Abstract
The PriA protein was identified in Escherichia coli as a factor involved in the replication of extrachromosomal elements such as bacteriophage φX174 and plasmid pBR322. Recent data show that PriA plays an important role in chromosomal replication, by promoting reassembly of the replication machinery during reinitiation of inactivated forks. A gene encoding a product 32% identical to the E.coli PriA protein has been identified in Bacillus subtilis. To characterise this protein, designated PriABs, we constructed priABs mutants. These mutants are poorly viable, filamentous and sensitive to rich medium and UV irradiation. Replication of pAMβ1-type plasmids, which is initiated through the formation of a D-loop structure, and the activity of the primosome assembly site ssiA of plasmid pAMβ1 are strongly affected in the mutants. The purified PriABs protein binds preferentially to the active strand of ssiA, even in the presence of B.subtilis SSB protein (SSBBs). PriABs also binds stably and specifically to an artificial D-loop structure in vitro. These data show that PriABs recognises two specific substrates, ssiA and D-loops, and suggest that it triggers primosome assembly on them. PriABs also displays a single-stranded DNA-dependent ATPase activity, which is reduced in the presence of SSBBs, unless the ssiA sequence is present on the ssDNA substrate. Finally, PriABs is shown to be an active helicase. Altogether, these results demonstrate a clear functional identity between PriAEc and PriABs. However, PriABs does not complement an E.coli priA null mutant strain. This host specificity may be due to the divergence between the proteins composing the E.coli and B.subtilis PriA-dependent primosomes.
INTRODUCTION
The Escherichia coli PriA protein was characterised as being required for replication of bacteriophage φX174 and plasmid ColE1 (1–3). In these extra-chromosomal elements PriA promotes the initiation of replication through its specific binding to DNA, followed by the ordered assembly of several other proteins, PriB, PriC, DnaT, DnaC, DnaB replicative helicase and DnaG primase. This particular nucleoprotein complex has been referred to as the φX174-type primosome (4,5 and references therein; for recent reviews see 3,6). This primosome can be sequentially assembled on two distinct DNA sites, specifically bound by PriA. One, designated pas (primosome assembly site), was characterised in φX174 and ColE1. PriA binds to the pas in the single-stranded DNA (ssDNA) form, when it folds into a particular structure not bound by the SSB protein (7). The second type of PriA binding site is a D-loop structure (8,9). This three-stranded molecule is an early intermediate of the replication of ColE1-type plasmids (10). The cellular function of PriA has emerged more recently, following the identification of its gene. PriA is not essential in E.coli, suggesting that initiation of DNA replication promoted by this protein is accessory (11–14). However, disruption of the priA gene decreases cell viability, causes sensitivity to rich medium, filamentation, UV sensitivity, deficiency in recombination and constitutive induction of the SOS response. These phenotypes led to the hypothesis that the cellular role of the φX174-type primosome is to restart stalled DNA replication, as well as to repair some types of DNA damage by linking DNA recombination to replication (6,15–17). The structural nature of the DNA specifically recognised by PriA supports this proposal: the three-stranded DNA molecules generated by recombinational repair of the DNA triggers the ordered cascade of primosomal proteins, inaugurated by PriA, to recruit the DNA replication machinery (18,19). To better indicate its cellular function, the φX174-type primosome has been renamed the replication restart primosome (17).
In addition to specific binding of DNA, PriA is also a 3′→5′ helicase, translocating along ssDNA in the direction opposite to the replication fork helicase. This DNA melting activity was shown to be dispensable for the central role of PriA in E.coli (20). Nevertheless, it has recently been proposed that PriA helicase activity would generate the ssDNA needed for the loading of DnaB when the forked substrate specifically targeted by PriA is double stranded (21–24).
Current knowledge about primosomal proteins in Bacillus subtilis is less detailed. Counterparts of the E.coli replicative helicase and the primase are known in B.subtilis (25,26), but there are no obvious homologues of the PriB, PriC, DnaT and DnaC primosomal proteins (27). Characterisation of the sole primosome assembly site isolated so far in Gram-positive bacteria, the ssiA sequence carried by the plasmid pAMβ1, has pointed to the existence of a φX174-type primosome in B.subtilis (28). Three B.subtilis essential proteins, DnaB, DnaD and DnaI, which are not encoded in the E.coli genome, are required for chromosomal replication (29) and for ssiA activity (28). A potential PriA analogue was tentatively identified more recently in B.subtilis on the basis of sequence homology (27,30,31). Masai et al. purified this protein from an insoluble fraction and, following renaturation, showed it to be a DNA-dependent ATPase displaying helicase activity and able to bind to an artificial D-loop structure (31). Nevertheless, this initial characterisation did not show that the protein was involved in replication restart in B.subtilis. In this report we address precisely this question in order to establish the existence of a PriA-dependent primosome in B.subtilis. We present in vivo evidence demonstrating functional analogy between this B.subtilis protein and E.coli PriA. We also report purification of this protein, which we designate PriABs, in a soluble form, with which we confirm and extend a previous in vitro study (31). More particularly, we report that PriABs displays a much stronger affinity for ssDNA than its E.coli counterpart. Altogether, this study confirms the existence of a PriA-dependent primosome in B.subtilis, built from a conserved initiator. Finally, we show that PriABs does not substitute for PriAEc in vivo, suggesting a host specificity for this protein which may be due to the divergence between the primosomal partners acting after PriA in the two bacteria.
MATERIALS AND METHODS
Bacterial strains and growth media
The strains used in this study are listed in Table 1. Bacillus subtilis strains are all derivatives of strain 168. They were cultivated either in LB medium or in minimal medium (Spizizen’s minimal salts) (32) supplemented with 0.1% d-glucose, 0.01% l-tryptophan, 0.1% casamino acids, 18 mg l–1 ammonium iron(III) citrate (∼17% iron; Merck), as indicated in the text, and, when required, with 0.6 µg ml–1 erythromycin (Em), 4 µg ml–1 chloramphenicol (Cm) and 0.5 or 1 mM IPTG. Competent cells were prepared as described in Bron (32). The restriction map of the priABs chromosomal region in strains PPBJ65, PPBJ69, PPBJ117 and PPBJ120 was verified by Southern analysis. Strain CBB294 is a derivative of PPBJ120 disrupted for priABs but carrying a mutation suppressing the lack of PriABs (dnaB75) (33).
Table 1. Plasmids, bacteriophages and strains used in this study.
| Strains, plasmids and phages | Description | Reference |
|---|---|---|
| E.coli strains | ||
| MiT898 | ΔendA araD139 Δ[ara-leu] galU galK hsdM hsdS rpsL Δ[lacIOPZYA] X74 | (56) |
| B834(DE3) | hsdS gall cIts857 ind1 metSam7 nin5 lacUV5-T7 gene 1 | (57) |
| TG1 | supE hsdΔ5 thi Δ(lac-proAB) F′[traD36 proAB+ lacIq lacZΔM15] | (38) |
| DM4000 | (lac-pro)XIII hisG4 argE3 ara14 xyl5 mtl1 rpsL31 sulA::Mu-d (Ap, lac, B::Tn9) | (49) |
| JC18983 | DM4000 priA2::Km | (49) |
| B.subtilis strains | ||
| 168 | trpC2 | C. Anagnostopoulos |
| PPBJ65 | 168 priABs::pAPJ12 | This work |
| PPBJ69 | 168 priAind::pAPJ13 | This work |
| PPBJ117 | 168 priA2Bs::pAPJ14 | This work |
| PPBJ120 | 168 priA1Bs::pAPJ11 | This work |
| CBB294 | 168 priA1Bs::pAPJ11 dnaB75 | (55) |
| Plasmids and phages | ||
| pCYB1 | pBR322-based vector (IMPACT system) | New England Biolabs |
| pTYB1 | pBR322-based vector (IMPACT system) | New England Biolabs |
| pVA798ΔRCR | pIP501 derivative | (48) |
| pMUTIN2 | pBR322 lacZ lacI EmR Pspac | (58) |
| pGB2 | pSC101-based vector | (35) |
| pADG6406-1 | pADG6406 ssiA+ carrying vector | (28) |
| pADG6406-2 | pADG6406 ssiA– carrying vector | This work |
| pAPJ11 | pMUTIN2 Pspac: (nt +689 to +1023 of priABs) | This work |
| pAPJ12 | pMUTIN2 Pspac: (nt +2079 to +2419 of priABs) | This work |
| pAPJ13 | pMUTIN2 Pspac: (nt –25 to +376 of priABs) | This work |
| pAPJ14 | pMUTIN2 Pspac: (nt +4 to +376 of priABs) | This work |
| pSMG3 | pCYB1::priABs | This work |
| pSMG19 | pTYB1::ssbBs | This work |
| pAPJ2 | pC194 and pBSSK– joined by HindIII site | This work |
| pAPJ9 | pAPJ2 carrying the ssiA active strand on ssDNA | This work |
| pAPJ10 | pAPJ2 carrying the ssiA inactive strand on ssDNA | This work |
| pAPJ41 | pGB2, Ptac-priABs | This work |
| pAPJ43 | pGB2, Ptac-priAEc | This work |
| M13-ssiA+ | M13mp19 carrying the ssiA active strand on ssDNA | This work |
| M13-ssiA– | M13mp19 carrying the ssiA inactive strand on ssDNA | This work |
Plasmid constructions and preparations were done in E.coli strain MiT898. Escherichia coli strains were grown on Luria broth supplemented with 25 mg ml–1 thymine or minimal medium M63 (34) supplemented with 0.2% d-glucose. Spectinomycin (Spc) (60 µg ml–1), ampicillin (100 µg ml–1), kanamycin (Km) (50 µg ml–1) and IPTG (for concentrations see Table 3) were added when required.
