Abstract
Background and Aims
Satellite DNAs (satDNAs) are repetitive sequences composed by tandemly arranged, often highly homogenized units called monomers. Although satDNAs are usually fast evolving, some satDNA families can be conserved across species separated by several millions of years, probably because of their functional roles in the genomes. Tyba was the first centromere-specific satDNA described for a holocentric organism, until now being characterized for only eight species of the genus Rhynchospora Vahl. (Cyperaceae). Here, we characterized Tyba across a broad sampling of the genus, analysing and comparing its evolutionary patterns with other satDNAs.
Methods
We characterized the structure and sequence evolution of satDNAs across a robust dadated phylogeny based on Hybrid Target-Capture Sequencing (hyb-seq) of 70 species. We mined the repetitive fraction for Tyba-like satellites to compare its features with other satDNAs and to construct a Tyba-based phylogeny for the genus.
Key Results
Our results show that Tyba is present in the majority of examined species of the genus, spanning four of the five major clades and maintaining intrafamily pairwise identity of 70.9% over 31 Myr. In comparison, other satellite families presented higher intrafamily pairwise identity but are phylogenetically restricted. Furthermore, Tyba sequences could be divided into 12 variants grouped into three different clade-specific subfamilies, showing evidence of traditional models of satDNA evolution, such as the concerted evolution and library models. Besides, a Tyba-based phylogeny showed high congruence with the hyb-seq topology. Our results show structural indications of a possible relationship of Tyba with nucleosomes, given its high curvature peaks over conserved regions and overall high bendability values compared with other non-centromeric satellites.
Conclusions
Overall, Tyba shows a remarkable sequence conservation and phylogenetic significance across the genus Rhynchospora, which suggests that functional roles might lead to long-term stability and conservation for satDNAs in the genome.
Keywords: Holocentromere, repetitive DNA, satellite DNA, phylogenetic signal, Rhynchospora Vahl
INTRODUCTION
Repetitive DNA has become a key element in genomic studies since it was discovered that these sequences compose a large fraction of most eukaryotic genomes (Gemmell, 2021). The satellite DNAs (satDNAs) are one of the most well-studied types of repetitive DNA, being composed by tandemly arranged, usually highly homogenized units called monomers (Garrido Ramos, 2017). Monomer consensus sequences are used to characterize similar satDNA into families (Novák et al., 2017; Oliveira et al., 2021). One of the key features of these sequences is the remarkably fast rates of sequence evolution, in terms of abundance and/or nucleotide sequence, resulting in accumulation of changes in short evolutionary times (Macas et al., 2002; Lower et al., 2018). This rapid diversification presents major challenges to the study of satDNA evolution (Plohl et al., 2012).
Although different theories have been proposed to explain satDNA evolution (Lower et al., 2018), the most prominent models are the concerted evolution and library models, which are often complementary to each other (Plohl et al., 2012; Garrido-Ramos, 2015; Camacho et al., 2022). The concerted evolution model follows the idea that the monomers of a satDNA array accumulate mutations in an independent manner and that these mutations are homogenized and fixed along the array following a molecular drive process (Dover, 2002; Plohl et al., 2012; Garrido-Ramos, 2015). The library model postulates that related species share a library of satDNA families of varying abundances caused by random expansion or contraction of arrays of these different satDNAs (Salser et al., 1976; Fry and Salser, 1977).
These two models try to explain the fast changes of satDNA repeats in sequence and abundance, although there are cases of long-term satDNA sequence conservation, usually observed in different taxonomic levels, such as families (Mravinac et al., 2002), orders (Robles et al., 2004) and even phyla (Petraccioli et al., 2015). The maintenance of ‘relic’ satDNA families might be indicative of functional roles for these sequences in eukaryotic genomes (Plohl et al., 2012). This functionality might be related to expression of non-coding RNA sequences, associated with nucleotypic and/or structural effects (Mravinac et al., 2005; Plohl et al., 2008). Sequence-based nucleosome-prediction models using conserved satDNA monomers suggest that they might have a role in nucleosome formation (Tsoumani et al., 2013; Escudero et al., 2019). These models analyse sequence properties such as dinucleotide periodicity and curvature patterns to predict the ‘bendability’ values of a DNA sequence, which can be an indicator of interaction with histones (Liu et al., 2011; Zhang et al., 2013).
Satellite DNAs are usually an integral part of the heterochromatin, frequently being found in functional regions such as the telomere and the centromere (Achrem et al., 2020). With regard to the centromere, most eukaryotes present monocentric chromosomes, in which the centromere is restricted to a single region, usually composed by long arrays of repetitive sequences, especially satDNAs (Plohl et al., 2014). These centromere-specific satDNAs present varying degrees of sequence conservation (Melters et al., 2013). For example, within the Poaceae family, the CentO centromeric satDNA found in Oryza was later shown to have high similarity with CentC, present in the centromeres of maize (Cheng et al., 2002; Zhong et al., 2002). In contrast, centromeric satDNAs of some Solanum species were shown to be chromosome specific (Gong et al., 2012; Zhang et al., 2014). In holocentric species, which present a dispersed centromere along each chromatid (Bureš et al., 2013), it was believed for a long time that centromere-specific repetitive sequences were not present (Marques and Pedrosa-Harand, 2016). For example, well-studied holocentric organisms, such as Luzula elegans and Caenorhabditi elegans, did not present repeats associated with CENH3, even after detailed genomic characterizations (Subirana and Messeguer, 2013; Heckmann et al., 2011). This changed with the discovery of the 172-bp satDNA Tyba in the sedge species Rhynchospora pubera (Cyperaceae). Tyba showed a line-like distribution along a groove positioned at the outer part of chromatids in R. pubera chromosomes co-localizing with the CENH3 protein, being the first centromere-specific satDNA described for a holocentric species (Marques et al., 2015). Tyba sequences were later confirmed to be present in eight other Rhynchospora species (Rhynchospora alba, R. breviuscula, R. cephalotes, R. ciliata, R. colorata, R. exaltata, R. tenerrima and R. tenuis), while being absent in Rhynchospora globosa (Rocha et al., 2016; Ribeiro et al., 2017; Costa et al., 2021; Hofstatter et al., 2022). This already suggests an old origin for this satellite DNA, given the estimated distance (~30 Myr; Buddenhagen, 2016) between the clade containing R. cephalotes + R. exaltata and the clade containing the other species that presented Tyba. Whole-genome sequencing of Rhynchospora species revealed that Tyba-based holocentromeres also impact genomic architecture, epigenome organization and karyotype evolution (Hofstatter et al., 2022). This suggests that the presence of Tyba at centromeres might have an adaptive role and might influence species diversification.
Rhynchospora is a cosmopolitan genus composed of ~400 species with a North American centre of diversity, with preliminary divergence time estimates placing the origin of the genus between 38 and 49 Mya (Buddenhagen, 2016; Thomas, 2020; Silva Filho et al., 2021). The presence of Tyba across evolutionarily distant species, such as R. pubera and R. cephalotes, coupled with its holocentromeric localization, makes this sequence an interesting case to study the evolution and dynamics of centromeric DNA in non-monocentric organisms in a macroevolutionary context. Here, we have performed a genus-wide investigation about the tempo and mode of Tyba evolution in Rhynchospora, comparing it with other satDNAs found in the genus. We mined repetitive DNA information from filtered off-target next generation sequencing (NGS) reads (Costa et al., 2021) from 70 species of Rhynchospora representing the major clades of the genus and used a robust phylogenetic framework to serve as the background for studying Tyba evolution. We aimed to answer the following questions.
How widely distributed is Tyba in Rhynchospora when compared with other satDNAs?
How conserved is Tyba across the whole genus?
What could be the reasons/mechanisms for spread of Tyba across the genus Rhynchospora?
MATERIALS AND METHODS
Sequence data acquisition and filtering
All target-capture sequencing data analysed here were obtained from the thesis by Buddenhagen (2016). Because we used off-target reads from target-capture sequencing, we opted to exclude from our analysis the data of any species that showed a percentage of annotated repeats smaller than the one obtained for R. cephalotes by Costa et al. (2021), which was the species with the least amount of classified repeats (~5%) that still presented good correlation values with genome-skimming data. In total, 77 accessions, representing 70 Rhynchospora Vahl species (~20% of the genus), were selected for our satellite mining analysis. From the same dataset, we collected data from six Carex L. species, two Chorizandra R. Br. species, Exocarya sclerioides Benth., Hypolytrum nemorum (Vahl) Spreng. and Scirpodendron ghaeri (Gaertn.) Merr. to serve as the outgroup (Supplementary data Table S1). All sequences used were deposited in GenBank under project number PRJNA672127.
