation and re‐entry in division are associated with a switch to glycolysis. This observation echoes similar ones in animals where the acquisition of pluripotency has been linked to a switch to glycolysis, supporting the concept of metabolic reprogramming of cell fate (Shyh‐Chang & Ng, 2017).
To verify whether this activation of sugar usage through glycolysis is indeed required for LR initiation and to rule out an effect of exogenous auxin, we induced LR initiation by gravistimulation (Lavenus et al, 2015) in the presence of 2‐deoxy‐d‐glucose (2D) a nonmetabolizable glucose analog blocking glycolysis. 2D was applied locally by an agar block positioned over the root bend (Fig 2D). Whereas in mock‐treated root bends, an LR primordium developed, none were formed upon 2DG treatment where the pericycle cells facing the xylem pole showed no divisions (Fig 2E and F). Auxin accumulation in the pericycle acts as a morphogenetic trigger for LR formation and is one of the earliest markers of LR initiation (Dubrovsky et al, 2008). To monitor auxin accumulation upon glycolysis inhibition, we stained root bends treated with 2DG for DR5:GUS. We did not detect DR5 activity in the bend upon 2DG treatment (Fig 2E and F). These indicate that inhibition of glycolysis blocks LR initiation at a very early stage.
Together these data show that glycolysis and glycolysis‐dependent metabolic activations are required to form an LR.
The TOR complex is activated upon lateral root induction
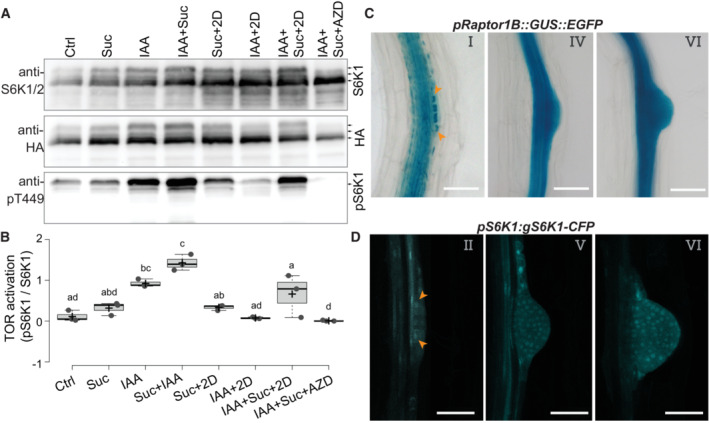
Lateral root formation is an auxin‐induced process regulated by glucose (Gupta et al, 2015) and requires carbohydrate catabolism (our results). The TOR complex (TORC) is required for shoot and root meristems activation (Xiong et al, 2013; Pfeiffer et al, 2016; Li et al, 2017) and is activated by glucose in shoots and roots (Xiong et al, 2013; Pfeiffer et al, 2016; Li et al, 2017) and additionally by auxin in the shoot (Xiong et al, 2013; Li et al, 2017; Schepetilnikov et al, 2017), we hypothesized that re‐activation of cell proliferation during LR induction could lead to TORC activation. We monitored the phosphorylation of the canonical TORC substrate S6K1 in roots upon treatment with sucrose and IAA (Fig 3A and B). Treatment with either sucrose or IAA led to an upregulation of S6K1 phosphorylation, while co‐treatment had a synergistic effect that was fully suppressed by treatment with the TORC inhibitor AZD8055 (Montané & Menand, 2013). Treating roots with IAA led to a glycolytic switch (Fig 2B and C); thus, we sought to check to which extent IAA's effects on TORC activation depend on glycolysis. For this, we repeated the sucrose and IAA treatments in the presence of 2D. Simultaneous provision of external 2D and sucrose did not lower the levels of pS6K1, possibly due to an excess of sucrose. Inhibition of glycolysis without additional sucrose led to a block of S6K1 phosphorylation induced by IAA, indicating that TORC activation by auxin depends on carbohydrate catabolism in roots. This glycolysis‐dependent promotion of TOR activity by auxin could be a specificity of heterotrophic tissues that allows a systemic integration of developmental progression with shoot photosynthetic capacity. Such coupling has been reported for the light‐dependent regulation of alternative splicing in roots triggered by shoot‐photosynthesized sugars and compromised when TOR levels or its activity is reduced (Riegler et al, 2021).
Figure 3. Auxin inducible S6K1 phosphorylation via TOR depends on the activation of glycolysis in the primary root.
-
ARepresentative western blot of root tissues of pUB10:S6K1‐3xHA treated by the indicated combination of auxin (IAA, 10 μM), sucrose (Suc, 110 mM), 2‐deoxy‐d‐glucose (2D, 20 mM) and AZD8055 (AZD, 10 μM) and probed with anti‐S6K1/2, anti‐HA or anti‐S6K1‐T449P. Blot is one of three biological replicates.
-
BQuantification of the relative S6K activation. Box plots show three biological replicates, and comparison between samples was performed by one‐way ANOVA and post‐hoc Tukey HSD Test (α = 0.05); different letters indicate significant differences.
-
CRepresentative DIC images showing RPT1B promoter expression at different stages of LR development in 10 DAG seedlings. Orange arrowheads indicate the accumulation of reporter signals in the dividing lateral root founder cells. Scale bars: 50 μm.
-
DRepresentative confocal section showing S6K1 expression in different stages of LR development in 10 DAG pS6K1:gS6K1‐CFP seedlings. Orange arrowheads indicate the accumulation of reporter signals in the dividing lateral root founder cells. Scale bars: 50 μm.
Source data are available online for this figure.
The previous results do not identify in which cells TOR is activated. To pinpoint in which tissues the TORC is present and active, we first used a reporter for the TORC subunit RAPTOR1B and detected its expression in the stele, LR founder cells of the pericycle, and LR primordia (Fig 3C). This expression pattern is similar to the one reported for the other TORC subunit LST8 (Moreau et al, 2012). It suggests that TORC is present in the stele, the activated pericycle, founder cells, and forming LR. We also monitored in which cells S6K1 is expressed using a CFP‐tagged genomic clone. This reporter specifically marked the actively dividing LR founder cells (Fig 3D), confirming an earlier report (Zhang et al, 1994). These results suggest that TORC is turned on in the activated pericycle cells but do not exclude that it could also be activated in the stele tissue important for pericycle activation, such as the protoxylem.
The TOR complex is required for LR formation
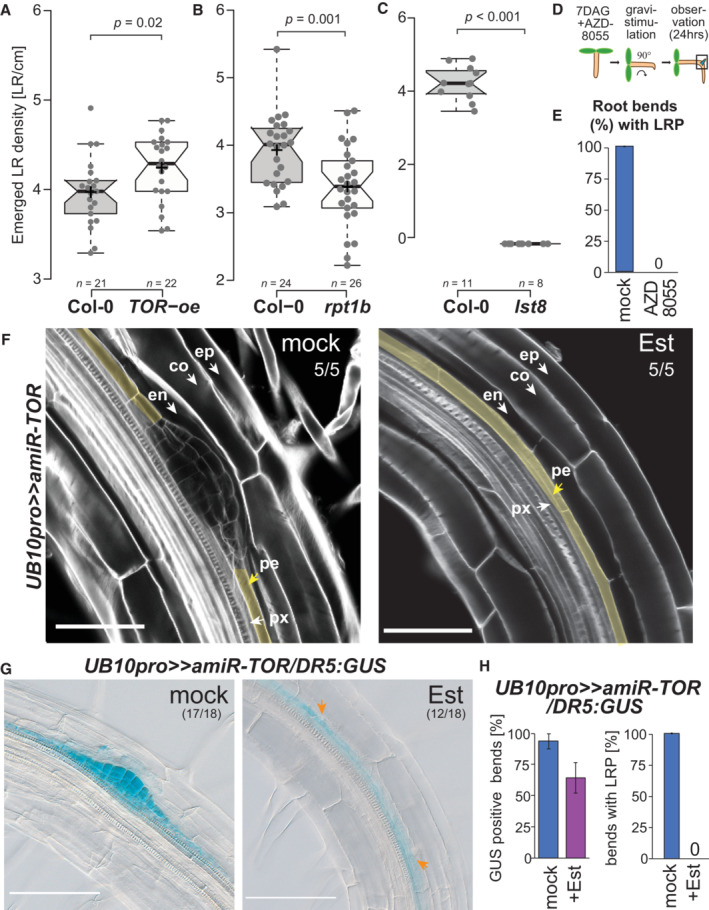
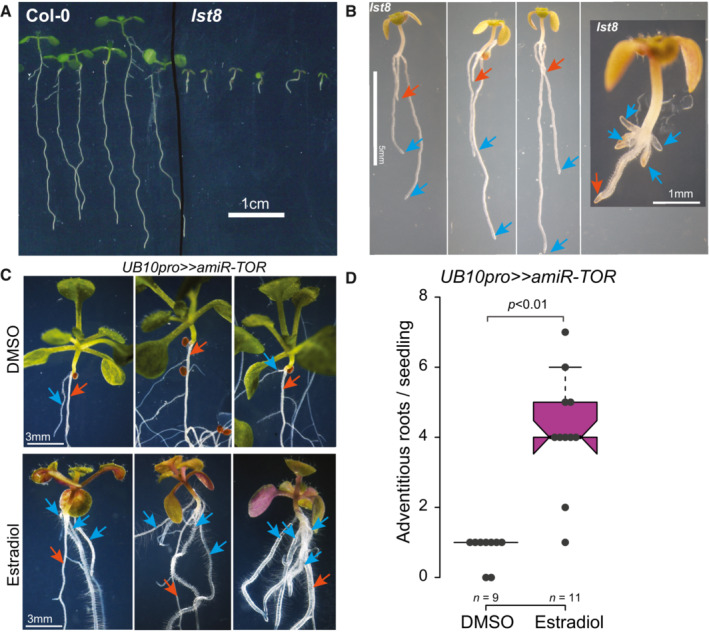
The glucose‐TOR signaling and the phytohormone jasmonate were recently shown to determine the branching angle of LR (Sharma et al, 2022). We looked at LR formation in plants with altered TORC levels or activity to test whether TORC is necessary for forming LR. We first used a TOR overexpression line (TOR‐oe; Deprost et al, 2007) and observed longer primary roots and increased density of emerged LR, indicating that elevated TOR levels can promote LR formation (Fig 4A; Appendix Fig S5). TOR‐null mutants are embryo‐arrested (Menand et al, 2002); we thus first quantified LR density in raptor1b (rpt1b) and lst8, two viable mutants that lead to reduced TOR activity (Moreau et al, 2012; Salem et al, 2017). Lateral root density was reduced in rpt1b and lst8, indicating that the full TORC activity might be necessary for proper LR formation (Fig 4B and C; Appendix Fig S5). Notably, the development of hypocotyl‐born adventitious roots was increased in the lst8 mutant (Fig EV1). To further confirm that TORC activity is required for LR formation, we treated wild‐type seedlings with AZD8055 (AZD). Lateral root initiation occurs in a region of the root where the endodermal Casparian stripe severely limits the apoplastic diffusion (Vermeer et al, 2014); we thus had to treat seedlings with 10 μM AZD, a concentration that fully inhibits primary root growth (Montané & Menand, 2013). To circumvent this shortcoming and be able to monitor the effect of TORC inhibition on LR formation, we turned to gravistimulation‐induced LR formation (Fig 4D, Lavenus et al, 2015). In these conditions, AZD treatment led to a complete block of LR formation (Fig 4E). To test the contribution of TOR itself to LR formation, we designed a ß‐estradiol (Est) inducible artificial miRNA against TOR that we first expressed from the UBIQUITIN promoter (UB10pro>>amiR‐TOR). After 24 h of Est treatment, TOR mRNA abundance was reduced to < 25% of that of the DMSO control indicating efficient knockdown of TOR mRNA (Appendix Fig S6). When LR formation was induced by gravistimulation in these conditions, we did not observe any division of the pericycle in the bend. At the same time, we observed stage III LR primordia in DMSO‐treated plants (Fig 4F). Notably, we observed that upon TOR knockdown, while LR formation was inhibited, AR formation from the hypocotyl was not (Fig EV1). Together, these data indicate that chemical and genetic inhibition of TOR kinase leads to a block of LR initiation and promotes AR formation in the hypocotyl.