Table 3. PriABs does not complement an E.coli priA null mutant.
| E.coli strain | priAEc status | Plasmid | IPTG (µM) | Phenotype | ||||
|---|---|---|---|---|---|---|---|---|
| Name | Induced protein | c.f.u. × 108 per OD600a | Growth on rich mediumb | UV resistancec | pBR322 replicationd | |||
| DM4000 | + | pGB2 | none | 0 | 2.4 | + | 0.6 | + |
| JC18983 | 2 | pGB2 | none | 0 | 0.7 | – | 0.0014 | – |
| JC18983 | 2 | pAPJ43 | PriAEc | 0 | 3.0 | + | ND | ND |
| JC18983 | 2 | pAPJ41 | PriABs | 0 | 0.6 | – | 0.0006 | – |
| 33 | 0.3 | – | 0.0004 | – | ||||
| 100 | 0.01 | – | 0.0010 | – | ||||
| DM4000 | + | pAPJ41 | PriABs | 0 | 3.1 | + | 0.7 | + |
| 100 | 3.3 | + | 0.5 | + | ||||
| 300 | 2.6 | + | 0.5 | ND | ||||
c.f.u., colony forming units; ND, not determined.
aStrains were grown exponentially at 37°C with the indicated dose of IPTG in minimal medium supplemented with spectinomycin. At OD600 = 0.4–0.6 cells were serially diluted, plated on minimal media without IPTG and with spectinomycin and c.f.u. were determined after 48 h incubation at 37°C.
bCells grown in minimal medium without IPTG and with spectinomycin were streaked onto LB plates supplemented with spectinomycin and the indicated dose of IPTG, and incubated at 37°C for 48 h. + and – indicate the presence and absence of colonies, respectively.
cAs in footnote a except that after plating, cells were irradiated at 20 J m–2. Results are expressed as the fraction of surviving cells.
dAbility of pBR322 to replicate was measured by its ability to transform competent cells to tetracyclin resistance. +, ∼106 transformants µg–1; –, <100 transformants µg–1.
Plasmids and M13 derivatives
The plasmids used in this study are listed in Table 1. Plasmids of the pAPJ series were constructed by inserting various PCR fragments digested with EcoRI and BamHI between the EcoRI and BamHI sites of pMUTIN2. These PCR fragments were generated using chromosomal DNA of strain 168 as template and the following oligonucleotides as primers: pAPJ11, OPP8 and OPP9; pAPJ12, OPP10 and OPP11; pAPJ13, OPP12 and OPP14; pAPJ14, OPP13 and OPP14. OPP8, 5′-CCGGAATTCGCTCTGTAACCATCAAAACCC-3′ (+689 to +710); OPP9, 5′-CGCGGATCCGAAGCGGCCCTTGAAGCGG-3′ (+1023 to +1002); OPP10, 5′-CCGGAATTCGCATGAAATGGCGCACCAGG-3′ (+2079 to +2098); OPP11, 5′-CGCGGATCCTTACATCATCATATAAGG-3′ (+2419 to +2401); OPP12, 5′-CCGGAATTCTCAAAACAAACCGGAGAGCGC-3′ (–25 to –3); OPP13, 5′-CCGGAATTCAATTTTGCAGAAGTCATCGTTG-3′ (+4 to +26); OPP14, 5′-CGCGGATCCGGAAGATCGGCTCCGTGTGC-3′ (+376 to +355). The priABs sequence is italicised and the EcoRI and BamHI sites are underlined. Arbitrary coordinates for the priABs sequence retained in the oligonucleotides are indicated in parentheses, giving the value +1 to the A of the proposed translational start of the ORF (27). The sequence of the insert in pAPJ13 has been checked to ascertain the absence of mutation.
For construction of pSMG3, the PriABs coding sequence was PCR amplified from chromosomal DNA of strain 168 with OPP17 and OPP18 as primers and inserted into the NdeI and SapI sites of pCYB1 (both blunted by Klenow filling in): OPP17, 5′-TGAATTTTGCAGAAGTCATCG-3′; OPP18, 5′-CATCATCATATAAGGATTCATATC-3′. This generated a triple fusion protein, PriABs–intein–chitin binding domain (CBD), expressed under the control of E.coli transcriptional (the Ptac promoter, inducible by IPTG) and translational signals. The sequence of priABs carried by pSMG3 has been verified.
For construction of pSMG19, the SSBBs coding sequence was PCR amplified from chromosomal DNA of strain 168 with OSMG18 and OSMG19 as primers, digested with NdeI and SapI and inserted in the sames sites of pTYB1: OSMG18, 5′-GAATTCCATATGCTTAACCGAGTTGTATTAG-3′; OSMG19, 5′-GAATTCGCTCTTCCGCAGAATGGAAGATCATCATCCGAGATG-3′ (sequences underlined in OSMG18 and OSMG19 represent a NdeI and a SapI site, respectively). This generated a triple fusion protein SSBBs–intein–CBD, expressed under the control of transcriptional and translational signals of bacteriophage T7. The sequence of ssbBs carried by pSMG19 has been verified.
pAPJ41 is a derivative of the pGB2 vector (35) which allows inducible expression of PriABs in E.coli and which does not need PriAEc for replication. This plasmid was constructed in several steps. First, the chromosome of strain PPBJ69 was digested with SwaI, ligated and transformed into E.coli to isolate a plasmid, pAPJ19, which contains the whole priABs ORF and additional 3′-flanking sequences. Second, the 3′ end of the priABs–intein–CBD ORF in pSMG3 (SacI–BamHI) was exchanged for the 3′ end of the priABs ORF of pAPJ19 (SacI–BglII) to generate plasmid pSMG4. Finally, to obtain pAPJ41 the Eco47III–PstI restriction fragment of pSMG4 carrying the laciq gene and the priABs ORF placed under the control of the Ptac promoter was cloned in the pGB2 vector between the SmaI and PstI sites located in the polylinker. pAPJ43 is almost identical to pAPJ41 except that it carries the PriAEc coding sequence in place of PriABs. It was constructed in two steps. First, the NdeI–PvuI fragment of the PriAEc-expressing plasmid described in Nurse et al. (36) was exchanged with a similarly cleaved fragment of pSMG3 to give plasmid pAPJ42. Then the MluI–HindIII fragment of pAPJ42 carrying the whole Ptac–priAEc artificial gene was exchanged for the corresponding MluI–HindIII fragment of pAPJ41 to give pAPJ43.
The two M13mp19 derivatives carrying the ssiA sequence in both orientations, M13-ssiA+ and M13-ssiA–, were constructed in two steps. The ssiA sequence (145 nt, coordinates 4712–4856 in pAMβ1) (37) was generated by PCR using pIL253 as template and OPP1 and OPP3 as primers, and inserted in both orientations in the SmaI site of the polylinker of plasmid pAPJ2, giving plasmids pAPJ9 and pAPJ10: OPP1, 5′-TAATTATTAGGGGGAGAAGGAGAGAG-3′; OPP3, 5′-CCTATAAAAGATAGAAAATTAAAAAATC-3′. The sequence of ssiA has been verified by DNA sequencing. The activity of ssiA has been verified in B.subtilis by showing that the ssDNA of plasmid pAPJ9 is efficiently converted into double-standed DNA (dsDNA) in vivo. Finally, the SalI–EcoRI fragments carrying ssiA from pAPJ9 and pAPJ10 were cloned into M13mp19 similarly cut, to give M13-ssiA+ and M13-ssiA–, respectively.
Plating efficiency and UV survival tests
Bacillus subtilis strains were grown to mid-log phase in minimal medium containing Em with and without IPTG (1 mM) as indicated (see legend to Fig. 2 and Table 2). To measure the plating efficiency, cultures were diluted appropriately and plated on minimal medium and on LB similarly supplemented with Em and IPTG, and incubated for 16–40 h at 37°C. To measure UV survival, minimal medium plates were irradiated immediately after plating with a 2 J m–2 dose of UV for different time periods and incubated for 16–40 h at 37°C. The tests with E.coli were performed similarly. Strains were grown in minimal medium containing Spc, supplemented or not with IPTG, as indicated (see Table 3), and spread on plates not supplemented with IPTG.
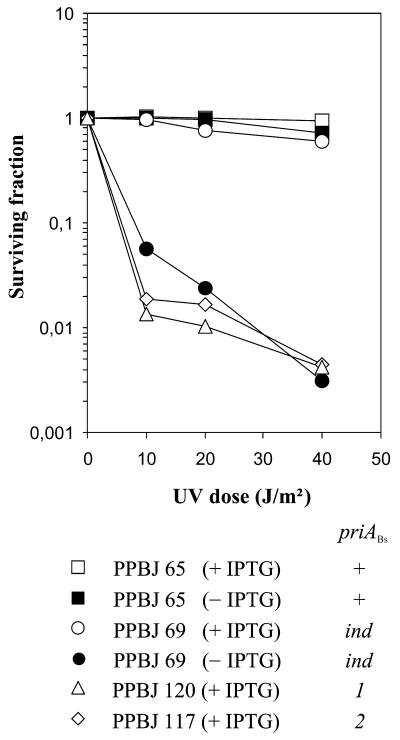
Figure 2.
UV sensitivity of B.subtilis priA mutant strains. Strains were grown in minimal medium supplemented with Em in the presence or absence of IPTG (1 mM), as indicated. Appropriate dilutions were plated on the same medium and immediately irradiated with different doses of UV. After incubation for 2 days at 37°C, the fraction of surviving cells was determined and plotted against the UV dose. Each point is the mean value of two independent determinations.
