Abstract
The RNA-recognition motif (RRM) is a common and evolutionarily conserved RNA-binding module. Crystallographic and solution structural studies have shown that RRMs adopt a compact α/β structure, in which four antiparallel β-strands form the major RNA-binding surface. Conserved aromatic residues in the RRM are located on the surface of the β-sheet and are important for RNA binding. To further our understanding of the structural basis of RRM-nucleic acid interaction, we carried out a high resolution analysis of UP1, the N-terminal, two-RRM domain of heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1), whose structure was previously solved at 1.75–1.9 Å resolution. The two RRMs of hnRNP A1 are closely related but have distinct functions in regulating alternative pre-mRNA splice site selection. Our present 1.1 Å resolution crystal structure reveals that two conserved solvent-exposed phenylalanines in the first RRM have alternative side chain conformations. These conformations are spatially correlated, as the individual amino acids cannot adopt each of the observed conformations independently. These phenylalanines are critical for nucleic acid binding and the observed alternative side chain conformations may serve as a mechanism for regulating nucleic acid binding by RRM-containing proteins.
INTRODUCTION
Human heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) is an abundant nuclear protein that has been implicated in many cellular processes, including packaging of nascent RNA polymerase II transcripts in the nucleus (1,2), regulating alternative 5′ splice site selection in pre-mRNA splicing (3–7), mediating the splicing-inhibitory effects of certain exonic splicing silencers (8–11), nuclear–cytoplasm transport (12), RNA annealing (13) and telomere length regulation (14). The protein consists of 320 amino acids and has two RNA-recognition motif (RRM) consensus sequences at the N-terminus, followed by a glycine-rich C-terminal region. Both the N-terminal domain, known as UP1, and the C-terminal domain are required for the function of hnRNP A1 in alternative splicing (15). Furthermore, two functional RRMs are required for function, as substituting a pair of conserved phenylalanines in either RRM with a pair of aspartic acids abolishes the activity of the protein in alternative splicing (15). The two RRMs of hnRNP A1 are closely related in sequence, yet they have distinct functions in pre-mRNA splicing. Domain swap experiments have shown that the alternative splicing function of hnRNP A1 requires an intact RRM2 at the C-terminal position, whereas the N-terminal RRM can be either RRM1 or RRM2 (16). Despite lacking the C-terminal domain, UP1 binds efficiently to hnRNP A1 high affinity binding sites determined by SELEX with the intact protein (17) and to single-stranded human telomeric DNA repeats, indicating that the two RRMs confer the binding specificity and are sufficient for high affinity binding (14,18–21). The C-terminal domain is capable of binding RNA independently, but it also engages in homophilic protein–protein interactions, resulting in cooperative RNA binding, which appears to be important for hnRNP complex assembly (2) and splicing silencing (8–11). Moreover, the C-terminal domain comprises signals involved in bi-directional nuclear–cytoplasmic transport of hnRNP A1 (22).
The RRM is the most common and best-characterized RNA-binding motif (23,24). RRM-containing proteins have been found in many pre-mRNA-binding proteins and a host of other RNA- and single-stranded (ss)DNA-binding proteins in all three kingdoms of life. The RRM is ∼70–90 amino acids in length and usually it can be readily identified by its highly conserved RNP1 octamer and RNP2 hexamer submotifs. These submotifs consist of characteristic arrays of aromatic amino acids and a number of interspersed, mostly hydrophobic, amino acids that are well conserved. The two sequence motifs are separated by ∼30 amino acids. The three-dimensional structures of about 20 RRMs have been determined (25). The RRM structure has a compact, globular α/β-fold. Four antiparallel β-strands form a consecutive sheet and two α-helices are packed on one side of the β-sheet. The exposed side of the β-sheet is the major RNA-binding surface. In particular, the conserved RNP1 and RNP2 aromatic residues located in the two central β-strands are involved in stacking interactions with the bases of the bound single-stranded nucleic acid. Though many of the protein–RNA interactions are through amino acids located on the β-sheet of the RRM core structure, the specificity for the RNA ligands usually requires residues outside the conserved structural elements. For example, loops connecting the β-strands or regions outside the conserved RRM domain can play prominent roles in RNA recognition (21,26–28). In addition, many RNA-binding proteins have multiple RRMs. To date, six structures of proteins with two tandem RRMs have been determined (21,29–36). The most notable difference among these structures is the spatial arrangement of the two RRMs with respect to each other. For example, the two RRMs in hnRNP A1 are held together by two pairs of Arg:Asp interactions (35,36), whereas the two RRMs in the other structures have different and more flexible spatial arrangements (29–34). Many of the multi-RRM proteins require contiguous RRMs for binding to their physiological target sites, and the RRMs act synergistically and function as an inseparable RNA-binding entity (17,21,37). These observations provide a rationale for how RRM-containing proteins can bind specifically to a wide spectrum of RNA targets.