Figure 4. TORC is required in the pericycle for lateral root formation.
-
A–CBox plots of emerged LR in TOR‐oe (A), rpt1b (B), and lst8 (C) at 14 DAG. Comparison between samples was performed by one‐way ANOVA and post‐hoc Tukey HSD Test (α = 0.05); different letters indicate significant differences. The number of roots scored is indicated.
-
DSchematic of the experimental setup used for scoring LR formation by gravistimulation upon inhibition of TOR by AZD8055 (10 μM).
-
EProportion of bends developing lateral root primordia after transfer to AZD8055 containing media and gravistimulation for 24 h (n = 10).
-
FRepresentative confocal images of calcofluor counterstained bends of 7DAG UB10pro>>amiR‐TOR seedlings following a 24 h pre‐treatment with mock (DMSO) or ß‐Estradiol (10 μM) and subsequent 24 h gravistimulation (ep: epidermis, co: cortex, en: endodermis, pe: pericycle, px: protoxylem). Numbers indicate the proportion of root bends with the depicted phenotype. Scale bar: 50 μm.
-
GRepresentative DIC images of bends in 7 DAG UB10pro>>amiR‐TOR/DR5:GUS seedlings stained for GUS activity after a 24 h pre‐treatment with mock (DMSO) or ß‐Estradiol (Est, 10 μM) and subsequent 24 h gravistimulation. The proportion of root bends showing the depicted phenotype is indicated. Scale bar: 100 μm.
-
HFraction of bends developing lateral root primordia and stained for GUS activity in primary root vasculature of UB10pro>>amiR‐TOR/DR5:GUS, n = 18 bends.
Source data are available online for this figure.
Figure EV1. Impairment of the TOR machinery causes the increased formation of adventitious roots.
-
APrimary root growth in 14‐day‐old lst8 seedlings is reduced compared to Col‐0.
-
BClose‐ups of 14‐day‐old lst8 seedlings with numerous adventitious roots (blue arrows) on the hypocotyl; the primary root is indicated with a red arrow.
-
CTOR knockdown induces the formation of adventitious roots. 14‐day‐old UB10pro>>amiR‐TOR seedlings transferred at 8 DAG to Est develop more adventitious roots from the hypocotyl (blue arrows) than DMSO‐treated seedlings. The red arrows indicate the primary root.
-
DDistribution of the number of hypocotyl‐borne adventitious roots produced in 14‐day‐old UB10pro>>amiR‐TOR seedlings transferred at 8 DAG to Est or DMSO. The number of biological replicates is indicated. Unpaired t‐test.
Source data are available online for this figure.
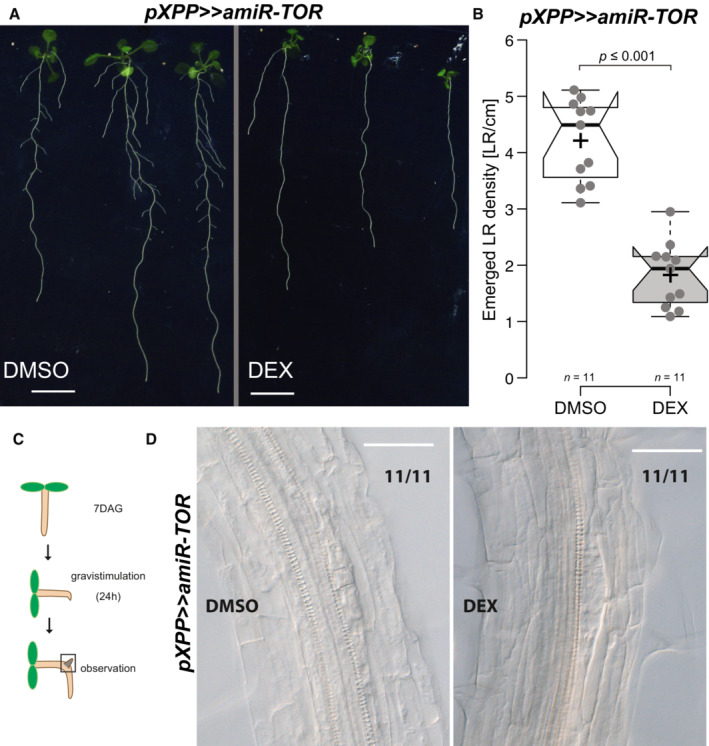
As an exogenous application of sugar or auxin can promote LR formation and increase TOR activity, we checked whether the block in LR formation induced by knocking down TOR could be reversed by treatments with auxin and/or sucrose. Neither sole nor combined applications of sucrose or auxin could rescue the inhibition of LR formation induced by the TOR knockdown (Appendix Fig S7). Reducing the abundance or activity of TOR blocks LR formation early, indicating an essential role for TOR. To determine whether TOR was required, particularly in the LR founder cells, we knocked down TOR in the xylem pole pericycle (XPP) cells from which LR founder cells derive (Parizot et al, 2008). For this, we drove the expression of the amiR‐TOR from the XPP‐specific promoter (Andersen et al, 2018) using a dexamethasone (Dex) inducible expression system (XPPpro>>amiR‐TOR). Impaired TORC function leads to starch accumulation in foliage (Caldana et al, 2013). We observed that Est‐treated UB10pro>>amiR‐TOR hyperaccumulated starch in their leaves (Appendix Fig S8). Similarly, we observed intense starch accumulation around the leaf vasculature in DEX‐treated XPPpro>>amiR‐TOR, suggesting TOR knockdown (Appendix Fig S8). In the root, induction of amiR‐TOR in the XPP cells led to a severe reduction in the number and density of emerged LR formed compared with mock‐induced plants (Fig EV2A and B). However, induction of LR by gravistimulation did not impair initiation events (Fig EV2C and D). Given that the promoter driving the expression of the amiR‐TOR switches off as soon as LR initiation starts (Andersen et al, 2018), this indicates that sustained downregulation of TOR is required in the pericycle to suppress the initiation of LR formation. The severe reduction in the number and density of emerged LR may be a consequence of the re‐activation of the XPPpro>>amiR‐TOR in the newly formed LR primordium.
Figure EV2. Xylem‐pole‐pericycle specific knockdown of TOR expression impairs the emergence of LR primordia.
-
APhenotype of pXPP>>amiR‐TOR seedlings grown on DMSO or 30 μM Dexamethasone (DEX) at 14 DAG. Scale bar: 5 mm.
-
BDensity of emerged LR in pXPP>>amiR‐TOR upon control or DEX treatment in 14‐day‐old seedlings.
-
CSchematic of the experimental setup used for scoring LR initiation by gravistimulation for 24 h, upon control or DEX treatment.
-
DRepresentative DIC images of root bends of 7DAG pXPP>>>>amiR‐TOR seedlings raised on (DMSO) or DEX and subsequent 24 h gravistimulation. Numbers indicate the proportion of root bends with the depicted phenotype. Scale bar: 50 μm.
Source data are available online for this figure.
To determine whether auxin accumulation was compromised in the TOR knockdown, we crossed the pDR5::GUS to the UB10pro>>amiR‐TOR line. In control conditions, we observed pDR5::GUS accumulation in the apical region of the stage III LR‐primordia 24 h after gravistimulation. Upon TOR knockdown, we detect in ~ 60% of the LR a faint DR5::GUS signal in the pericycle and the protoxylem (Fig 4G and H). Collectively, these data suggest that while TOR is required for LR initiation, TOR knockdown does not cause complete inhibition of auxin signaling.