Table 2. Plating efficiency of priA mutants on minimal and rich medium.
| Strain | priABs genotypea | c.f.u. × 108 per OD650b | ||
|---|---|---|---|---|
| IPTG | Minimal medium | Rich medium | ||
| PPBJ120 | 1 | + | 0.03 | <0.0001 |
| PPBJ117 | 2 | + | 0.07 | <0.0001 |
| PPBJ65 | + | + | 3.3 | 4.3 |
| PPBJ65 | + | – | 2.6 | 1.9 |
| PPBJ69 | ind | + | 1.9 | 1.5 |
| PPBJ69 | ind | – | 0.19 | 0.06 |
aSee Figure 1D.
bThe strains were grown to mid-log phase in minimal medium in the presence (+) or absence (–) of IPTG (1 mM). Appropriate dilutions of the cultures were plated on either rich or minimal medium containing IPTG. c.f.u., colony forming units. Values are the average of between two and four independent determinations, except for PPBJ69 in rich medium + IPTG, which was tested only once.
DNA manipulation and analysis
Standard techniques were used for DNA manipulation and cloning in E.coli (38). Total DNA from exponentially growing B.subtilis cells was extracted as described (28). After agarose gel electrophoresis, plasmid DNA was revealed by Southern blotting with α-32P-radiolabelled probe generated with a nick translation kit (Roche) with purified plasmid DNA in the presence of [α-32P]dATP (ICN). The different plamid species were revealed and quantified with a Storm apparatus (Molecular Dynamics) and ImageQuant software.
DNA sequencing of PCR products or plasmid templates was done with the PRISM sequencing kit (Applied Biosystems) and resolved on an automated DNA sequencer (Applied Biosystem 377A).
M13 ssDNA was prepared from TG1 cells as described (38). pAPJ9 ssDNA was similarly prepared from TG1 cells containing the helper phage M13K07 (38).
DNA probes used for the electrophoretic mobility shift assays were prepared by several means. The 174 nt long ssiA+/– ssDNA probes were excised by restriction from M13-ssiA+ and M13-ssiA– ssDNA with the use of the following oligonucleotides complementary to restriction sites flanking ssiA: OSMG42, 5′-CGTCGACCTGCAGCATGCA-3′; OSMG43, 5′-GCCGATGAATTCGATCCT-3′; OSMG44, 5′-GCCGATGAATTCGATTAAT-3′. The underlined sequences represent a PstI site in OSMG42 and an EcoRI site in OSMG43 and OSMG44. The OSMG42/OSMG43 pair was used to excise ssiA+ from the M13-ssiA+ ssDNA template and, similarly, the OSMG42/OSMG44 pair to excise ssiA– from M13-ssiA–. The ssDNA (50 nM) was heated at 65°C for 10 min in a 1 ml solution containing 100 mM NaCl, 20 mM Tris, pH 7.5, 1 mM DTT and 10 mM MgCl2 and the complementary pair of oligonucleotides (500 nM) were allowed to anneal by cooling the mixture slowly to 37°C. The DNA was then digested to completion with PstI and EcoRI. The 174 nt fragments released were purified from the larger bacteriophage DNA fragment and smaller oligonucleotides by gel filtration on Superose 6 (Pharmacia). They were then treated with shrimp phosphatase (Pharmacia) before 5′-end-labelling using [γ-32P]ATP (ICN) and T4 polynucleotide kinase. Finally, both fragments were purified by electrophoresis on 5% (w/v) polyacrylamide gels and recovered by passive elution in buffer E (10 mM Tris, pH 8, 1 mM EDTA, 0.2% SDS, 0.3 M NaCl) overnight at 30°C. The size and uniformity of the fragments was verified on a denaturing polyacrylamide gel using a sequence size ladder.
dsDNA and branched DNA molecules were prepared by annealing the following purified oligonucleotides, which are identical to those used by McGlynn et al. (8) for the study of PriAEc and RecG binding to D-loop and bubble structures: OSMG27, 5′-GACGCTGCCGAATTCTACCAGTGCCTTGCTAGCATCTTTGCCCACCTGCAGGTTCACCC-3′; OSMG27comp, 5′-GGTGAACCTGCAGGTGGGCAAAGATGTCCTAGCAAGGCACTGGTAGAATTCGGCAGCGTC-3′; OSMG28, 5′-GGGTGAACCTGCAGGTGGGCGGTGCTCATCGTAGGTTAGTTGGTAGAATTCGGCAGCGTC-3′; OSMG29, 5′-AAAGATGTCCTAGCAAGGCAC-3′. The D-loop was made by annealing OSMG27, OSMG28 and OSMG29 mixed at a molar ratio of 1:2:3, respectively. The bubble was made by annealing OSMG27 and OSMG28 and the dsDNA with OSMG27 and OSMG27comp at a molar ratio of 1:2. Annealing was performed by heating the oligonucleotides in buffer A (10 mM Tris, pH 8, 1 mM EDTA, 100 mM NaCl) for 5 min at 95°C, then they were left for 10 min at 65°C, followed by slow cooling to room temperature. In each combination, the OSMG27 oligonucleotide was 5′-end-labelled with [γ-32P]ATP (ICN) and T4 polynucleotide kinase prior to annealing. The expected synthetic DNA substrates were purified by elution in buffer E after separation from free oligonucleotides by native electrophoresis in a 5% polyacrylamide gel. The Ost4 oligonucleotide was used as a ssDNA probe, prepared as for the other DNA substrates: Ost4, 5′-GCCAAGCTTGCATGCCTGCAGGTCGACTCTAGAGGATCCCCGGGTACCGAGCTCGAATTCACTGGCCGTCGTTTTACAACGTCGTGACTG-3′.
For each probe, the concentrations of DNA substrates used in the gel mobility assay were estimated by monitoring the specific activity of the labelled oligonucleotide after end labelling and the final activity of the purified substrate.
Purification of PriABs, PriAEc and SSBBs
PriABs and SSBBs were expressed and purified using the IMPACT system (New England Biolabs). PriABs and SSBBs proteins fused to the intein–CBD tag were overproduced in strains MiT898 and B834 (DE3), respectively. To limit protein aggregation, cell growth was carried out at 25°C. Optimal conditions for PriABs production were IPTG induction for 3 h at the end of exponential growth, and for SSBBs production overnight growth without induction. Cells were harvested, resuspended in HEN500-T buffer (20 mM HEPES pH 7.6, 0.1 mM EDTA, 500 mM NaCl, 0.1% Triton X-100) and broken by sonication (Bioblock Vibracell 72408 sonicator, used as recommended by the supplier). The lysate was centrifugated at 4°C for 1 h at 20 000 g, the supernatant loaded onto chitin beads and the protein separated from intein by addition of 30 mM DTT and incubation overnight at 4°C. The protein was eluted and further purified by conventional chromatography. In the case of PriABs, the protein in HEN100-D buffer (20 mM HEPES pH 7.6, 0.1 mM EDTA, 100 mM NaCl, 1 mM DTT), was loaded onto a Hi-Trap SP-Sepharose column (Pharmacia) and eluted with a linear NaCl gradient in HED buffer. The fractions containing PriABs were loaded onto a Hi-Trap heparin column (Pharmacia) and the protein bound was eluted with HEN100-D. It was diluted twice with 100% glycerol and stored at –20°C. The yield of PriABs was ∼2 mg protein l–1 of culture and its purity was estimated to be 95%. In the case of SSBBs, the protein eluted from the chitin beads was further purified by successive chromatography on Hi-Trap Q-Sepharose and heparin columns (Pharmacia). The eluted proteins were finally treated by heat (at 85°C for 5 min) to eliminate by precipitation the contaminants which co-purify with SSBBs. We have shown that such a heat treatment does not modify SSBBs binding activity to ssDNA, as shown previously for SSB of E.coli (10). We have also observed by gel filtration on a Superose 12 column (Pharmacia) that purified SSBBs is a tetrameric protein, like its E.coli counterpart. The yield of SSBBs was ∼0.5 mg protein l–1 and its purity was estimated to be 95%.
The same purification procedure was used for PriAEc as for PriABs, except that expression was in strain JC19008 carrying plasmid pSMG24 (39). The yield of purified soluble protein was similar in both cases.
Immunodetection of PriABs
Immunisation against PriABs and serum preparation in the rabbit was entrusted to Eurogentec. Prior to injection, PriABs protein was further purified to homogeneity by electrophoresis on a SDS–polyacrylamide gel. The antibodies directed against PriABs were purified from serum by the method described in Pringle et al. (40) after coupling PriABs to Affi-Gel10 as recommended by the supplier (Bio-Rad). The relative levels of PriABs in the different strains used were determined by immunoblot analysis with the purified antibodies. Cells were grown exponentially in LB medium suplemented with IPTG. Cell lysates were prepared by lysozyme treatment of the harvested cells followed by brief sonication. The same amounts of total cellular proteins of each strain were then fractionated by SDS–PAGE on 8% gels and transferred to a Hybond PVDF membrane (Amersham) by electroblotting using a semi-dry transfer system. PriABs immunodetection was carried out as described in the ECL+ kit (Amersham). Purified anti-PriABs antibodies were diluted 1/500 for the hybridisation step. Protein G–horseradish peroxidase (Bio-Rad) was used to reveal anti-PriABs with a Storm apparatus (Molecular Dynamics) and quantification was with ImageQuant software.