Crystallographic and NMR studies have shown that large conformational changes are often associated with nucleic acid binding. For example, the loop connecting β2 and β3 in the N-terminal RRM of U1A exhibits large conformational differences between the apo- and RNA-bound structures (28,38); upon binding of UP1 to single-stranded telomeric DNA, the loop connecting the two RRMs becomes ordered and interacts directly with the ssDNA, and the two RRMs also undergo moderate movement (21). The domain movement is much more pronounced in the structure of the RNA-bound sex-lethal protein (31,33). On the other hand, few conformational differences between the amino acids in the RNP1 and RNP2 submotifs have been found among all the RRM structures studied so far, and a similar mode of protein–RNA interaction involving RNP1 and RNP2 amino acids has been observed for the various RRMs. It is conceivable, however, that key residues involved in RNA binding may adopt alternative conformations, which would suggest a mechanism of regulating RNA binding or an alternative mode of RNA interaction. For example, such a mechanism may contribute to the RNA binding and functional differences between two otherwise closely related RRMs, such as those in hnRNP A1. Nevertheless, all RRM structural analyses carried out to date have failed to resolve alternative side chain conformations, possibly due to resolution limits.
Here we report the 1.1 Å crystal structure of the two-RRM domain (UP1) of hnRNP A1. The crystal structures of UP1 and its complex with human telomeric ssDNA were previously determined at lower resolutions (21,35,36). The high resolution data in the present study allowed us to model alternative side chain conformations of a number of residues, capturing their more transient and dynamic conformational states. In particular, the atomic resolution structure revealed for the first time that the critical RNA-interacting phenylalanine residues could undergo concerted conformational changes.
MATERIALS AND METHODS
Data collection and processing
Details of protein production, purification and crystallization of recombinant human UP1 have been described (39). The crystals belong to spacegroup P21, with cell constants of a = 37.71 Å, b = 43.45 Å, c = 55.06 Å and β = 93.7°, and there is one UP1 molecule per asymmetric unit. Diffraction data were collected at 100°K on a Brandeis 2 × 2 CCD detector at the X12C beamline of the National Synchrotron Light Source, Brookhaven National Laboratory. To collect high resolution data, the detector to crystal distance was set to 78 mm and the 2θ angle was set to 9.97°. Other parameters used for data collection were: X-ray wavelength 0.90 Å; oscillation range 0.75° per image; exposure time 3.0 min. Data were processed using the HKL suite of programs (40) and the data statistics are shown in Table 1.
Table 1. Statistics from the crystallographic analysis.
| Data statistics | ||
| No. of observed/unique reflections | 180601/60334 | |
| Resolution (Å) | 50.00–1.10 | |
| Completeness (%) | 83.6 (52.0 in the last shell) | |
| Rmerge | 0.057 (0.236 in the last shell) | |
| <I/σ> | 11.0 | |
| Refinement statistics | ||
| No. of atoms (non-hydrogen) | 1545 (protein 1315; solvent 230) | |
| R factor | 0.146 (F > 4σ)/0.155 (all data) | |
| Rfree | 0.185 (F > 4σ)/0.194 (all data) | |
| r.m.s. deviations | ||
| bond lengths | 0.008 Å | |
| bond angle distances | 0.027 Å | |
| chiral volume restraints | 0.039 Å3 | |
| planarity restraints | 0.41 Å | |
| Mean B factors | ||
| main chain atoms | 15.8 Å2 | |
| side chain atoms | 21.9 Å2 | |
| solvent atoms | 36.2 Å2 |
Structure refinement
The 1.9 Å crystal structure of UP1 (36) was used as the starting model for refinement. Several rounds of conventional refinement were first carried out using CNS (41). Anisotropic refinement was then carried out with SHELXL-97 (42). CNS refinements were carried out against the scattering amplitudes (Fo) and SHELXL refinements against the intensities (Fo2). The data were randomly divided into two sets: a working set composed of 90% of the data and a test set composed of the remaining 10% for cross-validation purposes. After the refinement was completed, working and reference data sets were merged and used to calculate electron density maps. The programs O (43) and XtalView (44) were used for model rebuilding with σA and difference electron density maps. Most of the water molecules were placed with the help of the program SHELXWAT in SHELX-97, the rest being placed manually by inspection of the electron density maps.
