Abstract
Nanopores are label-free single-molecule analytical tools that show great potential for stochastic sensing of proteins. Here, we described a ClyA nanopore functionalized with different nanobodies through a 5–6 nm DNA linker at its periphery. Ty1, 2Rs15d, 2Rb17c, and nb22 nanobodies were employed to specifically recognize the large protein SARS-CoV-2 Spike, a medium-sized HER2 receptor, and the small protein murine urokinase-type plasminogen activator (muPA), respectively. The pores modified with Ty1, 2Rs15d, and 2Rb17c were capable of stochastic sensing of Spike protein and HER2 receptor, respectively, following a model where unbound nanobodies, facilitated by a DNA linker, move inside the nanopore and provoke reversible blockade events, whereas engagement with the large- and medium-sized proteins outside of the pore leads to a reduced dynamic movement of the nanobodies and an increased current through the open pore. Exploiting the multivalent interaction between trimeric Spike protein and multimerized Ty1 nanobodies enabled the detection of picomolar concentrations of Spike protein. In comparison, detection of the smaller muPA proteins follows a different model where muPA, complexing with the nb22, moves into the pore, generating larger blockage signals. Importantly, the components in blood did not affect the sensing performance of the nanobody-functionalized nanopore, which endows the pore with great potential for clinical detection of protein biomarkers.
Keywords: nanopores, nanobodies, protein detection, functionalization, specificity, modularity, general applicability
Nanopores can stochastically sense single molecules in real time and have been used to detect various analytes, such as metal ions,1,2 biomolecules,3,4 nucleic acids,5−7 and polypeptides.8,9 In particular, protein sensing10−12 by this technique holds additional advantages over other existing techniques such as enzyme-linked immunosorbent assays (ELISA) and mass spectrometry, as it can be exploited for real-time protein characterization13 and quantification,14 and it can also provide insight into protein unfolding kinetics,15 conformational changes,16,17 and ligand binding affinity.18,19 Besides that, nanopores can readily be integrated into small portable devices,20 which makes them very suitable for application in point-of-care diagnostics.
To date, a variety of nanopore-based strategies have been explored for protein sensing. Protein detection can be achieved directly by monitoring the current modulations induced by their translocation through (or binding inside) the lumen of the pore. The key to this strategy is to choose a pore with appropriate geometry that can accommodate the target. In the past decade, nanopores with large lumen volume such as Fragaceatoxin C (FraC),21 Cytolysin (ClyA),22 and pleurotolysin (PlyAB)23,24 have been exploited for the investigation of folded proteins. For instance, ClyA, with a relatively large (∼6 × 6 × 10 nm) cylindrical internal lumen, has shown the ability to capture and characterize different folded proteins25 and distinguish the interaction of peptides or DNA ligands with the protein.22 Although these biological nanopores have been proven effective, their fixed sizes and the limited types available in nature restrict their general application for the sensing of a broader variety of folded proteins of variable sizes.
In comparison, binder-assisted indirect detection of target proteins outside the nanopores has been emerging to be a more generic strategy for folded protein sensing.14,26−29 These approaches variously enable nanopores to detect large proteins that do not fit inside the nanopore, and by exploiting specific binding interactions to target proteins, they enhance the specificity of protein sensing compared to the naked nanopores. These strategies involve either capturing the target protein near the nanopore entrance, provoking a change in current, or transmitting the binding interactions occurring outside of a nanopore to the interior of the pore, leading to an altered ionic flow passing through the pore. So far, a variety of binders such as biotins,10 aptamers,14 peptides,26 and protein domains29 have been chemically or genetically functionalized on nanopores, which have been widely used for protein detection or protein–ligand binding studies.
In one example, Movileanu and Thakur established a platform for the investigation of protein–protein interaction where the protein domain (RNase barnase, Bn) containing a flexible 12-amino-acid peptide adaptor at the N-terminal was fused on the monomeric pore t-FhuA.29 Upon the binding of the cognate ligand protein (Barstar, Bs) to the protein binder, the adaptor was pulled away from the pore opening, which provoked distinguishable unblocking current events. This nanopore sensor showed the capacity of detecting and quantifying protein analytes in the presence of a background amount of serum; however, it had several drawbacks that limited its application in protein sensing. First, constructing nanopores with genetically encoded protein ligands is laborious, which makes this approach not optimal for a general platform for detecting proteins. In addition, the preparation of the nanopore requires protein refolding in urea and detergent, which may render the ligand non-functional.
In another example, Bayley etal. demonstrated that an aptamer-modified α-hemolysin (α-HL) nanopore, where a 15-mer DNA aptamer (TBA) was hybridized to an oligonucleotide that was covalently attached to a cysteine near the rim of the pore, allowed the detection of thrombin.14 The anchoring of the DNA adapter on the pore endows it with modularity; thus by changing the aptamer, various analytes can be detected using the same nanopore construct. Nevertheless, it is worth noting that given the diversity of the aptamers’ structures and lengths, different aptamers cannot be expected to behave in the same manner every time without significant experimentation; thus it is difficult to create a generic system that employs different aptamer binders for different desired targets. Particularly, for those platforms that use aptamers to increase sensing specificity, it can be problematic when employing them in biological samples such as blood, as some aptamers can be quickly degraded by nucleases.
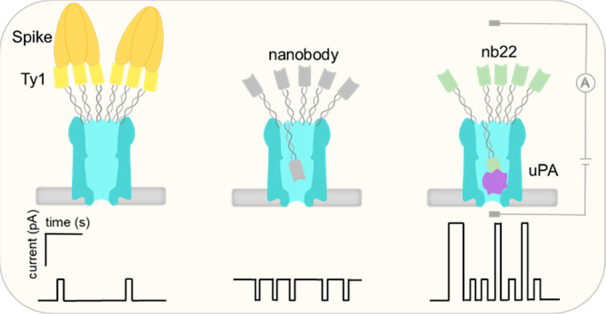
Here, we developed a modular nanopore sensor using site specifically tethered nanobodies as recognition elements, which allowed for the stochastic sensing of protein targets in complex samples. Nanobodies, derived from heavy-chain-only antibodies, have emerged as a rapidly growing family of strong protein binders.30−32 Nanobodies have similar sizes (12–14 kDa)33 and are nuclease-tolerant; in addition, they can be easily produced using a bacterial expression system and can readily be equipped with customized tags without affecting their function.34−37 These properties make nanobodies more favorable for the indirect protein sensing compared to aptamers. In this study, nanobodies, designed as replaceable modules, were immobilized on a ClyA dodecamer via DNA duplex formation. By simply changing the modules, four different nanobody-functionalized nanopores were constructed and all nanopore constructs showed the capacity for protein detection. In particular, benefiting from the multivalent interaction between trimeric SARS-CoV-2 Spike protein and multimerized Ty1 nanobodies, this approach enabled us to detect proteins in the picomolar range in the presence of blood. We expect this strategy to be a generic method that allows highly specific and sensitive detection of various proteins and protein-containing analytes (e.g., viruses, bacteria) in complex biofluids.