Gel mobility shift assays
For some experiments reported (see Figs 4 and 5) the reaction mixture (40 µl) contained 10 mM HEPES pH 7.5, 3 mM DTT, 200 mM NaCl, 0.2 mg/ml bovine serum albumin (BSA) and γ-32P-labelled DNA, at the concentrations specified in the figure legends. The amounts of purified PriABs and SSBBs (expressed in nM) are indicated in the figures. Reaction mixtures were incubated at 30°C for 15 min and analysed by gel electrophoresis through a 5 or 4% (80:1) polyacrylamide gel as indicated in the figure legends, following addition of 10 µl of 50% glycerol (supplemented with 0.04% xylene cyanol and 1 mg ml–1 BSA). In the case of the binding experiments performed with SSBBs and PriABs, SSBBs was pre-incubated for 15 min at 30°C with the ssDNA substrates prior to addition of PriABs, which was then allowed to further interact for 15 min at 30°C. For other experiments (see Fig. 6), various amounts of PriABs and PriAEc were incubated with labelled DNA substrates (0.1 nM) in 20 µl of R buffer (50 mM HEPES, 1 mM DTT, 1 mM EDTA, 0.1 mg ml–1 BSA, 50 mM NaCl, 12.5% glycerol, pH 7.4) at 30°C for 10 min. At the end of incubation, 5 µl of loading buffer (50% glycerol, 0.4% cyanol, 0.1 mg ml–1 BSA) was added and the samples were loaded on a 5% polyacrylamide gel (30:1) containing 5% glycerol and 0.25× TBE. Three different electrophoresis buffers were used: TAM (6 mM Tris, 5 mM Na acetate, 2 mM Mg acetate) (see Fig. 5), TEG (25 mM Tris, 0.19 M glycine, 1 mM EDTA) (see Figs 4 and 5) or 0.25× TBE (90 mM Tris–borate, 2 mM EDTA) (see Fig. 6). Electrophoresis was at 4°C at 12 V cm–1 for 2–4 h with circularisation of the buffer. Following electrophoresis, gels were dried under vacuum, revealed with a Storm apparatus (Molecular Dynamics), the radioactivity quantified with ImageQuant software and the apparent Kd determined according to Riggs et al. (41).
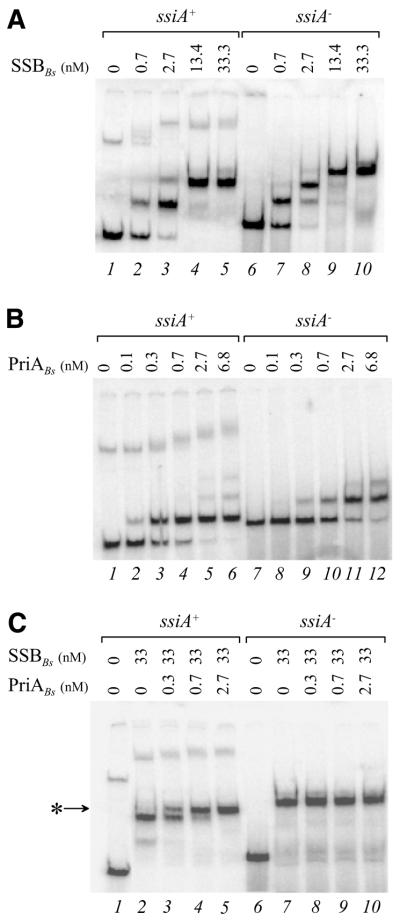
Figure 4.
PriABs binds to the active strand of ssiA in the presence of SSBBs. Protein–DNA complexes were generated at 30°C for 30 min and electrophoresed on a 4% non-denaturing polyacrylamide gel in TGE buffer at 4°C. A 174 nt long ssDNA fragment containing ssiA+ or ssiA– sequence was 5′ radiolabelled and used as a DNA binding substrate. Final protein concentrations of each binding experiment (expressed in nM) are indicated above each lane. SSBBs and PriABs refer to B.subtilis SSB and PriA proteins, respectively. (A) Binding of SSBBs. (B) Binding of PriABS. (C) Binding of PriABs to ssiA strands covered by SSBBs. The star indicates the PriABs supershift observed with ssiA+ substrate covered by SSBBs.
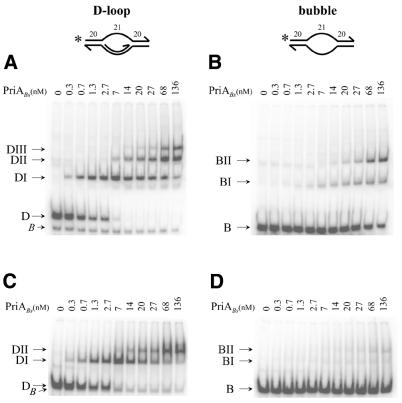
Figure 5.
PriABs binds preferentially to a D-loop structure. The numbering on the schematic representation of the D-loop and bubble substrates indicates the size (in nt) of the dsDNA and ssDNA part, the star indicates the position of the 32P radiolabelling present at the 5′ end. Purified PriABs protein at the indicated concentrations (expressed in nM) was added to the substrates, incubated at 30°C for 30 min and the protein–DNA complexes were resolved on a 5% non-denaturing polyacrylamide gel run at 4°C in TGE buffer (A and B) or in TAM buffer (C and D). (A and C) PriABs binding to the D-loop. (B and D) PriABs binding to the bubble.
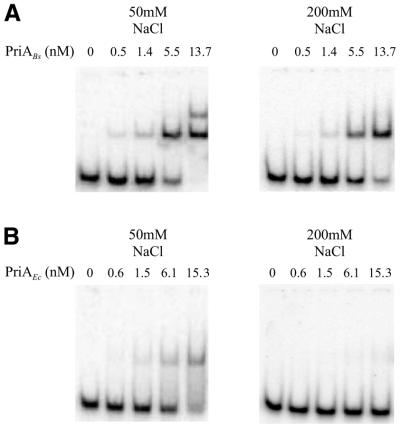
Figure 6.
PriABs binds more stably to ssDNA than PriAEc. Protein–DNA complexes were electrophoresed on a 5% non-denaturing polyacrylamide gel containing 5% glycerol in 0.25× TBE buffer at 4°C. The 90 nt long Ost4 oligonucleotide was 5′ radiolabelled and used as a DNA binding substrate with the indicated concentrations of PriABs (A) or PriAEc (B) (expressed in nM). Binding assays were performed in buffer containing either 50 (left) or 200 mM NaCl (right).
ATPase assay
ATPase activity was assayed by linking ATP hydrolysis to the oxidation of NADH as described previously (42). The dependence of the ATPase reaction on ssDNA cofactor was examined by the above method at 37°C in a buffer containing 50 mM HEPES pH 7.5, 50 mM NaCl, 5 mM MgCl2, 0.2 mg ml–1 BSA. The concentrations of DNA (expressed in nM nt), ATP and proteins PriABs and SSBBs (in nM) used in the assays are indicated in the figure legends, as well as the time course of the reaction. Kinetic experiments were performed in a UV/VIS spectrometer Lambda 20 (Perkin Elmer). Values for the Michaelis–Menten constants kcat and Km for ATP at saturating amounts of ssDNA were derived by fitting data directly to the Michaelis–Menten equation.
Helicase assay
The same standard forked DNA used for study of the Thermus aquaticus helicase (43) was used to assay the helicase activity of PriABs. It was similarly prepared by annealing the following two purified oligonucleotides, after labelling of the 5′ end of oligonucleotide OPP210 with T4 polynucleotide kinase (NEB): OPP210, 5′-T30CGAGCACCGCTGCGGCTGCACC-3′; OPP211, 5′-GGTGCAGCCGCAGCGGTGCTCG T30-3′.
Helicase assays were performed with the indicated amount of PriABs added to 1 nM DNA substrate in 20 µl of reaction buffer composed of 20 mM Tris pH 7.5, 50 mM NaCl, 3 mM MgCl2, 4 mM DTT, 20 µg ml–1 BSA, with or without addition of ATP (5 mM) as indicated. After 30 min incubation at 30°C each reaction was stopped with 5 µl of S solution (3% SDS, 100 mM EDTA, 40% glycerol, 0.1% xylene cyanol) and run through a 12% polyacrylamide gel in 1× TBE. Following electrophoresis, gels were dried under vacuum, revealed with a Storm apparatus (Molecular Dynamics) and the radioactivity present in the forked substrate and in the ssDNA product quantified with ImageQuant software to calculate the level of helicase activity expressed as a percentage of ssDNA generated in the assay.
RESULTS
Bacillus subtilis encodes a homologue of the E.coli primosomal PriA protein
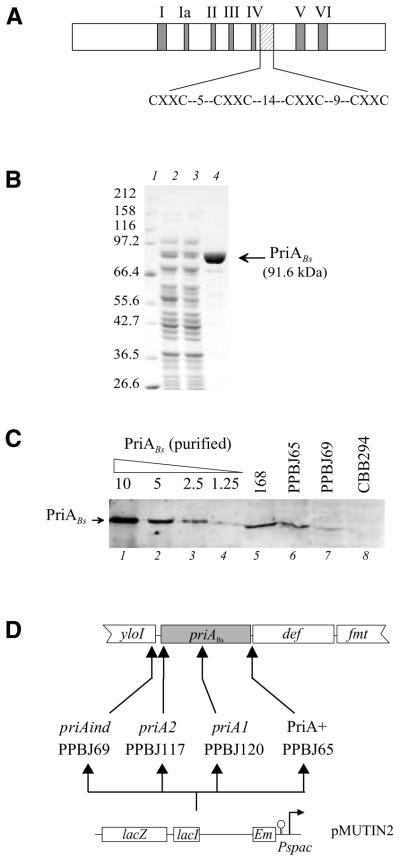
The sequence of a B.subtilis ORF encoding a homologue of PriAEc has been reported (27,30). This ORF, designated priABs, is located at 140° on the B.subtilis map. On the basis of sequence analysis, priABs is the second ORF of an operon including 12 ORFs. The level of homology of PriABs with the E.coli protein is highly significant, with 32% identity and 65% similarity distributed along the two proteins. However, PriABs contains 70 additional amino acids, clustered in the first third of the protein. Two distinct functional regions present in PriAEc are conserved in the B.subtilis protein. One corresponds to the seven canonical motifs typical of many helicases and the other to a cysteine-rich region, which may be organised in two consecutive small zinc finger-like motifs (Fig. 1A).