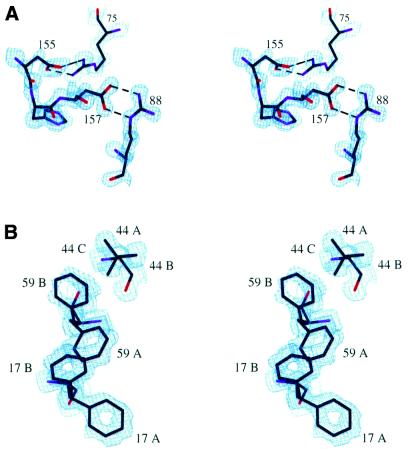
The high resolution data allowed us to refine the non-hydrogen atoms anisotropically. The solvent atoms were restrained to be weakly anisotropic with an estimated standard deviation of 0.1 Å2. Hydrogen atoms were generated automatically by the program and were included in the refinement using a riding model (42). Residues Phe17, Phe59 and Val44 exhibited correlated disorder, and this collective behavior was taken into consideration in the refinement using SHELX-97. Possible side chain conformations were generated and included using PART numbers and their occupancies were treated as free variables and refined using the FVAR and SUMP commands. Five permissible combinations of Phe17, Phe59 and Val44 were generated (namely AAA, AAB, AAC, ABA and BBA, in which the first, second and third letters denote the conformation of Phe17, Phe59 and Val44, respectively) and each combination was assigned a starting occupancy value. These occupancy values were treated as free variables in the refinement, with their sum constrained to be 1. For example, the occupancy of Phe17 in the A conformation is the sum of the occupancy values of AAA, AAB, AAC and ABA and the occupancy (or frequency of appearance) of the Phe17B/Phe59B/Val44A conformation corresponds to the value for the BBA combination. The positions and thermal motions of the relevant conformations (PARTs) were correlated using the EXYZ and EADP commands. Hydrogen atoms were not included for these residues. The final model contains 1315 protein atoms and 230 water molecules. Final R and Rfree values were 0.155 and 0.194, respectively, for all the data to 1.1 Å resolution. Figure 1A and B shows the electron density map around two regions of the protein: (i) amino acids involved in salt bridges between RRM1 and RRM2 are shown in Figure 1A; (ii) the conserved RNP1 and RNP2 aromatic residues of RRM1 are shown in Figure 1B. The maps shown are σA maps, i.e. (2mFo – DFc) × exp(iαC) maps in which m is the figure of merit and D is the Luzzati parameter (45). σA maps were used to minimize model bias. Detailed refinement statistics are provided in Table 1. The PROCHECK program (46) was used to check the stereochemical quality of the refined model. The Ramachandran plots showed that 94.4% of the residues were in the most favored region and 5.6% were in the additional allowed region. Figures were prepared with the program O (43) and structural alignments were done with the ALIGN program (47).
Figure 1.
Electron density maps. (A) A stereo view of the electron density map showing the Arg:Asp salt bridges that mediate the interactions between RRM1 and RRM2. The amino acids are shown as a stick model. Red, purple and black colors on the stick model are used to indicate oxygen, nitrogen and carbon atoms, respectively. Dashed lines indicate salt bridges. The map is a σA map contoured at the 2σ level. (B) A stereo view of the electron density map showing correlated alternative side chain conformations of Phe17, Phe59 and Val44. The amino acid side chains are numbered and shown as a stick model. Different side chain conformations are denoted with the suffixes A, B and C, as described in the text. The electron density map is a σA map contoured at the 1.2σ level.
RESULTS
Overall structure
The refined UP1 structure consists of 163 amino acids (Lys8–Ser91 and Thr103–Leu181) out of a total of 196 in the expressed UP1 fragment, plus 230 water molecules. Three regions were not modeled because of poor electron density: the first seven residues at the N-terminus, the last 16 residues at the C-terminus and the internal linker residues Arg92–Leu102. These regions were disordered in the original 1.75 and 1.9 Å structures (35,36) and they remained uninterpretable even in the present high resolution analysis. The linker and many of the C-terminal residues become ordered when the protein forms a complex with telomeric ssDNA (21), as they are involved in direct interaction with the bound single-stranded oligonucleotide.
UP1 comprises two tandem RNA-binding motifs: RRM1 (amino acids 15–89) and RRM2 (amino acids 106–180). Each RRM independently adopts the characteristic RRM fold. The two RRMs share 35% sequence identity and 55% amino acid similarity (16,21). The root mean square (r.m.s.) deviation from superimposition of the Cα positions of the two RRMs is 1.37 Å in the present high resolution structure. In our 1.9 Å resolution structure the r.m.s. deviation was 1.49 Å (36). The r.m.s. deviation of the Cα positions between the 1.1 and 1.9 Å structures is 0.177 Å, showing excellent agreement in the overall structure.