Results and Discussion
Functionalization of a ClyA Nanopore with Nanobodies
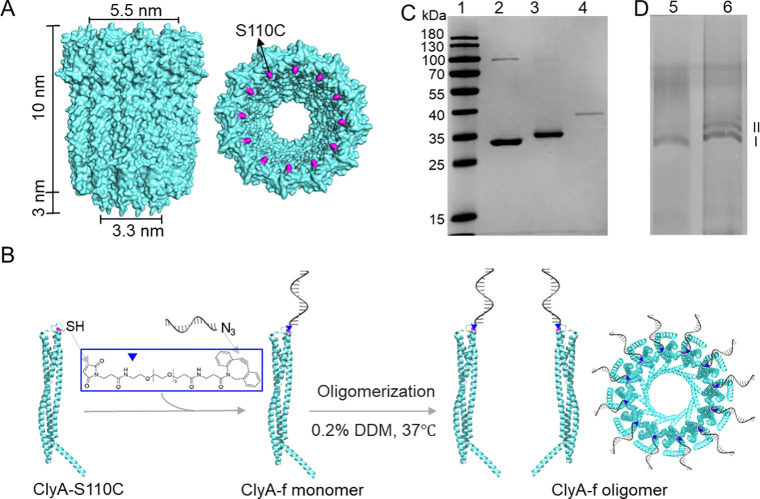
To specifically detect proteins with various sizes, we designed a ClyA nanopore functionalized with multiple nanobodies via a 16 base pair DNA duplex linker to the wide end of the pore. We hypothesized that the binding of target proteins to the nanobodies would alter the ionic flux through the nanopore, thus inducing a distinguishable current signal indicating protein detection. In prior studies,14,25 the conjugations of oligos with protein nanopores are achieved through the formation of a disulfide bond, which, however, is a less efficient reaction and produces a more unstable conjugate in complex biological environments. Hence, it is advantageous to first react the cysteine-containing proteins with a bifunctional cross-linker to incorporate a DBCO moiety and then use the highly efficient DBCO-azide click reaction to achieve the conjugation. To enable site-specific attachment of the DNA linker to ClyA, we used a mutated ClyA-AS38 variant bearing a serine to a cysteine substitution at position 110 (named as ClyA-S110C, Figure 1A). Then, a 16nt DNA oligonucleotide with an azide group at the 3′ end (f-azide) was attached to ClyA-S110C by using a maleimide-PEG4-DBCO linker (Figure 1B). With the addition of a 20-fold excess of the linkers to the ClyA-S110C, the band of the product was entirely upshifted compared to ClyA-S110C in the SDS-PAGE gel, indicating a high yield of ClyA-DBCO products (Figure 1C). Subsequently, the purified ClyA-DBCO was reacted with a 1.5-fold excess of f-azide, leading to a full yield of ClyA-f constructs (Figure 1C). Furthermore, after the self-assembly in the presence of detergent to form oligomerized pores, ClyA-S110C and ClyA-f dodecamers38 (Figure 1D, band I) were extracted from the blue native polyacrylamide gel.39 On the basis of the high conjugation efficiency of ClyA-f monomers and homogeneity of the oligomerization, we could assume that each ClyA-f dodecamer displays approximately 12 oligos ready for nanobody attachment.
Figure 1.
Attachment of ssDNA to ClyA nanopore. (A) Side view (left) and top view (right) of the ClyA structure (PDB: 6mrt). Serine (colored purple) at position 110 was genetically mutated to cysteine to enable site-specific chemical modification. (B) Schematic model showing the conjugation strategy of attaching ssDNA to a ClyA nanopore. A 16-mer oligonucleotide, named f, is conjugated to a ClyA monomer via a maleimide-PEG4-DBCO linker, where the maleimide reacts with the −SH group on the protein and DBCO is clicked to the azide group on the oligo. ClyA-f monomers then oligomerize to a ClyA-f oligomer in the presence of 0.2% n-dodecyl-beta-maltoside (DDM) at 37 °C. (C) SDS-PAGE analysis of the conjugation efficiency. Lane 1: protein ladder, lane 2: ClyA-S110C monomer, lane 3: after reaction of ClyA-S110C with maleimide-PEG4-DBCO (ClyA-DBCO), lane 4: after reaction of purified ClyA-DBCO with f-azide (ClyA-f). (D) Native polyacrylamide gel analysis of the oligomerization of ClyA-f. Lane 5: ClyA-f after oligomerization, lane 6: ClyA-S110C after oligomerization.
To allow nanobodies to anchor on ClyA nanopores, nanobodies were produced with an azide group preceding a histidine-tag at the N-terminal through unnatural amino acid incorporation by amber codon suppression37 and conjugated to a complementary strand of oligo f containing a DBCO group (f′-DBCO) at the 5′ end, via click chemistry. As a proof-of-concept, Ty1 nanobody, which can reversibly bind the receptor binding domain (RBD) of SARS-CoV-2 Spike proteins, was conjugated with f′ (Figure S1A). The binding activity of the oligo-attached nanobody was examined using a biolayer interferometry (BLI) assay, which showed that the attachment of the oligo did not significantly affect the binding affinity of the Ty1 nanobody to the RBD (Figure S1B). Furthermore, to test the feasibility of the attachment of nanobodies to ClyA, the ClyA-f monomers were incubated with a 5-fold excess of the Ty1-f′ conjugates and analyzed by SDS-polyacrylamide gel. It displayed that ClyA-f had an obvious mobility shift due to nanobody attachment (Figure S1C) and suggested that the attachment efficiency was up to 100%. Eventually, the nanobody-functionalized ClyA nanopores (ClyA-f-nb) were prepared by incubating the ClyA-f dodecamers with the respective nanobody-f′ modules.
Characterization of the Nanobody-Functionalized ClyA Nanopore
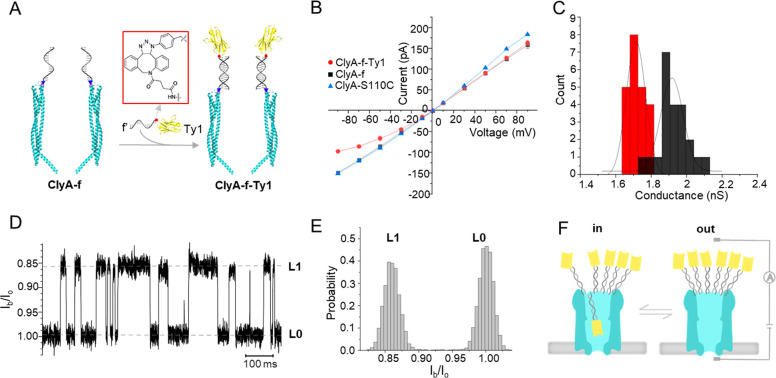
First, we conducted the electrical characterization of ClyA-S110C, ClyA-f, and Ty1-modified ClyA (ClyA-f-Ty1, Figure 2A) at different applied potentials using a single-channel recording system, to investigate the effect of the attachment of ssDNA and nanobodies. At applied potentials of ±35 mV, the current traces of ClyA-f were similar to that of ClyA-S110C, and no specific signal caused by the attached oligos entering the nanopore was observed (Figure S2). However, the current through the open ClyA-f pore was slightly smaller than that of ClyA-S110C when a positive potential larger than +35 mV was applied to the trans side (Figure 2B). It indicates that the electrophoretic force drives the oligos, attached to the cis side, into the lumen of ClyA and partially blocks it. Nevertheless, the conductive behavior of ClyA-f was not affected by the attachment of ssDNA at negative bias (Figure 2B). On the contrary, the attachment of Ty1 nanobodies had no effect on the ClyA-f-Ty1 pore at a positive potential (+35 mV), whereas the pore was partially blocked compared to ClyA-f when a negative potential (−35 mV) was applied (Figure S2). The blocked pore current at different applied potentials indicated that the current of ClyA-f-Ty1 was smaller than that of the non-nanobody-attached ClyA-f at negative potentials ranging from −10 to −90 mV (Figure 2B). As a result, the conductance of ClyA-f-Ty1 (1.71 ± 0.01 nS, n = 22) at −35 mV was smaller than that of ClyA-f (1.92 ± 0.01 nS, n = 22) (Figure 2C).
Figure 2.
Functionalization of the ClyA nanopore with Spike nanobody Ty1 and electrical characterization of the nanopore. (A) Schematic model showing the strategy of functionalizing the ClyA nanopore with the Ty1 nanobody, where Ty1-f′ was immobilized on the ClyA-f nanopore by DNA strand hybridization. (B) I–V curves of ClyA-S110C (blue triangle), ClyA-f (black square), and ClyA-f-Ty1 (red circle) at applied potential ranging from −90 to 90 mV (three independent experiments). (C) Histogram showing conductance distribution of the ClyA-f nanopore with (red) and without (black) Ty1 nanobody molecules. The conductance was calculated using the sum of the absolute current value under 35 and −35 mV applied potential divided by the sum of the absolute voltage value (n = 22). (D) Representative current traces of ClyA-f-Ty1 under an applied potential of −20 mV. Io is the open pore current and Ib is the blocked pore current. (E) All-point histogram of the current traces shown in D, which demonstrated a well-defined distribution of the blockade signals. (F) Schematic model interpreting the reversible conformation change between blocked (left) and open (right) states of ClyA-f-Ty1 at an applied potential of −20 mV, which corresponded to the movement of one of the Ty1 nanobodies in and out of the vestibule of the pore. All of the experiments were performed in 150 mM NaCl, 50 mM Tris-HCl, pH 7.5.
