Abstract
It has been almost 30 years since the invention of SELEX (Systematic Evolution of Ligands by Exponential Enrichment) methodology and the description of the first aptamers. In retrospect over the past 30 years, advances in aptamer development and application have demonstrated that aptamers are potentially useful reagents that can be employed in diverse areas within analytical chemistry, biotechnology, biomedicine, and molecular biology. While often touted as artificial antibodies with an ability to be selected for any target, aptamer development, unfortunately, lags behind development of analytical methodologies that employ aptamers, hindering deeper integration into the application of analytical tool development. This perspective covers recent advances in SELEX methodology for improving efficiency of the SELEX procedure and enhancing affinity and specificity of the selected aptamers – what we view as a critical barrier in the future role of aptamers in analytical chemistry. We discuss post-selection modifications that can be used for enhancing performance of the selected aptamers in analytical device by including understanding intermolecular interaction forces in the binding domain. While highlighting promising properties of aptamers that enable several analytical advances, we provide discussion on the challenges of penetration of aptamers in the analytical field.
Keywords: aptamers, SELEX, K d , affinity, specificity, sensors, analytical devices
Graphical Abstract
1. Introduction
Nucleic acid aptamers are short, single strands of RNA or DNA molecules with a stable three-dimensional (3D) structure selected by a well-known process, SELEX (Systematic Evolution of Ligands by Exponential Enrichment).1,2 In this process, an initial oligonucleotide library (~1012 –1015 sequences), tailored by flanking primer-binding regions on 5’ and 3’ ends of a randomized region (30- to 50-mer nucleotide), is designed for selection.3 Typically, selection is performed in a binding buffer containing common monovalent and divalent ions (Na+, K+, Mg2+, Ca2+, etc.) to screen the negatively charged phosphate backbone of the oligonucleotides. As such, secondary or tertiary oligonucleotide structural motifs, including G-quadruplexes, stem-loops, pseudoknots, kissing complexes, three-way junctions, bulges, and hairpins,4–8 are promoted to recognize target molecules through intermolecular interactions including hydrogen bonding, electrostatic interactions, van der Waals forces, π–π stacking, etc.9–12 Subsequently, the bound sequences are separated from the unbound sequences, eluted, and amplified by polymerase chain reaction (PCR). Iterative rounds of selection-amplification cycles are performed to enrich the oligonucleotide pool until desired target binding activity dominate the pool.
When considering aptamers as affinity reagents, intuitively, the counterpart – antibodies – come to mind, of which a rich analytical history exists. Discovered almost a century later than antibodies, aptamers have not yet fully transitioned from proof-of-concept demonstrations to clinical applications. As diagnostic and research reagents, antibodies have provided clinicians and researchers an extraordinarily powerful and ubiquitous tool in a variety of medical and scientific disciplines through the use of, for example, enzyme linked immunosorbent assays (ELISAs) among others. This in part due to the capability of antibodies to bind specific epitopes present on carbohydrates, nucleic acids, proteins or cell membranes.13–15
When compared to antibodies, synthetic affinity ligands such as aptamers are relatively straight forward and inexpensive to synthesize, display prolonged shelf life, are stable under non-physiological conditions, provide high batch-to-batch reproducibility, and offer flexible modification for improved functionality. Given these merits, aptamers display additional advantages in diagnostics for specific and sensitive detection of target molecules over their analogue antibodies especially when the analytes are small molecules (<~1700 Da). For instance polydisperse polysaccharides in animal tissue,16 glycosaminoglycans (GAGs), are nonimmunogenic and thus it has been challenging to harvest antibodies that display high affinity and specificity against GAGs. On the contrary, RNA aptamers against two unmodified GAGs: heparosan and chondroitin, respectively, were selected successfully through bead-based SELEX.17 Furthermore, aptamers for small molecules recognition have been selected and used for monitoring a variety of other small molecules.18–24Since the invention of SELEX in the early 1990s hundreds of aptamers have been selected. Typically, aptamers have been categorized based on their binding partner in the following five categories of targets: proteins, small molecules, cells, viruses and bacteria. It should be noted here, however, that “cell,” “bacteria,” and “virus” binding aptamers is a misnomer, as the aptamer is likely specific to some entities (e.g., protein, carbohydrate, etc.) expressed on the particle surface. Thus, identifying these entities is crucial for improving specificity of the aptamers towards these categories of targets and wider applications of the aptamers. Nonetheless, aptamers are typically classified into these categories. Here we list the most well-studied aptamers based on the number of articles employing the listed aptamers (Table S1). Readers are encouraged to read references 25–29 for more comprehensive lists of aptamers. 25–29
Model aptamers, such as the thrombin-binding, streptavidin-binding, ATP-binding, and cocaine-binding aptamers, are significant for demonstrating proof-of-concept aptamer-based analytical methods. However, with the overwhelming number of publications utilizing such a small set of aptamers, questions arise such as what challenges hinder or prevent the analytical aptamer community from integrating hundreds of existing aptamers into analytical platforms for real-world applications?
2. Aptamer Selection Strategies
In 1990, the SELEX method was described for the first time by Tuerk and Gold while exploring the interaction between the bacteriophage T4 DNA polymerase and its messenger mRNA.1 When randomizing one eight-base region of the mRNA, two different sequences displaying equivalent binding constant towards the T4 DNA polymerase were selected from a random pool of approximate 65,536 species. Concurrently the term aptamers, borrowed from the Latin word “aptus” and the Greek word “mers”, were first introduced by Ellington and Szostak who isolated RNA molecules capable of creating binding “pockets” for organic dyes.2 From then on, aptamers have been known as short single-stranded nucleic acid sequences that are selected through a directed evolution process for binding to a specific target with high affinity. One critical hurdle to the broader penetration of aptamers in analytical chemistry is that of specific – or how well the aptamer binds a specific target analyte. The following discusses how the SELEX procedure has advanced to tackle this very issue.
2.1. A Brief Introduction to Conventional Aptamer Selection: SELEX
In order to yield aptamers with high specificity and affinity to target molecules, the SELEX method involves steps of iterative binding, partitioning, dissociation, and amplification. Despite that more than 32 variants of SELEX process have been described in the past three decades, the fundamental steps of the procedure remain in most variations. Among these steps, immobilizing target molecules on a solid phase such as magnetic beads, glass coverslips, agarose-based resin, etc. is a common strategy that is used to enhance efficiency of the selection of aptamers for small molecules including drugs, toxins, antibiotics and molecular markers.30
When performing SELEX, the choice of nucleic acid library is the first of many steps that can be tailored to address the downstream application of the selected aptamer. In the first decade of implementing SELEX technology, RNA libraries were used more frequently than DNA libraries, as the general belief was more structural motifs could be generated from RNA.31 Despite RNA libraries having been wildly used in the early stage of SELEX, their stability and additional cost on extra modifications largely restricting wide applications of RNA aptamers. As a result, DNA libraries are being preferentially used in SELEX studies since 2008.32 In the initial library, ~1012 – 1015 nucleic acids containing a randomized region and two primer-binding sequences are exposed to the target. Through positive selection, the unbound nucleic acids are removed from the bound and those bound sequences are eluted for counter selection. During this process, structurally similar non-target compounds are introduced to incubate with the eluted bound sequences to effectively discriminate nonspecific nucleic acids. The recovered sequences are amplified by PCR (DNA SELEX) or reverse-transcription PCR (RNA SELEX). Enriched PCR products are applied in the next cycle of selection. To obtain the selected nucleic acids to display high affinity and specificity towards the target, iterative SELEX cycles are performed (Figure S1). Ultimately, the success of the conventional SELEX process depends heavily on the isolation of the bound nucleic acids from the unbound.
2.2. Improving Performance of Aptamers Through New Selection Strategies
The major disadvantages of conventional SELEX rest with tedious selection cycles, requirements for large quantities of reagents, or the selected aptamers display low affinity and specificity. On the basis of the fundamental steps of conventional SELEX, current SELEX technology has evolved significantly by integrating advancements achieved in disciplines such as synthetic chemistry, material science, engineering, molecular biology, and bioinformatics to improve efficiency of the SELEX procedure and enhance performance of the selected aptamers.
The limited chemical diversity originating from the finite combinations of nucleotides brings challenges for conventional SELEX. A selected aptamer normally comprises 10–40 nucleotides of 4 different nucleotide species.33 In contrast, the binding sites of an antibody consist of 110–130 amino acids, and each amino acid could be one of the 20 different amino acids.34 In view of the possible combinations, structure variations formed by aptamers are far from comparable to their proteinaceous counterpart antibodies. Perhaps the less chemical diversity of aptamers lowers the success rate of developing high performance aptamers.35 Improving the SELEX method typically focuses on four aspects: reduce the time for aptamer development, 36,37,38,39,40 incorporate novel designs and functions prior to the SELEX process for improving chemical diversity, 41, 42 increase the selection stringency,43, 44 and improve the process throughput of aptamers (Table S2). 45, 46, 47
Presumably, through increasing the interactions of DNA/RNA libraries with targets, higher affinity aptamers can be selected. To this end, various strategies have focused on endowing initial DNA/RNA libraries with more protein-like properties to enhance interactions with targets, since aptamers are lack of variety of physiochemical properties such as charge, hydrophobicity, pKa that proteins present.48 A novel evolutionary engineering method ExSELEX based on inducing hydrophobic interactions between DNA and targets was reported by Hirao and co-workers (Figure S2a).49 In this approach, randomized sequences comprising five bases: four natural bases and one artificial hydrophobic base, 7-(2-thienyl)imidazo[4,5-b]pyridine (Ds) were used as the improved library, from which a Ds-containing DNA aptamer was selected to display significantly higher affinity (Kd = 75 pM) comparing to the known DNA aptamer selected from natural DNA libraries. In addition to adding an extra artificial hydrophobic base in the initial library, base modification has also been conducted for initial library design by Gold et al (Figure S2b).50 In their report, DNA libraries incorporated one of four dUTPs modification at the 5-position was used as initial libraries for selecting modified aptamers. As a result, modified DNA SELEX was able to select aptamers against 13 “difficult” proteins whereas unmodified DNA SELEX failed.
When evaluating performance of the selected aptamers, specificity is another critical parameter. Because of the negatively charged phosphate backbone, aptamers may nonspecifically attract cationic molecules. Strategies using salt gradients, denaturing detergents, and cation-chelating additives have been implemented in the elution buffer to minimize nonspecific interactions.51–53 It is suspected that nonspecific binding of other molecules can also be contributed to interactions with the two primer-binding sequences that flank the randomized DNA/RNA sequences. To alleviate this, primer-free libraries were employed in some SELEX procedures, however, the complexity and cost by introducing restriction and ligation enzymes largely prevent researchers from pursuing this path.54–56 Along with minimizing nonspecific interactions, counter selection using structurally similar targets to incubate with initial nucleic acid libraries prior to/simultaneous with successive SELEX rounds is performed.57–59 For instance, a RNA aptamer that can distinguish mutated p53 against wild-type (WT) p53 was isolated via contrast SELEX.60 Positive selection and counter selection were performed simultaneously in a pool of the agarose beads and magnetic beads conjugated with WT and mutant proteins, respectively.