Alternative side chain conformations
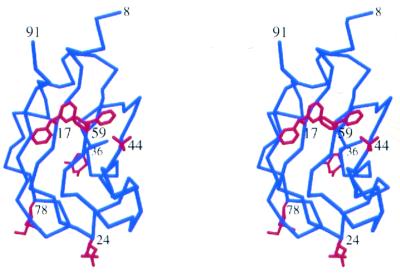
The high resolution analysis revealed that six amino acids in UP1 have alternative side chain conformations. Interestingly, all six amino acids are located in RRM1. These residues constitute 3% of the total residues in UP1. The locations of these residues (Phe17, Glu24, Gln36, Val44, Phe59 and Lys78) in the protein are shown in Figure 2.
Figure 2.
A stereo view of the location of residues with alternative side chain conformations. The Cα chain of RRM1 is shown in blue and the multi-conformational side chains are shown in red.
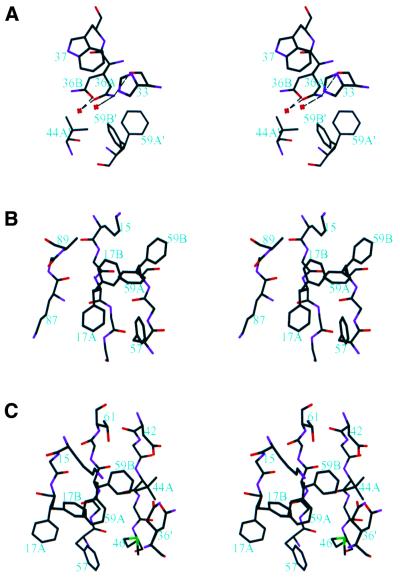
Three residues are located in loop regions and are exposed to the solvent: Glu24 in loop 1, Gln36 in loop 2 and Lys78 in loop 5. Although Glu24 and Lys78 are best modeled with alternative side chain conformations, the electron densities for the alternative conformations are not ideal, suggesting that these residues have considerable conformational flexibility. Gln36 has a better defined alternative conformational state and it is located in the vicinity of Phe17′, Phe59′ and Val 44′ (the prime is used to denote amino acids in a symmetry-related molecule) (Fig. 3A). The major and minor conformations of Gln36, designated the A and B conformations, have occupancies of 0.55 and 0.45, respectively. The two conformations are related by rotations around the Cα–Cβ and Cγ–Cδ bonds. The amide plane in Gln36A makes edge-on interactions with His33, Trp37 and Phe59B′. Nɛ2 of Gln36A interacts with the carbonyl oxygen of His33 and a water molecule via hydrogen bonds, and Oɛ1 of Gln36A hydrogen bonds to another water molecule. The amide plane of Gln36B makes edge-on interactions with Trp37 and Phe59B′. In addition, Nɛ2 of Gln36B contacts Cγ1 of Val44A′/Cγ2 of Val44B′ and Oɛ1 hydrogen bonds to a water molecule. The Gln36A conformation is slightly more preferred, perhaps because it makes ten intramolecular and three intermolecular contacts to other amino acids and three hydrogen bonds to water molecules. Gln36B makes nine intramolecular and three intermolecular contacts with other amino acids and one hydrogen bond to a water molecule. It appears that other conformations for Gln36 are not possible in the crystal lattice, because of potential steric conflicts with neighboring residues, namely Trp37, Phe59′, Met46′ and Val44′.
Figure 3.
Stereo views showing amino acids with alternative side chain conformations. Amino acids are shown as sticks. Carbon, nitrogen, oxygen and sulfur atoms are shown in black, purple, red and green, respectively. (A) Gln36 and its surrounding amino acids. Gln36 has two conformations, Gln36A and Gln36B, with occupancies of 0.55 and 0.45, respectively. Amino acids from a neighboring molecule in the crystal lattice are indicated with a prime after the amino acid number. Red crosses denote water molecules and dashed lines indicate hydrogen bonds. (B) Phe17 and its surrounding amino acids. Phe17 adopts two conformations, Phe17A and Phe17B, with occupancies of 0.65 and 0.35, respectively. (C) Phe59, Val44 and their surrounding amino acids. Phe59 adopts two conformations, Phe59A and Phe59B (occupancies of 0.57 and 0.43, respectively). Val44 has three conformations of which only Val44A (occupancy of 0.61) is labeled. The other two conformations, Val44B and Val44C, are shown but not labeled. They have similar occupancies and are related to the A conformation by rotating –120° and 120° around the Cα–Cβ bond.