When lowering the applied potential to −20 mV, we observed transient and reversible blockade signals (Figure 2D). These signals are composed of two current levels (L0 and L1), where L0 is similar to the one expected for the open pore current. The current blockade percentage ((Io – Ib)/Io (or ΔI/Io, where Io is the open pore current and Ib is the blocked pore current) of L1 events was 14.2 ± 0.3% (n = 3) and the average dwell time of these events was 21.1 ± 1.1 ms (n = 3). By fitting the all-point histogram of the current traces using the Gaussian function and calculating the proportion of the area under the curve, we found that the open probability of ClyA-f-Ty1 at −20 mV was 51% (Figure 2E). With the increase of the applied potential from −10 mV to −40 mV, the blockade probability of the pore and the dwell time of the blockade signals (tin) increased significantly, whereas the interval time of the pore remaining open (tout) greatly decreased (Figure S3). In particular, at a potential of −50 mV and above, the ClyA pore was almost permanently blocked. However, by reversing the applied potential, the ClyA-f-Ty1 nanopore can return to an unblocked state.
It is known that ClyA-AS generates a strong electroosmotic flow (EOF),40 which under negative applied potentials induces the capture of a variety of proteins.19 Given the small size of a nanobody (with a diameter of 2.5 and a height of 4 nm41), which is smaller than the lumen of the ClyA, it is likely that L1 is caused by a single nanobody (attached on ClyA via a 5.5 nm flexible dsDNA linker) entering the pore. The blockade probability increased with the potential, suggesting that the nanobody remained longer inside the nanopore as the EOF was increased. To further confirm our interpretation, we observed the irreversible opening of the pore after adding 5 U of DNase I to the cis side of the chamber in the presence of Mg2+ for about 30 min at −20 mV (Figure S4), presumably a result of dsDNA linker cleavage. This result confirmed that the nanobodies were successfully attached on the ClyA nanopore by DNA duplex formation, and it offered additional evidence for the interpretation of the blockade signals. Herein, we used “in” and “out” to define the position of the nanobodies either inside or outside the nanopore vestibule (Figure 2F), and tin and tout to represent the time that the nanobodies stay inside and outside of the nanopore, respectively.
In theory, the lumen size of ClyA (inner diameter of ∼5.5 nm at the rim, ∼3.3 nm at the constriction site, and a height of ∼13 nm) could accommodate multiple nanobodies with an approximate size of 2.5 × 2.5 × 4 nm3. However, in our study, more than 99% of the blockade signals were single-level events. Occasionally, we also observed two-level blockade signals (Figure S5). This suggests that the single-level blockade reflects a single-molecule event, and the occasional stepwise jump from 14% to 35% blockage is attributed to the repositioning of a second nanobody into the ClyA lumen. We further verified that the blockades were caused by a single nanobody entering the nanopore by testing the ClyA functionalized with fewer Ty1 nanobodies. Here we observed the same blockade level as the fully equipped ClyA-f-Ty1, but with much lower frequency (Figure S6). It is possible that once a nanobody is located inside the channel, the sterical hindrance and the electrostatic repulsion, caused by the attached dsDNA or a reduced EOF flow, exclude the entry of another nanobodies. These results demonstrated attachment of nanobodies to the ClyA nanopore via a flexible oligonucleotide linker that enables the coupled nanobody to dynamically move in and out of the nanopore, partially blocking the ionic current when in the nanopore (Figure 2F). Further, the results demonstrated the ability to control the dynamics between the in and the out states through applied voltage.
Real-Time Detection of SARS-CoV-2 Spike Protein
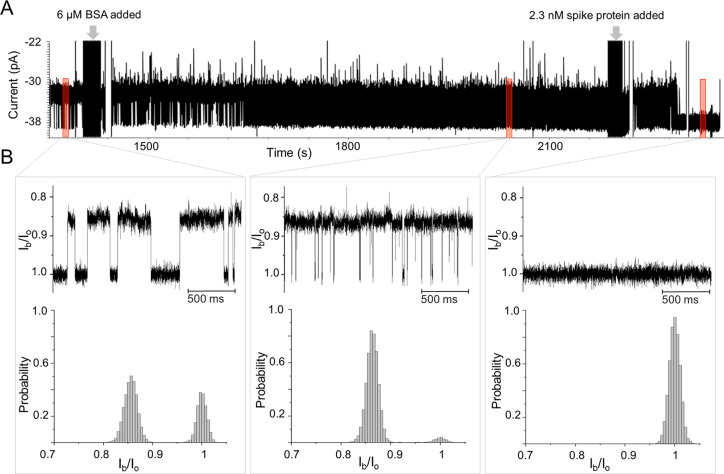
Since our aim eventually is to detect Spike proteins that are in a complex biological environment, we used bovine serum albumin (BSA) to investigate the effect of molecular crowding on the blockade signals. We observed that the addition of BSA to the cis side of the ClyA-f-Ty1 nanopore led to no additional blockade signals (Figure 3). In addition, both the tout and the open probability of ClyA-f-Ty1 decreased with the increasing concentration of BSA (Figure S7). Particularly, in the presence of 6 μM BSA and at a bias of −20 mV, the tout of ClyA-f-Ty1 decreased from 42.9 ± 38.9 ms to 4.6 ± 0.4 ms and the probability of ClyA-f-Ty1 remaining open-state decreased from 14.2 ± 7.5% to 2.1 ± 0.8% (n = 4). These results suggested that the presence of BSA drastically increased the capture of Ty1 by ClyA. Previous studies showed that molecular crowding can greatly enhance the capture of macromolecules to nanopores.9,42,43 Given that BSA possesses dimensions of 14 × 4 × 4 nm and a pI of 4.7 in aqueous solution,44 it is very likely that BSA generates a similar crowding effect that pushes the equilibrium of the ClyA-f-Ty1 toward the closed substate. Importantly, we found that the addition of BSA greatly minimized the pore-to-pore variance of nanobody-modified ClyA pores (Figure S7). Therefore, for further sensing applications, 6 μM BSA was added to the cis side of the chamber to minimize the background signal.
Figure 3.
Detection of Spike protein by ClyA-f-Ty1 nanopores. (A) Current traces of ClyA-f-Ty1 before and after the sequential addition of 6 μM BSA and 2.3 nM Spike protein. (B) Enlarged representative current traces from A (top), and all-point histograms of the current distribution (bottom). From left to right: before and after the sequential addition of 6 μM BSA and 2.3 nM Spike proteins. The experiments were performed in 150 mM NaCl, 50 mM Tris-HCl, pH 7.5.