Phylogenetic analyses and molecular dating
To anchor our findings on a phylogenetic backbone, we used the robust RaxML topology constructed by Buddenhagen (2016) with 256 target loci obtained by hybrid target-capture sequencing (hyb-seq). Although the original sampling contained 115 Rhynchospora accessions, the reads of some of these were not sufficient for the RepeatExplorer analysis, yielding poor annotations. Therefore, we pruned the original tree, leaving only 77 Rhynchospora accessions and the 11 outgroup species. This was done with the drop.tip function implemented in the package phytools (Revell, 2012) in R (R Core Team, 2022). This pruned tree was then submitted to a molecular clock analysis. Divergence times were estimated on BEAST v.1.8.3 (Drummond and Rambaut, 2007) through the CIPRES Science Gateway, using the pruned tree as a fixed topology. For calibration, we used the same points defined by Buddenhagen (2016), following a normal distribution with a 10% standard deviation. An uncorrelated relaxed lognormal clock (Drummond and Rambaut, 2007) and birth–death speciation model (Gernhard, 2008) were applied. Two independent runs of 100 000 000 generations were performed, sampling every 10 000 generations. After removing 25% of samples as burn-in, the independent runs were combined, and a maximum clade credibility (MCC) tree was constructed using TreeAnnotator v.1.8.2 (Rambaut and Drummond, 2013). To verify the effective sampling of all parameters and assess convergence of independent chains, we examined their posterior distributions in Tracer. The MCMC sampling was considered sufficient at effective sampling sizes (ESS) ≥ 200.
Satellite DNA mining
In order to prepare the target-capture sequencing data for satDNA mining, we first had to filter out all the reads containing the enriched targeted regions. For this, we followed the protocol presented by Costa et al. (2021) to acquire the unenriched off-target portion of the genomic libraries. The off-target datasets of each Rhynchospora species were uploaded to the RepeatExplorer pipeline (Novák et al., 2013) hosted at the web-based platform Galaxy (https://repeatexplorer-elixir.cerit-sc.cz/). RepeatExplorer uses a graph-based clustering algorithm to group sequences based on similarity, facilitating the identification of high-copy sequences of a genome. These clusters of sequences are then identified by cross-checking against repetitive element databases (Novák et al., 2013). Concurrently, the TAREAN (Tandem Repeat Analyzer) tool checks the clusters for predictions of tandem arrangement, building consensus sequences for these clusters (Novák et al., 2017).
Our dataset was submitted to three different run strategies in RepeatExplorer: (1) individual species clustering (ISC) analysis for each of the 77 Rhynchospora accessions in order to characterize the consensus of the satDNAs of each species; (2) a comparative clustering (CC) analysis with reads from all the Rhynchospora accessions, in order to identify shared satellite DNAs; and (3) a Tyba-like clustering (TLC) analysis, a comparative analysis using only reads that were mapped to a database of previously published Tyba using the Geneious read mapper implemented in Geneious v.7.1.9 (low-sensitivity preset; Kearse et al., 2012). Given that the genome sizes of most analysed species were unknown, we input all reads left after the filtering of target regions (off-target reads) for the ISC analysis, and the same number of reads (170 000) for each species was used to build the combined dataset for the CC analysis. This amount was decided based on the species that had the smallest number of reads analysed in the individual analysis (R. glaziovii, 173 544; Supplementary data Table S1). For the TLC analysis, we used all the 157 078 reads that were mapped to the Tyba database. The run parameters of the individual and comparative RepeatExplorer analysis were the same as those described by Costa et al. (2021).
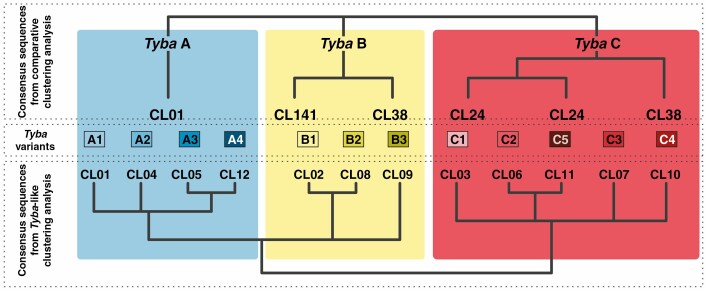
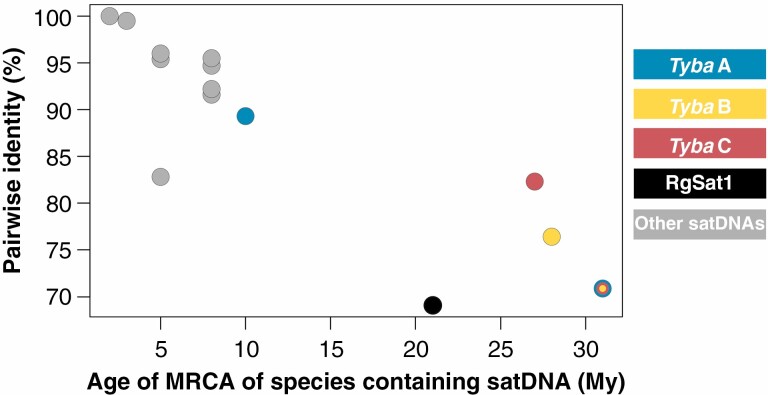
The CC and TLC analysis were used to divide Tyba into subfamilies and variants, respectively (Fig. 1). The consensus sequences of clusters annotated as Tyba in the CC analysis were aligned using the Geneious alignment tool, with default settings (Kearse et al., 2012). The alignment was used to produce an ‘approximately maximum-likelihood’ (AML) phylogenetic tree using FastTree, as implemented in Geneious v.7.1.9 (Kearse et al., 2012), forming monophyletic clades that were considered different subfamilies of the satDNA (Fig. 1). The similarity between the shared satDNA found in the CC analysis (including non-Tyba satDNA families) was also assessed by a dotplot constructed with DOTTER (Sonnhammer and Durbin, 1995). The consensus sequences found in the TLC analysis were also used to construct an AML tree, and each of the clusters could be mapped to one of the subfamilies, being considered variants (Fig. 1). The consensus sequences found in the ISC analysis were mapped to the Tyba variants and non-Tyba shared satDNAs to estimate the sequence diversity of each shared satDNA. To assess the relationship between sequence divergence and age of the shared satDNAs, the pairwise identity (as a percentage) of sequences mapped to each shared satDNA was estimated with Geneious and plotted against the time of divergence between species that contained each of the shared satDNAs.
Fig. 1.
Classification of satDNA family Tyba into subfamilies (uncovered in the species comparative clustering analysis) and variants [based on the Tyba-like (TL) reads in the clustering analysis]. Approximately maximum-likelihood (AML) trees show the phylogenetic relationships based on consensus sequence similarity. Coloured rectangles reflect the similarities between consensus sequences obtained in both analyses and are used to delimit sequences into subfamilies A (blue), B (yellow) and C (red). The clusters TL CL01, CL04, CL05 and CL12 formed a monophyletic clade and were all annotated as being similar to subfamily Tyba A (blue), being considered sequence variants A1–A4. Likewise, TL CL03, CL06, CL07, CL10 and CL11 formed a monophyletic clade that was annotated as being similar to Tyba C (red), thus being considered variants C1–C5. Although TL CL02, CL08 and CL09 were annotated as Tyba B, only CL02 and CL08 were monophyletic, with CL09 being in a polytomy with CL02 + CL08 and the clusters annotated as Tyba A. However, the most similar satellite to CL09 is CL02 (75% pairwise identity), which is further evidence that it should be considered a variant of Tyba B (yellow). Thus, CL02, CL08 and CL09 were considered to be variants B1–B3.
Satellite DNA-based phylogenetic analysis
We also performed a phylogenetic analysis based on the satDNA family Tyba and a separate analysis based on the other satDNAs found in the genus. For this, we separated all reads annotated as Tyba from the reads identified as other satDNAs (‘non-Tyba’ satDNAs) in the ISC analysis and adopted an Alignment and Assembly Free (AAF) methodology (Fan et al., 2015). AAF constructs phylogenies directly from unassembled genome sequence data, bypassing both genome assembly and alignment. Thus, it calculates the statistical properties of the pairwise distances between genomes, allowing it to optimize parameter selection and perform bootstrapping.
We also used the individual Tyba consensus from the ISC analysis to estimate the phylogenetic signal (Pagel’s λ; Pagel, 1999) of monomer length and the GC content of Tyba sequences. For this, we use the phytools package (Revell, 2012) implemented in R (R Core Team, 2022). For species that had more than one Tyba variant, we used the consensus sequence of the most abundant Tyba cluster (Supplementary data Table S2). This analysis presents a measure of similarity between phylogenetically close species regarding the studied characteristics. In this case, a value of λ closer to one would mean that closely related species would have more similar Tyba variants, whereas λ closer to zero would mean that closely related species had less similar Tyba variants than expected (Pagel, 1999).