Figure 1.
The B.subtilis PriA protein. (A) Schematic representation of the B.subtilis PriA protein. The double signature which characterises PriAEc and which is conserved in PriABs is presented: (i) the seven conserved motifs (I–VI) of the 3′→5′ helicase subfamily into which PriAEc is classified (filled boxes); (ii) the two putative zinc finger domains (hatched box); the spacing between the conserved cysteines is indicated. (B) Purification of PriABs. The PriABs protein was purified in E.coli using the IMPACT system. Proteins were resolved by 8% SDS–PAGE and stained with Coomassie brilliant blue. Lane 1, molecular weight standards; lane 2, soluble proteins of E.coli cells induced to express the fusion PriABs–intein–CBD; lane 3, flow-through of the chitin column; lane 4, elution from the chitin column after DTT cleavage. The purity obtained at this step has been estimated to be >95%. (C) Immunodetection of PriABs in B.subtilis. Protein preparations were resolved by 8% SDS–PAGE, transferred to PVDF membrane and probed with purified anti-PriABs antibodies. Lanes 1–4, the indicated amounts (in ng) of purified PriABs; lanes 5–8, total proteins extracted from an equal amount of cells grown in the presence of IPTG from strain 168 and derivatives PPBJ65, PPBJ69 and CBB294 (a derivative of PPBJ120 carrying an extragenic mutation suppressing the lack of PriABs, designated dnaB75) (33). The priABs allele present in each strain is indicated. (D) Schematic representation of the priABs chromosomal region in B.subtilis strains used in this study. priABs and flanking ORFs are represented by boxes. yloI is an ORF of unknown function; def and fmt are homologues of the E.coli def and fmt genes. In the lower part a schematic map of the pMUTIN2 integrative vector carrying the IPTG-inducible promoter Pspac is presented. Vertical arrows below the priABs ORF indicate the insertion sites of pMUTIN2 derivatives. The names of the resulting strains with their associated priA allele are indicated.
To show that priABs is transcribed and translated, its putative product has been overproduced in E.coli and purified (Fig. 1B) and specific antibodies directed against this protein have been prepared and purified (see Materials and Methods). Whole cellular protein extracts from B.subtilis were analysed by western blotting using the anti-PriABs antibodies (Fig. 1C). A protein identical in size with the purified PriABs was detected in the B.subtilis wild-type strain (Fig. 1C, lane 5). The specifity of the signal was demonstrated by its disappearance in strain CBB294, in which priABs has been disrupted (see below; Fig. 1C, lane 8). Therefore, PriABs is expressed in B.subtilis. The number of PriABs molecules per cell is between 50 and 100, as deduced from the western blot analysis.
Bacillus subtilis priA mutants are poorly viable, sensitive to rich medium and UV irradiation
To study the role of PriABs in cell physiology, we constructed priABs mutants. For this purpose we disrupted priABs by transforming B.subtilis 168 cells with non-replicative EmR plasmids carrying internal fragments of priABs (pAPJ11 and pAPJ14). As a control, we used a plasmid carrying the 3′ end of priABs, and thus expected to preserve the integrity of priABs upon insertion (pAPJ12). As E.coli null priA mutants are viable on minimal medium but not on rich medium (13), disruption was carried out on minimal medium. EmR transformants were obtained with the disrupting plasmids, giving strains PPBJ117 and PPBJ120, which carried priABs alleles designated priA2Bs and priA1Bs, respectively (Fig. 1D). However, colonies were much smaller in size than upon transformation with the control plasmid. As expected from the size of the colonies, these strains grow slowly in minimal medium (doubling time >160 min) and microscopic examination of the bacteria revealed that they were filamentous. Measurements of the plating efficiencies of these strains showed that they were 40–100-fold less viable than the control strain on minimal medium and were sensitive to rich medium (Table 2), as well as to UV irradiation (Fig. 2). A strain in which priABs is under the control of the Pspac promoter was also constructed (PPBJ69 carrying the priAind allele) (Fig. 1D). This strain displayed wild-type phenotypes in the presence of IPTG (see Table 2 and Fig. 2), although its level of PriABs was 3-fold lower than in the control strains, as shown by western blot analysis (Fig. 1C, compare lanes 5 and 6 with 7). In the absence of IPTG, however, this strain displayed phenotypes similar to those of disrupted strains: filamentation, small colonies and poor viability in minimal medium, as well as sensitivity to rich medium (Table 2) and sensitivity to UV irradiation (Fig. 2).
PriABs is required for primosome assembly on ssiA and D-loops in vivo
In E.coli, PriA was initially characterised as being required for two distinct modes of replication displayed by extrachromosomal elements. The first relies on a pas sequence required for replication of the ssDNA circular intermediate generated during rolling circle replication of bacteriophage φX174 (10). The second depends on a D-loop structure synthesised at an early step of the θ replication mode of ColEI-type plasmids (10). Interestingly, these two schemes of replication have been characterised in B.subtilis and have been shown to rely on identified primosomal proteins of this bacterium (28). Therefore, we tested PriABs dependence for these two modes of extrachromosomal replication. The pas-mediated conversion of ssDNA to dsDNA was measured in a plasmid rolling circle assay with the use of the pas sequence ssiA from plasmid pAMβ1 (28). D-loop-mediated replication was measured using an appropriate pAMβ1 derivative (28,44–46). The experiments were carried out in strains harbouring the conditional priAind allele, allowing modulation of priABs expression with the use of IPTG (Fig. 1).
To test the activity of ssiA, we used derivatives of the rolling circle plasmid pC194, which produce a ssDNA intermediate which is not efficiently converted to the dsDNA form in B.subtilis (47). This circular ssDNA molecule is detected by Southern blotting following electrophoresis of total DNA prepared from B.subtilis cells harbouring such plasmids (Fig. 3A). In the PriA+ strain PPBJ65, conversion of ssDNA to the dsDNA form is promoted by ssiA in the active orientation (ssiA+) but not in the inactive orientation (ssiA–) (Fig. 3A, lanes 3 and 1). In contrast, in the priAind strain, conversion is inefficient, irrespective of the ssiA orientation. Conversion was inefficient both in the absence (Fig. 3A) and presence of IPTG (not shown), indicating that the diminished level of PriABs in the induced priAind strain is not sufficient to support ssiA activity on a multicopy extrachromosomal element, while it appears sufficient for chromosomal replication.
Figure 3.
PriABs is required for ssiA activity and pAMβ1-type plasmid replication. (A) Bacillus subtilis PPBJ65 (PriABs+) and PPBJ69 (priAind) strains harbouring pC194-derived plasmid pADG6406-1 (ssiA+) or pADG6406-2 (ssiA–) were grown overnight in LB supplemented with IPTG, Em and Cm, then diluted 100-fold in fresh medium without IPTG and cultivated for ∼5 h. Total DNA was extracted and analysed by Southern hybridisation using 32P-labelled pC194 DNA as probe. (B) Bacillus subtilis PPBJ65 (PriABs+) and PPBJ69 (priAind) strains harbouring pAMβ1-derived plasmid pVA798ΔRCR were grown to mid-log phase without IPTG and their total DNA was extracted and analysed by Southern hybridisation, using 32P-labelled pVA798ΔRCR as probe. ssDNA and dsDNA, single-stranded and double-stranded DNA, respectively.
pAMβ1-type plasmids replicate by a θ mechanism that involves an early D-loop intermediate, in which the ssiA+ sequence is present on the ssDNA portion of the molecule (44–46). In the priAind mutant grown without IPTG the copy number of plasmid pVA798ΔRCR (a pAMβ1-type plasmid; 48) was ∼10-fold lower than in PriABs+ cells and the plasmid accumulated ssDNA (Fig. 3B). The ssDNA corresponded to the plasmid lagging strand template, as demonstrated by the use of strand-specific probes (not shown). A similar replication defect was observed when the cells were grown in the presence of IPTG (not shown), presumably reflecting the low PriABs level in the cells. The defect of pVA798ΔRCR replication in the absence of PriABs led to loss of the plasmid from priAind cells upon prolonged growth without IPTG and to its inability to become established in the priA1Bs strain (not shown). We conclude that PriABs is required for replication of pAMβ1-type plasmids and acts presumably by promoting primosome assembly on the D-loop intermediate. The ssiA sequence unmasked on the D-loop was previously shown not to be essential for pAMβ1 replication, suggesting a ssiA-independent mechanism(s) of primosome assembly (28). In priAind mutants pAMβ1 derivatives lacking ssiA exhibited a reduced copy number and accumulated ssDNA (not shown). We conclude that a ssiA-independent mechanism(s) of primosome loading during pAMβ1 replication is dependent on PriABs and assume that PriABs, like PriAEc, triggers primosome assembly on D-loops.
PriABs binds preferentially to the primosome assembly site ssiA
Analysis of ssDNA conversion to dsDNA indicated that PriABs acts in vivo at ssiA. In order to test whether PriABs recognises the active strand of ssiA, we carried out gel shift analyses with the purified protein. For these experiments two 174 nt long ssDNA substrates, carrying either the active or inactive strand of ssiA (designated ssiA+ and ssiA–, respectively), were prepared and radiolabelled (see Materials and Methods). Somewhat surprisingly, the two substrates behaved differently in non-denaturing gels: the ssiA+ strand migrated faster than the ssiA– strand (Fig. 4A, lanes 1 and 6) and was accompanied by a minor, slowly migrating product (Fig. 4A, lane 1). In contrast, the two strands appeared identical in size in a gel under denaturing conditions (data not shown). This suggests that the two strands might fold into different secondary or tertiary structures. Binding experiments conducted with B.subtilis SSB protein (SSBBs; see Materials and Methods for purification procedure) led to the same conclusion. Both strands were efficiently recognised by SSBBs, but the binding patterns were clearly different (Fig. 4A). The faster migrating form of ssiA+ gave primarily two shifted bands while ssiA– gave an additional band at saturating amounts of SSBBs. The slower ssiA+ form gave multiple shifted bands. This indicates that a part of the ssiA+ sequence may be poorly accessible to a SSBBs tetramer.