2.3. The Downstream Application of the Aptamer and Properties of the Target Analyte Should Drive Selection Strategy
To increase the probability of selecting suitable aptamers for analytical application, the downstream analytical device for incorporating the selected aptamers should be considered prior to perform the SELEX process. An example demonstrating that functionalities of the aptamer for the downstream application should be contemplated prior to SELEX is the selection of structure-switching aptamers for developing biosensors. Structure-switching aptamers provide a means to convert a binding event into a change in aptamer structure or conformation which can ultimately provide analytical readout using optical61–65 and electrochemical methods.66–71 To select aptamers able to induce detectable signal upon target binding, Xiao and coworkers developed an alternative selection strategy relying on binding-induced release of aptamers from duplexed nucleic acids. More specifically, the authors reported a generalizable approach for isolating small-molecule-binding aptamers with intrinsic dye-displacement functionality.72 In their approach, an aptamer against 3,4-methylenedioxypyrovalerone (MDPV) was isolated by displacing diethylthiatricarbocyanine (Cy7) that stacked in DNA three-way junctions (TWJs) from a randomized TWJs-structured DNA library pool, which resulted in a rapid change in absorbance due to the displacement of Cy7. Based on the small-molecule-binding aptamer, they successfully designed a colorimetric sensor for the detection of the small molecule MDPV.
3. Post-Selection Modifications as Strategy to Improve Performance of Aptamer in Analytical Device
As described above, the performance requirements of aptamers to be employed in analytical devices lie in the sensitivity and specificity of the aptamer-target interaction. While selection strategies continue to improve these features, there are also several strategies that can be employed post selection to improve the performance of aptamers for use in analytical devices. Key to the success of such strategies is the understanding of how interaction forces between aptamer and target affect binding. To this end, reliable analytical approaches, such as nuclear magnetic resonance spectroscopy (NMR) and X-ray crystallography and docking and molecular simulations, have been used to provide quantitative information about aptamer-target interactions.
3.1. Understanding Intermolecular Interactions in the Binding Domain as a Method for Improving Aptamer Performance
Formation of a specific and tightly bound aptamer-target complex represents a delicate balance, as it requires coordination of the following three participants: the binding buffer, the nucleic acid sequence, and the properties of the target analyte. The optimal binding buffer condition can be obtained by controlling the composition and concentration of monovalent and divalent salts, pH, and buffer identity. In the optimal binding buffer, secondary or tertiary structure motifs of the selected DNA/RNA aptamers can be promoted to interact with the target analytes including small molecules, macromolecules, cells, etc.73,74,83–85,75–82 However, the post selection use environment must be considered as often times solution conditions are not identical. In fact, this caveat was pointed out in a 2009 Annual Review in Analytical Chemistry article by Cho, Lee, and Ellington.86 The authors state “…knowledge of the sequence of an aptamer is often taken as an indication that only this sequence itself is necessary for experimental repetition. In fact, it is not only the sequence, but many other variables—such as buffer conditions, temperature, purification method, and conformational state—that affect the function of an aptamer. If an analytical researcher does not know or heed these important variables, then their experiments may be meaningless.”
An in-depth understanding of the intermolecular interactions between the binding domain of the selected aptamer and the target analyte is useful for improving aptamer performance in the downstream analytical device. If the binding domain of a selected aptamer is known, one can incorporate the binding domain into rational designs to improve aptamer performance. Tan and co-workers recently reported a smart strategy to improve aptamer specificity (Figure 1a).87 In their work, a structure-switchable aptamer (SW-Apt), achieved by conjugating pH-responsive i-motifs with tyrosine kinase-7 (PTK7)-binding aptamer sgc8, is capable of distinguishing identical target antigens on both tumor and healthy cells. At slightly acidic pH (tumor microenvironment), the i-motifs of the SW-Apt fold into quadruplex structures, enabling the recognition loops to bind receptors; whereas at physiological pH (healthy cells), the i-motifs of the SW-Apt are unstructured, preventing the recognition loops from binding to receptors. Furthermore, in another rational design method (Figure 1b),88 we introduced structural approaches to enhance the observed binding affinity and sensitivity of the aminoglycoside targeting RNA aptamer. Specifically we bimolecularly engineered several aptamers based on the parent RNA aptamer to create largest changes in aptamer conformation and thus signaling based on our sensor mechanism. Since the binding domain is known, by conserving the bases involved in the binding of tobramycin and destabilizing the secondary structure through deletion of bases in the parent aptamer, we demonstrated a semi-rational design strategy for improving the analytical performance.
Figure 1.
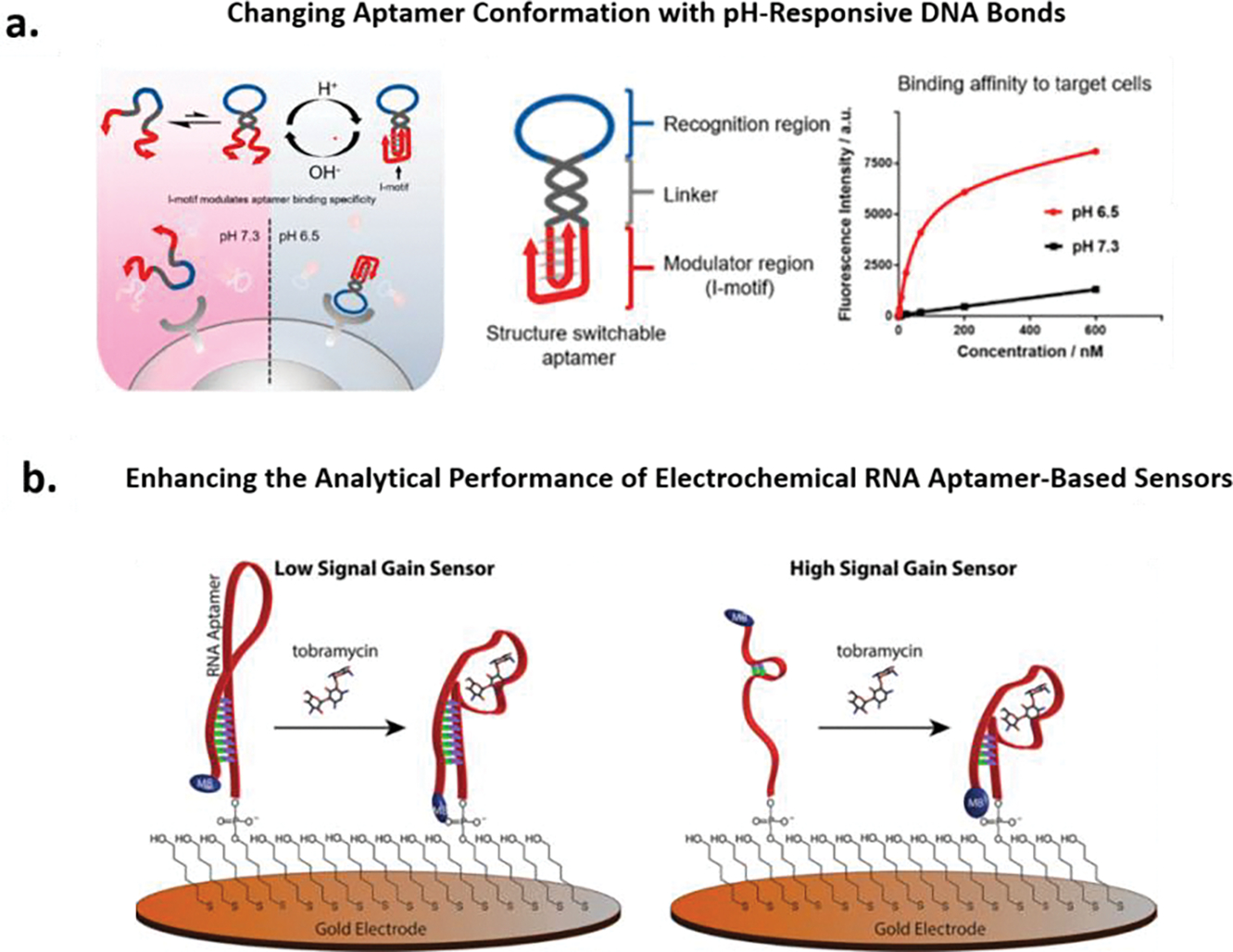
(a). Schematic representation of a structure-switchable aptamer (SW-Apt), achieved by conjugating pH-responsive i-motifs with tyrosine kinase-7 (PTK7)-binding aptamer sgc8. At acidic pH 6.5 (red), the i-motifs of the SW-Apt fold into quadruplex structures, enabling the recognition loops to bind receptors; whereas at physiological pH 7.3 (black), the i-motifs of the SW-Apt is unstructured, preventing the recognition loops from binding to receptors. Reproduced with permission from Ref 87. Copyright (2018) American Chemical Society. (b). Shown are the electrochemical structure-switching aptamer-based sensors for the detection of aminoglycoside antibiotics, low signal gain sensor and high signal gain sensor are shown on the left and right, respectively. High gain sensor was achieved by rationally engineering the secondary structure of the aptamer to create large conformation changes upon target binding. Reproduced with permission from Ref 88. Copyright (2014) American Chemical Society.
3.2. Semi-Rational Post-Selection Strategies to Improve Aptamer Performance Downstream
The analytical approaches introduced above have been used in post-selection strategies to provide quantitative information about aptamer-target interactions and the binding domain. Reportedly, ~400 publications have performed abovementioned three techniques, however, comparing with thousands of papers published on aptamer, characterization of the aptamer-target structures and binding mechanism is still lacking. Quantitative information about aptamer-target interactions and the binding domain could guide post-selection modifications to improve aptamer performance downstream.
In many cases, the selected full aptamers including primer regions, are not needed, as only the binding domain is essential for target recognition.89–97 Truncating the nonessential binding component of an aptamer may lead to improved specificity, sensitivity and cost-effectiveness of the oligonucleotide sequence, as nonspecific interactions could potentially be reduced.98–100 In an example of the benefits of truncation, Soontornworajit and co-workers designed a successful truncation strategy based on oligonucleotide hybridization combined with competitive antibody binding (Figure S3a).101 Tailored by the immunoassay results, which does not require knowledge of the binding site, the parent platelet derived growth factor (PDGF-BB) aptamer, comprising 86 nucleotides (nt) was reduced to truncated aptamers comprising 36–40 nt. The truncated aptamers displayed picomolar Kd values, among which, the 36 nt truncated aptamer displayed the highest affinity towards PDGF-BB, with a 150-fold increase compared with the parent aptamer. In another rational design reported by Xiao and co-workers (Figure S3b),102 structure-switching functionality was introduced to both the cocaine and ATP aptamer via exonuclease III (Exo III)-directed truncation. The parent cocaine aptamer pre-folds into a three-way-junction (TWJ) structure while the parent ATP aptamer pre-folds into a stem-loop structure. As Exo III-directed truncation halts four bases prior to the target-binding domain, the resulting truncated aptamers displayed a target-induced structure-switching functionality, which can be used in the downstream application of constructing structure-switching aptamer-based biosensors. The strength of this technique is that it does not require knowledge of the binding domain.