The other three multi-conformational residues are located on the RNA-binding surface: Phe17 on β1, Val44 on β2 and Phe59 on β3. Phe17 and Phe59 are part of the RNP2 and RNP1 submotifs, respectively. These aromatic residues are highly conserved among RRMs (23). They interact with RNA or ssDNA directly in the structure of RRM–RNA and RRM–ssDNA complexes. In the UP1–TR2 structure (21), in which TR2 is a ssDNA containing two human telomeric repeats, (TTAGGG)2, Val44 interacts with G6 via van der Waals interactions and it also forms part of a shallow hydrophobic channel where TR2 binds. Val44 is not conserved in other RRMs and the corresponding residues are not always involved in interacting with the nucleic acid. For example, the corresponding amino acid in the RRM2 of UP1 is Glu135, which does not interact with TR2 (21).
The side chain of Phe17 has two conformations, denoted Phe17A and Phe17B (Fig. 3B). Phe17A is the major conformation, with an occupancy of 0.65, Phe17B having an occupancy of 0.35. The two conformations are related by a rotation about the Cα–Cβ bond. Both conformers are in hydrophobic environments. Phe17A makes close contacts with Phe57, Phe59A, the aliphatic portion of Lys87 and the peptide plane of Gly19. The packing of the phenylalanine rings has a characteristic edge-on pattern (48) and the peptide plane of Gly19 is slightly inclined with respect to the Phe17A ring plane (Fig. 3B). Phe17B contacts Phe59B, Ala89 and the aliphatic portion of Lys15. Phe17A has a higher occupancy than Phe17B, possibly because Phe17A makes more intramolecular contacts than does Phe17B: Phe17A makes a total of 15 intramolecular contacts, while Phe17B makes a total of 12. On the other hand, the side chain of Phe17B contacts four water molecules, whereas Phe17A contacts none.
The two side chain conformations of Phe59 have occupancies of 0.57 (Phe59A) and 0.43 (Phe59B) and are related by a rotation about the Cα–Cβ bond (Fig. 3C). Phe59A contacts Met46, Phe17A and Phe57. Phe59B contacts Val44A, Asp42, Thr61, Lys15 and Phe17B. Phe59A makes a total of 14 intramolecular contacts, compared with 11 for Phe59B. An additional interaction stabilizing the B conformation is provided by an intermolecular contact with Gln36A′ of a symmetry-related molecule. Furthermore, Phe59A interacts with two water molecules, whereas only one such interaction is found for Phe59B.
There are three possible conformations for valine and all three conformers are observed for Val44 (Fig. 3C). The three conformers are denoted Val44A, Val44B and Val44C and they have occupancies of 0.61, 0.19 and 0.20, respectively. The high preference for Val44A appears to be due to the existence of the B conformation of Phe59. Val44A makes a total of eight intramolecular contacts; two of them are with Phe59B. Val44B and Val44C each makes five contacts. Both the Val44B and Val44C conformations are incompatible with the Phe59B conformation because of steric hindrance. In addition, Val44A and Val44B each make an intermolecular contact with Gln36A′. Val44A also contacts two water molecules, while Val44B and Val44C each contact one water molecule.
Correlated side chain conformations of Phe17, Phe59 and Val44
Each of the three residues, Phe17, Phe59 and Val44, can adopt more than one conformation, as described above. However, not all of the conformations can be independently adopted by these residues because of potential steric clashes. Permissible combinations are: (i) Phe17A/Phe59A/Val44A,B,C; (ii) Phe17A/Phe59B/Val44A; (iii) Phe17B/Phe59B/Val44A (Figs 1B and 3C). These three combinations have occupancies, or frequencies of appearance, of 0.57, 0.08 and 0.35, respectively (see Materials and Methods). The first combination is the same as that observed in our previous studies (21,36). In the second combination, Phe17 remains in the major conformation and Phe59 rotates away from Phe17. Interestingly, this was also observed in a previous study (35), although this combination is not energetically favored due to the absence of interactions between the two phenylalanines. To avoid steric clashes, Val44 is forced to adopt the A conformation whenever Phe59 is in the B conformation. In the third combination, the two phenylalanine residues show a concerted motion, rotating together away from the major conformation (Figs 1B and 3C). In this combination Val44 is again restricted to the A conformation.