TOR inhibition moderately affects the transcriptional auxin response associated with LR formation
While the reduction of TOR abundance or inhibition of its activity blocks LR formation, it does not fully block auxin signaling in these cells, suggesting that it is required either downstream or parallel to the auxin‐induced LR formation developmental program. To obtain a genome‐wide picture of the effects of TOR knockdown during the early phase of LR formation, we compared by RNA‐seq the transcriptomes of roots 6 h after the auxin‐induced synchronous pericycle activation (Himanen et al, 2002) in the inducible UB10pro>>amiR‐TOR background (Fig 5A). Transcriptome analysis identified 1,141 auxin‐responsive genes in control conditions (Appendix Fig S9). Upon TOR knockdown, the expression of these genes was barely changed (Fig 5B). We verified that inhibition of TOR activity by treatment with AZD8055 led to similar effects (Appendix Fig S10). Although no morphological sign of LR initiation in the pericycle could be observed upon TOR knockdown or inhibition, the auxin‐induced transcriptional response was globally unchanged. The SLR/IAA14 protein is a central regulator of LR initiation that controls the auxin‐dependent expression of genes in the pericycle essential for LR initiation (Fukaki et al, 2002). We thus examined in detail how this set of IAA‐induced genes behaved upon TOR knockdown. For this, we took advantage of an existing dataset that profiles the response of the pericycle in similar conditions upon inhibition of SLR‐dependent auxin signaling in the pericycle (Ramakrishna et al, 2019) and identified 475 SLR‐dependent genes responsive to auxin. These genes behaved the same in TOR knockdown and controls, indicating that although LR formation is inhibited, the transcriptional SLR‐dependent response to auxin in the pericycle is globally unaffected when TOR levels are reduced. Together these data suggest that upon TOR reduction, founder cells still perceive and respond transcriptionally to auxin but appear unable to transform this response into an LR initiation event.
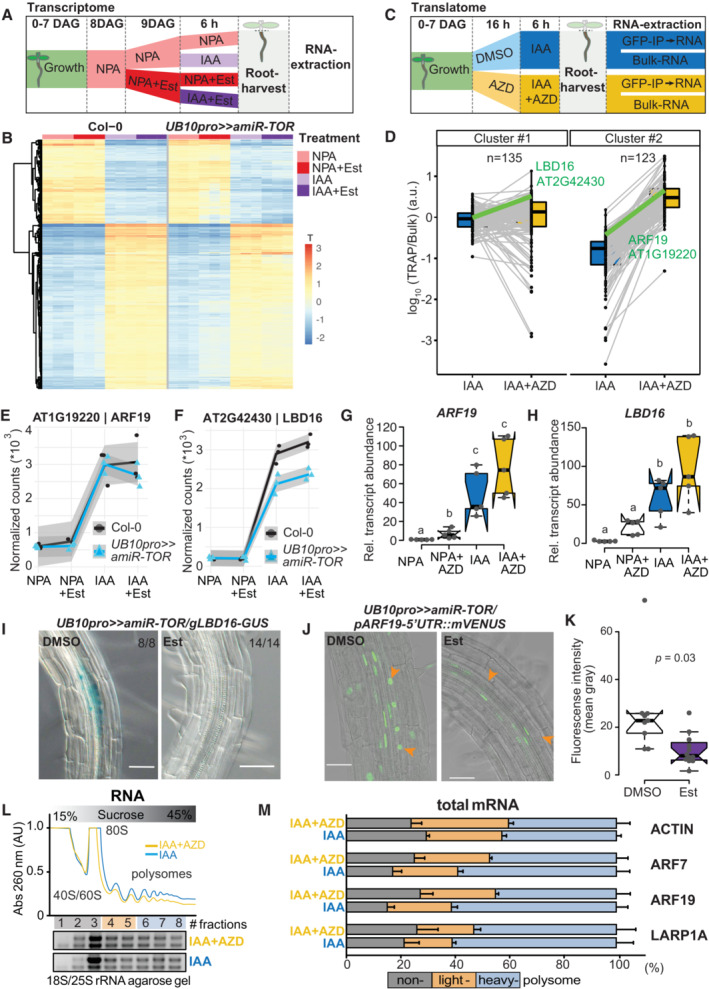
Figure 5. Effect of TOR on the auxin‐induced lateral root transcriptome and translatome.
-
ASchematic of the experimental setup used to profile the impact of TOR knockdown on the transcriptome during LR formation. IAA, NPA, and Est were all used at 10 μM concentration.
-
BHeatmap from a k‐means clustering analysis for 1,141 IAA‐dependent transcripts (log fold change > 1 & FDR < 0.05). n = 3 biological replicates.
-
CSchematic of the experimental setup used to profile the impact of TOR inhibition on the translatome during LR formation. IAA and AZD8055 were used at 10 μM.
-
DTranslational response (reads associated to ribosomes (TRAP) divided by reads in Bulk RNA) of 258 auxin‐induced genes. K‐means revealed two clusters, with mild (#1) to strong (#2) shifts in translation response upon TOR inhibition. The profiles of ARF19 and LBD16 are highlighted in green. n = 3 biological replicates.
-
E, FAbundance of ARF19 (E) and LBD16 (F) transcripts in RNAseq samples. mRNA accumulation in response to auxin is comparable for both, whether TOR is knocked down. n = 3 biological replicates.
-
G, HRelative expression levels (normalized to ACTIN) of ARF19 (G) and LBD16 (H) measured by RT–qPCR upon TOR activity inhibition with AZD8055. Comparison between samples was performed by one‐way ANOVA. Different letters indicate significant differences based on a post‐hoc Tukey HSD Test (n = 5 biological replicates, α = 0.05).
-
IDistribution of GUS‐staining in UB10pro>>amiR‐TOR/gLBD16‐GUS seedlings 24 h after bending is absent if previously treated for 24 with Est. The proportion of root bends with the depicted phenotype is indicated.
-
JRepresentative confocal images of bends of 7 DAG UB10pro>>amiR‐ TOR/pARF19‐5′UTR::mVENUS seedlings following a 24 h pre‐treatment with mock (DMSO) or ß‐ Estradiol and subsequent 24 h gravistimulation. Scale bar: 50 μm, n = 9 root bends.
-
KSignal (mean gray values) in the nuclei of the pericycle cells of UB10pro>>amiR‐TOR/pARF19‐5′UTR::mVENUS. Significant differences between DMSO and Est‐treated roots based on paired t‐test, n = 9 root bends.
-
L, MTotal lysates prepared from lateral roots treated or not with IAA and AZD were fractionated through sucrose gradients, and the relative redistribution (percentage of total) of ACTIN, ARF7, ARF19, and LARP1 mRNAs in every 8 fractions were studied by RT–qPCR analysis. (L) Polysome profiles. 40S, small ribosomal subunit; 60S, large ribosomal subunit; 80S, mono‐ribosome; polysomes, polyribosomes. AU is arbitrary units of RNA absorbance at A260 nanometers. (M) RT–qPCR analysis of mRNA redistribution through a sucrose gradient (8 fractions collected). Translation efficiency was computed as a percentage of mRNA in non‐polysome fractions (40/60/80S; fractions 1–3) against both lights (fractions 4–5) and heavy polysomes (fractions 6–8). The plot is representative of three independently performed experiments with similar results. Data are mean ± SEM.
Source data are available online for this figure.
TOR affects the translation of auxin‐responsive transcription factors
The contrast between the mild effect of TOR knockdown on the root transcriptome and the strong block of LR formation prompted us to investigate the effects of TOR inhibition on the translatome. To this end, we performed targeted purification of polysomal mRNA (TRAP‐Seq; Vragović et al, 2015) using a transgenic line ubiquitously expressing a GFP‐tagged RPL18 (Mustroph et al, 2009) 6 h after the synchronous pericycle activation by auxin treatment upon inhibition of TOR activity (AZD8055 treatment, AZD). To correct for the abundance of mRNA, bulk RNA‐Seq was performed on the same samples and used to normalize the reads purified with the ribosomes (Fig 5C). This TRAP/Bulk ratio measures the fraction of mRNA associated with ribosomes, be it polysomes or monosomes, and provides an indication of the degree of translation of a particular mRNA. The bulk RNA‐seq data analysis identified 271 transcripts upregulated upon IAA treatment. Although this number is reduced compared with the transcriptome analysis due to the absence of NPA pretreatment, 80% of these genes were also differentially expressed in the UB10pro>>amiR‐TOR transcriptome upon IAA treatment (Appendix Fig S11). Clustering these transcripts according to their TRAP/Bulk ratio between IAA and IAA + AZD conditions revealed two clusters. Cluster #1 consists of genes with moderate change in TRAP/Bulk ratio comparing IAA to IAA + AZD, whereas the change was more important for cluster #2 (Fig 5D). Examining these two clusters, we selected two candidates for further characterization, LBD16 (cluster #1) and ARF19 (cluster #2), both involved in LR initiation (Okushima et al, 2007).