Multivalent interaction has been widely exploited to improve binding affinity and enhance sensing sensitivity.45,46 It was reported that the binding affinity between Spike and Ty1 was dramatically increased by multimerization of the nanobody.47 Given the dodecamer structure and well-defined distance, the ClyA nanopore is predicted to be an optimal scaffold for multimerizing nanobodies, to allow multivalent recognition of Spike protein. To test the feasibility of this sensing system, SARS-CoV-2 Spike protein was added to the ClyA-f-Ty1 nanopore at a final concentration of 2.3 nM in the presence of 6 μM BSA. After incubating with Spike for about 1 min, we observed that the frequency of the blockade signals started to decrease and the tout increased (Figures 3, S8). Following that, the current traces gradually flattened, which was in accordance with the ClyA-f-Ty1 nanopore recovering to an open state (∼-38 pA) (Figures 3, S8). The increased open pore probability suggested that the attached nanobodies underwent a conformational transition from inside to outside the pore. Under a negative applied potential, the movement of the nanobodies is driven by the combined effect of the electroosmotic flow and electrophoresis. However, once outside, the nanobodies can capture a Spike protein that due to its size will retain the nanobodies outside the pore. In the presence of 6 μM BSA, the addition of 2.3 nM Spike led to an increase of open probability of the pore from 3.9% to 98.9%, whereas in the absence of BSA, the open probability only increased to 61% (Figure S9). Possibly, the molecular crowding caused by the BSA raised the local concentration of Spike near the ClyA pore opening, which increased the probability of the nanobody capturing Spike.9,42,43
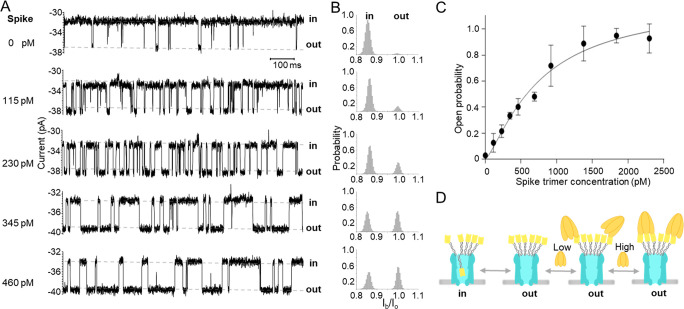
To make a calibration curve for Spike detection and to further investigate the binding kinetics of the trimeric Spike with the attached Ty1 nanobodies, we tested the response of the ClyA-f-Ty1 nanopore to different concentrations of Spike protein. At lower concentrations (0–460 pM), the open probability of ClyA-f-Ty1 increased with increasing Spike concentration over the entire range (Figure 4A,B). We also found that the average time of Ty1 locating outside the pore (tout) increased linearly with increasing concentration of Spike protein, while the time of Ty1 lodging inside the nanopore (tin) was independent of the concentration (Figure S10). This confirmed that the increased open probability was indeed caused by Spike protein associating with Ty1 nanobodies. As the Spike is a trimeric protein that can interact with three Ty1 nanobodies,48 any of the 12 Ty1 on a ClyA nanopore being occupied by one Spike would modulate the ionic flow. Thus, this makes our platform highly sensitive, capable of detecting Spike at picomolar concentration. When further increasing the Spike concentration, we found that the open probability was positively correlated with the concentration, reaching a plateau at around 2 nM (Figure 4C). The data can be fitted by the Hill–Langmuir equation with a Hill coefficient greater than 1 (n = 1.3), indicating that the binding between the trimeric Spike and the Ty1 was cooperative. The dissociation constant (Kd) obtained from the fitting was 760.6 pM, which was significantly lower than that of the monomeric RBD binding to Ty1 (Kd = 9 nM). These data were consistent with the fact that cooperative binding between multiple ligands and the same receptor can create a much stronger binding affinity,45,49 suggesting that the binding between the trimeric Spike and multimerized Ty1 was multivalent.
Figure 4.
Open probability of ClyA-f-Ty1 correlates positively with Spike trimer protein concentration. (A) Representative current traces of ClyA-f-Ty1 before and after the addition of increasing concentration of Spike trimer protein. (B) All-point histograms were displayed to show the current distribution before and after the addition of increasing concentration of Spike protein. (C) Curve regression of the open probability in the function of Spike concentrations. The curve was fitted by using the Hill–Langmuir equation Y = Bmax*Xh/(Kdh + Xh) (n = 1.3, Kd = 760.6 pM) (n = 3, the data are shown as mean ± standard deviation). (D) The schematic model showing the dynamics of the interaction between ClyA-f-Ty1 and Spike protein. Ty1 nanobodies dynamically move in and out of the ClyA nanopore under applied potential. Spike protein reversibly interacts with the Ty1 nanobodies attached on the nanopore, presumably in a multivalent fashion. The experiments were performed in 150 mM NaCl, 50 mM Tris-HCl, pH 7.5 in the presence of 6 μM BSA.
Moreover, we observed that in the presence of 6 μM BSA and 2.3 nM Spike, the histogram of the tout showed two peaks (Figure S8) with an average interevent time of 5.0 ± 1.3 ms and 20.2 ± 0.0 s (n = 3), respectively. The interevent duration of the first peak was similar to that observed without Spike protein (4.5 ± 1.3 ms, n = 3), indicating that those events were likely attributed to a single unbound Ty1 nanobody moving in and out of the nanopore. The events of the second peak are most likely caused by the nanobodies engaging with Spike. The tout caused by Spike at an intermediate concentration (690 pM) ranges from a few milliseconds to tens of seconds (Figure S11), probably reflecting that 12 Ty1 nanobodies can interact with the trimeric Spike protein in many different combinations. The short interevent time likely reflects a state where Spike only bound one nanobody, whereas the longer durations may reflect a state where one Spike trimer complex bound multiple nanobodies, creating avidity (Figure 4D).
Detection of SARS-CoV-2 Spike Protein in Blood
For sensing application in the clinic, it is crucial that the sensing efficiency and specificity of the sensor are not affected by blood components such as proteins, red and white blood cells, and platelets. To test the influence of blood components using our ClyA-f-Ty1 pore sensor, 1 μL (final concentration: 0.2% v/v) of defibrinated sheep blood was added to the cis side of the chamber in the presence of BSA (Figure S12A). Higher blood concentrations could not be tested, as they disrupted the integrity of the lipid bilayer. With 0.2% v/v whole blood, the conductive behavior of the ClyA-f-Ty1 nanopore was only slightly affected by the blood, and the membrane remained stable (Figure S12B–D). No obvious blood-induced blockade was observed, except for very few transient blockade signals with a current blockage of 31.5 ± 0.1% (Figure S12D, level 2). However, the dwell time of those events were very short (∼0.6 ms), suggesting it might be due to transient collision by proteins or platelets in the blood. Moreover, the changes of the open probability, the dwell time, and the interevent time of the ClyA-f-Ty1 nanopore before and after the addition of blood were negligible (Figure S12E–H).
After the addition of 2.3 nM Spike, the nanopore largely transited to an open state due to binding to the Ty1 nanobodies. Some large and second-long blockade events were also observed in this state (Figure S13). Likely, in the absence of Spike proteins, the steric hindrance of nanobodies on ClyA excluded blood components from entering the pore, whereas upon Spike protein binding to the nanobodies, the pore remained open so that some large proteins in blood occasionally entered the nanopore. It is worth noting that, unlike other approaches, in this system the protein does not need to enter the nanopore to be detected. This is important, because the applied potential required for protein detection in this assay was just −20 mV, which was much lower than that required for capturing proteins into nanopores.22 The lower voltage reduces the chances of capturing unwanted background contaminants in the nanopore. Furthermore, the coupled nanobodies at the entrance to the nanopore further prevent capture and interference of unwanted proteins and contaminants (e.g., background proteins in blood) in the nanopore, which drastically increases the selectivity of the nanopore for the target proteins.
General Applicability of Nanobody-Functionalized Nanopores as Protein Sensors
Nanobodies have similar characteristics in sizes and shapes.33 It is expected, therefore, that a variety of nanobodies can provoke similar transient blockage signals when immobilized on a ClyA nanopore, thus allowing the detection of variable-sized protein targets. Taking advantage of the modularity of our approach, we constructed ClyA nanopores with nanobodies 2Rs15d (ClyA-f-15d), 2Rb17c (ClyA-f-17c), and nb22 (ClyA-f-nb22), respectively. Among these nanobodies, 2Rs15d and 2Rb17c50 recognize the N-terminal half and C-terminal half of human epidermal growth factor receptor 2 (HER2) proteins that are highly expressed in breast cancer, and nb2251 recognizes murine urokinase-type plasminogen activator (muPA), which is a biomarker associated with cancer progression. All of the three nanobodies were successfully conjugated with the oligo f′, and these nanobodies could be functionalized on ClyA with high attachment efficiency (Figure S14). Due to their similarity in size, shape, and surface charge (Figure S15), we assume that these nanobodies have a similar effect on the electrical behavior of ClyA as Ty1. Indeed, all of the three nanobody-conjugated ClyA nanopores induced similar blockage signals at an applied potential of −20 mV (Figure S16, Figure S17). In the presence of 6 μM BSA, the blockade percentages caused by 2Rs15d, 2Rb17c, and nb22 were 11.7 ± 0.1%, 14.2 ± 0.4%, and 13.7 ± 0.1%, respectively. To verify the protein sensing capability, the recombinant soluble protein Her2-hFc (96 kDa) was added to the ClyA-f-15d and ClyA-f-17c pores, respectively. Similar to the phenomenon observed for the interaction of Spike with ClyA-f-Ty1, both nanobody-functionalized nanopores showed significantly increased open probability after the addition of Her2-hFc as a result of the target protein binding to the coupled nanobodies (Figure S16).