Sequence conservation
Given that the CC analysis included reads from all analysed species in a single run, it provides a broad view of satellite DNAs shared by two or more species. In contrast, with the ISC analysis each run contained reads from a single species, making it possible to find a greater number of satDNAs for each species. To determine whether these satDNAs were species specific or were part of the shared satDNAs, we mapped all the consensus sequences found in the ISC analysis to the consensus sequences of the shared satDNAs revealed in the CC analysis. For this, we uded the BowTie2 mapper (high-sensitivity preset, End-to-End, 60% pairwise identity) (Langmead and Salzberg, 2012). Given that we used a 60% pairwise identity threshold, ISC consensus sequences mapped to the same shared satDNA were considered to be from the same satDNA family. The alignments produced from the mapping were used to calculate the pairwise identity between all consensus sequences mapped to each shared satDNA. The age of shared satDNAs was estimated based on the age of the most common recent ancestor (MRCA) between the species that possessed the satDNAs, and a plot against pairwise identity was constructed using R.
Curvature/bendability analysis of shared satDNAs
Consensus sequences of the Tyba variants (mapped to their respective subfamily consensus sequences) and of the other shared satDNAs, including the non-centromeric satDNA previously found in Rhynchospora globosa (RgSat) (Ribeiro et al., 2017)) were used to estimate bendability and curvature plots based on the DNA sequence using the bend.it server (http://pongor.itk.ppke.hu/dna/bend_it.html). It uses the DNase I-based bendability parameters of Brukner et al. (1995) and the consensus bendability scale (Gabrielan and Zohary, 2004). We also used the DNA curvature analysis website by Gohlke (https://www.lfd.uci.edu/~gohlke/dnacurve/) based on nucleosome positioning (Goodsell and Dickerson, 1994) to produce three-dimensional models of the Tyba subfamilies sequences by opening the Helix Coordinate PDB file in the Geneious software.
RESULTS
Phylogenetic framework
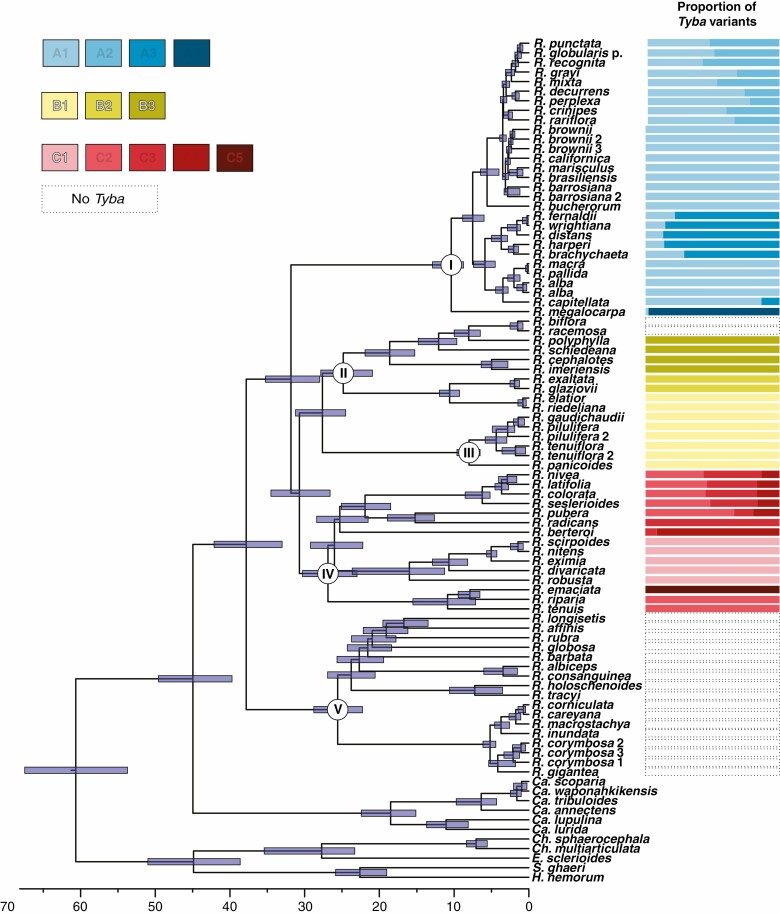
The phylogenetic reconstruction of Rhynchospora based on nuclear sequences showed five main clades (Fig. 2). According to our dating analysis, the crown age of the genus was estimated at 37.8 Mya [95% confidence interval (CI) = 33–42.1]. Clade V was the first to diverge, with crown age of ~25.6 Mya (95% CI = 22.2–28.8). Later, clade I (which comprises species traditionally classified as part of Eurhynchosporae; Gale, 1944) diverged from the remaining clades (31.8 Mya, 95% CI = 28–35.2), with clade IV diverging later from the rest at ~30.9 Mya (95% CI = 26.6–34.5). The remaining species split into clade II and clade III (species formerly recognized as Pleurostachys, recently synonymized as Rhynchospora section Pleurostachys; Thomas, 2020) at 27.6 Mya (95% CI = 24.5–31.2).
Fig. 2.
Phylogenetic relationships and molecular dating of Rhynchospora species. Coloured bars (according to the key at the top left of the figure) represent the proportion of different Tyba variants found in the genome of each species. Bars at the nodes of the tree represent the 95% confidence interval of the molecular dating analysis, with the axis scale representing divergence time (in millions of years). Circles with roman numerals in the nodes delimit the five clades discussed.
Tyba is a diverse satDNA family with several variants
The number of analysed reads of our ISC analyses ranged from 173 544 reads in R. glaziovii to 2 719 919 in R. pubera (Supplementary data Table S1). General satDNA abundance varied from 0.02% in R. emaciata to 9.14% in one of the accessions of R. corymbosa (Supplementary data Table S3). From the 537 satDNA consensus sequences found in the individual clustering analysis of all species, 133 were found to have ≥60% sequence similarity with one of the previously published Tyba sequences, with monomer length varying from 170 to 175 bp. These Tyba-like sequences were found in most species of clades I–IV, with only two species (R. biflora and R. racemosa) of clade II showing no trace of Tyba. Satellite DNAs from species of clade V did not show significant similarity with Tyba. We confirmed that these species from clades II and V did not present Tyba by being unable to find Tyba-like sequences in the full set of raw reads (Fig. 2).
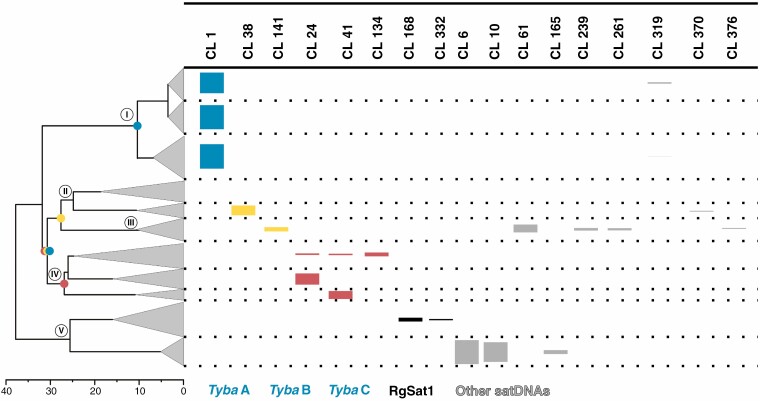
In our CC analysis, we found 23 clusters that were annotated by TAREAN as satellite DNAs. From these, 17 clusters were found to have a significant amount of reads in more than one species, being considered shared satDNAs (Fig. 3; Supplementary data Table S2). Six clusters of shared satDNAs (CL1, CL24, CL38, CL41, CL134 and CL141) were automatically annotated as having reads similar to Tyba, which was confirmed by the dotplot analysis (Supplementary data Fig. S1). The consensus sequences of these satDNAs were found to have a 61.7% pairwise identity. Based on the high similarity and RepeatExplorer annotation, we determined that these satDNAs belonged to the same satDNA family. An AML phylogeny showed that these six Tyba-like satDNAs could be divided into three groups based on sequence similarity (subfamilies A, B and C; see Fig. 1). Along with Tyba, we also annotated two further shared satDNA clusters that were found to be of the same family (Supplementary data Fig. S1) and presented high sequence similarity to RgSat, a non-centromeric satDNA found by Ribeiro et al. (2017). The clusters identified as RgSat also presented moderate similarity to CL06 (48.4% pairwise identity) and to the Tyba A subfamily sequences (46.0%), but these values were below our threshold for being considered to be from the same family.