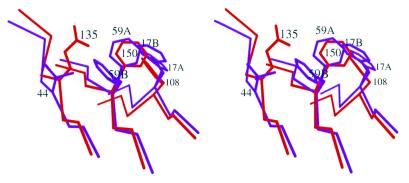
It is important to keep in mind that Phe17 and Phe59 are key residues involved in RNA/ssDNA binding. In all the RRM–nucleic acid complex structures solved so far the phenylalanines are exclusively in the A conformation. It is also clear that not all RRMs have suitable environments that would allow the alternative conformations of the phenylalanines observed here. For example, the corresponding phenylalanines in RRM2 of the present structure (Phe108 and Phe150) do not show alternative side chain conformations. There are two reasons that may account for the absence of an alternative side chain conformation of Phe150. (i) The distance between Phe150 and Glu135, which corresponds to Val44 in RRM1, is shorter than that between the corresponding residues in RRM1 (Fig. 4). The distance between the Cα atoms of Phe150 and Glu135 is 5.08 Å, while it is 5.48 Å between the Cα atoms of Phe59 and Val44. As a consequence, the Cβ atom of Glu135 is too close to Phe150 for it to adopt a B conformation similar to Phe59B (Fig. 4). (ii) A potential B conformation of Phe150 would clash with the bulky side chain of Glu135, especially the Cδ and Oɛ1 atoms, in the observed conformation (Fig. 4). Unlike the situation with the Cβ atom, these clashes can be avoided if Glu135 takes an alternative side chain conformation. The alternative side chain conformations in RRM1 and the absence of alternative conformations in RRM2 are not due to crystal contacts because: (i) no crystal contacts were found for Phe17; (ii) although Phe59B and Val44A/B also interact with Gln36 of a neighboring molecule, Phe59A and Val44A are the energetically favored conformations, as reflected by their higher occupancies compared with the other conformations; (iii) Glu135 is not involved in crystal contacts. Therefore, crystal contacts cannot account for the difference in conformational state between RRM1 and RRM2.
Figure 4.
Stereo diagram of the superimposition of the two RRMs showing why the RRM2 phenylalanines cannot adopt the alternative conformations observed in RRM1. Phe17, Phe59, Val44 (colored purple) and their counterparts in RRM2 (colored red) are shown as a stick model. Three β-strands (β1, β2 and β3) where these residues are located are also shown with the same color code. Glu135 in RRM2 is displaced from the corresponding position of Val44 in RRM1, creating clashes if Phe150 adopts the B conformation.
DISCUSSION
The 1.1 Å resolution structure of UP1 presented here shows that six residues in RRM1 have alternative side chain conformations. Among these residues, three, Phe17, Phe59 and Val44, are known to be important for nucleic acid binding. Interestingly, these alternative conformations of the side chains are correlated. Spatially correlated side chain disorder, analogous to that observed in the present structure, was previously reported in the high resolution crystal structure of crambin (49). In the case of crambin, conformational correlation of three residues over a range of 8–10 Å has been observed. It has been suggested that the spatial correlation may have resulted from short-range (4–8 Å), liquid-like correlated motion, as revealed in the analysis of diffuse X-ray scattering of insulin and lysozyme crystals (50,51). The correlated conformational change observed in the present structure may arise from similar underlying dynamics. The longest conformational correlation length in the present UP1 structure is 14.1 Å, which spans three residues, namely Phe17, Phe59 and Val44, and corresponds to the distance between the Cγ2 atom of Val44A/Cγ1 atom of Val44C and the Cɛ2 atom of Phe17A.
The corresponding phenylalanines in RRM2 do not show alternative conformations analogous to those in RRM1. As mentioned earlier, Phe150 cannot adopt an alternative conformation similar to that of Phe59B because Glu135 is shifted toward Phe150 by 0.4 Å as compared with the corresponding Val44 position with respect to Phe59 in RRM1. This difference between RRM1 and RRM2 is interesting in the context of hnRNP A1 function in pre-mRNA splicing. Despite the high sequence conservation and structural similarity between RRM1 and RRM2 of hnRNP A1, domain swapping and duplication experiments showed that the two RRMs have distinct functions (16). Efficient alternative splice site switching activity requires the presence of one copy of RRM2 preceding the C-terminal glycine-rich domain, whereas the N-terminal RRM can be either RRM1 or a copy of RRM2 (16). Based on previous structural results, it has been noted that RRM2 is necessary to maintain the correct positioning of the two RRMs via two pairs of salt bridges (21,36). However, restoring the two salt bridges to the RRM1-duplication variant of hnRNP A1 is not sufficient for gain of function in alternative splicing, suggesting that some unique features of RRM1 may interfere with proper interaction with RNA in the context of alternative splicing (16). It is possible that the alternative side chain conformations of the RNA-binding phenylalanines in the RRM1-duplication variant are incompatible with the type of RNA binding seen with the wild-type protein.