Similar to the post-selection strategy truncation, site-directed mutagenesis is a common method for identifying certain crucial nucleotides primarily involved in the aptamer-target interactions.103–105 Very recently, Willner and co-workers examined eight mutants of ATP-binding aptamers, where the thymine bases within the binding domain were substituted with cytosine bases (Figure S4a).106 Among the eight mutants, the ATP-aptamer mutant 7 displayed improved affinity (Kd = 15 ± 1 μM) over the parent ATPaptamer (Kd = 31 ± 3 μM), presumably due to more favorable hydrogen bonding and π−π interactions between the mutant ATP-binding aptamer and the target ATP. In addition to identifying crucial nucleotides on the aptamer to improve binding affinity, alternatively, Juncker and co-workers tuned binding affinity of aptamers by introducing single-base mismatches into complementary strands through an induced fit binding pathway (Figure S4b).107 In this pathway, a duplexed aptamer (DA) actively senses the ligand from the duplexed state. Subsequentially, target binding catalytically dissociates the DAs. To systematically and comprehensively profile DAs target-binding landscapes, aptamer-complementary element scanning (ACE-Scan) was used to study the binding kinetics of thousands of surface-assembled DAs. A combined 5′ and 3′ enantio heat map was obtained for identifying mutant DAs that displayed higher dissociation rates (koff) towards target binding. These two examples, however, typically require knowledge of the binding domain, as changes to this domain severely hinder binding.108
When the downstream application of selected aptamers is for ability to function in complex media, stability of the selected aptamers are major concerns, as nuclease degradation create practical barriers for the in vivo application of aptamers, particularly for RNA.109–114 Perhaps replacing the RNA sequence with a deoxy version is the simplest way one can envision to improve stability of RNA. Indeed, there was an attempt to increase the stability of RNA aptamer by employing the DNA version of the aptamer. Although it was claimed that the function of the RNA aptamer was retained in the DNA version,115 it was demonstrated later that the DNA dopamine aptamer responded to both dopamine and other structurally related catecholamine neurotransmitters.116 Given that simple replacement approach is not feasible for improving stability, various chemical modifications have been utilized to improve the stability of aptamers, among which, 3’-end capping with inverted thymidine has been shown to combat 3’-exonuclease in human serum.117 Other existing chemical modifications able to improve the stability include using LNA (locked nucleic acid) to substitute ribonucleotide, introducing 3’-biotin to the 3’-terminal of the aptamers,118–120 modifying the sugar rings with 2’-fluoro (2’-F) or 2’-amino (2’-NH2) ribose groups,121,122 replacing phosphodiester linkages of DNA with phosphorothioate analogs or triazole linkages,123–125 etc.
4. Analytical Application of Aptamers: Highlight the Advantages of using Aptamers as well as the Hurdles
Functioning as affinity reagents, both aptamers and antibodies have received great attention in diverse fields, including affinity isolation, biomarker discovery, food safety and environmental monitoring, and in particular clinical diagnostics. However, the analytical application of aptamers is still in its infancy while antibody-based methods dominate in clinical diagnostics. Nonetheless, since the invention of aptamers in the early 1990s, the field has witnessed significant advances in analytical methodologies that employ aptamers. In fact, we believe that aptamer-based methodologies can tackle challenges not accessible with antibody-based detection, particularly small molecule dynamics monitoring.
4.1. Illustrative Examples of Analytical Device That Outperform Antibody-Based Approach
As noted above, we believe that a particular promising application of aptamer-based analytical devices is the ability to monitor small molecule dynamics. For example, aptamer-based sensors capable of measuring real-time pharmacokinetic parameters of drugs can provide feedback information to ensure delivery of optimal drug dose for individual patients. Thus motivated, Soh and co-workers developed a microfluidic electrochemical detector for in vivo continuous monitoring (MEDIC) of doxorubicin (DOX) and kanamycin in human whole blood and in live rats for up to 4 h (Figure 2a).126 To achieve this detection, the authors developed a continuous-flow diffusion filter of the MEDIC device to protect the aptamer sensor surfaces from fouling in whole blood. Furthermore, the authors employed a kinetic differential measurement (KDM) to improve accuracy of real-time measurements by minimizing current drift (presumable from surface fouling) and enhancing signal-to-noise ratios. Expanding of this example, Plaxco and co-workers developed electrochemical aptamer-based (E-AB) sensors capable of real-time measurement of four drugs (i.e. doxorubicin, kanamycin, gentamicin, and tobramycin) in the bloodstream of awake, ambulatory animals (Figure 2b).127 Of note, the aptamer sequences employed are the same as those used in the MEDIC device. In contrast, however, to the complex continuous-flow diffusion filter adopted in the MEDIC invented by Soh and co-workers, the sensors were encased in a biocompatible polysulfone membrane with 0.2-μm pores to reduce fouling of the sensors placed in the jugular veins of anesthetized rats. As such, target molecules were able to reach the sensor surface while larger entities such as cells could not. The membrane-protected E-AB sensors achieved nanomolar precision and 3 s temporal resolution. It should be noted, the specificity afforded by the aptamer for DOX is limited because it also binds the analog daunomycin.128 In addition, the aminoglycosides used all bind to the same aptamer due to their structural similarity. These specificity limitations highlight the need for better selection techniques to be developed that can select aptamers capable of discriminating analogs or potentially metabolites of a target if needed. Ultimately, the sensor can only be as specific and sensitive as the aptamer that is used.
Figure 2.
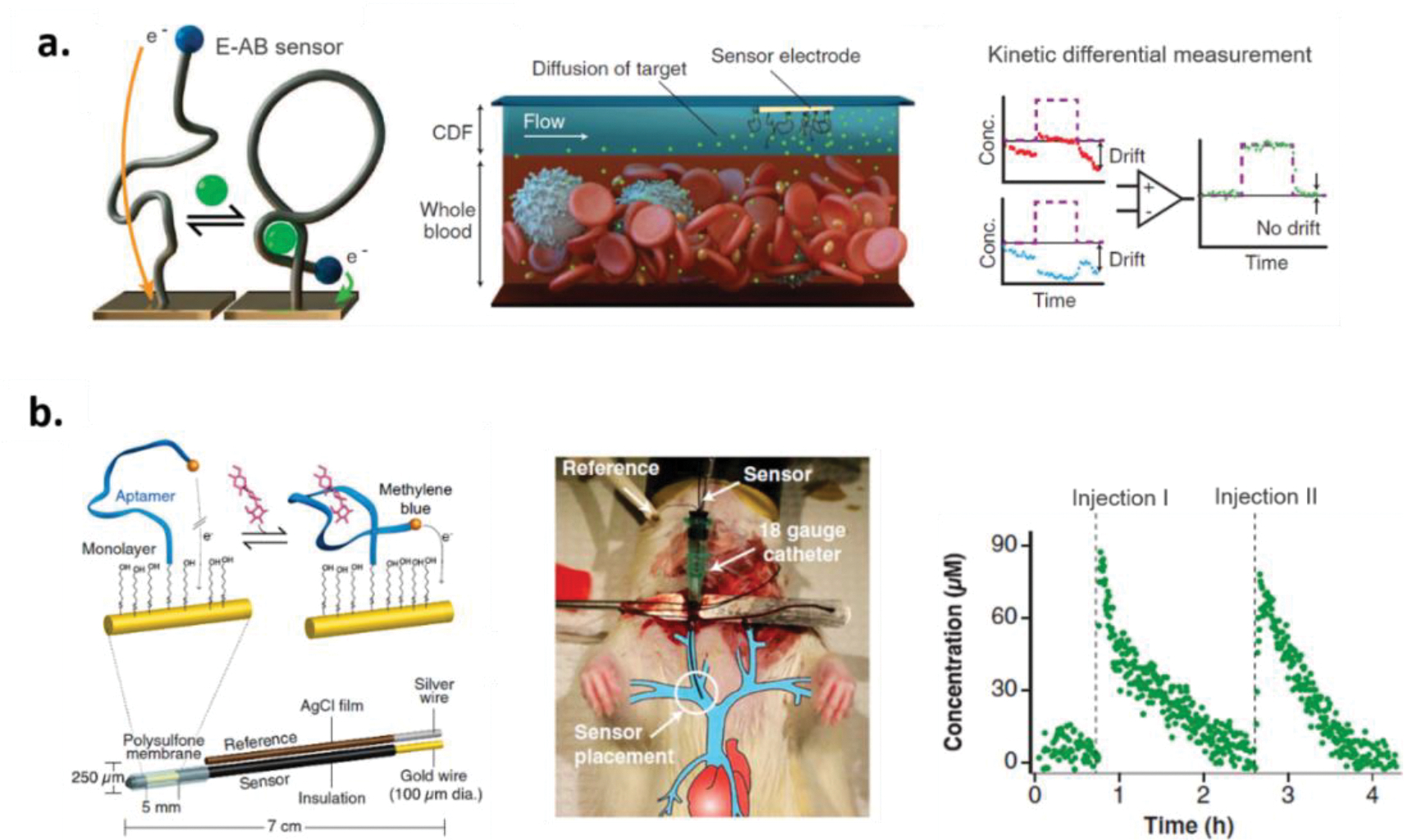
(a). Schematic representation of real-time measurement of doxorubicin and kanamycin in whole blood. The microfluidic electrochemical detector for in vivo continuous monitoring was developed by including an electrochemical aptamer-based sensor for the measurement and a continuous-flow diffusion filter to protect the aptamer probes from fouling. To minimize current drift and enhance signal-to-noise ratios, the kinetic differential measurement (KDM) was conduct for improving accuracy of real-time measurements. Reproduced with permission from Ref 126. Copyright (2013) The American Association for the Advancement of Science. (b). Schematic representation of real-time, continuous measurement of drugs in the bloodstream of awake, ambulatory animals. To protect the sensor from fouling, a biocompatible polysulfone membrane with 0.2-μm pores was used to encase the sensor. The resultant device was emplaced in one of the external jugulars of a rat using an 18-gauge catheter. Two serial injections of the antibiotic tobramycin in the blood of an anesthetized rat was demonstrated. Reproduced with permission from Ref 127. Copyright (2017) The National Academy of Sciences.
Apart from being employed in aptamer-based sensors for real-time measurements of drugs, RNA aptamers have found unique application as “light-up” RNAs, which mimic naturally existing green fluorescent proteins (GFP). The RNA aptamer 24–2 which binds the fluorophore 3,5- difluoro-4-hydroxybenzylidene imidazoline (DFHBI) was selected by Jaffrey and co-workers.129 Resembling enhanced GFP with fluorescence emission comparable to fluorescent proteins, the RNA-fluorophore complexes, termed Spinach, can be fused with structure-switching nucleic acids for in vivo imaging (Figure 3a). To improve the thermal stability and emission brightness of Spinach, systematic mutagenesis was conducted.130 When fused to the CGG repeat-containing RNA, the developed Spinach2 displayed improved fluorescence compared to the original Spinach (Figure 3b). Followed by Spinach and Spinach2, a 49-nt RNA aptamer, named Broccoli, was selected against the fluorophore (Z)-4-(3,5-difluoro-4-hydroxybenzylidene)-1,2-dimethyl-1Himidazol-5(4H)-one (DFHBI-1T) using a fluorescence-activated cell sorting (FACS)-based directed evolution approach (Figure 3c).131 With this direct cell-based selection, Broccoli demonstrates highly efficient cellular performance over Spinach2 for cell imaging, on account that Broccoli displays low magnesium dependence, higher thermostability, ability to fold without a tRNA scaffold, and shorter in size compared with 96-nt-long Spinach2. These recently developed “light-up” RNAs largely enable living-cell imaging for metabolites, proteins, and RNA, however, the living cells used in previous studies were mainly bacteria.132–134
Figure 3.