Fig. 3.
Shared satDNAs in analysed Rhynchospora. Each line represents one of the clades of the simplified Rhynchospora phylogeny on the left (based on the tree presented in Fig. 1). Clusters annotated as satDNA by RepeatExplorer are presented in the upper part, and their relative abundance in a given species is represented by the height of the rectangles. Rectangle colours are according to the key below. Small coloured circles at the nodes of the simplified tree represent possible points of origin of the different Tyba subfamilies.
In the TLC analysis, the 276 052 input reads were divided into 12 different clusters, each assigned to one of the main Tyba subfamilies by similarity hits to the custom repeat database (see Fig. 1; Supplementary data Table S2). An AML phylogenetic tree with the consensus sequence of the 12 Tyba-like clusters (TL-CLs) showed that these could also be divided into different clades. Each of these clades was assigned to one of the Tyba subfamilies discovered in the shared satDNA analysis based on the RepeatExplorer annotation. Thus, each of the TL-CLs was treated as one variant of a specific subfamily of Tyba (Fig. 1).
Tyba variants are clade specific within Rhynchospora
The CC analysis showed that Tyba was present among species of clades I, II, III and IV of Rhynchospora, with one of each Tyba subfamily being dominant in one specific clade, while subfamily B was present in both clades II and III (Fig. 3). No other satDNA was found in the CC analysis in more than one major clade. The RgSat satellite was found in clusters CL168 and CL332 (69.5% pairwise identity) and was present in only two species of clade V (R. globosa and R. rubra).
The TLC analysis mainly corroborated the results from the CC analysis (Fig. 3), with each clade presenting only one dominant Tyba subfamily (Fig. 2). Species of clades II and III showed only one variant from subfamily B per species. In contrast, in clades I and IV, a few subclades presented more than one variant from the same subfamily (A and C, respectively; Fig. 2).
Tyba sequences present high phylogenetic signal
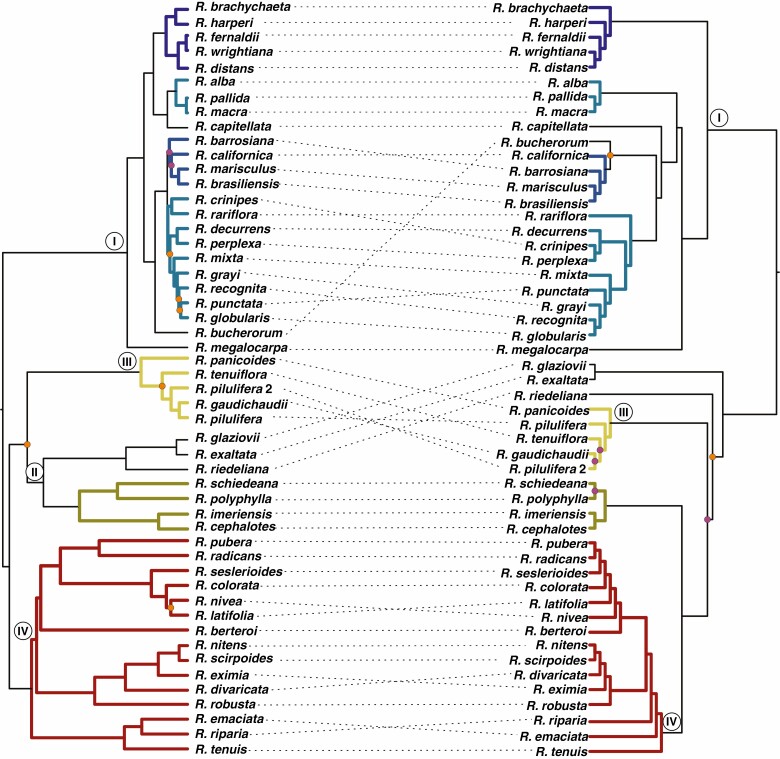
Although discrepancies between AAF Tyba-like and hyb-seq topologies were observed at higher levels, we could retrieve a large number of monophyletic subclades congruent with the hyb-seq tree (Fig. 4). Most of the relationships inside clade I were well recovered in the AAF topology, with a few discrepancies, such as R. capitellata and R. bucherorum (Fig. 4). The only subclade from clade III that was not monophyletic in the AAF analysis was R. tenuis (R. emaciata + R. riparia). Support values of most nodes of the AAF tree were >95%, with a few lower supports at shallow levels and at the base of the clade containing species from clades II, III and IV (Fig. 4). In contrast, the AAF phylogeny based on the non-Tyba satDNAs had more incongruences when compared with the nuclear phylogeny (Supplementary data Fig. S2). Clades I and III were recovered as monophyletic, whereas species from the remaining clades were scattered throughout the phylogeny.
Fig. 4.
Comparison of phylogenetic trees recovered from nuclear markers (left) and AAF of Tyba-like reads (right). Clades coloured in shades of blue (clade I), yellow (clade II) and red (clade III) are monophyletic in both trees. Circles at tree nodes refer to support values between 75 and 95% (orange) and <75% (purple). Nodes without circles presented maximum support.
To investigate whether Tyba sequence variability reflected phylogenetic relationships, we also assessed Pagel’s λ for the GC content (as a percentage) and monomer size (in base pairs) of the most abundant Tyba consensus sequence of each species. We found a high phylogenetic signal for both GC (λ = 0.92, P < 0.001) content and monomer size (λ = 0.93, P < 0.001), indicating that Tyba sequences from closely related species have similar nucleotide composition and monomer size, whereas distant species do not share similar values of these traits.
Tyba sequences are remarkably different from other satDNAs
In order to determine sequence divergence within different satDNA families, we mapped all the 532 satDNA consensus sequences found in the ISC analysis to the 17 shared satDNA consensus sequences found in the CC analysis. In this way, we could identify whether/which satDNAs from each species could be considered to be from the same satDNA family. In order to calculate intrafamily sequence divergence, we calculated the pairwise identity for all the resulting alignments. These identity values were plotted against the age of the MRCA of the species that shared that satDNA (Fig. 5). Tyba subfamilies presented lower pairwise identity than most of the other shared satDNAs (89.3, 76.4 and 82.3% for Tyba A, B and C, respectively), but were persistent through a longer evolutionary time (10, 28 and 27 Mya for Tyba A, B and C, respectively). Overall, all consensus satDNA sequences from the ISC analysis assigned to the Tyba family preserved a pairwise identity of 70.9%, with the MRCA of the species that contained Tyba having originated 31 Mya. The other shared satDNAs had high pairwise identity values (from 82.8% for CL10 to 100% for CL370), but were present only in species that diversified more recently, from 2 Mya (CL370) to 8 Mya (CL61) (Fig. 5). The only non-Tyba satDNA that was preserved through a relatively long evolutionary time was RgSat. Although it was present in only two species, those species shared a MRCA at ~21 Mya. However, the pairwise identity between the satDNAs mapped to the RgSat consensus was 69.1%. Overall, the Tyba subfamilies were older than most of the other satDNAs, while maintaining moderately high pairwise identity values (Fig. 5).
Fig. 5.
Plot of pairwise identity through evolutionary time for the shared satDNAs recovered in the comparative clustering analysis. Dots represent the shared satDNAs, with colours reflecting the key on the right side. The multicoloured dot refers to the whole Tyba family.
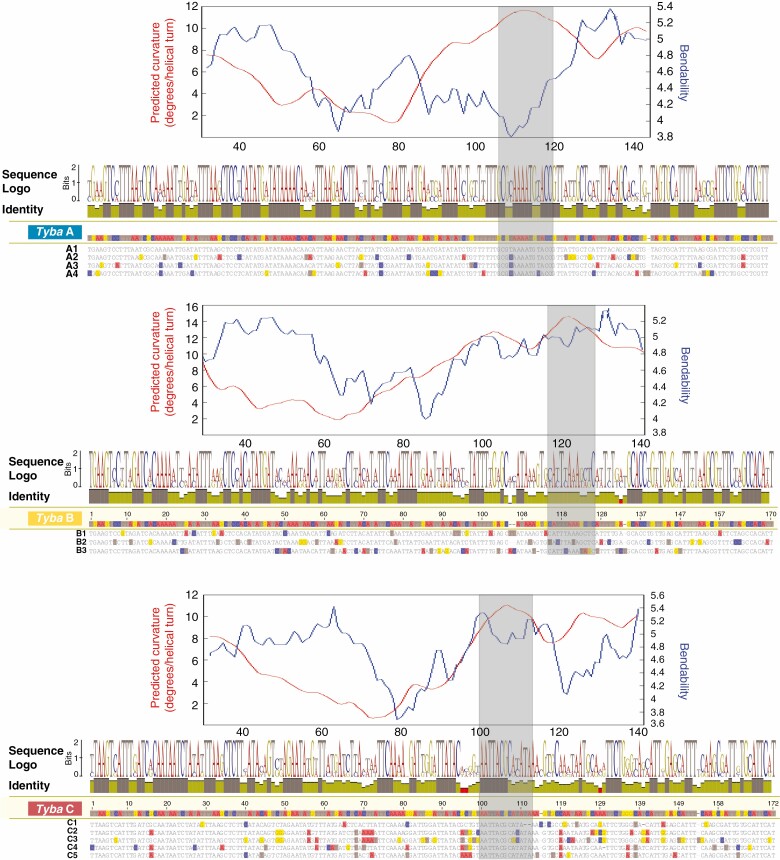
To check sequence conservation of the Tyba variants, the consensus sequences of the 12 variants were mapped to the consensus sequence of the three Tyba subfamilies, with identity values being calculated in a 2 bp sliding window. Overall, Tyba A had a smaller number of mutations among its variants, with a high identity throughout most of the sequence (Fig. 6). Both Tyba B and Tyba C variants showed a larger number of regions bearing low identity, but also presented highly conserved motifs, such as the ones highlighted by grey blocks in Fig. 6.