PriABs binding to the ssiA substrates indicated a 7-fold preference for ssiA+ (Kd = 0.45 nM) over ssiA– (Kd = 3.1 nM) (Fig. 4B). PriABs generated one complex with the ssiA+ substrate at low concentration and up to three complexes at higher concentrations. The slow migrating form of ssiA+ was also shifted by PriABs to poorly resolved multiple bands. These experiments revealed that PriABs can bind stably to the inactive ssiA strand. This interaction does not depend on the presence of the ssiA sequence since it occurs with any ssDNA molecule longer than 41 nt, but not with dsDNA, to which PriABs binds poorly (data not shown).
Finally, we carried out a binding experiment between PriABs and ssiA strands preincubated with an excess of SSBBs. Under these conditions only the ssiA+ substrate was shifted by PriABs (Fig. 4C). Both fast and slow migrating forms of ssiA+ were shifted, indicating that they have related structures (Fig. 4C).
PriABs binds specifically to a D-loop structure
Analysis of pAMβ1-related plasmids indicated that PriABs acts in vivo at D-loops (see above). We therefore investigated whether PriABs binds to the D-loop and bubble structures used previously to reveal PriAEc binding (8). As shown in Figure 5, PriABs bound preferentially to the D-loop (Kd = 1.5 nM) in comparison to a bubble (Kd = 150 nM). Three complexes (DI, DII and DIII) (Fig. 5A) appeared consecutively with increasing amounts of PriABs added to the D-loop, whereas only two were observed with the bubble (BI and BII) (Fig. 5B). The appearance of DI at low protein concentrations, followed by that of DII, DIII, BI and BII at higher concentrations, clearly demonstrates the preferential binding of PriABs to the D-loop structure. A small amount of unbound contaminant bubble structure in the D-loop preparation (Fig. 5A, band B) provided an internal control which confirmed the preference for the D-loop. Furthermore, complexes of PriABs with the bubble substrate (BI and BII) were much less stable than those generated with the D-loop, since they almost completely disappeared when electrophoresed under destabilising conditions (i.e. in the presence of magnesium; Fig. 5D), whereas complexes with the D-loop remained stable (Fig. 5C). These results confirmed and detailed what was previously reported with PriABs purified differently from an insoluble form (31).
PriABs binds strongly to ssDNA
The above gel shift experiments indicated that PriABs displays a high affinity for ssDNA, which would clearly distinguish PriABs from its functional homologue PriAEc. To compare the two proteins with respect to their ssDNA binding activity, we performed gel shift experiments with the ssDNA substrate Ost4, a 90 nt long oligonucleotide. The PriAEc protein was purified by a procedure similar to that used for PriABs (see Materials and Methods). As shown in Figure 6, PriABs binds with a better affinity than PriAEc to the ssDNA probe (compare Fig. 6A and B, left; Kd = 4 and 20 nM, respectively). PriABs generated two discrete retarded bands, while a smear from the retarded band to the free DNA was observed with PriAEc. Such a gel shift pattern is strongly indicative that PriAEc binding to ssDNA is unstable. Accordingly, increasing the ionic strength of the binding buffer nearly eliminated the band shift induced by PriAEc, while PriABs still bound to the substrate efficiently under those conditions (compare Fig. 6A and B, right).
PriABs is a ssDNA-dependent ATPase displaying helicase activity
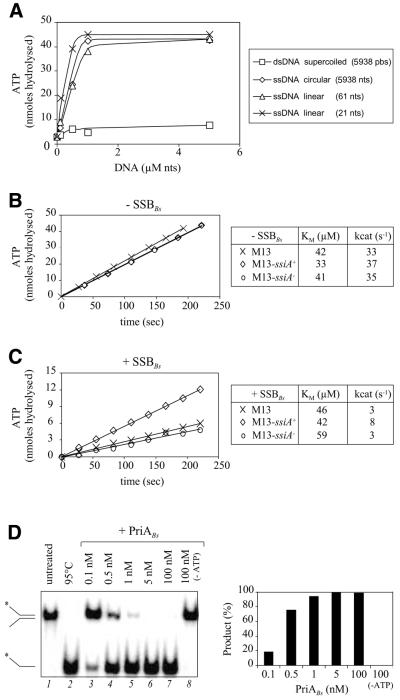
PriAEc is a ssDNA-dependent ATPase fueling its helicase activity (3). We have observed that PriABs induces ATP hydrolysis in the presence of naked ssDNA, but not dsDNA (Fig. 7A), indicating that it has a similar activity. Stable binding of PriABs to ssDNA is not required for this activity, because a 21 nt long oligonucleotide efficiently triggered PriABs ATPase activity (Fig. 7A) but did not form a stable complex with PriABs as judged by gel shift experiments (data not shown). The presence of ssiA+ or ssiA– in naked M13 circular ssDNA did not affect the ATPase activity of PriABs (Fig. 7B). However, addition of SSBBs protein prior to PriABs, which reduced ATPase activity with all substrates, had less effect with the ssiA+ than with the ssiA– substrate (Fig. 7C). We conclude that PriABs is a ssDNA-dependent ATPase and suggest that SSBBs limits the accessibility of PriABs to ssDNA, unless a specialised DNA sequence, such as ssiA, is present in the ssDNA. The ATPase activity of PriABs is linked to the translocase/helicase activity already reported for PriABs (31). As illustrated in Figure 7D, we have also confirmed that our PriABs preparation displayed helicase activity: provided that ATP was included in the reaction, PriABs efficiently unwound a Y-shaped DNA molecule used in the helicase assay (43).
Figure 7.
PriABs is a ssDNA-dependent ATPase displaying helicase activity. (A) PriABs (55 nM) was incubated at 37°C for 30 min in the presence of increasing amounts of DNA at a fixed concentration of ATP (45 µM). The calculated amount of ATP hydrolysed is plotted against DNA concentration. Supercoiled dsDNA and ssDNA were prepared from the pAPJ9 phagemid as described in Materials and Methods. Purified oligonucleotides used for construction of the D-loop substrate were used as linear ssDNA substrates (cf. Fig. 5). (B and C) ssDNA-dependent ATPase activity of PriABs was measured at 37°C in the presence of constant amounts (1 µM nt) of three ssDNA substrates, in the absence (B) or presence (C) of SSBBs protein. In the experiment conducted with SSBBs, ssDNA substrates were incubated with this protein (0.5 monomer/nt) for 30 min at 37°C prior to addition of PriABs. A representative kinetic experiment at 0.1 mM ATP is shown; similar experiments were carried out at different ATP concentrations in order to calculate the kcat and Km values of the enzyme for ATP. These values are reported in the adjacent table and are the average of two independent experiments. M13, M13- mp19 ssDNA; M13-ssiA+, M13-mp19 ssDNA carrying the ssiA+ sequence; M13-ssiA–, M13-mp19 ssDNA carrying the ssiA– sequence. (D) PriABs displays helicase activity. (Left) Native gel analysis of PriABs unwinding activity. Lanes 1 and 2 contain, respectively, the synthetic forked DNA substrate and the labelled ssDNA liberated following heating at 95°C for 5 min, which are represented schematically on the left. Lanes 3–7 contain reactions performed with increasing amounts of PriABs (indicated in nM at the top of the gel). Lane 8 contains a reaction performed with the same amount of PriABs as in lane 7, but without ATP in the reaction buffer. (Right) Quantitation of PriABs unwinding activity. Helicase activity is expressed as the percentage of the liberated ssDNA strand quantified in each sample lane in the gel presented in (A) (see Materials and Methods).
PriABs does not complement an E.coli priA null mutant
The results presented above show that PriABs is functionally equivalent to PriAEc in vivo and in vitro, as was previously proposed by Masai et al. (31). A question raised by this identity is whether one PriA can substitute for the other in vivo. We tested this hypothesis with PriABs in E.coli. For this purpose we cloned priABs in pGB2, a plasmid which does not depend on PriAEc for replication, and placed it under the control of E.coli translational and IPTG-inducible transcriptional signals to give plasmid pAPJ41 (see Materials and Methods). Another plasmid, pAPJ43, carrying the priAEc coding sequence under the same expression signals, was constructed as a control. The priAEc null mutant strain JC18983 and the wild-type isogenic strain DM4000 (49) were transformed by the two plasmids and tested for several phenotypes associated with the lack of PriAEc: viability, growth on rich medium, UV sensitivity and replication of the ColE1-type plasmid pBR322. As expected, the priAEc-carrying plasmid corrected the phenotypes of the priA mutant even without IPTG induction (Table 3) (only viability and sensitivity to rich medium were tested for this plasmid). In contrast, the priABs-carrying plasmid did not correct any of the mutant phenotypes, either non-induced or induced with low IPTG concentrations (Table 3). At higher IPTG concentrations induction of PriABs was toxic (Table 3). The toxicity associated with PriABs expression was not observed in the wild-type strain (Table 3). These results show that PriABs cannot substitute for PriAEc in vivo.
DISCUSSION
We report a detailed analysis of a B.subtilis protein proposed to be the counterpart of the E.coli PriA primosomal protein on the basis of sequence similarities (27,30,31), confirming and extending a previous biochemical analysis of this protein (31). Several lines of in vivo and in vitro evidence demonstrate that this protein is indeed PriA.