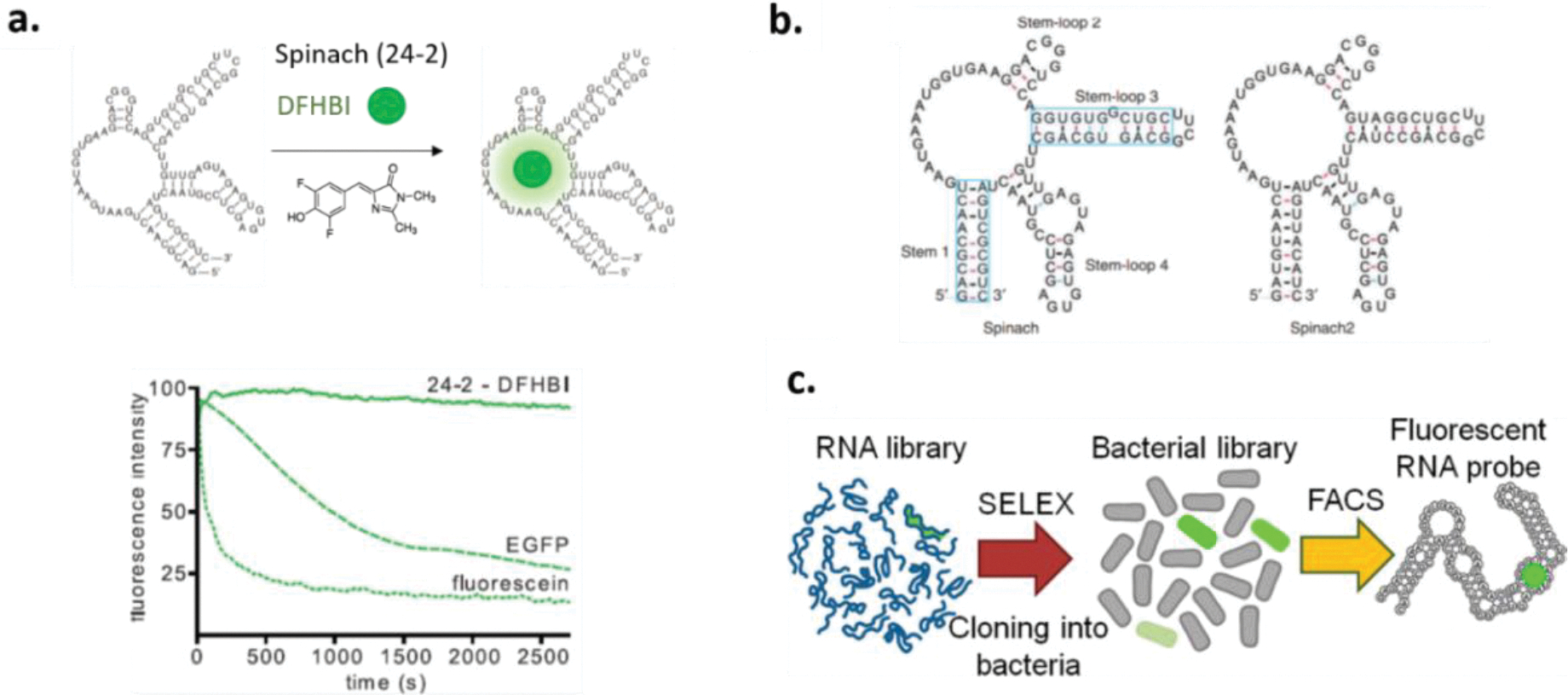
(a). Schematic representation of formation of the 24–2–DFHBI (3,5- difluoro-4-hydroxybenzylidene imidazolinone) complex, termed Spinach. Photobleaching curves for 24–2–DFHBI, enhanced green fluorescent protein (EGFP), and fluorescein demonstrates that Spi nach displays fluorescence emission comparable to fluorescent proteins. Reproduced with permission from Ref 129. Copyright (2011) The American Association for the Advancement of Science. (b). Schematic representation of secondary structure of Spinach and Spinach2. Systematic mutagenesis was conducted on Spinach stem 1 and stem-loop 3 (boxed) to generate Spinach2. Reproduced with permission from Ref 130. Copyright (2013) Springer Nature. (c). Shown is fluorescence-activated cell sorting (FACS)-based directed evolution approach for generating the Broccoli aptamer that displays highly efficient cellular performance. Reproduced with permission from Ref 131. Copyright (2014) American Chemical Society.
4.2. Reading Between the Lines – The Challenges of Aptamers in Analytical Field
While the above examples are excellent and demonstrate some of the potential promise of using aptamers in analytical chemistry, there still remain challenges that transcend the analytical transduction methodologies presented. For example, typically when an aptamer is examined in the lens of a specific analytical method or device, many times this is done for a proof-of-concept demonstration that the aptamer functions under specific conditions. However, critical readers may ask: “why don’t we see more analytical permeation of aptamers in the field?” or: “How do we move beyond proof of concept?” or: “Why do I keep reading about the same 4 aptamers?”
While contemplating the answer for such a question, inspecting the success of the most popular aptamer - thrombin aptamers might provide insight. The proof-of-concept analytical applications of thrombin aptamers have been demonstrated through various examples including designing thrombin biosensors involving various transducers,135–140 using duplexed aptamers based on thrombin aptamers for exploring the ligand binding dynamics,107,141,142 improving signal-to-background ratio and sensitivity of the thrombin aptamer by using split thrombin aptamer fragments,143–146 programming multiprotein nanoarrays with defined nanometer spacing and precision employing thrombin aptamers,147 constructing target-specific imaging platforms based on thrombin aptamers for in vivo imaging,148–150 etc. Given these across-the-board applications, we believe the secret of the success of the thrombin aptamer is two-fold: 1). thrombin aptamers are proven to work via a multidisciplinary array of methods; 2). the binding domain of thrombin aptamers and intermolecular forces between thrombin aptamers and the target are well characterized and the specificity is well documented.
According to the Aptagen online database (https://www.aptagen.com/aptamer-index), there are more than 500 aptamers selected to date. These aptamers are proven to work under specific conditions (e.g., buffer condition, temperature, etc.). Among them, it is highly possible that some of the selected aptamers are better aptamers display high specificity and affinity that can be applied in the multidisciplinary array of analytical device. However, the challenges of aptamers in analytical field lie in figuring out promising aptamers that can pass the test from multidisciplinary examination and concentrating the efforts of the field to rigorously characterize and further improve performance of the promising aptamers. It is up to the aptamer field to decide which are the promising aptamers worth being taken to the next level.
5. Conclusion
The aptamer community is going to celebrate 30 years of the SELEX technology. In retrospect over the past thirty years, advances in the field of aptamer demonstrates that aptamers have the potential to be powerful reagents for use in diverse areas within analytical chemistry as well as biotechnology, biomedicine, and molecular biology. Aptamers have the potential to provide additional advantages over their affinity counterparts, antibodies, as noted from the very beginning of their invention. It is our perspective that one area aptamers can shine in the future of analytical chemistry, when coupled with the right transduction mechanism, is in the specific monitoring of small molecule dynamics whether that will be in the fields of, for example, biomedicine, therapeutic monitoring, or chemistry of the brain applications. And while the analytical methodology employing aptamers continues to grow, we see a lag in the application of such affinity reagents beyond proof-of-concept examples. Our perspective highlights potential challenges in using aptamer which include challenges like poor specificity or sensitivity. Even some of the best illustrative examples of the analytical usage of aptamers rely on aptamers that exhibit limited specificity. On the contrary, there are shining examples of aptamers that display exceptional affinity (the theophylline aptamer151 comes to mind). This often-cited example, however, raises a question that is often encountered in this field – why are some aptamer sequences more prolific than others? This is most likely a result of the fact that the top five or so aptamers have been rigorously characterized by a wide range of researchers. This characterization ranges from structural biochemical characterization, careful analysis of binding specificity and the nature of the biomolecular interactions, and in some cases these aptamers have been subjected to rational post-selection modification for improved performance in analytical devices. Rigorous characterization, however, is not always complete. An example of such is the recent publication declaring the arsenic aptamer does not indeed specifically bind arsenic, even though it has been used in >20 publications describing analytical applications.152
In short, aptamers still hold a lot of promise for use as affinity reagents in the development of analytical devices and methods. These methods have advanced considerably, even demonstrating the ability to detect specific small molecules in the blood stream of a living animal. What will propel aptamers in their future role in analytical chemistry? The field as a whole needs to elevate more rigorously-characterized sequences. This evaluation is a multidisciplinary effort and is critical to the further success of aptamers.