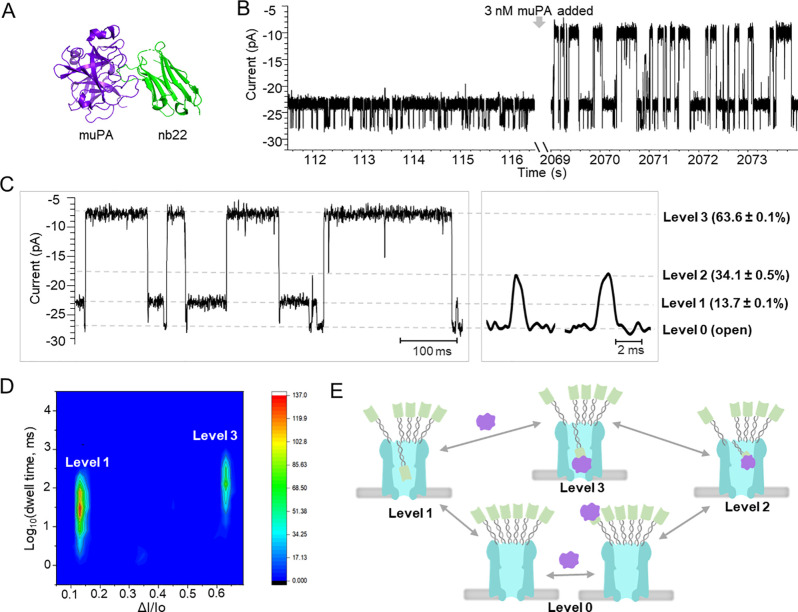
Furthermore, we tested the feasibility of ClyA-f-nb22 for protein sensing. Interestingly, after target protein muPA (48 kDa, pI 8.53, Figure 5A) was added to ClyA-f-nb22 pores, at a potential of −15 mV, we observed a new class of blockade events (levels 2 and 3) in addition to the open-pore level (level 0) and events provoked by the nanobody (level 1) (Figure 5B–D). At −15 mV, the new level 3 blockade showed a current blockade of 63.6 ± 0.1% and was relatively long in duration, at 45.5 ± 1.5 ms, while level 2 blocked to 34.1 ± 0.5% and was very short in duration, at 1.6 ± 0.4 ms (Figures 5C, S18). Level 3 blockades were not observed before adding muPA (Figure 5B) or when muPA was added to ClyA-f (Figures S19, S20) or ClyA-f-Ty1 nanopores (Figures S21, S22), suggesting that they were not caused by free muPA protein itself nor by nonspecific interaction between nanobodies and the proteins. Most likely, level 3 blockades reflect the entries of nb22:muPA complexes inside the nanopore. In addition, as the applied potential increased from −5 mV to −15 mV, the dwell time of the level 3 events increased by around 1.5 orders of magnitude (Figure S23), which was consistent with the fact that positively charged muPA:nb22 complexes tend to reside in the nanopore a longer time at higher negative potential. These results further confirmed that muPA complexing with nb22 enters the ClyA pore, provoking the level 3 blockade events.
Figure 5.
ClyA nanopore functionalized with nanobody nb22 for the detection of muPA. (A) The crystal structure of muPA (purple) in complex with the nb22 nanobody (green) (PDB: 5LHR) in a cartoon presentation. The binding affinity of nb22 to muPA measured by SPR:51kon = (4.6 ± 0.8) × 105 M–1 s–1, koff = (7.8 ± 2.2) × 10–5 s–1, KD = 0.2 ± 0.03 nM. (B) Representative current traces of ClyA-f-nb22 before and after adding 3 nM muPA under −15 mV applied potential. (C) Enlarged representative current traces after adding 3 nM muPA at −15 mV. The signals consisted of three blockade levels with current blocking percentages of 13.7 ± 0.1%, 34.1 ± 0.5%, and 63.6 ± 0.1%, respectively. (D) Heatmap of the blockade events observed after the addition of 3 nM muPA with the logarithm of the dwell time against current blockade percentage. (E) The schematic model demonstrated the conformation changes of ClyA-f-nb22 in response to muPA proteins. The experiments were performed in 150 mM NaCl, 50 mM Tris-HCl, pH 7.5 with the presence of 6 μM BSA.
In comparison, level 2 blockades did not significantly change with applied voltage. Given the short dwell time and non-voltage dependency (Figure S23), level 2 blockades may reflect the transient collision of the nb22:muPA complex with the ClyA nanopore rather than full entry into the nanopore. Based on the above analysis, we built a model (Figure 5E) displaying the conformational transitions of ClyA-f-nb22 in response to muPA, which corresponded to the observed multiple current levels. During the single-channel recording experiments, we did not observe signatures induced by the release of the nb22:muPA complex, and we found that the frequency of the level 3 events was not dependent on the concentration of muPA. It might be attributed to the high binding affinity (KD = 0.2 nM51) between nb22 and muPA and the low off rate (koff = 7.8 × 10–5 s–1) of the nb22:muPA complex. Taken together, these results demonstrate the ability to detect smaller target analytes inside the nanopore through binding to the coupled nanobodies.
Conclusions
In this study, we constructed a generic nanopore sensor, which can be easily functionalized with different nanobody recognition units, allowing for specific and sensitive detection of proteins. We have demonstrated that four different nanobody-functionalized ClyA nanopores provoked distinctive current changes upon the binding to specific proteins with sizes ranging from tens to hundreds of kilodaltons, showing the feasibility for protein sensing. Detection of various-sized proteins follows different mechanisms, where the engagement of large proteins retains the nanobodies outside of the pore, increasing the current through the open pore, whereas small proteins complexing with the nanobodies were dragged into the pore, generating larger blockade signals. These findings suggest that various proteins regardless of their sizes, shapes, and charges can be detected by the nanobody-functionalized nanopore platform. In addition, the multimerization of nanobodies on the ClyA nanopore enabled the detection of the trimeric Spike proteins at picomolar concentrations, which implies that multivalent interaction between the nanobodies and the target protein strongly increases the sensitivity of protein quantification. Importantly, protein sensing by the present approach requires very low applied potential, and the attached nanobodies on the nanopore as gatekeepers prevent the entry of noncognate proteins, which greatly reduces the interference of other components in blood and increases the sensing selectivity. This nanopore sensor shows great potential for highly sensitive detection of biomarkers, bacteria, or viruses in biofluids obtained from patients.
Experimental Section
Materials
All the chemicals, except as specifically stated, were purchased from Sigma-Aldrich. Unnatural amino acid (UAA) 4-azido-l-phenylalanine (pAzF) used in this study was synthesized in-house following a reported protocol.52 All the DNA oligos were purchased from IDT.
Expression and Purification of pAzF-Modified Nanobodies
The DNA encoding for Ty1,48 nb22,51 2Rs15d, and 2Rb17c50 nanobodies were cloned into PET22b (+) plasmid (Addgene), respectively, with a pelB leader sequence at the N-terminal and a hexahistidine tag (6xHis) at the C-terminal. An amber stop codon (TAG) was added before the 6xHis to incorporate UAA into the nanobody. The production of pAzF-modified nanobody was conducted by following an established protocol.37 First, the constructed plasmid was transformed into BL21 E. coli cells. Cells were cultured in 1 L of TB medium supplemented with 100 mL of salt buffer (0.17 M KH2PO4, 0.72 M K2HPO4), 1 mL of 2 M MgCl2, 1 mL of 100 mg/mL ampicillin, 1 mL of 50 mg/mL spectinomycin, 10 mL of 10% glucose, and 250 mg of 4-azido-l-phenylalanine at 37 °C at 200 rpm. When the OD600 reached 0.6–0.9, IPTG with a final concentration of 1 mM was added. The protein induction was completed at 25 °C by overnight shaking. The cells were harvested by centrifuging at 4 °C and 4500 rpm for 15 min, which were then resuspended in 24 mL of cold TES buffer (0.2 M Tris, pH 8, 0.5 mM EDTA, 0.5 M sucrose). The suspension was incubated at 4 °C and 200 rpm (horizontal rotator) for 6 h, followed by the addition of 48 mL of 1/4 TES buffer and incubation at 4 °C and 200 rpm overnight. Subsequently, the cell suspension was centrifuged at 4 °C and 12000g for 30 min. The supernatant was collected and supplemented with 5 mM MgCl2, followed by a further purification by FPLC (GE Healthcare) using a 5 mL HisTrap column (GE Healthcare). Binding buffer and elution buffer used here were 20 and 500 mM imidazole, respectively; both were supplemented with 20 mM sodium phosphate pH 7.4, 500 mM NaCl. The purity of the protein was analyzed on 4–12% SDS-PAGE gel.