Fig. 6.
Bendability (blue)/curvature (red) propensity, sequence logos, identity plot and alignment of Tyba variants assigned to each Tyba subfamily. Heights of nucleotides in the sequence logos are proportional to the number of times that base appeared in the alignment. Dark brown values in the identity plot represent high sequence identity, while yellow values represent moderate and red low sequence identity. Base pairs that are different from the consensus sequence are coloured in the alignment. Grey rectangles represent the approximate region where the curvature peak was estimated.
Curvature and bendability for the Tyba subfamilies and the other shared satDNAs were calculated based on DNase I parameters. Curvature peaks were found in regions that were fairly conserved in the variants of subfamilies A and C, which was not the case for the overall less-conserved variants of subfamily B (Fig. 6). Likewise, bendability values were higher at conserved regions separated by a less-conserved region between them, which presented the lowest bendability values (Fig. 6). The other shared satDNAs presented varied curvature/bendability profiles. Most sequences presented multiple curvature peaks, and bendability values did not follow the same pattern observed in Tyba (Supplementary data Fig. S2). The RgSat sequence, in contrast, presented a single thin curvature peak and overall low bendability values, with only one peak (Supplementary data Fig. S2).
DISCUSSION
The holocentromeric satDNA Tyba is conserved across the Rhynchospora phylogeny
We demonstrated that the holocentromeric satellite DNA Tyba is evolutionarily persistent and a relatively old family of satDNA in the genus Rhynchospora, especially when compared with the other shared satDNAs of the genus. Usually, satDNAs have a fast rate of evolution and only species that are closely related share satellite families (Lower et al., 2018). In more extreme cases, sequence divergence between centromeric satDNAs of different chromosomes in the same karyotypes have been reported (Zhang et al., 2014).
The library model for satDNA evolution postulates that, depending on the homogenization rates (under concerted evolution), it is possible to find a low number of copies of a satDNA family in a phylogenetically distant species, whereas that family can be dominant on genomes in other closely related species (Plohl et al., 2012; Garrido-Ramos, 2015). Here, Tyba was found in four of the five main clades of Rhynchospora, and its monomer sequence could be classified into three subfamilies, with each clade presenting a single type. We could find different variants of Tyba in different abundances across closely related species, especially in clades I and IV. Although many species presented only one Tyba variant, it is important to note that we used reads in a way comparable to a genome-skimming analysis (Costa et al., 2021). This means that we had a small coverage for most of the analysed species, which, in turn, could mask low-abundant Tyba variants in the RepeatExporer analysis (Novák et al., 2017).
The propelling mechanisms of Tyba satDNA evolution
Different mechanisms seem to explain the diversification of Tyba holocentromeric satDNA in Rhynchospora. The high phylogenetic signal of GC content and monomer length and the moderate congruence between our Tyba-like AAF phylogeny and the hyb-seq phylogeny are evidence of efficient clade/subclade-specific sequence homogenization of Tyba. Although the AAF phylogeny and the phylogenetic signal analysis used different types of data (raw NGS reads and TAREAN consensus sequences, respectively), both analyses showed that closely related species have more similar sequences of Tyba than distant species. Concerted evolution can lead to fast divergence between the satellites of reproductively isolated organisms, making these sequences potentially informative phylogenetic markers (Kuhn et al., 2012; Lorite et al., 2017; Belyayev et al., 2019).
Our results suggest that both concerted evolution and the library model explain Tyba evolution in Rhynchospora. It has been demonstrated that mechanisms of concerted evolution can lead to the homogenization of different satDNA subfamilies that might have composed a genomic library of a group of species (Plohl et al., 2010; Ahmad et al., 2020). According to this view, all three subfamilies of Tyba might have existed in the satDNA library of the ancestor of all species that possess Tyba, as has been speculated for other phylogenetically conserved satDNA families (Quesada del Bosque et al., 2011, 2014; Lorite et al., 2017). However, ~30 Myr of evolutionary time might have led to the observed clade-specific homogenization. One of the results of this clade-specific homogenization is that the clade-specific homogenized sequences can become useful phylogenetic markers (Kuhn et al., 2012; Lorite et al., 2017; Belyayev et al., 2019). Although homogenization can occur independently from natural selection, it is possible that favourable characteristics could lead to evolutionary success of individuals presenting specific centromeric sequences, which could lead to the selection for particular sequence variants within a centromeric satDNA array (Pérez-Gutiérrez et al., 2012; Lower et al., 2018).
Upon its discovery, two variants of Tyba were identified in R. pubera (Marques et al., 2015), and only one of these two was found in the closely related R. tenuis (Ribeiro et al., 2017), both positioned in clade IV. In our analysis, we could observe a total of 12 different variants within three subfamilies in each of the major Rhynchospora clades. Thus, patterns of both the library model and concerted evolution could be found in clade IV. Younger lineages present up to three different Tyba variants in their composition, whereas older lineages have only one dominant variant, which is congruent with the idea that concerted evolution is more efficient over long evolutionary times (Pérez-Gutiérrez et al., 2012).
We found both variants previously discovered in R. pubera (Marques et al., 2015) and a third, new variant in this species. Comparatively, Tyba sequences from species of clades II and III appear to have had a faster homogenization process, with only a single variant being dominant in each species, but the same variant being shared with other species of the clade or even between clades suggests its older origin. This could lead to clade III and a subclade of clade II appearing as monophyletic in the AAF topology. A number of factors have been proposed to explain why homogenization rates might vary between satDNA sequences, such as array length, organization and genomic location (Navajas-Pérez et al., 2009; Kuhn et al., 2012; Lorite et al., 2017). The two variants of the oldest non-Tyba shared satDNA found in our CC analysis, RgSat, showed only 69% pairwise similarity. This satellite, first found in R. globosa, presents a traditional block-like distribution at the terminal regions of metaphase chromosomes and a clustered disposition at the chromocenters of interphasic nuclei (Ribeiro et al., 2017). In contrast, Tyba has been confirmed cytologically to be co-localized with the holocentromeres of species from clades II and IV (Marques et al., 2015; Rocha et al., 2016; Ribeiro et al., 2017; Costa et al., 2021) and was shown to have a dispersed distribution in interphasic nuclei (Marques et al., 2015; Ribeiro et al., 2017). Thus, the fact that a dispersed satDNA presents such high rates of homogenization, even when compared with a traditional block-like satDNA, is remarkable.
The Eurhynchosporae clade (formerly a tribe; Gale, 1944), corresponding to clade I in our phylogeny, presents an interesting case for satDNA evolution in a fast-diversifying group. This clade, which appears here as the first lineage to diverge among the clades that contain Tyba, was shown to present an increase in the diversification rate (Larridon et al., 2021). In situations of rapid diversification, sequence evolution is often not rapid enough for sequences to diverge significantly between species (Giarla and Esselstyn, 2015). According to this view, species within a recent diversification event tend also to present with similar dominant satDNA variants (Quesada del Bosque et al., 2013; Dogan et al., 2021). Although the A1 variant is widespread among clade I in our analyses, the other three Tyba A variants appear in specific subclades at comparable abundance, in a classic library model display.