Supplementary Material
6. References
- (1).Tuerk C; Gold L Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science (80-.). 1990, 249 (4968), 505–510. 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- (2).Ellington AD; Szostak JW In Vitro Selection of RNA Molecules That Bind Specific Ligands. Nature 1990, 346 (6287), 818–822. 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- (3).Cowperthwaite MC; Ellington AD Bioinformatic Analysis of the Contribution of Primer Sequences to Aptamer Structures. J. Mol. Evol. 2008, 67 (1), 95–102. 10.1007/s00239-008-9130-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Tok JB-H; Cho J; Rando RR RNA Aptamers That Specifically Bind to a 16S Ribosomal RNA Decoding Region Construct. Nucleic Acids Res. 2000, 28 (15), 2902–2910. 10.1093/nar/28.15.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Bock LC; Griffin LC; Latham JA; Vermaas EH; Toole JJ Selection of Single-Stranded DNA Molecules That Bind and Inhibit Human Thrombin. Nature 1992, 355, 564–566. 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- (6).Boiziau C; Dausse E; Yurchenko L; Toulmé JJ DNA Aptamers Selected against the HIV-1 Trans-Activation-Responsive RNA Element Form RNA-DNA Kissing Complexes. J. Biol. Chem. 1999, 274 (18), 12730–12737. 10.1074/jbc.274.18.12730. [DOI] [PubMed] [Google Scholar]
- (7).Tuerk C; MacDougal S; Gold L RNA Pseudoknots That Inhibit Human Immunodeficiency Virus Type 1 Reverse Transcriptase. Proc. Natl. Acad. Sci. 2006, 89 (15), 6988–6992. 10.1073/pnas.89.15.6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Riccitelli NJ; Lupták A Computational Discovery of Folded RNA Domains in Genomes and in Vitro Selected Libraries. Methods 2010, 52 (2), 133–140. 10.1016/j.ymeth.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Mairal T; Cengiz Özalp V; Lozano Sánchez P; Mir M; Katakis I; O’Sullivan CK Aptamers: Molecular Tools for Analytical Applications. Anal. Bioanal. Chem. 2008, 390 (4), 989–1007. 10.1007/s00216-007-1346-4. [DOI] [PubMed] [Google Scholar]
- (10).Troisi R; Napolitano V; Spiridonova V; Krauss IR; Sica F Several Structural Motifs Cooperate in Determining the Highly Effective Anti-Thrombin Activity of NU172 Aptamer. Nucleic Acids Res. 2018, 46 (22), 12177–12185. 10.1093/nar/gky990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Lang PT; Pettersen EF; Case DA; Mukherjee S; Thomas V; Kuntz ID; Brozell SR; Rizzo RC; James TL; Meng EC DOCK 6: Combining Techniques to Model RNA-Small Molecule Complexes. RNA 2009, 15 (6), 1219–1230. 10.1261/rna.1563609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Unruh JR; Gokulrangan G; Lushington GH; Johnson CK; Wilson GS Orientational Dynamics and Dye-DNA Interactions in a Dye-Labeled DNA Aptamer. Biophys. J. 2005, 88 (5), 3455–3465. 10.1529/biophysj.104.054148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Lipman NS; Jackson LR; Trudel LJ; Weis-Garcia F Monoclonal Versus Polyclonal Antibodies: Distinguishing Characteristics, Applications, and Information Resources. ILAR J. 2005, 46 (3), 258–268. 10.1093/ilar.46.3.258. [DOI] [PubMed] [Google Scholar]
- (14).Chow CK; Allan BW; Chai Q; Atwell S; Lu J Therapeutic Antibody Engineering to Improve Viscosity and Phase Separation Guided by Crystal Structure. Mol. Pharm. 2016, 13 (3), 915–923. 10.1021/acs.molpharmaceut.5b00817. [DOI] [PubMed] [Google Scholar]
- (15).Lee HJ; Pardridge WM Monoclonal Antibody Radiopharmaceuticals: Cationization, Pegylation, Radiometal Chelation, Pharmacokinetics, and Tumor Imaging. Bioconjug. Chem. 2003, 14 (3), 546–553. 10.1021/bc0256648. [DOI] [PubMed] [Google Scholar]
- (16).Pomin VH; Mulloy B Glycosaminoglycans and Proteoglycans. Pharmaceuticals 2018, 11 (27), 1–9. 10.3390/ph11010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Kizer M; Li P; Cress BF; Lin L; Jing TT; Zhang X; Xia K; Linhardt RJ; Wang X RNA Aptamers with Specificity for Heparosan and Chondroitin Glycosaminoglycans. ACS Omega 2018, 3 (10), 13667–13675. 10.1021/acsomega.8b01853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Santos-Cancel M; Simpson LW; Leach JB; White RJ Direct, Real-Time Detection of Adenosine Triphosphate Release from Astrocytes in Three-Dimensional Culture Using an Integrated Electrochemical Aptamer-Based Sensor. ACS Chem. Neurosci. 2019, 10 (4), 2070–2079. 10.1021/acschemneuro.9b00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Borman RP; Wang Y; Nguyen MD; Ganesana M; Lee ST; Venton BJ Automated Algorithm for Detection of Transient Adenosine Release. ACS Chem. Neurosci. 2017, 8 (2), 386–393. 10.1021/acschemneuro.6b00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Mariottini D; Idili A; Nijenhuis MAD; De Greef TFA; Ricci F DNA-Based Nanodevices Controlled by Purely Entropic Linker Domains. J. Am. Chem. Soc. 2018, 140 (44), 14725–14734. 10.1021/jacs.8b07640. [DOI] [PubMed] [Google Scholar]
- (21).Swensen JS; Xiao Y; Ferguson BS; Lubin AA; Lai RY; Heeger AJ; Plaxco KW; Soh HT Continuous, Real-Time Monitoring of Cocaine in Undiluted Blood Serum via a Microfluidic, Electrochemical Aptamer-Based Sensor. J. Am. Chem. Soc. 2009, 131 (12), 4262–4266. 10.1021/ja806531z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Wang L; Musile G; McCord BR An Aptamer-Based Paper Microfluidic Device for the Colorimetric Determination of Cocaine. Electrophoresis 2018, 39 (3), 470–475. 10.1002/elps.201700254. [DOI] [PubMed] [Google Scholar]
- (23).Liu Y; Yan K; Zhang J Graphitic Carbon Nitride Sensitized with CdS Quantum Dots for Visible-Light-Driven Photoelectrochemical Aptasensing of Tetracycline. ACS Appl. Mater. Interfaces 2016, 8 (42), 28255–28264. 10.1021/acsami.5b08275. [DOI] [PubMed] [Google Scholar]
- (24).Jalalian SH; Karimabadi N; Ramezani M; Abnous K; Taghdisi SM Electrochemical and Optical Aptamer-Based Sensors for Detection of Tetracyclines. Trends Food Sci. Technol. 2018, 73, 45–57. 10.1016/j.tifs.2018.01.009. [DOI] [Google Scholar]
- (25).Kaur H Recent Developments in Cell-SELEX Technology for Aptamer Selection. Biochim. Biophys. Acta - Gen. Subj 2018, 1862 (10), 2323–2329. 10.1016/j.bbagen.2018.07.029. [DOI] [PubMed] [Google Scholar]
- (26).Akki SU; Werth CJ Critical Review: DNA Aptasensors, Are They Ready for Monitoring Organic Pollutants in Natural and Treated Water Sources? Environ. Sci. Technol. 2018, 52 (16), 8989–9007. 10.1021/acs.est.8b00558. [DOI] [PubMed] [Google Scholar]
- (27).Li F; Yu Z; Han X; Lai RY Electrochemical Aptamer-Based Sensors for Food and Water Analysis: A Review. Anal. Chim. Acta 2019, 1051, 1–23. 10.1016/j.aca.2018.10.058. [DOI] [PubMed] [Google Scholar]
- (28).Yüce M; Ullah N; Budak H Trends in Aptamer Selection Methods and Applications. Analyst 2015, 140 (16), 5379–5399. 10.1039/c5an00954e. [DOI] [PubMed] [Google Scholar]
- (29).Ruscito A; DeRosa MC Small-Molecule Binding Aptamers: Selection Strategies, Characterization, and Applications. Front. Chem 2016, 4, 1–14. 10.3389/fchem.2016.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Ozer A; Pagano JM; Lis JT New Technologies Provide Quantum Changes in the Scale, Speed, and Success of SELEX Methods and Aptamer Characterization. Mol. Ther. - Nucleic Acids 2014, 3, e183. 10.1038/mtna.2014.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Walter Gilbert. Origin of Life: The RNA World. Nature 1986, 319, 618. 10.1038/319618a0. [DOI] [Google Scholar]
- (32).Vorobyeva MA; Davydova AS; Vorobjev PE; Pyshnyi DV; Venyaminova AG Key Aspects of Nucleic Acid Library Design for in Vitro Selection. Int. J. Mol. Sci. 2018, 19 (2). 10.3390/ijms19020470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Ali MH; Elsherbiny ME; Emara M Updates on Aptamer Research. Int. J. Mol. Sci. 2019, 20 (10), 1–23. 10.3390/ijms20102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Schroeder Harry W. Jr and Lisa Cavacini P Structure and Function of Immunoglobulins. J Allergy Clin Immunolallergy Clin Immunol 2010, 125 (2), S41–S52. 10.1016/j.jaci.2009.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Mercier MC; Dontenwill M; Choulier L Selection of Nucleic Acid Aptamers Targeting Tumor Cell-Surface Protein Biomarkers. Cancers (Basel). 2017, 9 (69), 1–33. 10.3390/cancers9060069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Yang J; Bowser MT Capillary Electrophoresis-SELEX Selection of Catalytic DNA Aptamers for a Small-Molecule Porphyrin Target. Anal. Chem. 2013, 85 (3), 1525–1530. 10.1021/ac302721j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Spiga FM; Maietta P; Guiducci C More DNA-Aptamers for Small Drugs: A Capture-SELEX Coupled with Surface Plasmon Resonance and High-Throughput Sequencing. ACS Comb. Sci. 2015, 17 (5), 326–333. 10.1021/acscombsci.5b00023. [DOI] [PubMed] [Google Scholar]
- (38).Stoltenburg R; Reinemann C; Strehlitz B FluMag-SELEX as an Advantageous Method for DNA Aptamer Selection. Anal. Bioanal. Chem. 2005, 383 (1), 83–91. 10.1007/s00216-005-3388-9. [DOI] [PubMed] [Google Scholar]
- (39).Oh SS; Ahmad KM; Cho M; Kim S; Xiao Y; Soh HT Improving Aptamer Selection Efficiency through Volume Dilution, Magnetic Concentration, and Continuous Washing in Microfluidic Channels. Anal. Chem. 2011, 83 (17), 6883–6889. 10.1021/ac201269f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Tsao S-M; Lai J-C; Horng H-E; Liu T-C; Hong C-Y Generation of Aptamers from A Primer-Free Randomized SsDNA Library Using Magnetic-Assisted Rapid Aptamer Selection. Sci. Rep. 2017, 7 (1), 45478. 10.1038/srep45478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).King DJ; Ventura DA; Brasier AR; Gorenstein DG Novel Combinatorial Selection of Phosphorothioate Oligonucleotide Aptamers †. Biochemistry 1998, 37 (47), 16489–16493. 10.1021/bi981780f. [DOI] [PubMed] [Google Scholar]