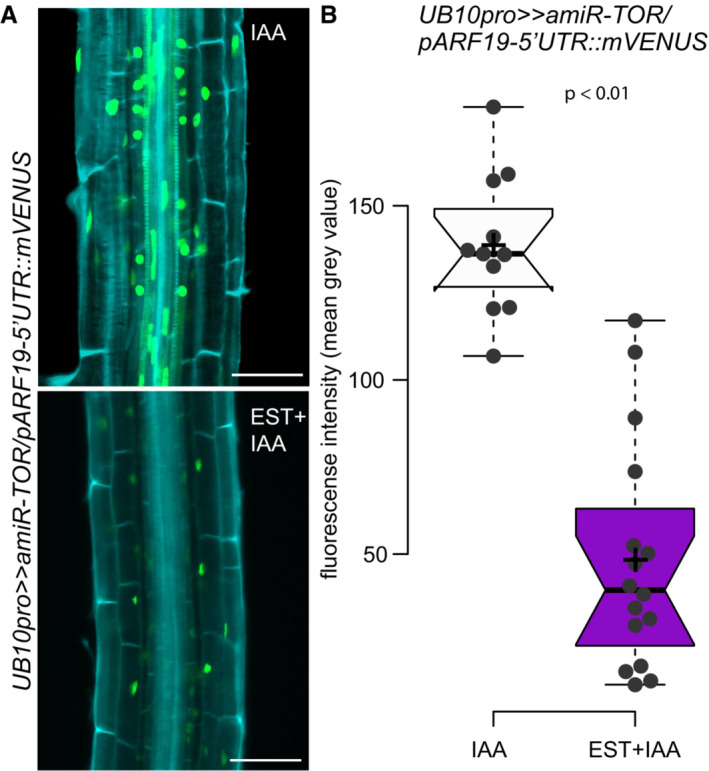
In the UB10pro>>amiR‐TOR transcriptome, these two genes were induced by IAA, and this induction was not affected by knocking down TOR (Fig 5E and F). This result was independently confirmed by RT–qPCR upon inhibition of TOR activity by AZD8055 (Fig 5G and H), indicating that transcription of these genes is not affected by TOR abundance or activity. In the translatome data, both genes had a higher TRAP/Bulk ratio upon inhibition of TOR activity. This suggests that the translation of these genes is different when TOR is inhibited. To verify this, we crossed the UB10pro>>amiR‐TOR line to translational reporters for ARF19 and LBD16 and monitored the effect of TOR knockdown on the expression of the reporters. For ARF19, the expression of the mVenus reporter is controlled by the ARF19 promoter, the 5′UTR, and the 1st intron (pARF19‐5′UTR::mVenus; Truskina et al, 2021). For LBD16, we used a GUS‐tagged genomic clone (Sheng et al, 2017). In both cases, whereas in control conditions, expression of the reporters could be detected in the cells of the LR primordium, upon TOR knockdown, their translation was severely reduced while still present in the neighboring cells (Fig 5I–K). This indicates that TOR controls the expression of these two genes during LR initiation at the translational level. Similar results for ARF19 accumulation were obtained in roots following synchronized LR‐induction via auxin treatment, confirming the comparability of gravitropic induction (single LR) and proliferation of numerous LRs using the lateral root inducible system (Fig EV3). To further confirm that TOR can regulate the expression of genes at the translation level, we looked at the association of the endogenous ARF19 transcript with ribosomes in wild‐type plants treated or not with AZD8055 during auxin‐induced LR formation. Comparing the polysome profile upon TOR inhibition to the control revealed a shift from heavy to light fractions indicative of a reduced ribosome processivity (Fig 5L and M). This shift was comparable with the one observed for LARP1, a transcript whose translation is TOR‐dependent (Scarpin et al, 2020). Note that the distribution among ribosomal fractions was similar under both conditions for the housekeeping gene ACTIN (Fig 5M). As ARF19 and ARF7 are jointly essential for LR initiation (Okushima et al, 2007), we also profiled the association of ARF7 mRNA, whose expression is not auxin‐induced, with ribosomes. Like ARF19, ARF7 mRNA shifted from heavy to light fractions upon TOR inhibition, indicating that its translation is controlled by TOR (Fig 5M). Together these data show that the translation of both key transcription factors mediating auxin signaling during LR initiation is modulated by TOR. Intriguingly, the 5′UTR of both ARF7 and ARF19 mRNA contains several upstream open reading frames (uORF) that require a TOR‐dependent translation re‐initiation step to allow expression of the main ORF (Schepetilnikov et al, 2013), providing a likely mechanism by which TOR could regulate the expression of these genes. Collectively, our data support a model in which TOR acts as a metabolic gatekeeper for LR formation by locally integrating the availability of shoot‐derived photoassimilates with the auxin‐mediated LR developmental program through control of the translation of key transcription factors. Such a model would ensure the integration and coordination of the developmental and metabolic cues required to form a new organ. Given the vast array of TOR outputs, it is likely that TOR may exert its gatekeeper role through additional mechanisms, such as promoting cell cycle progression via E2F as previously established (Xiong et al, 2013).
Figure EV3. Silencing TOR‐expression reduces ARF19 in TOR‐deficient pericycle cells after synchronized LR‐induction.
-
ARepresentative confocal images of mVenus accumulation in 7 DAG UB10pro>>amiR‐TOR/pARF19‐5′UTR::mVENUS seedlings. Seedlings were pre‐treated for 24 h with mock (DMSO) or 10 μM ß‐Estradiol to induce TOR knockdown and then transferred to 10 μM IAA to induce LR formation synchronously. Scale bar: 50 μm, n ≥ 11 individual roots.
-
BQuantification of mean gray values in the nuclei of the pericycle cells. Significant differences between mVENUS‐signal of 10 μM IAA treated seedlings either DMSO or Est pre‐treated roots based on paired t‐test.
Source data are available online for this figure.
TOR inhibits WOX11 expression in the root and controls noncanonical primary root branching
Given that ARF7 and ARF19 regulate the transcriptional activation of LBD16, and TOR controls their translation, we wondered how LBD16 transcripts still accumulate upon TOR inhibition or knockdown (Fig 5F).
The Arabidopsis root system is determined by the production of lateral roots, which originate from the growing root, and adventitious roots (ARs), which are formed from non‐root organs. WOX11 is a crucial trigger of AR formation (Liu et al, 2014; Sheng et al, 2017). In standard laboratory conditions, WOX11 does not contribute to LR formation. However, when plants are grown on soil or upon wounding, the primary root can produce both WOX11‐mediated roots and non‐WOX11‐mediated roots (Sheng et al, 2017). Interestingly WOX11‐mediated and non‐WOX11‐mediated root initiation converges on the activation of LBD16 (Sheng et al, 2017). This led us to examine whether WOX11 is involved in LBD16 regulation upon TOR knockdown. We observed that WOX11 transcription increases in the UB10pro>>amiR‐TOR mutant or upon AZD treatment (Fig 6A; Appendix Fig S12). To know in which tissue WOX11 is expressed upon TOR knockdown, we crossed a WOX11pro::GUS line to UB10pro>>amiR‐TOR. In control conditions, and as previously reported by Sheng et al, the WOX11pro::GUS signal was undetectable during LR formation; more specifically, WOX11 expression was absent in xylem‐pole pericycle cells. Upon TOR knockdown, we observed the WOX11pro::GUS signal in the pericycle (Fig 6B). Together, these data show that TOR inhibition or knockdown (Appendix Fig S12) promotes WOX11 expression. In these conditions, LBD16 transcription is promoted independently of ARF7/19 (Sheng et al, 2017; Fig 6C). Through its control of LBD16 translation, TOR would also control the WOX11‐dependent root branching. To test this hypothesis, we cut the root of the arf7/arf19 double mutant to induce the formation of only WOX11‐dependent side roots (Sheng et al, 2017). On 1/2MS medium 6 days after excision, we observed the expected formation of roots near the wounded region (Fig 6D and F). On 1/2MS medium containing AZD, no roots were formed near the cut side (Fig 6E and G), indicating that TOR activity is also required for the non‐canonical, WOX11‐dependent primary root branching (Fig 6H). WOX11‐mediated root initiation is a hallmark of root formation from autotrophic tissues (hypocotyl, leaves) that can be coopted for noncanonical primary root branching upon stress (Sheng et al, 2017). By inhibiting WOX11 expression, TOR prevents the activation of this fail‐safe branching mechanism in sink tissue (Fig 6H). In source tissue such as the hypocotyl, reduction of TOR abundance (amiR‐TOR) or its activity (lst8 mutant) does not prevent AR formation induced by WOX11 up‐regulation and may even promote it (Fig EV1). The role of TOR in AR formation has been reported in Arabidopsis (Deng et al, 2017). However, inhibition of TOR activity reduced AR formation, a difference from what we observed. The difference in response may arise from the conditions used. Deng et al scored AR formation in dark‐grown hypocotyl where the root was removed, while in our case, plants were grown in light with intact roots. These differences may hint at the existence of complex regulation of TOR roles in autotrophic tissues in response to different physiological status. How TOR levels modulate the expression of WOX11 and how, in the hypocotyl, TOR regulation of LBD16 is alleviated remain to be elucidated.
Figure 6. TOR is required for aARF7/ARF19 independent formation of adventitious roots via WOX11.
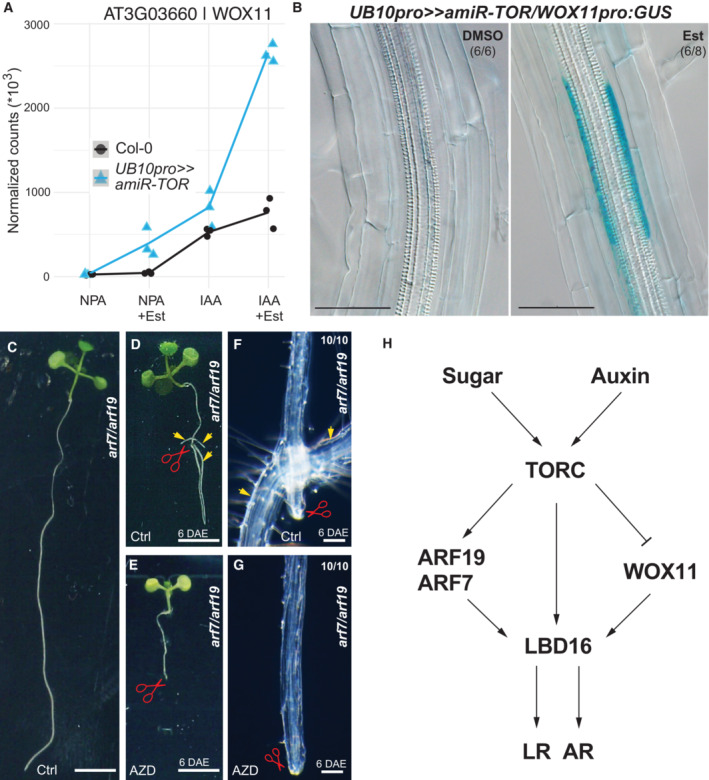
-
AAbundance of WOX11 transcripts in RNAseq samples. mRNA accumulation in response to auxin is elevated when TOR is knocked down. n = 3 biological replicates.
-
BDistribution of GUS‐staining in UB10pro>>amiR‐TOR/WOX11pro::GUS seedlings 6 days after transfer to 10 μM Est. The proportion of root bends with the depicted phenotype is indicated.
-
CThe arf7/arf19 mutant shows defective LR initiation.
-
DFormation of roots (arrows) close to the wound site of arf7/arf19 primary roots 6 days after excision (DAE).
-
EBlocking TOR‐activity via exposure to 10 μM AZD8055 (AZD) resulted in the loss of rooting capability on the wound site of arf7/arf19 primary roots at 6 DAE. Scale bars = 3 mm.
-
F, GClose‐ups indicate root formation location or absence of root formation on ctrl media and AZD‐containing media, respectively, in arf7/arf19 primary roots at 6 DAE. The proportion of root wound sites with the depicted phenotype is indicated. Scale bars = 100 μm.
-
HModel for TOR dependency of root branching in Arabidopsis.