The evolutionary persistence of Tyba might be related to its centromeric distribution
Given its apparent phylogenetic conservation through long evolutionary times, what factors might be responsible for such long evolutionary reach of Tyba? It has been proposed in several studies that this long-term conservation of a satDNA sequence might suggest structural/nucleotypic functional roles for these elements (Mravinac et al., 2005; Lower et al., 2018). Immunofluorescence in situ hybridization experiments and chromatin immunoprecipitation sequencing (ChIP-seq) with the centromeric protein CENH3 have shown that Tyba is located at holocentromeres of R. breviuscula, R. cephalotes, R. ciliata, R. pubera and R. tenuis (Marques et al., 2015; Rocha et al., 2016; Ribeiro et al., 2017; Costa et al., 2021; Hofstatter et al., 2022), which could indicate a functional association. Although we cannot ascertain that every satDNA identified here as Tyba, or derived from Tyba, has a holocentromeric distribution, it might have a conserved chromosomal location because Tyba has been shown to be centromere specific in phylogenetically distant species, such as R. cephalotes (Costa et al., 2021) and R. pubera (Marques et al., 2015). Furthermore, the remarkably conserved sequence composition, monomer length and high abundance might be yet another indicator that all Tyba satDNAs found here are indeed centromeric, given that centromeric satDNAs are often the most abundant in most organisms (Melters et al., 2013). More recently, sequencing of whole Rhynchospora genomes demonstrated the centromeric role of Tyba, which can impact evolutionary processes such as genomic architecture, epigenome organization and karyotype evolution (Hofstatter et al., 2022).
Although satDNA is a major constituent of eukaryotic centromeres, sequence conservation is extremely low in most cases (Plohl et al., 2014; Garrido-Ramos, 2017). In fact, centromeric satDNAs have been shown to differ among chromosomes of the same karyotype in some plant species (Gong et al., 2012; Iwata et al., 2013; Zhang et al., 2014). An investigation of putative centromeric satDNA of 282 species across eukaryotes showed that sequence similarity rapidly declines through divergence, reaching background noise levels after 50 Myr of divergence (Melters et al., 2013), which would make Tyba, at 31 Myr old, relatively old when compared with most centromeric satDNAs.
Our curvature/bendability analysis have shown the highest values of curvature and bendability at similar regions of the Tyba subfamilies sequences. In addition, this analysis also showed that Tyba sequences are more flexible than the sequence of RgSat, a confirmed non-centromeric satDNA. Other shared satDNAs presented variable patterns of curvature, with multiple peaks and mostly constant bendability values throughout the sequences. The curvature patterns of Tyba are similar to the proposed patterns of core DNA (tight sequences that wrap around a histone), in which large curvature at the ends of the sequence facilitate nucleosome formation (Liu et al., 2008). In the same way, high and stable bendability values for satellite sequences were also proposed to facilitate nucleosomal organization of centromeric proteins (Escudero et al., 2019). Our data are supported by genomic analyses that confirmed the centromeric role of Tyba satDNA (Hofstatter et al., 2022).
One important observation is that Tyba is not present in all Rhynchospora species, being absent from the whole of clade V and a few species from clade II, which emphasizes that Tyba is not necessary for holocentricity. For example, the clade V species R. globosa, which has meiotic evidence of its holocentricity (Arguelho et al., 2012), presented only satDNAs disposed in block-like patterns (Ribeiro et al., 2017). Although Tyba is probably not necessary for the holocentromere of Rhynchospora species, it is possible that its sequence confers advantages regarding nucleosome formation and positioning that might have resulted in its conservation for a long evolutionary time (Talbert and Henikoff, 2020).
It has also been shown that satDNAs can be associated with retrotransposons at functional parts of the centromere, as is the case with the satDNA CentO and retrotransposon CRR of rice (Cheng et al., 2002). A similar association was found in R. pubera, where the Ty3-Gypsy retroelement CRRh was shown to have a similar pattern to Tyba, spread throughout the holocentromeres (Marques et al., 2015). It has been proposed that an association with mobile elements could facilitate the spread of centromeric satDNAs. Most notably, the association with mobile elements seemed to have facilitated the spread and persistence of relic satDNA BIV160 for >500 Myr of evolution (Plohl et al., 2010). In the bivalve mollusc Crassostrea gigas, a close association of a Helitron transposable element with centromeric regions seemed to facilitate the spread of abundant satDNAs to different chromosomes (Tunjić-Cvitanić et al., 2021). In Rhynchospora, Hofstatter et al. (2022) discovered that 468 Tyba loci associated highly with fragments from a non-autonomous Helitron (TCR1) in R. pubera, suggesting that Tyba satellite spread could have been facilitated by TCR1 transposition activity. Thus, the association with mobile elements could favour the conservation of Tyba sequences across Rhynchospora holocentric chromosomes, providing the mobility and means of fixation for this satDNA.
Here, 404 satDNA consensus sequences found in the individual species clustering analysis (mean of approximately five per accession) could not be mapped to the Tyba and non-Tyba shared satDNAs, meaning that those were probably species specific. The oldest non-Tyba shared satDNA (RgSat, previously found in R. globosa by Ribeiro et al., 2017) was present only in two distantly related species (R. rubra and R. globosa) that shared a common ancestor ~21 Mya. In contrast, clades containing Tyba shared a common ancestor 31.8 Mya and maintained a pairwise identity >70%. Although the concept of evolutionarily old satDNAs might be uncommon, it is not exactly rare (see Plohl et al., 2010; Quesada del Bosque et al., 2013). The long-term conservation of these ‘relic’ sequences might indicate that this type of repeat could provide evolutionary advantages by playing a functional role (Plohl et al., 2012; Lower et al., 2018).
Here, we have unravelled the evolutionary history of the holocentromere-specific satellite Tyba (Marques et al., 2015, 2016; Ribeiro et al., 2018) in what is one of the largest samplings for satDNA-related analysis to date. We observed a high sequence conservation and phylogenetic significance for Tyba, especially when compared with other satDNAs observed in the genus. Tyba seems to have evolved by a combination of satDNA library diversification and concerted evolution. The sequence has been shown to persist throughout a long evolutionary time, with clade-specific variants, and was present in most of the genus, in striking contrast to the other satDNAs found in Rhynchospora. Our results suggest that such conservation could be linked to a functional role within the holocentromeres. Although we present a thorough outline of Tyba evolution, future studies with long-read sequencing techniques might help us to gain a better understanding of the organization of Tyba arrays, which could provide further clues about its intimate association with the centromeres of Rhynchospora.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Diogo Cabral-de-Melo (Universidade Estadual de São Paulo), Dr André Luís Laforga Vanzela (Universidade Estadual de Londrina), Dr Giovana Augusta Torres (Universidade Estadual de Lavras) and Dr Lyderson Facio Viccini (Universidade Federal de Juiz de Fora) for providing comments and suggestions for the manuscript.
Contributor Information
Lucas Costa, Laboratory of Plant Cytogenetics and Evolution, Department of Botany, Federal University of Pernambuco, Recife-PE, Brazil.
André Marques, Department of Chromosome Biology, Max Planck Institute for Plant Breeding Research, Cologne, Germany.
Christopher E Buddenhagen, AgResearch Ltd, Agroecology, Ruakura, New Zealand.
Andrea Pedrosa-Harand, Laboratory of Plant Cytogenetics and Evolution, Department of Botany, Federal University of Pernambuco, Recife-PE, Brazil.
Gustavo Souza, Laboratory of Plant Cytogenetics and Evolution, Department of Botany, Federal University of Pernambuco, Recife-PE, Brazil.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following.
Figure S1: dotplot of shared satDNAs found in the comparative clustering analysis of Rhynchospora species. Figure S2: bendability/curvature propensity plots of shared satDNAs RgSat, CL06, CL10, CL165, CL239, CL261 and CL319. Table S1: list of Rhynchospora species, voucher number, herbarium, number of reads before and after filtering target reads and total number of reads analysed in the individual clustering analysis into RepeatExplorer. Table S2: name, monomer length, GC content and consensus sequence of shared satDNAs uncovered in the comparative clustering analysis, Tyba variants uncovered in the Tyba-like clustering analysis and of the Tyba consensus uncovered in the individual species clustering analysis. Table S3: proportion of different repetitive DNA classes per species identified in individual clustering analyses.
FUNDING
This study was supported in part by the Coordenacão de Aperfeicoamento de Pessoal de Nıvel Superior–Brasil (CAPES; finance code 001), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior–Programa Institucional de Internacionalização [CAPES-PRINT; project number 88887.363884/2019-00 (L.C.)] and Conselho Nacional de Desenvolvimento Científico e Tecnologico [CNPq; grant number 141037/2018-0 (L.C.)].
CONFLICT OF INTEREST
All authors have declared that there are no conflicts of interest regarding this article.