Typical phenotypes associated with the lack of PriA in E.coli have been observed with B.subtilis priA mutant cells. These include poor viability, slow growth, filamentation and sensitivity to rich medium and UV. In E.coli these defects of priA mutants are thought to be due to a deficiency in the repair of arrested replication forks (for reviews see 6,16,17). We therefore propose that PriABs plays a similar role in replication fork reactivation in B.subtilis.
PriAEc is required for replication of several E.coli extrachromosomal elements (3). We report defects in two modes of extrachromosomal replication in B.subtilis priA mutants. One is the ssiA-dependent conversion of ssDNA to dsDNA. ssiA has been shown to act as a primosome assembly site in B.subtilis (28), similarly to the pas sequence of bacteriophage φX174 in E.coli (3). Moreover, we show that PriABs binds stably and specifically to the active strand of ssiA in vitro, as does the E.coli protein to the pas sequence of φX174 (5). PriABs still binds to ssiA in the presence of SSBBs protein. These results show that PriABs binds ssDNA carrying ssiA and suggest that it triggers primosome assembly at this site. Interestingly, ssiA appears to adopt a particular structure while its complementary strand does not. It has been shown that pas sites in E.coli are structurally distinct from their complementary strands and are resistant to melting by SSB and it is proposed that this feature determines their recognition by PriAEc (3). Similarly, we propose that the structure adopted by ssiA is refractory to melting by SSBBs and that this contributes to its specific recognition by PriABs in vivo.
Another mode of extrachromosomal replication dependent on PriABs is that of the pAMβ1-type plasmids, which is similar to that described for the E.coli ColE1-type replicons (44). It involves the formation of a D-loop structure, to which PriAEc binds specifically and promotes primosome assembly in vitro (8,9,18,19). PriABs protein efficiently binds an artificial D-loop structure in vitro (this study; 31). We observed that PriABs interaction with this three-stranded DNA molecule is more specific and much more stable than with a bubble structure. These combined in vivo and in vitro analyses suggest that PriABs triggers primosome assembly on such branched molecules, as reported for PriAEc. Such structures are thought to be targeted by PriA during DNA recombinational repair (3,6,16,17).
Another characteristic shared by the B.subtilis and E.coli PriA proteins is their low quantity in the cell, estimated to be 50–100 molecules per cell (50). We present evidence that this level can be reduced 3-fold in B.subtilis without the appearance of detectable cellular defects. However, the diminished quantity of PriABs is not high enough to sustain a normal level of PriABs-dependent extrachromosomal replication.
During the course of this study the purification of PriABs, a description of its binding to D-loop structures and its ssDNA-dependent ATPase and helicase activities have been reported (31). Our in vitro observations with a purified soluble form of PriABs confirm and extend this preliminary report. We have observed a more stable binding of PriABs than PriAEc to ssDNA. Probably associated with this property is the capability of PriABs to bind to bubble structures, although less stably and with a lower affinity than to the D-loop structure. SSBBs prevents PriABs binding to ssDNA, but not to the ssiA sequence (this study) nor to a forked structure (data not shown). Therefore, we propose that SSBBs protein participates in the specific targeting of PriABs-mediated primosome assembly to the DNA, as recently concluded for the E.coli primosomal restart machinery (24). PriA and SSB are two proteins highly conserved in bacteria and are two players involved in the early steps of replication fork re-activation. Therefore, an identical functional scaffold of replication restart appears conserved in these microorganisms in the initial stages.
Despite the strong similarities between the two bacterial PriA proteins, PriABs does not substitute for PriAEc in vivo, which shows its host specificity. We have observed that PriABs is toxic in E.coli priA cells, but not in isogenic wild-type cells. It is possible that PriABs competes in the mutant with the primosomal pathways that operate in the absence of PriAEc (51). Another possibility would be that production of PriABs adds yet another defect to the priA mutant cells, which are already affected in the metabolism of chromosomal DNA. The strong affinity of PriABs for ssDNA might be responsible for this toxicity. Nevertheless, the lack of toxicity of PriABs in wild-type cells suggests that it neither competes efficiently with the endogenous PriAEc for its regular chromosomal substrates nor titrates other protein partners.
The host specificity of the PriA protein raises the question of the protein content of the PriA-dependent primosome in B.subtilis. The E.coli PriA primosomal partners are PriB, PriC, DnaT, DnaC, DnaB (the replication fork helicase) and DnaG (the primase). Likely counterparts of the DnaB helicase and the DnaG primase are encoded respectively by dnaC and dnaG (formerly dnaE) in B.subtilis (25,26), but no obvious homologues of PriB, PriC and DnaT have been found in B.subtilis (27). We have suggested that three B.subtilis proteins, DnaB, DnaD and DnaI, initially identified as required for initiation of chromosome replication, are primosomal proteins. Indeed, they are required for the activity of the primosome assembly site ssiA and are involved in the replication of pAMβ1-type plasmids (28,52–54). Moreover, DnaI was shown to interact with DnaC in a two-hybrid assay and to co-localise with DnaB in the cell (55). Recently we isolated dnaB mutations that suppress the phenotypes of B.subtilis priA mutants. Interestingly, the in vivo defects of primosome assembly observed in a priA mutant were compensated for by a dnaB mutation, in a dnaD- and dnaI-dependent manner (33). Furthermore, we directly showed in vitro that purified PriA, DnaD and DnaB proteins specifically interact in this order on a forked DNA substrate, mimicking the product of recombinational repair of a stalled replication fork (39). Altogether, these genetic and biochemical observations suggest that PriABs, DnaB, DnaD and DnaI could act together to load the replication fork helicase DnaC onto DNA during replication fork restart. Therefore, the E.coli and B.subtilis restart primosomes have apparently diverged at the proteins acting between the PriA initiator and the replicative helicase.
Acknowledgments
ACKNOWLEDGEMENTS
We thank D. Mazel and J. Errington for providing DNA sequences of the priA region before publication, M. Farache for technical help, S. Sandler and K. J. Marians for providing the E.coli strains DM4000 and JJC18983 and the PriAEc expression plasmid and M. -A. Petit for critical reading of the manuscript. This work was supported, in part, by the Ministère de l’Education Nationale, de la Recherche et de la Technologie (Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires) and the European Commission (BIO4-CT98-0250). P.P. is on the CNRS staff.
REFERENCES
- 1.Wickner S. and Hurwitz,J. (1974) Conversion of φX174 viral DNA to double stranded form by purified E. coli proteins. Proc. Natl Acad. Sci. USA, 71, 4120–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schekman R., Weiner,J., Weiner,A. and Kornberg,A. (1975) Ten proteins required for conversion of φX174 single-stranded DNA to duplex form in vitro. J. Biol. Chem., 250, 5859–5865. [PubMed] [Google Scholar]
- 3.Marians K.J. (1999) PriA: at the crossroads of DNA replication and recombination. Prog. Nucleic Acid Res. Mol. Biol., 63, 39–67. [DOI] [PubMed] [Google Scholar]
- 4.Allen G.C. and Kornberg,A. (1993) Assembly of the primosome of DNA replication in Escherichia coli. J. Biol. Chem., 268, 19204–19209. [PubMed] [Google Scholar]
- 5.Ng J.Y. and Marians,K.J. (1996) The ordered assembly of the φX174-type primosome. I. Isolation and identification of intermediate protein-DNA complexes. J. Biol. Chem., 271, 15642–15648. [DOI] [PubMed] [Google Scholar]
- 6.Marians K.J. (2000) PriA-directed replication fork restart in Escherichia coli. Trends Biochem. Sci., 25, 185–189. [DOI] [PubMed] [Google Scholar]
- 7.Shlomai J.M. and Kornberg,A. (1980) An Escherichia coli replication protein that recognizes a unique sequence within a hairpin region in φX174 Proc. Natl Acad. Sci. USA, 77, 799–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGlynn P., Al-Deib,A.A., Liu,J., Marians,K.J. and Lloyd,R.G. (1997) The DNA replication protein PriA and the recombination protein RecG bind D-loops. J. Mol. Biol., 270, 212–221. [DOI] [PubMed] [Google Scholar]
- 9.Nurse P., Liu,J. and Marians,K.J. (1999) Two modes of PriA binding to DNA. J. Biol. Chem., 274, 25026–25032. [DOI] [PubMed] [Google Scholar]
- 10.Kornberg A. and Baker,T. (1992) DNA Replication, 2nd Edn. W.H.Freeman and Co., New York, NY.