Source data are available online for this figure.
These results shed light on the metabolic control of root branching. They reveal the central role TOR plays in an organ entirely dependent on the supply of carbohydrates from the source tissues in matching the availability of resources to the decision to form a new organ.
Materials and Methods
Reagents and Tools table
| Reagent/resource | Reference or source | Identifier or catalog number |
|---|---|---|
| Experimental models | ||
| DR5:GUS | Benková et al (2003) | |
| arf7/arf19 | Okushima et al (2007) | |
| LBD16‐SRDX | Goh et al (2012) | |
| slr | Fukaki et al (2002) | |
| pGATA23::shy2‐2‐GR and pGATA23::slr1‐GR | Ramakrishna et al (2019) | |
| TOR‐oe | Deprost et al (2007) | G548 |
| raptor1b‐1 | Salem et al (2017) | SALK_101990 |
| lst8 | Moreau et al (2012) | |
| 35Spro::GFP‐RPL18 | Mustroph et al (2009) | |
| pARF19‐5′UTR::mVenus | Truskina et al (2021) | |
| gLBD16‐GUS | Sheng et al (2017) | |
| p35S:HF‐GFP‐ RPL18 | Mustroph et al (2009) | N69096 |
| pWOX11::GUS | Liu et al (2014) | |
| Recombinant DNA | ||
| pUB10:S6K1‐3xHA | This study | |
| UB10pro>>amiR‐TOR | This study | |
| XPPpro>>amiR‐TOR | This study | |
| pRAPTOR1B::GUS:eGFP | This study | |
| pS6K1:gS6K1‐CFP | This study | |
| Antibodies | ||
| Anti‐S6K1 (phospho T449) | Abcam | ab207399 |
| Anti‐S6K1/2 | Agrisera AB | AS12‐1855 |
| Oligonucleotides and sequence‐based reagents | ||
| PCR primers | This study | Dataset EV2 |
| Chemicals, enzymes and other reagents | ||
| IAA | Sigma‐Aldrich | I2886 |
| Naphthylphthalamic acid (NPA) | Sigma‐Aldrich | N0640 |
| AZD8055 | MedChemExpress | HY‐10422 |
| 2‐Deoxyglucose | Sigma‐Aldrich | D9761 |
| ß‐estradiol | Sigma‐Aldrich | E2758 |
| amyloglucosidase | Roche | ROAMYGL |
| Ribitol | Sigma‐Aldrich | PHR3526 |
| N‐Methyl‐N‐trimethylsilyltrifluoracetamid | Sigma‐Aldrich | M7891 |
| SYBR‐Green Mix | Thermo Fisher | 4,368,577 |
| GFP‐Trap® | Chromotek | gtma |
| TRIzol | Invitrogen | 15,596,026 |
| Complete Protease Inhibitor EDTA‐free | Roche | 04693159001 |
| Software | ||
| R | https://www.r‐project.org/ | |
| TargetSearch | Cuadros‐Inostroza et al (2009) | |
| DeSeq2 | Love et al (2014) | |
| STAR | https://github.com/alexdobin/STAR | v2.5.2 |
| Fiji | Schindelin et al (2012) | |
| Other | ||
| Illumina NexSeq 500 | Illumina | |
| Ribo‐Zero Plus rRNA Depletion Kit | Illumina | |
Methods and Protocols
Plant material and growth conditions
Plants of Arabidopsis thaliana ecotype Colombia (Col‐0) were grown under fluorescent illumination (50 μE m−2 s−1) in long‐day conditions (16‐h light/8‐h dark) at 22°C. Seeds were surface sterilized (ethanol 70% and SDS 0.1%) and placed on ½ Murashige and Skoog (MS) medium adjusted to pH 5.7 containing 1% agar (Duchefa). Following stratification (4°C in the dark, > 24 h).
Construction of vectors and plant transformation
Unless specified otherwise, the plasmids were generated using the GreenGate modular cloning system (Lampropoulos et al, 2013). For pUB10:S6K1‐3xHA, the following modules were combined in pGGZ003: UBQ10 promoter (A), B‐dummy (B), S6K1 (AT3G08730, obtained by PCR on Col‐0 gDNA; 1,398 bp) (C), 3xHA (D), 35S terminator (E) and p35S:D‐ alaR:t35S (F). The ß‐Estradiol inducible amiR‐TOR line (UB10pro>>amiR‐TOR) was designed based on (Siligato et al, 2016), and two intermediate vectors (pAP039 and pAP043) were combined in pGGZ003. For pAP039 the following modules were combined in pGGM000: pGGA044 Olex TATA (A), B‐dummy (B), TOR amiRNA (TTTATAACAACAAGTTGGCGT, generated in this study, C), D‐dummy (D), RBCS terminator (E) 250 bp HA adapter (G). For pAP043, the following modules were combined in pGGN000: UBQ10 promoter (A) B‐dummy (B), CDS of chimeric TF XVE amplified from pLB12 (Brand et al, 2006) in two PCRs to domesticate an endogenous Eco31I site. (C), D‐dummy (D), UBQ10 terminator (E) 250 bp HA adapter (G). For XPPpro>>amiR‐TOR (pXPP::LhG4:GR/6xOP::amiR‐TOR), the intermediate module pAP097 was built consisting of HA‐adaptor, 6xOp (A), B‐dummy (B), TORamiRNA (C), D‐dummy (D), UB10 terminator (E) and HygrR (F) in pGGN000, and combined with pSW303 (in pGGM000) consisting of pXPP (A), B‐dummy (B), LhG4:GR (C), D‐dummy (D), RBCS terminator (E) FH‐adaptor (F). Both modules were combined in pGGZ003 to generate the final vector. For the pRAPTOR1B::GUS:eGFP transcriptional fusion, 1,360 bp upstream of the of RAPTOR1B (AT3G08850) were amplified by PCR, cloned into the pDONR221TM P1P2 by BP reaction (BP clonase, Thermofisher), and sub‐cloned into the destination vector pHGWFS7.0 by LR reaction (LR clonase, Thermofisher). The pS6K1:gS6K1‐CFP is an S6K1 genomic line with a C‐terminal CFP clone. It was generated by PCR amplification (DNA KOD Hot‐start DNA Polymerase, Novagen), cloned into the pENTRD‐TOPO Gateway vector using the manufacturer's protocol, and confirmed by sequencing. This clone was then used as a template to generate AscI‐S6K1p::S6K1g(No STOP)‐PacI fragments that were then ligated into the promoterless pBa002a vector to generate pBa002a/S6K1p::S6K1g‐CFP. The clone was confirmed by sequencing. The primers used for cloning and sequencing are listed in Dataset EV2. Agrobacterium tumefaciens (Agl‐0, GV3101 or ABI50) based plant transformation was carried out using the floral dip method (Clough & Bent, 1998). All plant lines examined were homozygous if not indicated otherwise. Homozygosity was determined by antibiotic resistance, and three independent lines were analyzed in the T3 generation.
Pharmacological treatments
For IAA treatments (10 μM), samples were treated for 6 h before sampling. TOR inhibitor AZD8055 (10 μM) was applied for 16 h before inducing LR formation by auxin for an additional 6 h. Similarly, seedlings were transferred for 16 h to 2‐Deoxyglucose (20 mM) containing media to block glycolysis before seedlings were treated for an additional 6 h with auxin to induce LR formation. UB10pro>>amiR‐TOR was expressed by transferring seedlings to plates containing ß‐Estradiol (10 μM) for 24 h.
Synchronous induction of lateral root induction
We used the previously described LR‐induciblesystem (Himanen et al, 2002). In brief, dense horizontal lanes of sterilized seeds were placed on sterile nylon membranes (SEFAR, Switzerland) and, 7 days after germination, were transferred to plates with fresh ½ MS medium containing 10 μM NPA (Naphthylphthalamic acid for 24 h before shifting to 10 μM IAA).
Starch quantification
Enzymatic quantification of starch was conducted according to (Smith & Zeeman, 2006) with few modifications. In brief, for each of the four replicates, rosettes of 4–6 individual plants at 15 DAG were snap‐frozen in liquid nitrogen, weight, and tissue lyzed before being boiled twice in 90% (v/v) ethanol. The decolored ground tissues were washed with 100% ethanol and once with water. Starch in the insoluble remains was hydrolyzed by adding 200 μl 0.2 M KOH and heating for 1 h at 95°C before neutralizing the solution via the application of 35 μl 1 M acetic acid and boiled for 30 min. Total starch in 50 μl of the resulting solution was then digested into glucose monomers via amyloglucosidase at 37°C overnight and further processed for starch quantification as described in (Smith & Zeeman, 2006).
GC–MS‐based metabolite profiling
Profiling of central carbon metabolism intermediates was performed using GC–MS according to metabolite extraction and analysis steps initially as described previously (Roessner et al, 2001). Briefly, 15–40 mg of the previously collected frozen root tissues was homogenized by tissue lyzer in liquid nitrogen and mixed with 360 μl ice‐cold methanol. 20 μg of Ribitol was added as an internal normalizing standard. After extraction (15 min, 70°C), 200 μl chloroform and 400 μl water were added, and samples were mixed vigorously before centrifugation. 200 μl of the upper methanol–water phase containing polar to semipolar metabolites was collected and dried in a vacuum concentrator. Derivatization followed thereafter, including methoximation of the concentrated residues followed by silylation. To this end, the residues were first re‐suspended in a methoxyamine‐hydrochloride/pyridine solution to methoxymize the carbonyl groups. Samples were then heated (90 min, 37°C) and subsequently silylated with N‐methyl‐N‐ trimethylsilyltrifloracetamide (37°C, 30 min). GC–MS analysis was performed on a gas chromatograph system equipped with a quadrupole mass spectrometer (GC‐MS‐QP2010, Shimadzu, Duisburg, Germany). For this, 1 μl of each sample was injected in split mode with a split ratio of 1:100, and the separation of derivatized metabolites was carried out on an RTX‐ 5MS column (Restek Corporation, Bellefonte, PA) using instrumental settings optimized by (Lisec et al, 2006).