LITERATURE CITED
- Achrem M, Szućko I, Kalinka A.. 2020. The epigenetic regulation of centromeres and telomeres in plants and animals. Comparative Cytogenetics 14: 265–311. doi: 10.3897/compcytogen.v14i2.51895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad SF, Singchat W, Jehangir M, et al. 2020. Dark matter of primate genomes: satellite DNA repeats and their evolutionary dynamics. Cells 9: 2714. doi: 10.3390/cells9122714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguelho EG, Michelan VS, Nogueira FM, et al. 2012. New chromosome counts in Brazilian species of Rhynchospora (Cyperaceae). Caryologia 65: 140–146. doi: 10.1080/00087114.2012.711675. [DOI] [Google Scholar]
- Belyayev A, Josefiová J, Jandová M, Kalendar R, Krak K, Mandák B.. 2019. Natural history of a satellite DNA family: from the ancestral genome component to species-specific sequences, concerted and non-concerted evolution. International Journal of Molecular Sciences 20: 1201. doi: 10.3390/ijms20051201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brukner I, Sánchez R, Suck D, Pongor S.. 1995. Sequence-dependent bending propensity of DNA as revealed by DNase I: parameters for trinucleotides. EMBO Journal 14: 1812–1818. doi: 10.1002/j.1460-2075.1995.tb07169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buddenhagen CE. 2016. A view of Rhynchosporeae (Cyperaceae) diversification before and after the application of anchored phylogenomics across the angiosperms. PhD Thesis, Florida State University, USA. [Google Scholar]
- Bureš P, Zedek F, Markova M.. 2013. Holocentric chromosomes. In: Greilhuber J, Dolezel J, Wendel JF, eds. Plant genome diversity, Vol. 2. Vienna: Springer, 187–204. [Google Scholar]
- Camacho JPM, Cabrero J, López-León MD, et al. 2022. Satellitome comparison of two oedipodine grasshoppers highlights the contingent nature of satellite DNA evolution. BMC Biology 20: 36. doi: 10.1186/s12915-021-01216-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Dong F, Langdon T, et al. 2002. Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. The Plant Cell 14: 1691–1704. doi: 10.1105/tpc.003079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa L, Marques A, Buddenhagen C, et al. 2021. Aiming off the target: recycling target capture sequencing reads for investigating repetitive DNA. Annals of Botany 128: 835–848. doi: 10.1093/aob/mcab063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan M, Pouch M, Mandáková T, et al. 2021. Evolution of tandem repeats is mirroring post-polyploid cladogenesis in Heliophila (Brassicaceae). Frontiers in Plant Science 11: 607893. doi: 10.3389/fpls.2020.607893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover G. 2002. Molecular drive. Trends in Genetics 18: 587–589. doi: 10.1016/s0168-9525(02)02789-0. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A.. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology 7: 214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudeiro A, Adega F, Robinson TJ, Heslop-Harrison JS, Chaves R.. 2019. Conservation, divergence, and functions of centromeric satellite DNA families in the Bovidae. Genome Biology and Evolution 11: 1152–1165. doi: 10.1093/gbe/evz061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Ives AR, Surget-Groba Y, Cannon CH.. 2015. An assembly and alignment-free method of phylogeny reconstruction from next-generation sequencing data. BMC Genomics 16: 522. doi: 10.1186/s12864-015-1647-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry K, Salser W.. 1977. Nucleotide sequences of HS-α satellite DNA from kangaroo rat dipodomys ordii and characterization of similar sequences in other rodents. Cell 12: 1069–1084. doi: 10.1016/0092-8674(77)90170-2. [DOI] [PubMed] [Google Scholar]
- Gabrielan E, Zohary D.. 2004. Wild relatives of food crops native to Armenia and Nakhichevan. Flora Mediterranea 14: 5–80. [Google Scholar]
- Gale S. 1944. Rhynchospora, section Eurhynchospora, in Canada, the United States and the West Indies. In: Gale S, eds. Contributions from the Gray Herbarium of Harvard University. Massachusetts: New England Botanical Club, 89–134. [Google Scholar]
- Garrido-Ramos MA. 2015. Satellite DNA in plants: more than just rubbish. Cytogenetic and Genome Research 146: 153–170. doi: 10.1159/000437008. [DOI] [PubMed] [Google Scholar]
- Garrido-Ramos M. 2017. Satellite DNA: an evolving topic. Genes 8: 230. doi: 10.3390/genes8090230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmell NJ. 2021. Repetitive DNA: genomic dark matter matters. Nature Reviews Genetics 22: 342–342. doi: 10.1038/s41576-021-00354-8. [DOI] [PubMed] [Google Scholar]
- Gernhard T. 2008. New analytic results for speciation times in neutral models. Bulletin of Mathematical Biology 70: 1082–1097. doi: 10.1007/s11538-007-9291-0. [DOI] [PubMed] [Google Scholar]
- Giarla TC, Esselstyn JA.. 2015. The challenges of resolving a rapid, recent radiation: empirical and simulated phylogenomics of Philippine shrews. Systematic Biology 64: 727–740. doi: 10.1093/sysbio/syv029. [DOI] [PubMed] [Google Scholar]
- Gong Z, Wu Y, Koblížková A, et al. 2012. Repeatless and repeat-based centromeres in potato: implications for centromere evolution. The Plant Cell 24: 3559–3574. doi: 10.1105/tpc.112.100511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodsell DS, Dickerson RE.. 1994. Bending and curvature calculations in B-DNA. Nucleic Acids Research 22: 5497–5503. doi: 10.1093/nar/22.24.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann S, Schroeder-Reiter E, Kumke K, et al. 2011. Holocentric chromosomes of Luzula elegans are characterized by a longitudinal centromere groove, chromosome bending, and a terminal nucleolus organizer region. Cytogenetic and Genome Research 134: 220–228. doi: 10.1159/000327713. [DOI] [PubMed] [Google Scholar]
- Hofstatter PG, Thangavel G, Lux T, et al. 2022. Repeat-based holocentromeres influence genome architecture and karyotype evolution. Cell 185: 3153–3168.e18. doi: 10.1016/j.cell.2022.06.045. [DOI] [PubMed] [Google Scholar]
- Iwata A, Tek AL, Richard MMS, et al. 2013. Identification and characterization of functional centromeres of the common bean. The Plant Journal 76: 47–60. doi: 10.1111/tpj.12269. [DOI] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn GCS, Küttler H, Moreira-Filho O, Heslop-Harrison JS.. 2012. The 1.688 repetitive DNA of Drosophila: concerted evolution at different genomic scales and association with genes. Molecular Biology and Evolution 29: 7–11. doi: 10.1093/molbev/msr173. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9: 357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larridon I, Spalink D, Jiménez‐Mejías P, et al. 2021. The evolutionary history of sedges (Cyperaceae) in Madagascar. Journal of Biogeography 48: 917–932. doi: 10.1111/jbi.14048. [DOI] [Google Scholar]
- Liu H, Wu J, Xie J, Yang X, Lu Z, Sun X.. 2008. Characteristics of nucleosome core DNA and their applications in predicting nucleosome positions. Biophysical Journal 94: 4597–4604. doi: 10.1529/biophysj.107.117028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Duan X, Yu S, Sun X.. 2011. Analysis of nucleosome positioning determined by DNA helix curvature in the human genome. BMC Genomics 12: 72. doi: 10.1186/1471-2164-12-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorite P, Muñoz-López M, Carrillo JA, et al. 2017. Concerted evolution, a slow process for ant satellite DNA: study of the satellite DNA in the Aphaenogaster genus (Hymenoptera, Formicidae). Organisms Diversity & Evolution 17: 595–606. doi: 10.1007/s13127-017-0333-7. [DOI] [Google Scholar]
- Lower SS, McGurk MP, Clark AG, Barbash DA.. 2018. Satellite DNA evolution: old ideas, new approaches. Current Opinion in Genetics & Development 49: 70–78. doi: 10.1016/j.gde.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macas J, Meszaros T, Nouzova M.. 2002. PlantSat: a specialized database for plant satellite repeats. Bioinformatics 18: 28–35. doi: 10.1093/bioinformatics/18.1.28. [DOI] [PubMed] [Google Scholar]
- Marques A, Pedrosa-Harand A.. 2016. Holocentromere identity: from the typical mitotic linear structure to the great plasticity of meiotic holocentromeres. Chromosoma 125: 669–681. doi: 10.1007/s00412-016-0612-7. [DOI] [PubMed] [Google Scholar]