Conjugation of Nanobody with f′-Oligo
Oligo f′ ordered with an amine group at the 5′ end (NH2-C6-5′ ATCCGCGGGTGTCGGG 3′) was first reacted with a 20-fold excess of NHS-DBCO in 60% DMSO at pH 8.0 and 25 °C overnight. After being purified by ethanol precipitation and subsequent reverse-phase HPLC, the DBCO-oligo was incubated with azide-modified nanobody in PBS at 25 °C overnight. The reaction was optimized by adding a different ratio of nanobody and f′-DBCO oligo. When the molar ratio was 5:1, the conjugation yield was above 70% (Figure S1A). Therefore, this ratio was applied for the conjugation of all four nanobodies. Subsequently, the nanobody-f′ conjugates were purified by ion-exchange chromatography and verified by either 16% denaturing urea polyacrylamide gel electrophoresis or SDS-PAGE.
Expression and Purification of the ClyA-S110C Nanopore
The ClyA-S110C construct was prepared by mutating the serine on position 110 to a cysteine in the cysteine-free variant ClyA-AS as previously reported.38 The constructed plasmid was transformed into an E. coli BL21 (DE3) electrocompetent cell by electroporation. Cells were cultured in 2× YT medium containing 100 μg/mL ampicillin at 37 °C and 200 rpm until the OD600 reached 0.8–1. Protein expression was induced by adding 0.5 mM IPTG and incubating at 20 °C and 200 rpm overnight. Cells were harvested by centrifugation at 6500 rpm and 4 °C for 15 min. The pellets were stored in a −80 °C freezer for at least 1 h and then thawed at 37 °C, followed by resuspension in 20 mL of lysis buffer (10 mM imidazole pH 8.0, 150 mM NaCl, 50 mM Tris·HCl, pH 7.5, 1 mM MgCl2, 5 mM TCEP) supplemented with 0.2 mg/mL of lysozyme. After incubating at 4 °C for 25 min on a rotator, the cells were further lysed by sonication. The lysate was then centrifuged at 6500 rpm and 4 °C for 30 min, and the supernatant was collected and incubated with Ni-NTA beads (Qiagen) at room temperature for 1 h on a rotator. Nonspecific binding protein was removed by at least 20 column volumes of wash buffer (10 mM imidazole pH 8.0, 150 mM NaCl, 50 mM Tris.HCl, pH 7.5), and the target protein was eluted from the beads in elution buffer (200 mM EDTA pH 7.5, 150 mM NaCl, 50 mM Tris·HCl, pH 7.5). The purity of the protein was analyzed on 4–12% SDS-PAGE gel.
Preparation of ClyA-f-nb Nanopore
The freshly purified ClyA-S110C was first incubated with a 20× molar excess of DBCO-PEG4-maleimide at a pH of 7.5 and 4 °C overnight, gently shaking. Unreacted DBCO-PEG4-maleimide was removed in standard buffer (150 mM NaCl, 50 mM Tris·HCl, pH 7.5) using a 3 kDa cutoff Amicon filter (Millipore). The purified ClyA-PEG4-DBCO was then incubated with a 1.5-fold excess of f-azide oligo at 4 °C overnight, gently shaking, to create the ssDNA-modified “ClyA-f” monomers. The f-azide oligo linker was prepared by reaction of an oligo with an amino modification at the 5′ end (NH2-C6-5′-CCCGACACCCGCGGAT-3′) with an azidobutyric acid NHS ester. SDS-PAGE gel was utilized to check the click reaction efficiency. The ClyA-f monomer was oligomerized in the presence of 0.2% n-dodecyl-β-d-maltoside (DDM) by incubating at 37 °C for 30 min. Subsequently, the oligomerized ClyA-S110C and ClyA-f were analyzed and purified by blue native polyacrylamide gel electrophoresis (BN-PAGE, Bio-Rad). Due to the negative charges of DNA oligos,14 ClyA-f oligomers migrated slightly faster than ClyA-S110C oligomers. According to the previous study,38 the lowest oligomeric bands of ClyA-S110C and ClyA-f were type-I nanopores (12-mer). Therefore, ClyA-S110C and ClyA-f dodecamers were obtained by slicing these bands from the gel. After eluting from the gel pieces using 30 μL of the standard buffer with the presence of 0.02% DDM, the ClyA-f oligomer solution was aliquoted into 5 μL/tube. The concentration of ClyA-f dodecamers eluted from the gel was too low to be measured by either Nanodrop or Bradford assay. Therefore, prior to the single-channel recording experiments, an excess of nanobody-f′ (∼40 pmol) was incubated with 5 μL of the ClyA-f oligomers at room temperature for at least 30 min to ensure each ClyA nanopore was modified with as many as possible nanobodies.
Single-Channel Recording Experiment (150 mM NaCl, 50 mM Tris-HCl, pH 7.5)
Electrical recordings were performed using a vertical planar lipid membrane setup as described previously.53 Briefly, a 1,2-diphytanoyl-sn-glycero-3-phosphocholine (DPhPC, purchased from Avanti Polar Lipids) lipid bilayer was formed on the aperture of the Teflon membrane on the chamber. After being connected to a patch-clamp amplifier (Axopatch 200B, Axon Instruments) using Ag/AgCl electrodes, both trans and cis sides of the chamber were filled with electrolyte buffer: 150 mM NaCl, 50 mM Tris-HCl, pH 7.5. ClyA nanopores were added into the cis side of the chamber, which was connected to the ground electrode. After pore insertion, extra ClyA was removed by several buffer exchanges. DNaseI (Sigma-Aldrich), BSA, muPA (kindly provided by Emil Oldenburg), Her2 (acquired from SinoBiological), and various concentrations of Spike proteins (SARS-CoV-2 S protein, purchased from ACROBiosystems) were all added to the cis side if not specifically stated. All recordings were conducted using a Bessel low-pass filter of 2 kHz and a sampling rate of 10 kHz. All electrical recording current traces were filtered by a Gaussian low-pass filter with a cutoff of 1 kHz prior to analysis. The data analysis software we used in this study is Clampfit (version 10.6.1.1, Molecular Devices).
Acknowledgments
We thank Emil Oldenburg for providing muPA protein. We also thank Junyi Su, Pernille Beth, and Marjan Omer for their help in nanobody production. This work was funded by the Villum Foundation to the Biomolecular Nanoscale Engineering Center (BioNEC) (grant 18333), the Danish National Research Foundation to the Center for Cellumar Signal Pattens (CellPAT) (grant DNRF 135), and Chinese Scholarship Council (CSC) grant.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.2c12733.