- (42).He W; Elizondo-Riojas M-A; Li X; Lokesh GLR; Somasunderam A; Thiviyanathan V; Volk DE; Durland RH; Englehardt J; Cavasotto CN; et al. X-Aptamers: A Bead-Based Selection Method for Random Incorporation of Druglike Moieties onto Next-Generation Aptamers for Enhanced Binding. Biochemistry 2012, 51 (42), 8321–8323. 10.1021/bi300471d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Liu Y; Wang C; Li F; Shen S; Tyrrell DLJ; Le XC; Li X-F Blotted on a Membrane. Anal. Chem. 2012, 84 (18), 7603–7606. 10.1021/ac302047e. [DOI] [PubMed] [Google Scholar]
- (44).Riley KR; Gagliano J; Xiao J; Libby K; Saito S; Yu G; Cubicciotti R; Macosko J; Colyer CL; Guthold M; et al. Combining Capillary Electrophoresis and Next-Generation Sequencing for Aptamer Selection. Anal Bioanal Chem 2015, 407 (6), 1527–1532. 10.1007/s00216-014-8427-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Lerga TM; Jauset-Rubio M; Skouridou V; Bashammakh AS; El-Shahawi MS; Alyoubi AO; O’Sullivan CK High Affinity Aptamer for the Detection of the Biogenic Amine Histamine. Anal. Chem. 2019, 91 (11), 7104–7111. 10.1021/acs.analchem.9b00075. [DOI] [PubMed] [Google Scholar]
- (46).Cho M; Xiao Y; Nie J; Stewart R; Csordas AT; Oh SS; Thomson JA; Soh HT Quantitative Selection of DNA Aptamers through Microfluidic Selection and High-Throughput Sequencing. Proc. Natl. Acad. Sci. 2010, 107 (35), 15373–15378. 10.1073/pnas.1009331107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Rabal O; Pastor F; Villanueva H; Soldevilla MM; Hervas-Stubbs S; Oyarzabal J In Silico Aptamer Docking Studies: From a Retrospective Validation to a Prospective Case Study’TIM3 Aptamers Binding. Mol. Ther. - Nucleic Acids 2016, 5, e376. 10.1038/mtna.2016.84. [DOI] [PubMed] [Google Scholar]
- (48).Wang T; Chen C; Larcher LM; Barrero RA; Veedu RN Three Decades of Nucleic Acid Aptamer Technologies: Lessons Learned, Progress and Opportunities on Aptamer Development. Biotechnol. Adv. 2019, 37 (1), 28–50. 10.1016/j.biotechadv.2018.11.001. [DOI] [PubMed] [Google Scholar]
- (49).Matsunaga KI; Kimoto M; Hirao I High-Affinity DNA Aptamer Generation Targeting von Willebrand Factor A1-Domain by Genetic Alphabet Expansion for Systematic Evolution of Ligands by Exponential Enrichment Using Two Types of Libraries Composed of Five Different Bases. J. Am. Chem. Soc. 2017, 139 (1), 324–334. 10.1021/jacs.6b10767. [DOI] [PubMed] [Google Scholar]
- (50).Gold L; Ayers D; Bertino J; Bock C; Bock A; Brody EN; Carter J; Dalby AB; Eaton BE; Fitzwater T; et al. Aptamer-Based Multiplexed Proteomic Technology for Biomarker Discovery. PLoS One 2010, 5 (12), e15004. 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Arnold S; Pampalakis G; Kantiotou K; Silva D; Cortez C; Missailidis S; Sotiropoulou G One Round of SELEX for the Generation of DNA Aptamers Directed against KLK6. Biol. Chem. 2012, 393 (5), 343–353. 10.1515/hsz-2011-0253. [DOI] [PubMed] [Google Scholar]
- (52).Kim K; Lee S; Ryu S; Han D Efficient Isolation and Elution of Cellular Proteins Using Aptamer-Mediated Protein Precipitation Assay. Biochem. Biophys. Res. Commun. 2014, 448 (1), 114–119. 10.1016/j.bbrc.2014.04.086. [DOI] [PubMed] [Google Scholar]
- (53).Rockey WM; Huang L; Kloepping KC; Baumhover NJ; Giangrande PH; Schultz MK Synthesis and Radiolabeling of Chelator-RNA Aptamer Bioconjugates with Copper-64 for Targeted Molecular Imaging. Bioorganic Med. Chem. 2011, 19 (13), 4080–4090. 10.1016/j.bmc.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Pan W; Xin P; Clawson GA Minimal Primer and Primer-Free SELEX Protocols for Selection of Aptamers from Random DNA Libraries. Biotechniques 2008, 44 (3), 351–360. 10.2144/000112689. [DOI] [PubMed] [Google Scholar]
- (55).Pan W; Clawson GA The Shorter the Better: Reducing Fixed Primer Regions of Oligonucleotide Libraries for Aptamer Selection. Molecules 2009, 14 (4), 1353–1369. 10.3390/molecules14041353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Jarosch F; Buchner K; Klussmann S In Vitro Selection Using a Dual RNA Library That Allows Primerless Selection. Nucleic Acids Res. 2006, 34 (12), e86. 10.1093/nar/gkl463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Latulippe DR; Szeto K; Ozer A; Duarte FM; Kelly CV; Pagano JM; White BS; Shalloway D; Lis JT; Craighead HG Multiplexed Microcolumn-Based Process for Efficient Selection of RNA Aptamers. Anal. Chem. 2013, 85 (6), 3417–3424. 10.1021/ac400105e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Tan Y; Guo Q; Xie Q; Wang K; Yuan B; Zhou Y; Liu J; Huang J; He X; Yang X; et al. Single-Walled Carbon Nanotubes (SWCNTs)-Assisted Cell-Systematic Evolution of Ligands by Exponential Enrichment (Cell-SELEX) for Improving Screening Efficiency. Anal. Chem. 2014, 86 (19), 9466–9472. 10.1021/ac502166b. [DOI] [PubMed] [Google Scholar]
- (59).Valenzano S; De Girolamo A; DeRosa MC; McKeague M; Schena R; Catucci L; Pascale M Screening and Identification of DNA Aptamers to Tyramine Using in Vitro Selection and High-Throughput Sequencing. ACS Comb. Sci. 2016, 18 (6), 302–313. 10.1021/acscombsci.5b00163. [DOI] [PubMed] [Google Scholar]
- (60).Chen L; Rashid F; Shah A; Awan HM; Wu M; Liu A; Wang J; Zhu T; Luo Z; Shan G The Isolation of an RNA Aptamer Targeting to P53 Protein with Single Amino Acid Mutation. Proc. Natl. Acad. Sci. 2015, 112 (32), 10002–10007. 10.1073/pnas.1502159112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Feagin TA; Maganzini N; Soh HT Strategies for Creating Structure-Switching Aptamers. ACS Sensors 2018, 3 (9), 1611–1615. 10.1021/acssensors.8b00516. [DOI] [PubMed] [Google Scholar]
- (62).Furuhata Y; Kobayashi M; Maruyama R; Sato Y; Makino K; Michiue T; Yui H; Nishizawa S; Yoshimoto K Programmable RNA Detection with a Fluorescent RNA Aptamer Using Optimized Three-Way Junction Formation. RNA 2019, 25 (5), 590–599. 10.1261/rna.069062.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Wang S; Zhang Y; Pang G; Zhang Y; Guo S Tuning the Aggregation/Disaggregation Behavior of Graphene Quantum Dots by Structure-Switching Aptamer for High-Sensitivity Fluorescent Ochratoxin A Sensor. Anal. Chem. 2017, 89 (3), 1704–1709. 10.1021/acs.analchem.6b03913. [DOI] [PubMed] [Google Scholar]
- (64).Tan Z; Feagin TA; Heemstra JM Temporal Control of Aptamer Biosensors Using Covalent Self-Caging to Shift Equilibrium. J. Am. Chem. Soc. 2016, 138 (20), 6328–6331. 10.1021/jacs.6b00934. [DOI] [PubMed] [Google Scholar]
- (65).Shi Y; Dai H; Sun Y; Hu J; Ni P; Li Z Fluorescent Sensing of Cocaine Based on a Structure Switching Aptamer, Gold Nanoparticles and Graphene Oxide. Analyst 2013, 138 (23), 7152–7156. 10.1039/c3an00897e. [DOI] [PubMed] [Google Scholar]
- (66).Li H; Arroyo-Currás N; Kang D; Ricci F; Plaxco KW Dual-Reporter Drift Correction To Enhance the Performance of Electrochemical Aptamer-Based Sensors in Whole Blood. J. Am. Chem. Soc. 2016, 138 (49), 15809–15812. 10.1021/jacs.6b08671. [DOI] [PubMed] [Google Scholar]
- (67).Lai RY; Plaxco KW; Heeger AJ Aptamer-Based Electrochemical Detection of Picomolar Platelet-Derived Growth Factor Directly in Blood Serum. Anal. Chem. 2007, 79 (1), 229–233. 10.1021/ac061592s. [DOI] [PubMed] [Google Scholar]
- (68).Wu Y; Midinov B; White RJ Electrochemical Aptamer-Based Sensor for Real-Time Monitoring of Insulin. ACS Sensors 2019, 4 (2), 498–503. 10.1021/acssensors.8b01573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).White RJ; Plaxco KW Exploiting Binding-Induced Changes in Probe Flexibility for the Optimization of Electrochemical Biosensors. Anal. Chem. 2010, 82 (1), 73–76. 10.1021/ac902595f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).White RJ; Rowe AA; Plaxco KW Re-Engineering Aptamers to Support Reagentless, Self-Reporting Electrochemical Sensors. Analyst 2010, 135 (3), 589–594. 10.1039/b921253a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Porchetta A; Vallée-Bélisle A; Plaxco KW; Ricci F Using Distal-Site Mutations and Allosteric Inhibition to Tune, Extend, and Narrow the Useful Dynamic Range of Aptamer-Based Sensors. J. Am. Chem. Soc. 2012, 134 (51), 20601–20604. 10.1021/ja310585e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Yu H; Yang W; Alkhamis O; Canoura J; Yang K-A; Xiao Y In Vitro Isolation of Small-Molecule-Binding Aptamers with Intrinsic Dye-Displacement Functionality. Nucleic Acids Res. 2018, 46 (8), e43. 10.1093/nar/gky026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Pinto A; Lennarz S; Rodrigues-Correia A; Heckel A; O’Sullivan CK; Mayer G Functional Detection of Proteins by Caged Aptamers. ACS Chem. Biol. 2012, 7 (2), 360–366. 10.1021/cb2003835. [DOI] [PubMed] [Google Scholar]
- (74).Soontornworajit B; Zhou J; Zhang Z; Wang Y Aptamer-Functionalized in Situ Injectable Hydrogel for Controlled Protein Release. Biomacromolecules 2010, 11 (10), 2724–2730. 10.1021/bm100774t. [DOI] [PubMed] [Google Scholar]
- (75).Ageely EA; Kartje ZJ; Rohilla KJ; Barkau CL; Gagnon KT Quadruplex-Flanking Stem Structures Modulate the Stability and Metal Ion Preferences of RNA Mimics of GFP. ACS Chem. Biol. 2016, 11 (9), 2398–2406. 10.1021/acschembio.6b00047. [DOI] [PubMed] [Google Scholar]
- (76).Bozza M; Sheardy RD; Dilone E; Scypinski S; Galazka M Characterization of the Secondary Structure and Stability of an RNA Aptamer That Binds Vascular Endothelial Growth Factor. Biochemistry 2006, 45 (24), 7639–7643. 10.1021/bi0521442. [DOI] [PubMed] [Google Scholar]
- (77).Mayne L; Lin CY; Christie SDR; Siwy ZS; Platt M The Design and Characterization of Multifunctional Aptamer Nanopore Sensors. ACS Nano 2018, 12 (5), 4844–4852. 10.1021/acsnano.8b01583. [DOI] [PubMed] [Google Scholar]