- Marques A, Ribeiro T, Neumann P, et al. 2015. Holocentromeres in Rhynchospora are associated with genome-wide centromere-specific repeat arrays interspersed among euchromatin. Proceedings of the National Academy of Sciences of the United States of America 112: 13633–13638. doi: 10.1073/pnas.1512255112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques A, Schubert V, Houben A, Pedrosa-Harand A.. 2016. Restructuring of holocentric centromeres during meiosis in the plant Rhynchospora pubera. Genetics 204: 555–568. doi: 10.1534/genetics.116.191213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melters DP, Bradnam KR, Young HA, et al. 2013. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biology 14: R10. doi: 10.1186/gb-2013-14-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravinac B, Plohl M, Mestrović N, Ugarković D.. 2002. Sequence of PRAT satellite DNA “frozen” in some coleopteran species. Journal of Molecular Evolution 54: 774–783. doi: 10.1007/s00239-001-0079-9. [DOI] [PubMed] [Google Scholar]
- Mravinac B, Plohl M, Ugarković E.. 2005. Preservation and high sequence conservation of satellite DNAs suggest functional constraints. Journal of Molecular Evolution 61: 542–550. doi: 10.1007/s00239-004-0342-y. [DOI] [PubMed] [Google Scholar]
- Navajas-Pérez R, Quesada del Bosque ME, Garrido-Ramos MA.. 2009. Effect of location, organization, and repeat-copy number in satellite-DNA evolution. Molecular Genetics and Genomics 282: 395–406. doi: 10.1007/s00438-009-0472-4. [DOI] [PubMed] [Google Scholar]
- Novák P, Neumann P, Pech J, Steinhaisl J, Macas J.. 2013. RepeatExplorer: a Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 29: 792–793. doi: 10.1093/bioinformatics/btt054. [DOI] [PubMed] [Google Scholar]
- Novák P, Ávila Robledillo L, Koblížková A, Vrbová I, Neumann P, Macas J.. 2017. TAREAN: a computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acids Research 45: e111–e111. doi: 10.1093/nar/gkx257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira MAS, Nunes T, Dos Santos MA, et al. 2021. High-throughput genomic data reveal complex phylogenetic relationships in Stylosanthes Sw (Leguminosae). Frontiers in Genetics 12: 727314. doi: 10.3389/fgene.2021.727314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel M. 1999. Inferring the historical patterns of biological evolution. Nature 401: 877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- Pérez-Gutiérrez MA, Suárez-Santiago VN, López-Flores I, Romero AT, Garrido-Ramos MA.. 2012. Concerted evolution of satellite DNA in Sarcocapnos: a matter of time. Plant Molecular Biology 78: 19–29. doi: 10.1007/s11103-011-9848-z. [DOI] [PubMed] [Google Scholar]
- Petraccioli A, Odierna G, Capriglione T, et al. 2015. A novel satellite DNA isolated in Pecten jacobaeus shows high sequence similarity among molluscs. Molecular Genetics and Genomics 290: 1717–1725. doi: 10.1007/s00438-015-1036-4. [DOI] [PubMed] [Google Scholar]
- Plohl M, Luchetti A, Meštrović N, Mantovani B.. 2008. Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene 409: 72–82. doi: 10.1016/j.gene.2007.11.013. [DOI] [PubMed] [Google Scholar]
- Plohl M, Petrović V, Luchetti A, et al. 2010. Long-term conservation vs high sequence divergence: the case of an extraordinarily old satellite DNA in bivalve mollusks. Heredity 104: 543–551. doi: 10.1038/hdy.2009.141. [DOI] [PubMed] [Google Scholar]
- Plohl M, Meštrović N, Mravinac B.. 2012. Satellite DNA evolution. In: Garrido-Ramos MA, ed. Genome dynamics. Basel: S. Karger, 126–152. doi: 10.1159/000337122. [DOI] [PubMed] [Google Scholar]
- Plohl M, Meštrović N, Mravinac B.. 2014. Centromere identity from the DNA point of view. Chromosoma 123: 313–325. doi: 10.1007/s00412-014-0462-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada del Bosque ME, Navajas-Pérez R, Panero JL, Fernández-González A, Garrido-Ramos MA.. 2011. A satellite DNA evolutionary analysis in the North American endemic dioecious plant Rumex hastatulus (Polygonaceae). Genome 54: 253–260. doi: 10.1139/g10-115. [DOI] [PubMed] [Google Scholar]
- Quesada del Bosque ME, López-Flores I, Suárez-Santiago VN, Garrido-Ramos MA.. 2013. Differential spreading of HinfI satellite DNA variants during radiation in Centaureinae. Annals of Botany 112: 1793–1802. doi: 10.1093/aob/mct233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada del Bosque MEQ, López-Flores I, Suárez-Santiago VN, Garrido-Ramos MA.. 2014. Satellite-DNA diversification and the evolution of major lineages in Cardueae (Carduoideae Asteraceae). Journal of Plant Research 127: 575–583. doi: 10.1007/s10265-014-0648-9. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2022. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. Available at: https://www.R-project.org/ [Google Scholar]
- Rambaut A, Drummond AJ.. 2013. TreeAnnotator v1. 7.0. Available as Part of the BEAST package. Available at: http://beast.bio.ed.ac.uk
- Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution 3: 217–223. doi: 10.1111/j.2041-210X.2011.00169.x. [DOI] [Google Scholar]
- Ribeiro T, Marques A, Novák P, et al. 2017. Centromeric and non-centromeric satellite DNA organisation differs in holocentric Rhynchospora species. Chromosoma 126: 325–335. doi: 10.1007/s00412-016-0616-3. [DOI] [PubMed] [Google Scholar]
- Ribeiro T, Buddenhagen CE, Thomas WW, Souza G, Pedrosa-Harand A.. 2018. Are holocentrics doomed to change? Limited chromosome number variation in Rhynchospora Vahl (Cyperaceae). Protoplasma 255: 263–272. doi: 10.1007/s00709-017-1154-4. [DOI] [PubMed] [Google Scholar]
- Robles F, de la Herrán R, Ludwig A, Ruiz Rejón C, Ruiz Rejón M, Garrido-Ramos MA.. 2004. Evolution of ancient satellite DNAs in sturgeon genomes. Gene 338: 133–142. doi: 10.1016/j.gene.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Rocha DM, Marques A, Andrade CGTJ, et al. 2016. Developmental programmed cell death during asymmetric microsporogenesis in holocentric species of Rhynchospora (Cyperaceae). Journal of Experimental Botany 67: 5391–5401. doi: 10.1093/jxb/erw300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salser W, Bowen S, Browne D, et al. 1976. Investigation of the organization of mammalian chromosomes at the DNA sequence level. Federation Proceedings 35: 23–35. [PubMed] [Google Scholar]
- Silva Filho PJS, Thomas WW, Boldrini II.. 2021. Redefining Rhynchospora section Tenues (Cyperaceae), a phylogenetic approach. Botanical Journal of the Linnean Society 196: 313–328. doi: 10.1093/botlinnean/boab002. [DOI] [Google Scholar]
- Sonnhammer ELL, Durbin R.. 1995. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167: GC1–G10. doi: 10.1016/0378-1119(95)00714-8. [DOI] [PubMed] [Google Scholar]
- Subirana JA, Messeguer X.. 2013. A satellite explosion in the genome of holocentric nematodes. PLoS One 8: e62221. doi: 10.1371/journal.pone.0062221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S.. 2020. What makes a centromere? Experimental Cell Research 389: 111895. doi: 10.1016/j.yexcr.2020.111895. [DOI] [PubMed] [Google Scholar]
- Thomas WW. 2020. Two new species of Rhynchospora (Cyperaceae) from Bahia, Brazil, and new combinations in Rhynchospora section Pleurostachys. Brittonia 72: 273–281. doi: 10.1007/s12228-020-09621-0. [DOI] [Google Scholar]
- Tsoumani KT, Drosopoulou E, Mavragani-Tsipidou P, Mathiopoulos KD.. 2013. Molecular characterization and chromosomal distribution of a species-specific transcribed centromeric satellite repeat from the olive fruit fly, Bactrocera oleae. PLoS One 8: e79393. doi: 10.1371/journal.pone.0079393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunjić-Cvitanić M, Pasantes JJ, García-Souto D, Cvitanić T, Plohl M, Šatović-Vukšić E.. 2021. Satellitome analysis of the Pacific oyster Crassostrea gigas reveals new pattern of satellite DNA organization, highly scattered across the genome. International Journal of Molecular Sciences 22: 6798. doi: 10.3390/ijms22136798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Koblížková A, Wang K, et al. 2014. Boom-bust turnovers of megabase-sized centromeric DNA in Solanum species: rapid evolution of DNA sequences associated with centromeres. The Plant Cell 26: 1436–1447. doi: 10.1105/tpc.114.123877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Talbert PB, Zhang W, et al. 2013. The CentO satellite confers translational and rotational phasing on cenH3 nucleosomes in rice centromeres. Proceedings of the National Academy of Sciences of the United States of America 110: E4875–E4883. doi: 10.1073/pnas.1319548110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong CX, Marshall JB, Topp C, et al. 2002. Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. The Plant Cell 14: 2825–2836. doi: 10.1105/tpc.006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.