Conjugation of Ty1 nanobody with oligo f′; current traces of ClyA-S110C, ClyA-f, ClyA-f-Ty1; voltage dependency of the blockade signals; verification of nanobody attachment by DNase; two-level blockade signals; current traces of ClyA with less Ty1; effect of BSA; analysis of signal caused by 2.3 nM Spike; detection of Spike in the absence of BSA; Spike concentration dependency; current traces after adding 690 pM Spike; effect of blood; Spike detection in the presence of blood; nanobody-oligo conjugation; crystal structure of nanobodies; Her2 protein detection; current traces of ClyA-f-nb22; analysis of blockade events caused by muPA; addition of muPA to ClyA-f; addition of muPA to ClyA-f-Ty1; analysis of level 2 and level 3 events caused by muPA (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Braha O.; Gu L.-Q.; Zhou L.; Lu X.; Cheley S.; Bayley H. Simultaneous Stochasticsensing of Divalent Metal Ions. Nat. Biotechnol. 2000, 18 (Ii), 1005–1007. 10.1038/79275. [DOI] [PubMed] [Google Scholar]
- Zeng T.; Li T.; Li Y.; Liu L.; Wang X.; Liu Q.; Zhao Y.; Wu H.-C. DNA-Based Detection of Mercury(II) Ions through Characteristic Current Signals in Nanopores with High Sensitivity and Selectivity. Nanoscale 2014, 6 (15), 8579–8584. 10.1039/C4NR02062F. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Dou L.; Zhang M.; Wang Y.; Jiang X.; Li X.; Wei L.; Chen Y.; Zhou C.; Geng J. Real-Time Sensing of Neurotransmitters by Functionalized Nanopores Embedded in a Single Live Cell. Mol. Biomed. 2021, 2 (1), 6. 10.1186/s43556-021-00026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boersma A. J.; Brain K. L.; Bayley H. Real-Time Stochastic Detection of Multiple Neurotransmitters with a Protein Nanopore. ACS Nano 2012, 6 (6), 5304–5308. 10.1021/nn301125y. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Zheng D.; Tan Q.; Wang M. X.; Gu L. Q. Nanopore-Based Detection of Circulating MicroRNAs in Lung Cancer Patients. Nat. Nanotechnol. 2011, 6 (10), 668–674. 10.1038/nnano.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao C.; Ying Y.-L.; Hu Z.-L.; Liao D.-F.; Tian H.; Long Y.-T. Discrimination of Oligonucleotides of Different Lengths with a Wild-Type Aerolysin Nanopore. Nat. Nanotechnol. 2016, 11 (8), 713–718. 10.1038/nnano.2016.66. [DOI] [PubMed] [Google Scholar]
- Deamer D.; Akeson M.; Branton D. Three Decades of Nanopore Sequencing. Nat. Biotechnol. 2016, 34 (5), 518–524. 10.1038/nbt.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefureac R.; Long Y. T.; Kraatz H. B.; Howard P.; Lee J. S. Transport of α-Helical Peptides through α-Hemolysin and Aerolysin Pores. Biochemistry 2006, 45 (30), 9172–9179. 10.1021/bi0604835. [DOI] [PubMed] [Google Scholar]
- Larimi M. G.; Mayse L. A.; Movileanu L. Interactions of a Polypeptide with a Protein Nanopore Under Crowding Conditions. ACS Nano 2019, 13 (4), 4469–4477. 10.1021/acsnano.9b00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahie M.; Chisholm C.; Chen M. Resolved Single-Molecule Detection of Individual Species within a Mixture of Anti-Biotin Antibodies Using an Engineered Monomeric Nanopore. ACS Nano 2015, 9 (2), 1089–1098. 10.1021/nn506606e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varongchayakul N.; Song J.; Meller A.; Grinstaff M. W. Single-Molecule Protein Sensing in a Nanopore: A Tutorial. Chem. Soc. Rev. 2018, 47, 8512–8524. 10.1039/C8CS00106E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waduge P.; Hu R.; Bandarkar P.; Yamazaki H.; Cressiot B.; Zhao Q.; Whitford P. C.; Wanunu M. Nanopore-Based Measurements of Protein Size, Fluctuations, and Conformational Changes. ACS Nano 2017, 11 (6), 5706–5716. 10.1021/acsnano.7b01212. [DOI] [PubMed] [Google Scholar]
- Yusko E. C.; Bruhn B. R.; Eggenberger O. M.; Houghtaling J.; Rollings R. C.; Walsh N. C.; Nandivada S.; Pindrus M.; Hall A. R.; Sept D.; Li J.; Kalonia D. S.; Mayer M. Real-Time Shape Approximation and Fingerprinting of Single Proteins Using a Nanopore. Nat. Nanotechnol. 2017, 12 (4), 360–367. 10.1038/nnano.2016.267. [DOI] [PubMed] [Google Scholar]
- Rotem D.; Jayasinghe L.; Salichou M.; Bayley H. Protein Detection by Nanopores Equipped with Aptamers. J. Am. Chem. Soc. 2012, 134 (5), 2781–2787. 10.1021/ja2105653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Larrea D.; Bayley H. Multistep Protein Unfolding during Nanopore Translocation. Nat. Nanotechnol. 2013, 8 (4), 288–295. 10.1038/nnano.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waduge P.; Hu R.; Bandarkar P.; Yamazaki H.; Cressiot B.; Zhao Q.; Whitford P. C.; Wanunu M. Nanopore-Based Measurements of Protein Size, Fluctuations, and Conformational Changes. ACS Nano 2017, 11 (6), 5706–5716. 10.1021/acsnano.7b01212. [DOI] [PubMed] [Google Scholar]
- Van Meervelt V.; Soskine M.; Singh S.; Schuurman-Wolters G. K.; Wijma H. J.; Poolman B.; Maglia G. Real-Time Conformational Changes and Controlled Orientation of Native Proteins Inside a Protein Nanoreactor. J. Am. Chem. Soc. 2017, 139 (51), 18640–18646. 10.1021/jacs.7b10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soskine M.; Biesemans A.; Maglia G. Single-Molecule Analyte Recognition with ClyA Nanopores Equipped with Internal Protein Adaptors. J. Am. Chem. Soc. 2015, 137 (17), 5793–5797. 10.1021/jacs.5b01520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernia S.; Van Der Heide N. J.; Galenkamp N. S.; Gouridis G.; Maglia G. Current Blockades of Proteins inside Nanopores for Real-Time Metabolome Analysis. ACS Nano 2020, 14 (2), 2296–2307. 10.1021/acsnano.9b09434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M.; Koren S.; Miga K. H.; Quick J.; Rand A. C.; Sasani T. A.; Tyson J. R.; Beggs A. D.; Dilthey A. T.; Fiddes I. T.; Malla S.; Marriott H.; Nieto T.; O’Grady J.; Olsen H. E.; Pedersen B. S.; Rhie A.; Richardson H.; Quinlan A. R.; Snutch T. P.; Tee L.; Paten B.; Phillippy A. M.; Simpson J. T.; Loman N. J.; Loose M. Nanopore Sequencing and Assembly of a Human Genome with Ultra-Long Reads. Nat. Biotechnol. 2018, 36 (4), 338–345. 10.1038/nbt.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G.; Willems K.; Soskine M.; Wloka C.; Maglia G. Electro-Osmotic Capture and Ionic Discrimination of Peptide and Protein Biomarkers with FraC Nanopores. Nat. Commun. 2017, 8 (1), 935. 10.1038/s41467-017-01006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meervelt V.; Soskine M.; Maglia G. Detection of Two Isomeric Binding Configurations in a Protein-Aptamer Complex with a Biological Nanopore. ACS Nano 2014, 8 (12), 12826–12835. 10.1021/nn506077e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G.; Willems K.; Bartelds M.; Van Dorpe P.; Soskine M.; Maglia G. Electro-Osmotic Vortices Promote the Capture of Folded Proteins by Plyab Nanopores. Nano Lett. 2020, 20 (5), 3819–3827. 10.1021/acs.nanolett.0c00877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G.; Voorspoels A.; Versloot R. C. A.; van der Heide N. J.; Carlon E.; Willems K.; Maglia G.. PlyAB Nanopores Detect Single Amino Acid Differences in Folded Haemoglobin from Blood. Angew. Chem. Int. Ed. 2022, 61 ( (34), ), 10.1002/anie.202206227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soskine M.; Biesemans A.; Moeyaert B.; Cheley S.; Bayley H.; Maglia G. An Engineered ClyA Nanopore Detects Folded Target Proteins by Selective External Association and Pore Entry. Nano Lett. 2012, 12 (9), 4895–4900. 10.1021/nl3024438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H.; Braha O.; Gu L. Q.; Cheley S.; Bayley H. Single-Molecule Observation of the Catalytic Subunit of CAMP-Dependent Protein Kinase Binding to an Inhibitor Peptide. Chem. Biol. 2005, 12 (1), 109–120. 10.1016/j.chembiol.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Wang S.; Haque F.; Rychahou P. G.; Evers B. M.; Guo P. Engineered Nanopore of Phi29 Dna-Packaging Motor for Real-Time Detection of Single Colon Cancer Specific Antibody in Serum. ACS Nano 2013, 7 (11), 9814–9822. 10.1021/nn404435v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howorka S.; Nam J.; Bayley H.; Kahne D. Stochastic Detection of Monovalent and Bivalent Protein-Ligand Interactions. Angew. Chemie - Int. Ed. 2004, 43 (7), 842–846. 10.1002/anie.200352614. [DOI] [PubMed] [Google Scholar]