We previously solved the crystal structure of UP1 bound to two repeats of human telomeric ssDNA, TTAGGGTTAGGG (21). Interestingly, in that structure, RRM1 and RRM2 from two different protein molecules bind to the same strand of DNA, with each RRM recognizing one of the two telomeric repeats. This particular mode of binding reflects the high similarity between the two RRMs and the repetitive nature of the hnRNP A1 high affinity binding sites in the telomeric sequence. Nevertheless, truncation derivatives of hnRNP A1 lacking one of the RRMs are still capable of residual RNA binding and each isolated RRM appears to have unique nucleic acid-binding properties (16,17,52,53). This difference between the RRMs can be at least partially attributed to several amino acids located on their RNA-binding surface. In view of the findings reported here, alternative side chain conformations may also contribute to the different properties of RRM1 and RRM2. Using the structure of the UP1–TR2 complex as a reference (21), we modeled how UP1 in the Phe17B/Phe59B/Val44A conformation may bind RNA/ssDNA. We found that UP1 in this conformation cannot bind TR2 in the same manner as seen previously (21), because of significant steric clashes between TR2 and Phe17B/Phe59B (Fig. 5). In particular, Phe59B is within 2.0 Å of G5 of TR2 and approximately parallel stacking between Phe17/Phe59 and purine bases is not maintained. A possible UP1 (Phe17B/Phe59B/Val44A)–TR2 binding mode can be modeled (data not shown), but the path of the nucleic acid is significantly different from that observed for UP1 in the standard conformation (21). It is not known at present whether the UP1 (Phe17B/Phe59B/Val44A)–nucleic acid interaction occurs in nature. Nevertheless, it can be said that the alternative side chain conformations would significantly alter the observed mode of nucleic acid binding by RRM-containing proteins, thus providing a potential mechanism for regulating RNA binding or perhaps providing an alternative mode of RNA binding.
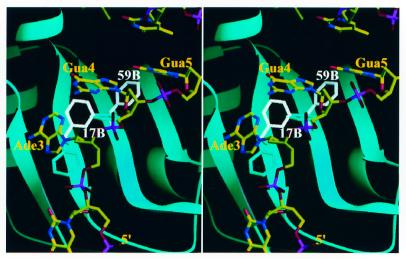
Figure 5.
Alternative conformations of Phe17 and Phe59 change protein–RNA interactions. The stereo diagram shows the superimposition of a human telomeric ssDNA (TR2) resembling the high affinity RNA-binding sequence of hnRNP A1 onto the 1.1 Å resolution structure of UP1. The protein structure is shown as a ribbon representation (cyan). Phe17, Phe59 and TR2 are shown as a stick model. Phe17A and Phe59A are shown in cyan; Phe17B and Phe59B are shown in white and labeled. The color code used for TR2 is red for oxygen, blue for nitrogen, yellow for carbon and magenta for phosphorus.
We have examined a number of crystal and NMR structures of RRM-containing proteins to see if any of the aromatic residues in the RNP1 and RNP2 submotifs also have alternative side chain conformations. Interestingly, the solution structure of RRM2 of Drosophila sex-lethal protein shows that Tyr214, which corresponds to Phe17 in UP1, is in a conformation similar to that of Phe17B in UP1 (54). In the crystal structure of sex-lethal with and without bound RNA, Tyr214 has a conformation similar to that of Phe17A in UP1. Phe256, which corresponds to Phe59 in UP1, also adopts different conformations between the NMR and crystal structures. However, the conformation of Phe256 in the NMR structure of sex-lethal differs from that of Phe59B in UP1. Therefore, the second RRM of sex-lethal may also serve as a prototype for studying the effect of RNA binding due to alternative side chain conformations of conserved aromatic residues in the RNP submotifs. Given that the RRM is the most common RNA-binding motif, it is likely that the observed alternative side chain conformations of the conserved aromatic residues also occur in other RRM-containing proteins. In future studies it will be of interest to determine the effects of alternative side chain conformation on RNA binding.
Atomic coordinates of the refined model have been deposited with the Protein Data Bank (PDB code 1L3K).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Bob Sweet at the X12C beamline of the National Synchrotron Light Source, Brookhaven National Laboratory, for help with data collection and Chris Waddling for advice on the use of SHELX in the region of correlated disorder. This work was supported in part by the W. M. Keck Foundation (R.M.X.) and by NIH grants CA13106 (A.R.K.) and GM55874 (R.M.X.).
PDB accession no. 1L3K
REFERENCES
- 1.Dreyfuss G., Matunis,M.J., Pinol-Roma,S. and Burd,C.G. (1993) hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem., 62, 289–321. [DOI] [PubMed] [Google Scholar]
- 2.McAfee J.G., Huang,M., Soltaninassab,S., Rech,J.E., Iyengar,S. and LeStourgeon,W.M. (1997) The packaging of pre-mRNA. In Krainer,A.R. (ed.), Eukaryotic mRNA Processing. IRL, Oxford, UK, pp. 68–102.