- 11.Lee E.H. and Kornberg,A. (1991) Replication deficiencies in priA mutants of Escherichia coli lacking the primosomal replication n′ protein. Proc. Natl Acad. Sci. USA, 88, 3029–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nurse P., Zavitz,K.H. and Marians,K.J. (1991) Inactivation of the Escherichia coli PriA DNA replication protein induces the SOS response. J. Bacteriol., 173, 6686–6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masai H., Asai,T., Kubota,Y., Arai,K. and Kogoma,T. (1994) Escherichia coli PriA protein is essential for inducible and constitutive stable DNA replication. EMBO J., 15, 5338–5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kogoma T., Cadwell,G.W., Barnard,K.G. and Asai,T. (1996) The DNA replication priming protein, PriA, is required for homologous recombination and double-strand break repair. J. Bacteriol., 178, 1258–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kogoma T. (1997) Stable DNA replication: interplay between DNA replication, homologous recombination and transcription. Microbiol. Mol. Biol. Rev., 61, 212–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cox M.M., Goodman,M.F., Kreuzer,K.N., Sherratt,D.J., Sandler,S.J. and Marians,K.J. (2000) The importance of repairing stalled replication forks. Nature, 404, 37–41. [DOI] [PubMed] [Google Scholar]
- 17.Sandler S.J. and Marians,K.J. (2000) Role of PriA in replication fork reactivation in Escherichia coli. J. Bacteriol., 182, 9–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J. and Marians,K.J. (1999) PriA-directed assembly of a primosome on D loop DNA. J. Biol. Chem., 274, 25033–25041. [DOI] [PubMed] [Google Scholar]
- 19.Liu J., Xu,L., Sandler,S.J. and Marians,K.J. (1999) Replication fork assembly at recombination intermediates is required for bacterial growth. Proc. Natl Acad. Sci. USA, 96, 3552–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zavitz K.H. and Marians,K.J. (1992) ATPase-deficient mutants of the Escherichia coli DNA replication protein PriA are capable of catalyzing the assembly of active primosomes. J. Biol. Chem., 267, 6933–6940. [PubMed] [Google Scholar]
- 21.Jones J.M. and Nakai,H. (1997) The φX174-type primosome promotes replisome assembly at the site of recombination in bacteriophage Mu transposition. EMBO J., 16, 6886–6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones J.M. and Nakai,H. (1999) Duplex opening by primosome protein PriA for replisome assembly on a recombination intermediate. J. Mol. Biol., 289, 503–516. [DOI] [PubMed] [Google Scholar]
- 23.Jones J.M. and Nakai,H. (2000) PriA and phage T4 gp59: factors that promote DNA replication on forked DNA substrates. Mol. Microbiol., 36, 519–527. [DOI] [PubMed] [Google Scholar]
- 24.Jones J.M. and Nakai,H. (2001) Escherichia coli PriA helicase: fork binding orients the helicase to unwind the lagging strand side of arrested replication forks. J. Mol. Biol., 312, 935–947. [DOI] [PubMed] [Google Scholar]
- 25.Sakamoto Y., Nakai,S., Moriya,S., Yoshikawa,H. and Ogasawara,N. (1995) The Bacillus subtilis dnaC gene encodes a protein homologous to the DnaB helicase of Escherichia coli. Microbiology, 141, 641–644. [DOI] [PubMed] [Google Scholar]
- 26.Wang L.F., Price,C.W. and Doi,R.H. (1985) Bacillus subtilis dnaE encodes a protein homologous to DNA primase of Escherichia coli. J. Biol. Chem., 260, 3368–3372. [PubMed] [Google Scholar]
- 27.Kunst F., Ogasawara,N., Moszer,I., Albertini,A.M., Alloni,G., Azevedo,V., Bertero,M.G., Bessieres,P., Bolotin,A., Borchert,S. et al. (1997) The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature, 390, 249–256. [DOI] [PubMed] [Google Scholar]
- 28.Bruand C., Ehrlich,S.D. and Jannière,L. (1995) Primosome assembly site in Bacillus subtilis. EMBO J., 14, 2642–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshikawa H. and Wake,R.G. (1993) Initiation and termination of chromosome replication. In Sonenshein,A.L., Hoch,J.A. and Losick,R. (eds), Bacillus subtilisand Other Gram Positive Bacteria: Biochemistry, Physiology and Molecular Genetics. American Society of Microbiology, Washington, DC, pp. 507–528.
- 30.Mazel D., Coic,E., Blanchard,S., Saurin,W. and Marlière,P. (1997) A survey of polypeptide deformylase function throughout the eubacterial lineage. J. Mol. Biol., 266, 939–949. [DOI] [PubMed] [Google Scholar]
- 31.Masai H., Deneke,J., Furui,Y., Tanaka,T. and Arai,K.I. (1999) Escherichia coli and Bacillus subtilis PriA proteins essential for recombination-dependent DNA replication: involvement of ATPase/helicase activity of PriA for inducible stable DNA replication. Biochimie, 81, 847–857. [DOI] [PubMed] [Google Scholar]
- 32.Bron S. (1990) Plasmids. In Harwood,C.R. and Cutting,S.M. (eds), Molecular Biological Methods for Bacillus. John Wiley & Sons, Chichester, UK, pp. 75–174.
- 33.Bruand C., Farache,M., McGovern,S., Ehrlich,S.D. and Polard,P. (2001) DnaB, DnaD and DnaI proteins are components of the Bacillus subtilis replication restart primosome. Mol. Microbiol., 42, 245–256 [DOI] [PubMed] [Google Scholar]
- 34.Miller J.H. (1992) A Short Course in Bacterial Genetics. A Laboratory Manual and Handbook for Escherichia coliand Related Bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 35.Churchward G., Belin,D. and Nagamine,Y. (1984) A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene, 31, 165–171. [DOI] [PubMed] [Google Scholar]
- 36.Nurse P., DiGate,R.J., Zavitz,K.H. and Marians,K.J. (1990) Molecular cloning and DNA sequence analysis of Escherichia coli priA, the gene encoding the primosomal protein replication factor Y. Proc. Natl Acad. Sci. USA, 87, 4615–4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swinfield T.J., Oultram,J.D., Thompson,D.E., Brehm,J.K. and Minton,N.P. (1990) Physical characterisation of the replication region of the Streptococcus faecalis plasmid pAMβ1. Gene, 87, 79–90. [PubMed] [Google Scholar]
- 38.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Marsin S., McGovern,S., Ehrlich,S.D., Bruand,C. and Polard,P. (2001) Early steps of Bacillus subtilis primosome assembly. J. Biol. Chem., 276, 45818–45825. [DOI] [PubMed] [Google Scholar]
- 40.Pringle J.R., Adams,A.E., Drubin,D.G. and Haarer,B.K. (1991) Immunofluorescence methods for yeast. Methods Enzymol., 194, 565–602. [DOI] [PubMed] [Google Scholar]
- 41.Riggs A.D., Suzuki,H. and Bourgeoss,S. (1970) Lac repressor-operator interaction. I. Equilibrium studies. J. Mol. Biol., 48, 67–83. [DOI] [PubMed] [Google Scholar]
- 42.Pullman M.E., Penefsky,H.S., Datta,A. and Racker,E. (1960) Partial resolution of the enzymes catalysing oxidative phosphorylation. J. Biol. Chem., 235, 3322–3329. [PubMed] [Google Scholar]
- 43.Kaplan D.L. and Steitz,T.A. (2000) DnaB from Thermus aquaticus unwinds forked duplex DNA with asymetric tail length dependence. J. Biol. Chem., 274, 6889–6897. [DOI] [PubMed] [Google Scholar]
- 44.Bruand C., Le Chatelier,E., Ehrlich,S.D. and Jannière,L. (1993) A fourth class of theta-replicating plasmids: the pAMβ1 family from gram-positive bacteria. Proc. Natl Acad. Sci. USA, 90, 11668–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jannière L., Bidnenko,V., McGovern,S., Ehrlich,S.D. and Petit,M.-A. (1997) Replication terminus for DNA polymerase I during initiation of pAMβ1 replication: role of the plasmid-encoded resolution system. Mol. Microbiol., 23, 525–535. [DOI] [PubMed] [Google Scholar]
- 46.Bidnenko V., Ehrlich,S.D. and Jannière,L. (1998) In vivo relations between pAMβ1-encoded type I topoisomerase and plasmid replication. Mol. Microbiol., 28, 1005–1016. [DOI] [PubMed] [Google Scholar]
- 47.Gros M.-F., te Riele,H. and Ehrlich,S.D. (1987) Rolling circle replication of single-stranded DNA plasmid pC194. EMBO J., 6, 3863–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pujol C., Chédin,F., Ehrlich,S.D. and Jannière,L. (1998) Inhibition of a naturally occurring rolling-circle replicon in derivatives of the theta-replicating plasmid pIP501. Mol. Microbiol., 29, 709–718. [DOI] [PubMed] [Google Scholar]
- 49.Sandler S.J., Samra,H.S. and Clark,A.J. (1996) Differential suppression of priA2::kan phenotypes in Escherichia coli K-12 by mutations in priA, lexA and dnaC. Genetics, 143, 5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shlomai J. and Kornberg,A. (1980) A prepriming DNA replication enzyme of Escherichia coli. I. Purification of protein n′: a sequence-specific, DNA-dependent ATPase. J. Biol. Chem., 255, 6789–6793. [PubMed] [Google Scholar]
- 51.Sandler S.J. (2000) Multiple pathways for restarting DNA replication forks in Escherichia coli K-12. Genetics, 155, 487–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koonin E.V. (1992) DnaC protein contains a modified ATP-binding motif and belongs to a novel family of ATPases including also DnaA. Nucleic Acids Res., 20, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bruand C. and Ehrlich,S.D. (1995) The Bacillus subtilis dnaI gene is part of the dnaB operon. Microbiology, 141, 1199–1200. [DOI] [PubMed] [Google Scholar]
- 54.Bruand C., Sorokin,A., Serror,P. and Ehrlich,S.D. (1995) Nucleotide sequence of the Bacillus subtilis dnaD gene. Microbiology, 141, 321–322. [DOI] [PubMed] [Google Scholar]
- 55.Imai Y., Ogasawara,N., Ishigo-oka,D., Kadoya,R., Daito,T. and Moriya,S. (2000) Subcellular localization of Dna-initiation proteins of Bacillus subtilis: evidence that chromosome replication begins at either edge of the nucleoids. Mol. Microbiol., 36, 1037–1048. [DOI] [PubMed] [Google Scholar]
- 56.Polard P., Ton-Hoang,B., Haren,L., Bétermier,M., Walczak,R. and Chandler,M. (1996) IS911-mediated transpositional recombination in vitro. J. Mol. Biol., 264, 68–81. [DOI] [PubMed] [Google Scholar]
- 57.Studier F.W. and Moffatt,B.A. (1986) Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol., 189, 113–130. [DOI] [PubMed] [Google Scholar]
- 58.Vagner V., Dervyn,E. and Ehrlich,S.D. (1998) A vector for systematic gene inactivation in Bacillus subtilis. Microbiology, 144, 3097–3104. [DOI] [PubMed] [Google Scholar]