GC–MS data processing
Raw GC–MS data files were first converted into an ANDI‐MS universal file format for spectrum deconvolution and compound identification. Baseline correction, peak identification, retention time (RT) alignment, and library matching with the reference collection of the Golm Metabolome Database (GMD, http://gmd.mpimp‐golm.mpg.de/) were obtained using the TargetSearch R package from Bioconductor (Cuadros‐Inostroza et al, 2009). Kovats retention indices used for library matching were calculated for deconvoluted mass spectra from measurements of an alkane mixture (Sigma‐Aldrich, St. Louis, MO). The Shimadzu GCMS solutions software (v2.72) interface was further used for the manual curation of annotation of some metabolites versus authentic standards analyzed under the above‐described analytical conditions. CSV output files (shoot and root data sets) from the data processing were exported with peak areas obtained for quantifier ions selected for deconvoluted spectra consistently detected in all analyzed samples. Peak areas (Dataset EV1) were scaled on a sample basis according to the extracted amount of root tissue and relative to the peak area obtained for the ribitol internal standard to correct for putative extraction and analytical performance variations across the different measurements. Finally, peak areas for the abovementioned compounds obtained in solvent/blank samples were subtracted as background signals from biological samples. For hierarchical clustering analysis of normalized relative peak levels, data were z‐score transformed, and clustering was conducted with Ward's clustering method.
S6K phosphorylation assay
For each sample, dissected and flash‐frozen roots of ca. 200 seedlings were used. Proteins were extracted from 40 mg root materials in 200 μl 1× MOPS buffer (0.1 M MOPS, 50 mM NaCl, 5% SDS, 10% glycerol, 4 mM EDTA [pH 7.5], 0.3% ß‐mercaptoethanol) supplemented with 1.5% phosphatase inhibitor cocktail 2 (Sigma‐Aldrich, St. Louis, MO). After adding extraction buffer, samples were briefly mixed and heated at 95°C for 7 min. Cellular debris was removed by centrifugation (10 min, 14,000 rpm, RT). Protein extracts were supplemented with 5x Laemmli buffer (Bromophenol blue [0.05%], 0.3 M Tris buffer [pH 6.8], 50% glycerol, 0.1 M DTT) and reheated for 5 min to 95°C. 20 μl protein extract was separated on a 12% SDS gel and transferred to a Nitrocellulose membrane (Sigma‐Aldrich, St. Louis, MO). Anti‐S6K1 (phospho T449) polyclonal antibody (No. ab207399, Abcam, Cambridge, UK) was used to detect S6K phosphorylation. S6K1/2 antibody (AS12‐1855, Agrisera AB, Vännäs, Sweden) was used to detect total S6K1, Ponceau‐S counterstain for confirmation of equal loading.
Histochemical analysis and microscopy
GUS activity was assayed at 37°C overnight following a modified version of the protocol (Weigel & Glazebrook, 2002): the initial washing with the staining buffer (without X‐Gluc) and vacuum steps were omitted. GUS staining was followed by fixation in a 4% HCl and 20% methanol solution (15 min at 65°C), followed by 7% NaOH in 60% ethanol (15 min, room temperature). Seedlings were subsequently cleared in successive ethanol baths for 10 min (40, 20, 10%), followed by a 20 min incubation in 25% glycerol and 5% ethanol. Finally, seedlings are mounted in 50% glycerol for imaging with DIC microscopy using an Axio Imager. M1 (Carl 478 Zeiss, Oberkochen, Germany) with a 20× objective. For starch staining, seedlings were collected in the morning of the indicated DAG, fixed and cleared as described above, then stained for 30 min with 2 ml of Lugol's Iodine solution according to (Caspar et al, 1985) and immediately scanned using a RICOH IM C3000 system (https://www.ricoh‐europe.com/). Calcofluor White counterstaining was performed with seedlings fixed for 30 min in 4% PFA in 1× PBS (RT), as described (Ursache et al, 2018). Root bend sections were cleared with ClearSee (Kurihara et al, 2015)for 1 day and imaged on a Leica SP8 confocal microscope with a 40×, NA = 1.3 oil immersion objective. Calcofluor White fluorescence was detected using the 405 nm excitation laser line and an emission range of 425–475 nm. mVENUS was detected using the 488 nm excitation laser line and an emission range of 500–550 nm.
Lateral root quantification
Seedlings were grown for 14 days on square dishes placed upright under a long‐day light regime as described above. Plates were subsequently scanned using a RICOH IM C3000 system (https://www.ricoh‐europe.com/), and primary root length was measured with fiji. Using a stereomicroscope (SteREO Discovery.V12, Zeiss, Jena), all emerged LR on each seedling were counted, and subsequently, the density of LR per cm of primary root was calculated.
Roots resection
Root cutting in the arf7/arf19 mutant was performed as described previously (Sheng et al, 2017) with the following modification, plants were cut at 8 DAG.
RNA‐seq analysis
Samples (Col‐0 or inducible UB10pro>>amiR‐TOR line) were prepared for harvesting using the synchronous induction of lateral root procedure. All samples were pre‐treated with 10 μM NPA for 24 h, then shifted to plates containing 10 μM NPA and 10 μM Estradiol or DMSO control for an additional 24 h before being shifted to either 10 μM IAA + 10 μM Estradiol or DMSO for LR induction or maintained on the same plates. Root tissue was harvested after 6 h. All sampling points were performed in triplicate. About 200 segments of the lower two‐thirds seedling roots were pooled for each sample. The total RNA of the 24 samples (2 genotypes × 4 treatments × 3 replicates) was extracted with the Universal RNA purification kit (EURx). Illumina NextSeq libraries were prepared from 2 μg of total RNA, and sequencing was performed on NextSeq 500 flow cells (12 samples per cell). Reads were mapped onto the Arabidopsis thaliana genome (TAIR10), and the number of reads per transcript was computed using STAR (version 2.5.2b). All the subsequent analysis was done with R (www.r‐project.org/) using the DESeq2 package (Love et al, 2014). Differentially expressed genes were identified using a grouping variable that combines the Treatment (NPA, NPA_Est, IAA, IAA_ESt) and Genotype (col‐0, UB10pro>>amiR‐TOR) variables at log2 fold change > 1 and false discovery rate < 0.05. The procedure was inspired by workflow. The 1,141 genes induced by IAA were defined as the differentially expressed genes (NPA vs. IAA) common that were insensitive to the effect of Estradiol in col‐0. The RNASeq data have been deposited to GEO (GSE199202) as part of the SuperSeries GSE199211.
RT–qPCR
Seedlings were pretreated at 7 DAG for 16 h either with 10 μM AZD8055 or DMSO control media before being transferred to DMSO, 10 μM IAA, 10 μM AZD8055 or 10 μM AZD8055 + 10 μM IAA. Root tissues of ca. 200 seedlings were then dissected after 6 h. All samplings were performed in quintuplicate or quadruplicate. Total RNA was extracted with the RNeasy Plant Mini Kit (Qiagen), and 2 μg RNA was reverse transcribed with RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher). Quantitative RT–PCR (RT–qPCR) was performed using gene‐specific primers (see Dataset EV2) in a total volume of 20 μl Absolute qPCR SYBR‐Green Mix (Thermo Fisher) on a qTOWER3 (Analytik Jena) apparatus according to the manufacturer's instructions.
TRAP‐Seq
The TRAP‐seq experiment was conducted according to Thellmann et al (2020). In brief, seedlings ubiquitously expressing a GFP‐tagged RPL18 ribosomal protein (p35S:HF‐GFP‐ RPL18, N69096; Mustroph et al, 2009) were pretreated at 7 DAG for 16 h either with 10 μM AZD8055 or DMSO control media before being transferred to DMSO, 10 μM IAA, 10 μM AZD8055 or 10 μM AZD8055 + 10 μM IAA. Root tissues were then dissected after 6 h. All samplings were performed in triplicate. For each sample, about 1,500 segments of the lower two‐thirds of the seedling roots were pooled, flash‐frozen in liquid nitrogen, and later homogenized with a polysome‐extraction buffer (Thellmann et al, 2020). The suspension was centrifuged for 15 min (16,000 g, 4°C). An aliquot of the homogenate was used for Bulk‐RNA‐ extraction by TRIzol reagent (Invitrogen). The remaining supernatant was incubated with GFP‐Trap® Magnetic Agarose beads (Chromotek, Munich, Germany). Ribosomal bound RNA was obtained by immunoprecipitation with magnetic anti‐GFP beads following the manufacturer's instructions and subsequently purified by TRIzol reagent (Invitrogen). Illumina NextSeq libraries were prepared from 2 μg of total RNA (bulk) or 100 ng (Ribosome bound) after depleting the rRNA via Ribo‐Zero Plus rRNA Depletion Kit (Illumina) and sequencing performed on NextSeq 500 flow cells (12 samples per cell). Reads were mapped onto the Arabidopsis thaliana genome (TAIR10), and the number of reads per transcript was computed using STAR (version 2.5.2b). All the subsequent analysis was done with R (www.r‐project.org/) using the DESeq2 package (Love et al, 2014). For each assay type (Bulk and TRAP), the variance was stabilized by a ‐log transform, and z‐score were derived for all transcripts genome‐wide. The log10 ratio of TRAP to Bulk signal was then computed for all transcripts. The auxin‐responsive genes used for k‐means clustering based on the ratio of TRAP to Bulk signal were identified in the Bulk set comparing DMSO to IAA treatment (|log2FC| > 1, FDR < 0.05). The TRAP‐Seq and Bulk RNA seq data have been deposited to GEO (GSE199203) as part of the SuperSeries GSE199211.
Polysome profile analysis
Root samples were frozen and ground in liquid nitrogen. The powder was resuspended in Polysome extraction buffer (100 mM HEPES‐KOH pH 8.0, 150 mM KCl, 25 mM Mg(OAc)2, 25 mM EGTA pH 8.0, 0.5% NP‐40, 250 mM sucrose, 5 mM dithiothreitol [DTT], Complete Protease Inhibitor EDTA‐free [Roche]), and the final lysate was cleared by high‐speed centrifugation for 15 min at 4°C. The equivalent of 100 a.u. (A260; measured on Nanodrop) was layered on top of the 15–45% (w/v) sucrose density gradients and then centrifuged at 29,000 rpm in a SW60‐Ti rotor for 3 h at 4°C. The polysome profiles were generated by continuous absorbance measurement at 260 nm using a Gradient Fractionation System (Biocomp Instruments), eight fractions were collected, and total RNA from individual fractions was extracted with Tri‐Reagent (Trizol) and reverse transcribed with High‐Capacity cDNA Reverse Transcription Kit (Applied Biosystem). Quantitative RT–PCR (RT–qPCR) was performed using gene‐specific primers (see File S2) in a total volume of 10 μl SYBR Green Master mix (Roche) on a LightCycler LC480 apparatus (Roche) according to the manufacturer's instructions.