- (78).Nakatsuka N; Abendroth JM; Cheung KM; Xu X; Zhao C; Zhu B; Rim YS; Yang Y; Weiss PS; Andrews AM; et al. Aptamer-Field-Effect Transistors Overcome Debye Length Limitations for Small-Molecule Sensing. Science (80-.). 2018, 362 (6412), 319–324. 10.1126/science.aao6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Sachan A; Ilgu M; Kempema A; Kraus GA; Nilsen-Hamilton M Specificity and Ligand Affinities of the Cocaine Aptamer: Impact of Structural Features and Physiological NaCl. Anal. Chem. 2016, 88 (15), 7715–7723. 10.1021/acs.analchem.6b01633. [DOI] [PubMed] [Google Scholar]
- (80).Feng J; Walter NG; Brooks CL Cooperative and Directional Folding of the PreQ1 Riboswitch Aptamer Domain. J. Am. Chem. Soc. 2011, 133 (12), 4196–4199. 10.1021/ja110411m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Wolter AC; Weickhmann AK; Nasiri AH; Hantke K; Ohlenschläger O; Wunderlich CH; Kreutz C; Duchardt-Ferner E; Wöhnert J A Stably Protonated Adenine Nucleotide with a Highly Shifted PKaValue Stabilizes the Tertiary Structure of a GTP-Binding RNA Aptamer. Angew. Chemie - Int. Ed. 2017, 56 (1), 401–404. 10.1002/anie.201609184. [DOI] [PubMed] [Google Scholar]
- (82).Haller A; Altman RB; Soulière MF; Blanchard SC; Micura R Folding and Ligand Recognition of the TPP Riboswitch Aptamer at Single-Molecule Resolution. Proc. Natl. Acad. Sci. 2013, 110 (11), 4188–4193. 10.1073/pnas.1218062110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Förster U; Weigand JE; Trojanowski P; Suess B; Wachtveitl J Conformational Dynamics of the Tetracycline-Binding Aptamer. Nucleic Acids Res. 2012, 40 (4), 1807–1817. 10.1093/nar/gkr835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Buck J; Wacker A; Warkentin E; Wöhnert J; Wirmer-Bartoschek J; Schwalbe H Influence of Ground-State Structure and Mg2+ Binding on Folding Kinetics of the Guanine-Sensing Riboswitch Aptamer Domain. Nucleic Acids Res. 2011, 39 (22), 9768–9778. 10.1093/nar/gkr664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Wurster SE; Bida JP; Her YF; Maher LJ Characterization of Anti-NF-ΚB RNA Aptamer-Binding Specificity in Vitro and in the Yeast Three-Hybrid System. Nucleic Acids Res. 2009, 37 (18), 6214–6224. 10.1093/nar/gkp670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Cho EJ; Lee J-W; Ellington AD Applications of Aptamers as Sensors. Annu. Rev. Anal. Chem. 2009, 2 (1), 241–264. 10.1146/annurev.anchem.1.031207.112851. [DOI] [PubMed] [Google Scholar]
- (87).Li L; Jiang Y; Cui C; Yang Y; Zhang P; Stewart K; Pan X; Li X; Yang L; Qiu L; et al. Modulating Aptamer Specificity with PH-Responsive DNA Bonds. J. Am. Chem. Soc. 2018, 140 (41), 13335–13339. 10.1021/jacs.8b08047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Schoukroun-Barnes LR; Wagan S; White RJ Enhancing the Analytical Performance of Electrochemical RNA Aptamer-Based Sensors for Sensitive Detection of Aminoglycoside Antibiotics. Anal. Chem. 2014, 86 (2), 1131–1137. 10.1021/ac4029054. [DOI] [PubMed] [Google Scholar]
- (89).Alsager OA; Kumar S; Zhu B; Travas-Sejdic J; McNatty KP; Hodgkiss JM Ultrasensitive Colorimetric Detection of 17-Estradiol: The Effect of Shortening Dna Aptamer Sequences. Anal. Chem. 2015, 87 (8), 4201–4209. 10.1021/acs.analchem.5b00335. [DOI] [PubMed] [Google Scholar]
- (90).Gu C; Xiang Y; Guo H; Shi H Label-Free Fluorescence Detection of Melamine with a Truncated Aptamer. Analyst 2016, 141 (14), 4511–4517. 10.1039/c6an00537c. [DOI] [PubMed] [Google Scholar]
- (91).Chinnappan R; AlAmer S; Eissa S; Rahamn AA; Abu Salah KM; Zourob M Fluorometric Graphene Oxide-Based Detection of Salmonella Enteritis Using a Truncated DNA Aptamer. Microchim. Acta 2018, 185 (1). 10.1007/s00604-017-2601-9. [DOI] [PubMed] [Google Scholar]
- (92).Lee EH; Lim HJ; Lee SD; Son A Highly Sensitive Detection of Bisphenol A by NanoAptamer Assay with Truncated Aptamer. ACS Appl. Mater. Interfaces 2017, 9 (17), 14889–14898. 10.1021/acsami.7b02377. [DOI] [PubMed] [Google Scholar]
- (93).Bhamidipati M; Cho HY; Lee KB; Fabris L SERS-Based Quantification of Biomarker Expression at the Single Cell Level Enabled by Gold Nanostars and Truncated Aptamers. Bioconjug. Chem. 2018, 29 (9), 2970–2981. 10.1021/acs.bioconjchem.8b00397. [DOI] [PubMed] [Google Scholar]
- (94).Dhiman A; Anand A; Malhotra A; Khan E; Santra V; Kumar A; Sharma TK Rational Truncation of Aptamer for Cross-Species Application to Detect Krait Envenomation. Sci. Rep. 2018, 8 (1), 1–8. 10.1038/s41598-018-35985-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Sun Y; Duan N; Ma P; Liang Y; Zhu X; Wang Z Colorimetric Aptasensor Based on Truncated Aptamer and Trivalent DNAzyme for Vibrio Parahemolyticus Determination. J. Agric. Food Chem. 2019, 67 (8), 2313–2320. 10.1021/acs.jafc.8b06893. [DOI] [PubMed] [Google Scholar]
- (96).Orava EW; Jarvik N; Shek YL; Sidhu SS; Gariépy J A Short DNA Aptamer That Recognizes TNFα and Blocks Its Activity in Vitro. ACS Chem. Biol. 2013, 8 (1), 170–178. 10.1021/cb3003557. [DOI] [PubMed] [Google Scholar]
- (97).Zhang H; Li XF; Le XC Tunable Aptamer Capillary Electrophoresis and Its Application to Protein Analysis. J. Am. Chem. Soc. 2008, 130 (1), 34–35. 10.1021/ja0778747. [DOI] [PubMed] [Google Scholar]
- (98).Wang X; Gao X; He J; Hu X; Li Y; Li X; Fan L; Yu HZ Systematic Truncating of Aptamers to Create High-Performance Graphene Oxide (GO)-Based Aptasensors for the Multiplex Detection of Mycotoxins. Analyst 2019, 144 (12), 3826–3835. 10.1039/c9an00624a. [DOI] [PubMed] [Google Scholar]
- (99).Li X; Zhang W; Liu L; Zhu Z; Ouyang G; An Y; Zhao C; Yang CJ In Vitro Selection of DNA Aptamers for Metastatic Breast Cancer Cell Recognition and Tissue Imaging. Anal. Chem. 2014, 86 (13), 6596–6603. 10.1021/ac501205q. [DOI] [PubMed] [Google Scholar]
- (100).Macdonald J; Houghton P; Xiang D; Duan W; Shigdar S Truncation and Mutation of a Transferrin Receptor Aptamer Enhances Binding Affinity. Nucleic Acid Ther. 2016, 26 (6), 348–354. 10.1089/nat.2015.0585. [DOI] [PubMed] [Google Scholar]
- (101).Vu CQ; Rotkrua P; Tantirungrotechai Y; Soontornworajit B Oligonucleotide Hybridization Combined with Competitive Antibody Binding for the Truncation of a High-Affinity Aptamer. ACS Comb. Sci. 2017, 19 (10), 609–617. 10.1021/acscombsci.6b00163. [DOI] [PubMed] [Google Scholar]
- (102).Wang Z; Yu H; Canoura J; Liu Y; Alkhamis O; Fu F; Xiao Y Introducing Structure-Switching Functionality into Small-Molecule-Binding Aptamers via Nuclease-Directed Truncation. Nucleic Acids Res. 2018, 46 (13), e81. 10.1093/nar/gky305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Sekhon SS; Lee SH; Lee KA; Min J; Lee BT; Kim KW; Ahn JY; Kim YH Defining the Copper Binding Aptamotif and Aptamer Integrated Recovery Platform (AIRP). Nanoscale 2017, 9 (8), 2883–2894. 10.1039/c6nr09408b. [DOI] [PubMed] [Google Scholar]
- (104).Zheng X; Hu B; Gao SX; Liu DJ; Sun MJ; Jiao BH; Wang LH A Saxitoxin-Binding Aptamer with Higher Affinity and Inhibitory Activity Optimized by Rational Site-Directed Mutagenesis and Truncation. Toxicon 2015, 101, 41–47. 10.1016/j.toxicon.2015.04.017. [DOI] [PubMed] [Google Scholar]
- (105).Zhang Z; Oni O; Liu J New Insights into a Classic Aptamer: Binding Sites, Cooperativity and More Sensitive Adenosine Detection. Nucleic Acids Res. 2017, 45 (13), 7593–7601. 10.1093/nar/gkx517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Biniuri Y; Albada B; Willner I Probing ATP/ATP-Aptamer or ATP-Aptamer Mutant Complexes by Microscale Thermophoresis and Molecular Dynamics Simulations: Discovery of an ATP-Aptamer Sequence of Superior Binding Properties. J. Phys. Chem. B 2018, 122 (39), 9102–9109. 10.1021/acs.jpcb.8b06802. [DOI] [PubMed] [Google Scholar]
- (107).Munzar JD; Ng A; Juncker D Comprehensive Profiling of the Ligand Binding Landscapes of Duplexed Aptamer Families Reveals Widespread Induced Fit. Nat. Commun. 2018, 9 (1), 343. 10.1038/s41467-017-02556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Schoukroun-Barnes LR; White RJ Rationally Designing Aptamer Sequences with Reduced Affinity for Controlled Sensor Performance. Sensors 2015, 15 (4), 7754–7767. 10.3390/s150407754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109).Zhang X; Battig MR; Chen N; Gaddes ER; Duncan KL; Wang Y Chimeric Aptamer − Gelatin Hydrogels as an Extracellular Matrix Mimic for Loading Cells and Growth Factors. Biomacromolecules 2016, 17 (3), 778–787. 10.1021/acs.biomac.5b01511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Kratschmer C; Levy M Effect of Chemical Modifications on Aptamer Stability in Serum. Nucleic Acid Ther. 2017, 27 (6), 335–344. 10.1089/nat.2017.0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Kuai H; Zhao Z; Mo L; Liu H; Hu X; Fu T; Zhang X; Tan W Circular Bivalent Aptamers Enable in Vivo Stability and Recognition. J. Am. Chem. Soc. 2017, 139 (27), 9128–9131. 10.1021/jacs.7b04547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Kovacevic KD; Gilbert JC; Jilma B Pharmacokinetics, Pharmacodynamics and Safety of Aptamers. Adv. Drug Deliv. Rev. 2018, 134, 36–50. 10.1016/j.addr.2018.10.008. [DOI] [PubMed] [Google Scholar]
- (113).Shigdar S; Macdonald J; O’Connor M; Wang T; Xiang D; Shamaileh H. Al; Qiao L; Wei M; Zhou SF; Zhu Y; et al. Aptamers as Theranostic Agents: Modifications, Serum Stability and Functionalisation. Sensors (Switzerland) 2013, 13 (10), 13624–13637. 10.3390/s131013624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Ni S; Yao H; Wang L; Lu J; Jiang F; Lu A; Zhang G Chemical Modifications of Nucleic Acid Aptamers for Therapeutic Purposes. Int. J. Mol. Sci. 2017, 18 (8), 1683. 10.3390/ijms18081683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Walsh R; Derosa MC Biochemical and Biophysical Research Communications Retention of Function in the DNA Homolog of the RNA Dopamine Aptamer. Biochem. Biophys. Res. Commun. 2009, 388 (4), 732–735. 10.1016/j.bbrc.2009.08.084. [DOI] [PubMed] [Google Scholar]