- Thakur A. K.; Movileanu L. Real-Time Measurement of Protein–Protein Interactions at Single-Molecule Resolution Using a Biological Nanopore. Nat. Biotechnol. 2019, 37 (1), 96–101. 10.1038/nbt.4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyldermans S. Nanobodies: Natural Single-Domain Antibodies. Annu. Rev. Biochem. 2013, 82 (1), 775–797. 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- Beghein E.; Gettemans J. Nanobody Technology: A Versatile Toolkit for Microscopic Imaging, Protein-Protein Interaction Analysis, and Protein Function Exploration. Front. Immunol. 2017, 8 (JUL), 1–14. 10.3389/fimmu.2017.00771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang W.; Ding C.; Zheng D.; Ma X.; Yi L.; Tong X.; Wu C.; Xue C.; Yu Y.; Zhou Q. Nanobody Conjugates for Targeted Cancer Therapy and Imaging. Technol. Cancer Res. Treat. 2021, 20, 153303382110101. 10.1177/15330338211010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z.; Yang Y.; Chen X.; Han Z.; Zhou J.; Li B.; He X. Positive Charge in the Complementarity-Determining Regions of Synthetic Nanobody Prevents Aggregation. Biochem. Biophys. Res. Commun. 2021, 572, 1–6. 10.1016/j.bbrc.2021.07.054. [DOI] [PubMed] [Google Scholar]
- Jullien D.; Vignard J.; Fedor Y.; Béry N.; Olichon A.; Crozatier M.; Erard M.; Cassard H.; Ducommun B.; Salles B.; Mirey G. Chromatibody, a Novel Non-Invasive Molecular Tool to Explore and Manipulate Chromatin in Living Cells. J. Cell Sci. 2016, 129 (13), 2673–2683. 10.1242/jcs.183103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier J.; Traenkle B.; Rothbauer U. Real-Time Analysis of Epithelial-Mesenchymal Transition Using Fluorescent Single-Domain Antibodies. Sci. Rep. 2015, 5 (1), 13402. 10.1038/srep13402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Audenhove I.; Van Impe K.; Ruano-Gallego D.; De Clercq S.; De Muynck K.; Vanloo B.; Verstraete H.; Fernández L. Á.; Gettemans J. Mapping Cytoskeletal Protein Function in Cells by Means of Nanobodies. Cytoskeleton 2013, 70 (10), 604–622. 10.1002/cm.21122. [DOI] [PubMed] [Google Scholar]
- Teodori L.; Omer M.; Märcher A.; Skaanning M.; Andersen V.; Nielsen J.; Oldenburg E.; Lin Y.; Gothelf K.; Kjems J. Site-Specific Nanobody-Oligonucleotide Conjugation for Super-Resolution Imaging. J. Biol. Methods 2022, 9 (1), e159. 10.14440/jbm.2022.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soskine M.; Biesemans A.; De Maeyer M.; Maglia G. Tuning the Size and Properties of ClyA Nanopores Assisted by Directed Evolution. J. Am. Chem. Soc. 2013, 135 (36), 13456–13463. 10.1021/ja4053398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soskine M.; Biesemans A.; Moeyaert B.; Cheley S.; Bayley H.; Maglia G. An Engineered ClyA Nanopore Detects Folded Target Proteins by Selective External Association and Pore Entry. Nano Lett. 2012, 12 (9), 4895–4900. 10.1021/nl3024438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems K.; Ruić D.; Lucas F.; Barman U.; Verellen N.; Hofkens J.; Maglia G.; Van Dorpe P. Accurate Modeling of a Biological Nanopore with an Extended Continuum Framework. Nanoscale 2020, 12 (32), 16775–16795. 10.1039/D0NR03114C. [DOI] [PubMed] [Google Scholar]
- Kijanka M.; Dorresteijn B.; Oliveira S.; van Bergen en Henegouwen P. M. Nanobody-Based Cancer Therapy of Solid Tumors. Nanomedicine 2015, 10 (1), 161–174. 10.2217/nnm.14.178. [DOI] [PubMed] [Google Scholar]
- Yao F.; Peng X.; Su Z.; Tian L.; Guo Y.; Kang X. F. Crowding-Induced DNA Translocation through a Protein Nanopore. Anal. Chem. 2020, 92 (5), 3827–3833. 10.1021/acs.analchem.9b05249. [DOI] [PubMed] [Google Scholar]
- Chau C. C.; Radford S. E.; Hewitt E. W.; Actis P. Macromolecular Crowding Enhances the Detection of DNA and Proteins by a Solid-State Nanopore. Nano Lett. 2020, 20 (7), 5553–5561. 10.1021/acs.nanolett.0c02246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright A. K.; Thompson M. R. Hydrodynamic Structure of Bovine Serum Albumin Determined by Transient Electric Birefringence. Biophys. J. 1975, 15 (2), 137–141. 10.1016/S0006-3495(75)85797-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valero J.; Civit L.; Dupont D. M.; Selnihhin D.; Reinert L. S.; Idorn M.; Israels B. A.; Bednarz A. M.; Bus C.; Asbach B.; Peterhoff D.; Pedersen F. S.; Birkedal V.; Wagner R.; Paludan S. R.; Kjems J. A Serum-Stable RNA Aptamer Specific for SARS-CoV-2 Neutralizes Viral Entry. Proc. Natl. Acad. Sci. U. S. A. 2021, 118 (50), e2112942118. 10.1073/pnas.2112942118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig P.-A.; Das H.; Liu H.; Kümmerer B. M.; Gohr F. N.; Jenster L.-M.; Schiffelers L. D. J.; Tesfamariam Y. M.; Uchima M.; Wuerth J. D.; Gatterdam K.; Ruetalo N.; Christensen M. H.; Fandrey C. I.; Normann S.; Tödtmann J. M. P.; Pritzl S.; Hanke L.; Boos J.; Yuan M.; Zhu X.; Schmid-Burgk J. L.; Kato H.; Schindler M.; Wilson I. A.; Geyer M.; Ludwig K. U.; Hällberg B. M.; Wu N. C.; Schmidt F. I.. Structure-Guided Multivalent Nanobodies Block SARS-CoV-2 Infection and Suppress Mutational Escape. Science (80-.) 2021, 371 ( (6530), ), 10.1126/science.abe6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moliner-Morro A.; Sheward D.; Karl V.; Perez Vidakovics L.; Murrell B.; McInerney G. M.; Hanke L. Picomolar SARS-CoV-2 Neutralization Using Multi-Arm PEG Nanobody Constructs. Biomolecules 2020, 10 (12), 1661. 10.3390/biom10121661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke L.; Vidakovics Perez L.; Sheward D. J.; Das H.; Schulte T.; Moliner-Morro A.; Corcoran M.; Achour A.; Karlsson Hedestam G. B.; Hällberg B. M.; Murrell B.; McInerney G. M. An Alpaca Nanobody Neutralizes SARS-CoV-2 by Blocking Receptor Interaction. Nat. Commun. 2020, 11 (1), 4420. 10.1038/s41467-020-18174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumbro E.; Alexander-Katz A. Influence of Binding Site Affinity Patterns on Binding of Multivalent Polymers. ACS Omega 2020, 5 (19), 10774–10781. 10.1021/acsomega.0c00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaneycken I.; Devoogdt N.; Van Gassen N.; Vincke C.; Xavier C.; Wernery U.; Muyldermans S.; Lahoutte T.; Caveliers V. Preclinical Screening of Anti-HER2 Nanobodies for Molecular Imaging of Breast Cancer. FASEB J. 2011, 25 (7), 2433–2446. 10.1096/fj.10-180331. [DOI] [PubMed] [Google Scholar]
- Kromann-Hansen T.; Louise Lange E.; Peter Sørensen H.; Hassanzadeh-Ghassabeh G.; Huang M.; Jensen J. K.; Muyldermans S.; Declerck P. J.; Komives E. A.; Andreasen P. A. Discovery of a Novel Conformational Equilibrium in Urokinase-Type Plasminogen Activator. Sci. Rep. 2017, 7 (1), 1–11. 10.1038/s41598-017-03457-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson M. B.; Brown D. B.; Vasquez C. A.; Ziller J. W.; Johnston K. M.; Weiss G. A. Synthesis and Explosion Hazards of 4-Azido-L-Phenylalanine. J. Org. Chem. 2018, 83 (8), 4525–4536. 10.1021/acs.joc.8b00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglia G.; Heron A. J.; Stoddart D.; Japrung D.; Bayley H. Analysis of Single Nucleic Acid Molecules with Protein Nanopores. Bone 2010, 23, 591–623. 10.1016/S0076-6879(10)75022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.