Statistical analysis
Plants were randomly assigned to groups and treatments during sowing. Samples size was not determined in advance; the largest possible number of plants per group was analyzed. No blinding measure was installed. Statistical analyses were performed with R. Normality was tested using the Shapiro–Wilk test; if not met, a nonparametric test was used. Levene's test tested the assumption on equality of variance; if not met, the assumption was relaxed in the test. The methods and P‐values are summarized in the figure legends. Data were plotted using R and Microsoft Excel.
Author contributions
Michael Stitz: Conceptualization; resources; data curation; formal analysis; validation; investigation; visualization; methodology; writing – original draft; writing – review and editing. David Kuster: Data curation; formal analysis; investigation; writing – review and editing. Maximillian Reinert: Data curation; investigation. Mikhail Schepetilnikov: Data curation; formal analysis; investigation; visualization. Béatrice Berthet: Data curation; formal analysis; investigation. Jazmin Reyes‐Hernández: Data curation; investigation. Denis Janocha: Resources; writing – review and editing. Anthony Artins: Resources. Marc Boix: Resources. Rossana Henriques: Resources. Anne Pfeiffer: Resources. Jan Lohmann: Resources. Emmanuel Gaquerel: Conceptualization; data curation; formal analysis; supervision; funding acquisition; visualization; methodology; writing – original draft; project administration; writing – review and editing. Alexis Maizel: Conceptualization; formal analysis; supervision; funding acquisition; visualization; methodology; writing – original draft; project administration; writing – review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Appendix S1
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Source Data for Expanded View
PDF+
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Acknowledgments
We thank A. Leibfried for critically reading the manuscript, M. Burow (DynaMo Center, Dept. of Plant and Environmental Sciences, University of Copenhagen, Denmark) for TOR‐oe seeds, S. Savaldi‐Goldstein (Faculty of Biology, Technion Haifa, Israel), for the p35S:HF‐ GFP‐RPL18 line, A. Bishopp (University of Nottingham, UK) for the pARF19‐5′UTR::mVenus line, Christian Meyer for sending the lst8 seeds—twice (INRA, Institute Jean‐Pierre Bourgin) and Lin Xu (Institute of Plant Physiology and Ecology, Shanghai, PRC) for the gLBD16‐GUS line and Lin Xu for providing the WOX11pro:GUS line. The authors gratefully acknowledge the data storage service SDS@hd supported by the Ministry of Science, Research and the Arts Baden‐Württemberg (MWK), the COS Metabolomics Core Technology Platform for GC/MS instrument access, the Cluster of Excellence Cellular Networks of the University of Heidelberg (CellNetworks) through grant EcTOP6 “Metabolism and Development” and the German Research Foundation (DFG) through grants INST 35/1314‐1 FUGG, INST 35/1503‐1 FUGG and FOR2581. Open Access funding enabled and organized by ProjektDEAL. Open Access funding enabled and organized by Projekt DEAL.
The EMBO Journal (2023) 42: e111273
Contributor Information
Emmanuel Gaquerel, Email: emmanuel.gaquerel@ibmp-cnrs.unistra.fr.
Alexis Maizel, Email: alexis.maizel@cos.uni-heidelberg.de.
Data availability
The datasets produced in this study are available in the following databases: RNA‐Seq data: Gene Expression Omnibus GSE199211 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE199211).
References
- Andersen TG, Naseer S, Ursache R, Wybouw B, Smet W, De Rybel B, Vermeer JEM, Geldner N (2018) Diffusible repression of cytokinin signalling produces endodermal symmetry and passage cells. Nature 555: 529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrivault S, Guenther M, Ivakov A, Feil R, Vosloh D, van Dongen JT, Sulpice R, Stitt M (2009) Use of reverse‐phase liquid chromatography, linked to tandem mass spectrometry, to profile the Calvin cycle and other metabolic intermediates in Arabidopsis rosettes at different carbon dioxide concentrations. Plant J 59: 826–839 [DOI] [PubMed] [Google Scholar]
- Atkinson JA, Rasmussen A, Traini R, Voß U, Sturrock C, Mooney SJ, Wells DM, Bennett MJ (2014) Branching out in roots: uncovering form, function, and regulation. Plant Physiol 166: 538–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini C, Pacurar DI, Perrone I (2014) Adventitious roots and lateral roots: similarities and differences. Annu Rev Plant Biol 65: 639–666 [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Blázquez MA, Nelson DC, Weijers D (2020) Evolution of plant hormone response pathways. Annu Rev Plant Biol 71: 327–353 [DOI] [PubMed] [Google Scholar]
- Brand L, Hörler M, Nüesch E, Vassalli S, Barrell P, Yang W, Jefferson RA, Grossniklaus U, Curtis MD (2006) A versatile and reliable two‐component system for tissue‐specific gene induction in Arabidopsis . Plant Physiol 141: 1194–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldana C, Li Y, Leisse A, Zhang Y, Bartholomaeus L, Fernie AR, Willmitzer L, Giavalisco P (2013) Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana . Plant J 73: 897–909 [DOI] [PubMed] [Google Scholar]
- Cao P, Kim S‐J, Xing A, Schenck CA, Liu L, Jiang N, Wang J, Last RL, Brandizzi F (2019) Homeostasis of branched‐chain amino acids is critical for the activity of TOR signaling in Arabidopsis . Elife 8: e50747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar T, Huber SC, Somerville C (1985) Alterations in growth, photosynthesis, and respiration in a starchless mutant of Arabidopsis thaliana (L.) deficient in chloroplast phosphoglucomutase activity. Plant Physiol 79: 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crookshanks M, Taylor G, Dolan L (1998) A model system to study the effects of elevated CO2 on the developmental physiology of roots: the use of Arabidopsis thaliana . J Exp Bot 49: 593–597 [Google Scholar]
- Cuadros‐Inostroza A, Caldana C, Redestig H, Kusano M, Lisec J, Peña‐Cortés H, Willmitzer L, Hannah MA (2009) TargetSearch – a Bioconductor package for the efficient preprocessing of GC‐MS metabolite profiling data. BMC Bioinformatics 10: 428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B, Vassileva V, Parizot B, Demeulenaere M, Grunewald W, Audenaert D, Van Campenhout J, Overvoorde P, Jansen L, Vanneste S et al (2010) A novel aux/IAA28 signaling cascade activates GATA23‐dependent specification of lateral root founder cell identity. Curr Biol 20: 1697–1706 [DOI] [PubMed] [Google Scholar]
- Dembinsky D, Woll K, Saleem M, Liu Y, Fu Y, Borsuk LA, Lamkemeyer T, Fladerer C, Madlung J, Barbazuk B et al (2007) Transcriptomic and proteomic analyses of pericycle cells of the maize primary root. Plant Physiol 145: 575–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng K, Dong P, Wang W, Feng L, Xiong F, Wang K, Zhang S, Feng S, Wang B, Zhang J et al (2017) The TOR pathway is involved in adventitious root formation in Arabidopsis and potato. Front Plant Sci 8: 793–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolaï M, Bedu M, Robaglia C, Meyer C (2007) The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep 8: 864–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrenel T, Caldana C, Hanson J, Robaglia C, Vincentz M, Veit B, Meyer C (2016) TOR signaling and nutrient sensing. Annu Rev Plant Biol 67: 261–285 [DOI] [PubMed] [Google Scholar]
- Dubrovsky JG, Sauer M, Napsucialy‐Mendivil S, Ivanchenko MG, Friml J, Shishkova S, Celenza J, Benkova E (2008) Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc Natl Acad Sci USA 105: 8790–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M (2002) Lateral root formation is blocked by a gain‐of‐function mutation in the SOLITARY‐ROOT/IAA14 gene of Arabidopsis . Plant J 29: 153–168 [DOI] [PubMed] [Google Scholar]
- Ge Y, Fang X, Liu W, Sheng L, Xu L (2019) Adventitious lateral rooting: the plasticity of root system architecture. Physiol Plant 165: 39–43 [DOI] [PubMed] [Google Scholar]
- Goh T, Joi S, Mimura T, Fukaki H (2012) The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development 139: 883–893 [DOI] [PubMed] [Google Scholar]
- Graf A, Schlereth A, Stitt M, Smith AM (2010) Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc Natl Acad Sci USA 107: 9458–9463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Singh M, Laxmi A (2015) Interaction between glucose and brassinosteroid during the regulation of lateral root development in Arabidopsis . Plant Physiol 168: 307–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K, Boucheron E, Vanneste S, de Almeida EJ, Inzé D, Beeckman T (2002) Auxin‐mediated cell cycle activation during early lateral root initiation. Plant Cell 14: 2339–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Schopfer P (2012) Photosynthetic sucrose acts as cotyledon‐derived long‐distance signal to control root growth during early seedling development in Arabidopsis . Proc Natl Acad Sci USA 109: 11217–11221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara D, Mizuta Y, Sato Y, Higashiyama T (2015) ClearSee: a rapid optical clearing reagent for whole‐plant fluorescence imaging. Development 142: 4168–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampropoulos A, Sutikovic Z, Wenzl C, Maegele I, Lohmann JU, Forner J (2013) GreenGate – a novel, versatile, and efficient cloning system for plant transgenesis. PLoS One 8: e83043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavenus J, Goh T, Roberts I, Guyomarc'h S, Lucas M, De Smet I, Fukaki H, Beeckman T, Bennett M, Laplaze L (2013) Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci 18: 450–458 [DOI] [PubMed] [Google Scholar]