- (116).Alvarez-martos I; Ferapontova EE Biochemical and Biophysical Research Communications A DNA Sequence Obtained by Replacement of the Dopamine RNA Aptamer Bases Is Not an Aptamer. Biochem. Biophys. Res. Commun. 2017, 489 (4), 381–385. 10.1016/j.bbrc.2017.05.134. [DOI] [PubMed] [Google Scholar]
- (117).Fine SL; Martin DF; Kirkpatrick P Pegaptanib Sodium. Nat. Rev. Drug Discov. 2005, 4, 187–188. 10.1038/nrd1677. [DOI] [PubMed] [Google Scholar]
- (118).Edwards SL; Poongavanam V; Kanwar JR; Roy K; Hillman KM; Prasad N; Leth-Larsen R; Petersen M; Marušič M; Plavec J; et al. Targeting VEGF with LNA-Stabilized G-Rich Oligonucleotide for Efficient Breast Cancer Inhibition. Chem. Commun. 2015, 51 (46), 9499–9502. 10.1039/c5cc02756j. [DOI] [PubMed] [Google Scholar]
- (119).Shi H; He X; Cui W; Wang K; Deng K; Li D; Xu F Locked Nucleic Acid/DNA Chimeric Aptamer Probe for Tumor Diagnosis with Improved Serum Stability and Extended Imaging Window in Vivo. Anal. Chim. Acta 2014, 812, 138–144. 10.1016/j.aca.2013.12.023. [DOI] [PubMed] [Google Scholar]
- (120).Darfeuille F; Hansen JB; Orum H; Di Primo C; Toulmé JJ LNA/DNA Chimeric Oligomers Mimic RNA Aptamers Targeted to the TAR RNA Element of HIV-1. Nucleic Acids Res. 2004, 32 (10), 3101–3107. 10.1093/nar/gkh636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (121).Padilla R; Sousa R Efficient Synthesis of Nucleic Acids Heavily Modified with Non- Canonical Ribose 2’-Groups Using a MutantT7 RNA Polymerase (RNAP). Nucleic Acids Res. 1999, 27 (6), 1561–1563. 10.1093/nar/27.6.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (122).Peng CG; Damha MJ G-Quadruplex Induced Stabilization by 2′-Deoxy-2′-Fluoro-d-Arabinonucleic Acids (2′F-ANA). Nucleic Acids Res. 2007, 35 (15), 4977–4988. 10.1093/nar/gkm520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (123).Pozmogova GE; Zaitseva MA; Smirnov IP; Shvachko AG; Murina MA; Sergeenko VI Anticoagulant Effects of Thioanalogs of Thrombin-Binding DNA-Aptamer and Their Stability in the Plasma. Bull. Exp. Biol. Med. 2010, 150 (2), 180–184. 10.1007/s10517-010-1099-5. [DOI] [PubMed] [Google Scholar]
- (124).El-Sagheer AH; Brown T Click Nucleic Acid Ligation: Applications in Biology and Nanotechnology. Acc. Chem. Res. 2012, 45 (8), 1258–1267. 10.1021/ar200321n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (125).Zaitseva M; Kaluzhny D; Shchyolkina A; Borisova O; Smirnov I; Pozmogova G Conformation and Thermostability of Oligonucleotide d(GGTTGGTGTGGTTGG) Containing Thiophosphoryl Internucleotide Bonds at Different Positions. Biophys. Chem. 2010, 146 (1), 1–6. 10.1016/j.bpc.2009.09.011. [DOI] [PubMed] [Google Scholar]
- (126).Ferguson BS; Hoggarth DA; Maliniak D; Ploense K; White RJ; Woodward N; Hsieh K; Bonham AJ; Eisenstein M; Kippin TE; et al. Real-Time, Aptamer-Based Tracking of Circulating Therapeutic Agents in Living Animals. Sci. Transl. Med. 2013, 5 (213), 1–9. 10.1126/scitranslmed.3007095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (127).Arroyo-Currás N; Somerson J; Vieira PA; Ploense KL; Kippin TE; Plaxco KW Real-Time Measurement of Small Molecules Directly in Awake, Ambulatory Animals. Proc. Natl. Acad. Sci. 2017, 114 (4), 645–650. 10.1073/pnas.1613458114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (128).Wochner A; Menger M; Orgel D; Cech B; Rimmele M; Erdmann VA; Glökler J A DNA Aptamer with High Affinity and Specificity for Therapeutic Anthracyclines. Anal. Biochem. 2008, 373 (1), 34–42. 10.1016/j.ab.2007.09.007. [DOI] [PubMed] [Google Scholar]
- (129).Paige JS; Wu KY; Jaffrey SR RNA Mimics of Green Fluorescent Protein. Science (80-.). 2011, 333 (6042), 642–646. 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (130).Strack RL; Disney MD; Jaffrey SR A Superfolding Spinach2 Reveals the Dynamic Nature of Trinucleotide Repeat–Containing RNA. Nat. Methods 2013, 10 (12), 1219–1224. 10.1038/nmeth.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (131).Filonov GS; Moon JD; Svensen N; Jaffrey SR Broccoli: Rapid Selection of an RNA Mimic of Green Fluorescent Protein by Fluorescence-Based Selection and Directed Evolution. J. Am. Chem. Soc. 2014, 136 (46), 16299–16308. 10.1021/ja508478x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (132).Karunanayake Mudiyanselage APKK; Yu Q; Leon-Duque MA; Zhao B; Wu R; You M Genetically Encoded Catalytic Hairpin Assembly for Sensitive RNA Imaging in Live Cells. J. Am. Chem. Soc. 2018, 140 (28), 8739–8745. 10.1021/jacs.8b03956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (133).Su Y; Hickey SF; Keyser SGL; Hammond MC In Vitro and in Vivo Enzyme Activity Screening via RNA-Based Fluorescent Biosensors for S-Adenosyl- l -Homocysteine (SAH). J. Am. Chem. Soc. 2016, 138 (22), 7040–7047. 10.1021/jacs.6b01621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (134).Kellenberger CA; Wilson SC; Sales-Lee J; Hammond MC RNA-Based Fluorescent Biosensors for Live Cell Imaging of Second Messengers Cyclic Di-GMP and Cyclic AMP-GMP. J. Am. Chem. Soc. 2013, 135 (13), 4906–4909. 10.1021/ja311960g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (135).Bai Y; Feng F; Zhao L; Wang C; Wang H; Tian M; Qin J; Duan Y; He X Aptamer/Thrombin/Aptamer-AuNPs Sandwich Enhanced Surface Plasmon Resonance Sensor for the Detection of Subnanomolar Thrombin. Biosens. Bioelectron. 2013, 47, 265–270. 10.1016/j.bios.2013.02.004. [DOI] [PubMed] [Google Scholar]
- (136).Collins CM; Yui S; Roberts CES; Kojic I Thrombin Detection Using a Piezoelectric Aptamer-Linked Immunosorbent Assay. Anal. Biochem. 2013, 443 (1), 97–103. 10.1016/j.ab.2013.08.019. [DOI] [PubMed] [Google Scholar]
- (137).He P; Liu L; Qiao W; Zhang S Ultrasensitive Detection of Thrombin Using Surface Plasmon Resonance and Quartz Crystal Microbalance Sensors by Aptamer-Based Rolling Circle Amplification and Nanoparticle Signal Enhancement. Chem. Commun. 2014, 50 (12), 1481–1484. 10.1039/c3cc48223e. [DOI] [PubMed] [Google Scholar]
- (138).Cho H; Baker BR; Wachsmann-Hogiu S; Pagba CV; Laurence TA; Lane SM; Lee LP; Tok JBH Aptamer-Based SERRS Sensor for Thrombin Detection. Nano Lett. 2008, 8 (12), 4386–4390. 10.1021/nl802245w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (139).Chang H; Tang L; Wang Y; Jiang J; Li J Graphene Fluorescence Resonance Energy Transfer Aptasensor for the Thrombin Detection. Anal. Chem. 2010, 82 (6), 2341–2346. 10.1021/ac9025384. [DOI] [PubMed] [Google Scholar]
- (140).Mir M; Vreeke M; Katakis I Different Strategies to Develop an Electrochemical Thrombin Aptasensor. Electrochem. commun. 2006, 8 (3), 505–511. 10.1016/j.elecom.2005.12.022. [DOI] [Google Scholar]
- (141).Krauss IR; Spiridonova V; Pica A; Napolitano V; Sica F Different Duplex/Quadruplex Junctions Determine the Properties of Anti-Thrombin Aptamers with Mixed Folding. Nucleic Acids Res. 2016, 44 (2), 983–991. 10.1093/nar/gkv1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (142).Krauss IR; Napolitano V; Petraccone L; Troisi R; Spiridonova V; Mattia CA; Sica F Duplex/Quadruplex Oligonucleotides: Role of the Duplex Domain in the Stabilization of a New Generation of Highly Effective Anti-Thrombin Aptamers. Int. J. Biol. Macromol. 2018, 107, 1697–1705. 10.1016/j.ijbiomac.2017.10.033. [DOI] [PubMed] [Google Scholar]
- (143).Liu J; Liu Y; Yang X; Wang K; Wang Q; Shi H; Li L Exciton Energy Transfer-Based Fluorescent Sensing through Aptamer-Programmed Self-Assembly of Quantum Dots. Anal. Chem. 2013, 85 (22), 11121–11128. 10.1021/ac403023p. [DOI] [PubMed] [Google Scholar]
- (144).Lin Z; Chen L; Zhu X; Qiu B; Chen G Signal-on Electrochemiluminescence Biosensor for Thrombin Based on Target-Induced Conjunction of Split Aptamer Fragments. Chem. Commun. 2010, 46 (30), 5563–5565. 10.1039/c0cc00932f. [DOI] [PubMed] [Google Scholar]
- (145).Chen J; Zeng L Enzyme-Amplified Electronic Logic Gates Based on Split/Intact Aptamers. Biosens. Bioelectron. 2013, 42 (1), 93–99. 10.1016/j.bios.2012.10.030. [DOI] [PubMed] [Google Scholar]
- (146).Liu X; Shi L; Hua X; Huang Y; Su S; Fan Q; Wang L; Huang W Target-Induced Conjunction of Split Aptamer Fragments and Assembly with a Water-Soluble Conjugated Polymer for Improved Protein Detection. ACS Appl. Mater. Interfaces 2014, 6 (5), 3406–3412. 10.1021/am405550j. [DOI] [PubMed] [Google Scholar]
- (147).Chhabra R; Sharma J; Ke Y; Liu Y; Rinker S; Lindsay S; Yan H Spatially Addressable Multiprotein Nanoarrays Templated by Aptamer-Tagged DNA Nanoarchitectures. J. Am. Chem. Soc. 2007, 129 (34), 10304–10305. 10.1021/ja072410u. [DOI] [PubMed] [Google Scholar]
- (148).Zhang J; Smaga LP; Satyavolu NSR; Chan J; Lu Y DNA Aptamer-Based Activatable Probes for Photoacoustic Imaging in Living Mice. J. Am. Chem. Soc. 2017, 139 (48), 17225–17228. 10.1021/jacs.7b07913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (149).Riccardi C; Russo Krauss I; Musumeci D; Morvan F; Meyer A; Vasseur JJ; Paduano L; Montesarchio D Fluorescent Thrombin Binding Aptamer-Tagged Nanoparticles for an Efficient and Reversible Control of Thrombin Activity. ACS Appl. Mater. Interfaces 2017, 9 (41), 35574–35587. 10.1021/acsami.7b11195. [DOI] [PubMed] [Google Scholar]
- (150).Nakatsuka MA; Mattrey RF; Esener SC; Cha JN; Goodwin AP Aptamer-Crosslinked Microbubbles: Smart Contrast Agents for Thrombin-Activated Ultrasound Imaging. Adv. Mater. 2012, 24 (45), 6010–6016. 10.1002/adma.201201484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (151).Jenison RD; Gill SC; Pardi A; Polisky B High-Resolution Molecular Discrimination by RNA. Science (80-.). 1994, 263 (5152), 1425–1429. 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- (152).Zong C; Liu J The Arsenic-Binding Aptamer Cannot Bind Arsenic: Critical Evaluation of Aptamer Selection and Binding. Anal. Chem. 2019, 91 (16), 10887–10893. 10.1021/acs.analchem.9b02789. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