- Lavenus J, Goh T, Guyomarc'h S, Hill K, Lucas M, Voß U, Kenobi K, Wilson MH, Farcot E, Hagen G et al (2015) Inference of the Arabidopsis lateral root gene regulatory network suggests a bifurcation mechanism that defines primordia flanking and central zones. Plant Cell 27: 1368–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Cai W, Liu Y, Li H, Fu L, Liu Z, Xu L, Liu H, Xu T, Xiong Y (2017) Differential TOR activation and cell proliferation in Arabidopsis root and shoot apexes. Proc Natl Acad Sci USA 114: 201618782–201618786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang B, Zhu X, Li R, Fu J, Cui H (2020) Novel regulators of sugar‐mediated lateral root development in Arabidopsis thaliana . Genes 11: 1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR (2006) Gas chromatography mass spectrometry‐based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Liu J, Sheng L, Xu Y, Li J, Yang Z, Huang H, Xu L (2014) WOX11 and 12 are involved in the first‐step cell fate transition during de novo root organogenesis in Arabidopsis . Plant Cell 26: 1081–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol 15: 550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M, Kenobi K, von Wangenheim D, VOβ U, Swarup K, De Smet I, Van Damme D, Lawrence T, Péret B, Moscardi E et al (2013) Lateral root morphogenesis is dependent on the mechanical properties of the overlaying tissues. Proc Natl Acad Sci USA 110: 5229–5234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana . Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C (2002) Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci USA 99: 6422–6427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montané M‐H, Menand B (2013) ATP‐competitive mTOR kinase inhibitors delay plant growth by triggering early differentiation of meristematic cells but no developmental patterning change. J Exp Bot 64: 4361–4374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau M, Azzopardi M, Clément G, Dobrenel T, Marchive C, Renne C, Martin‐Magniette M‐L, Taconnat L, Renou J‐P, Robaglia C et al (2012) Mutations in the Arabidopsis homolog of LST8/GβL, a partner of the target of rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. Plant Cell 24: 463–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidhara P, Weiste C, Collani S, Krischke M, Kreisz P, Draken J, Feil R, Mair A, Teige M, Müller MJ et al (2021) Perturbations in plant energy homeostasis prime lateral root initiation via SnRK1‐bZIP63‐ARF19 signaling. Proc Natl Acad Sci USA 118: e2106961118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJH, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey‐Serres J (2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis . Proc Natl Acad Sci USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis . Plant Cell 19: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parizot B, Laplaze L, Ricaud L, Boucheron‐Dubuisson E, Bayle V, Bonke M, De Smet I, Poethig SR, Helariutta Y, Haseloff J et al (2008) Diarch symmetry of the vascular bundle in Arabidopsis root encompasses the pericycle and is reflected in distich lateral root initiation. Plant Physiol 146: 140–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A, Janocha D, Dong Y, Medzihradszky A, Schöne S, Daum G, Suzaki T, Forner J, Langenecker T, Rempel E et al (2016) Integration of light and metabolic signals for stem cell activation at the shoot apical meristem. Elife 5: e17023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna P, Ruiz Duarte P, Rance GA, Schubert M, Vordermaier V, Vu LD, Murphy E, Vilches Barro A, Swarup K, Moirangthem K et al (2019) EXPANSIN A1‐mediated radial swelling of pericycle cells positions anticlinal cell divisions during lateral root initiation. Proc Natl Acad Sci USA 116: 8597–8602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegler S, Servi L, Scarpin MR, Godoy Herz MA, Kubaczka MG, Venhuizen P, Meyer C, Brunkard JO, Kalyna M, Barta A et al (2021) Light regulates alternative splicing outcomes via the TOR kinase pathway. Cell Rep 36: 109676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie A (2001) Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13: 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sairanen I, Novák O, Pěnčík A, Ikeda Y, Jones B, Sandberg G, Ljung K (2012) Soluble carbohydrates regulate auxin biosynthesis via PIF proteins in Arabidopsis . Plant Cell 24: 4907–4916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem MA, Li Y, Wiszniewski A, Giavalisco P (2017) Regulatory‐associated protein of TOR (RAPTOR) alters the hormonal and metabolic composition of Arabidopsis seeds, controlling seed morphology, viability and germination potential. Plant J 92: 525–545 [DOI] [PubMed] [Google Scholar]
- Santos Teixeira JA, Ten Tusscher KH (2019) The systems biology of lateral root formation: connecting the dots. Mol Plant 12: 784–803 [DOI] [PubMed] [Google Scholar]
- Scarpin MR, Leiboff S, Brunkard JO (2020) Parallel global profiling of plant TOR dynamics reveals a conserved role for LARP1 in translation. Elife 9: e58795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Dimitrova M, Mancera‐Martínez E, Geldreich A, Keller M, Ryabova LA (2013) TOR and S6K1 promote translation reinitiation of uORF‐containing mRNAs via phosphorylation of eIF3h. EMBO J 32: 1087–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Makarian J, Srour O, Geldreich A, Yang Z, Chicher J, Hammann P, Ryabova LA (2017) GTPase ROP2 binds and promotes activation of target of rapamycin, TOR, in response to auxin. EMBO J 36: 886–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütz LM, Louveaux M, Vilches Barro A, Bouziri S, Cerrone L, Wolny A, Kreshuk A, Hamprecht FA, Maizel A (2021) Integration of cell growth and asymmetric division during lateral root initiation in Arabidopsis thaliana . Plant Cell Physiol 62: 1269–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M, Sharma M, Jamsheer KM, Laxmi A (2022) Jasmonic acid coordinates with light, glucose and auxin signalling in regulating branching angle of Arabidopsis lateral roots. Plant Cell Environ 45: 1554–1572 [DOI] [PubMed] [Google Scholar]
- Sheng L, Hu X, Du Y, Zhang G, Huang H, Scheres B, Xu L (2017) Non‐canonical WOX11‐mediated root branching contributes to plasticity in Arabidopsis root system architecture. Development 144: 3126–3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Wu Y, Sheen J (2018) TOR signaling in plants: conservation and innovation. Development 145: dev160887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh‐Chang N, Ng H‐H (2017) The metabolic programming of stem cells. Genes Dev 31: 336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siligato R, Wang X, Yadav SR, Lehesranta S, Ma G, Ursache R, Sevilem I, Zhang J, Gorte M, Prasad K et al (2016) MultiSite gateway‐compatible cell type‐specific gene‐inducible system for plants. Plant Physiol 170: 627–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Zeeman SC (2006) Quantification of starch in plant tissues. Nat Protoc 1: 1342–1345 [DOI] [PubMed] [Google Scholar]
- Sulpice R, Pyl E‐T, Ishihara H, Trenkamp S, Steinfath M, Witucka‐Wall H, Gibon Y, Usadel B, Poree F, Piques MC et al (2009) Starch as a major integrator in the regulation of plant growth. Proc Natl Acad Sci USA 106: 10348–10353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thellmann M, Andersen TG, Vermeer JE (2020) Translating ribosome affinity purification (TRAP) to investigate Arabidopsis thaliana root development at a cell type‐specific scale. J Vis Exp 10.3791/60919 [DOI] [PubMed] [Google Scholar]
- Torres‐Martínez HH, Hernández‐Herrera P, Corkidi G, Dubrovsky JG (2020) From one cell to many: morphogenetic field of lateral root founder cells in Arabidopsis thaliana is built by gradual recruitment. Proc Natl Acad Sci USA 117: 20943–20949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truskina J, Han J, Chrysanthou E, Galvan‐Ampudia CS, Lainé S, Brunoud G, Macé J, Bellows S, Legrand J, Bågman A‐M et al (2021) A network of transcriptional repressors modulates auxin responses. Nature 589: 116–119 [DOI] [PubMed] [Google Scholar]
- Ursache R, Andersen TG, Marhavý P, Geldner N (2018) A protocol for combining fluorescent proteins with histological stains for diverse cell wall components. Plant J 93: 399–412 [DOI] [PubMed] [Google Scholar]
- Ursache R, De Jesus Vieira Teixeira C, Dénervaud Tendon V, Gully K, De Bellis D, Schmid‐Siegert E, Grube Andersen T, Shekhar V, Calderon S, Pradervand S et al (2021) GDSL‐domain proteins have key roles in suberin polymerization and degradation. Nat Plants 7: 353–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J, Han C, Gadeyne A, Eeckhout D, Matthijs C, Cannoot B, De Winne N, Persiau G, Van De Slijke E, Van de Cotte B et al (2019) Capturing the phosphorylation and protein interaction landscape of the plant TOR kinase. Nat Plants 5: 316–327 [DOI] [PubMed] [Google Scholar]
- Vanneste S, Rybel BD, Beemster GTS, Ljung K, Smet ID, Van Isterdael G, Naudts M, Iida R, Gruissem W, Tasaka M et al (2005) Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14‐mediated lateral root initiation in Arabidopsis thaliana . Plant Cell 17: 3035–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer JEM, von Wangenheim D, Barberon M, Lee Y, Stelzer EHK, Maizel A, Geldner N (2014) A spatial accommodation by neighboring cells is required for organ initiation in Arabidopsis . Science 343: 178–183 [DOI] [PubMed] [Google Scholar]
- Vilches Barro A, Stöckle D, Thellmann M, Ruiz‐Duarte P, Bald L, Louveaux M, von Born P, Denninger P, Goh T, Fukaki H et al (2019) Cytoskeleton dynamics are necessary for early events of lateral root initiation in Arabidopsis . Curr Biol 29: 2443–2454.e5 [DOI] [PubMed] [Google Scholar]
- Vragović K, Sela A, Friedlander‐Shani L, Fridman Y, Hacham Y, Holland N, Bartom E, Mockler TC, Savaldi‐Goldstein S (2015) Translatome analyses capture of opposing tissue‐specific brassinosteroid signals orchestrating root meristem differentiation. Proc Natl Acad Sci USA 112: 923–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Arabidopsis: A Laboratory Manual. New York, NY: CSHL Press; [Google Scholar]
- Xiong Y, McCormack M, Li L, Hall Q, Xiang C, Sheen J (2013) Glucose‐TOR signalling reprograms the transcriptome and activates meristems. Nature 496: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SH, Lawton MA, Hunter T, Lamb CJ (1994) atpk1, a novel ribosomal protein kinase gene from Arabidopsis. I. Isolation, characterization, and expression. J Biol Chem 269: 17586–17592 [PubMed] [Google Scholar]