- 3.Fu X.D., Mayeda,A., Maniatis,T. and Krainer,A.R. (1992) General splicing factors SF2 and SC35 have equivalent activities in vitro and both affect alternative 5′ and 3′ splice site selection. Proc. Natl Acad. Sci. USA, 89, 11224–11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayeda A. and Krainer,A.R. (1992) Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell, 68, 365–375. [DOI] [PubMed] [Google Scholar]
- 5.Mayeda A., Helfman,D.M. and Krainer,A.R. (1993) Modulation of exon skipping and inclusion by heterogeneous nuclear ribonucleoprotein A1 and pre-mRNA splicing factor SF2/ASF. Mol. Cell. Biol., 13, 2993–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caceres J.F., Stamm,S., Helfman,D.M. and Krainer,A.R. (1994) Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science, 265, 1706–1709. [DOI] [PubMed] [Google Scholar]
- 7.Yang X., Bani,M.R., Lu,S.J., Rowan,S., Ben-David,Y. and Chabot,B. (1994) The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc. Natl Acad. Sci. USA, 91, 6924–6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Del Gatto-Konczak F., Olive,M., Gesnel,M.C. and Breathnach,R. (1999) hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol. Cell. Biol., 19, 251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bilodeau P.S., Domsic,J.K., Mayeda,A., Krainer,A.R. and Stoltzfus,C.M. (2001) RNA splicing at human immunodeficiency virus type 1 3′ splice site A2 is regulated by binding of hnRNP A/B proteins to an exonic splicing silencer element. J. Virol., 75, 8487–8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caputi M., Mayeda,A., Krainer,A.R. and Zahler,A.M. (1999) hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J., 18, 4060–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu J., Mayeda,A. and Krainer,A.R. (2001) Exon identity established through differential antagonism exonic splicing silencer-bound hnRNP A1 and enhancer bound SR proteins. Mol. Cell, 8, 1351–1361. [DOI] [PubMed] [Google Scholar]
- 12.Pinol-Roma S. and Dreyfuss,G. (1992) Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature, 355, 730–732. [DOI] [PubMed] [Google Scholar]
- 13.Pontius B.W. (1993) Close encounters: why unstructured, polymeric domains can increase rates of specific macromolecular association. Trends Biochem. Sci., 18, 181–186. [DOI] [PubMed] [Google Scholar]
- 14.LaBranche H., Dupuis,S., Ben-David,Y., Bani,M.R., Wellinger,R.J. and Chabot,B. (1998) Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nature Genet., 19, 199–202. [DOI] [PubMed] [Google Scholar]
- 15.Mayeda A., Munroe,S.H., Caceres,J.F. and Krainer,A.R. (1994) Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J., 13, 5483–5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayeda A., Munroe,S.H., Xu,R.M. and Krainer,A.R. (1998) Distinct functions of the closely related tandem RNA-recognition motifs of hnRNP A1. RNA, 4, 1111–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burd C.G. and Dreyfuss,G. (1994) RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J., 13, 1197–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erlitzki R. and Fry,M. (1997) Sequence-specific binding protein of single-stranded and unimolecular quadruplex telomeric DNA from rat hepatocytes. J. Biol. Chem., 272, 15881–15890. [DOI] [PubMed] [Google Scholar]
- 19.Ishikawa F., Matunis,M.J., Dreyfuss,G. and Cech,T.R. (1993) Nuclear proteins that bind the pre-mRNA 3′ splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol. Cell. Biol., 13, 4301–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKay S.J. and Cooke,H. (1992) hnRNP A2/B1 binds specifically to single stranded vertebrate telomeric repeat TTAGGGn. Nucleic Acids Res., 20, 6461–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding J., Hayashi,M.K., Zhang,Y., Manche,L., Krainer,A.R. and Xu,R.M. (1999) Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev., 13, 1102–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pollard V.W., Michael,W.M., Nakielny,S., Siomi,M.C., Wang,F. and Dreyfuss,G. (1996) A novel receptor-mediated nuclear protein import pathway. Cell, 86, 985–994. [DOI] [PubMed] [Google Scholar]
- 23.Birney E., Kumar,S. and Krainer,A.R. (1993) Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res., 21, 5803–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burd C.G. and Dreyfuss,G. (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science, 265, 615–621. [DOI] [PubMed] [Google Scholar]
- 25.Perez-Canadillas J.M. and Varani,G. (2001) Recent advances in RNA-protein recognition. Curr. Opin. Struct. Biol., 11, 53–58. [DOI] [PubMed] [Google Scholar]
- 26.Scherly D., Boelens,W., Dathan,N.A., van Venrooij,W.J. and Mattaj,I.W. (1990) Major determinants of the specificity of interaction between small nuclear ribonucleoproteins U1A and U2B″ and their cognate RNAs. Nature, 345, 502–506. [DOI] [PubMed] [Google Scholar]
- 27.Gorlach M., Burd,C.G. and Dreyfuss,G. (1994) The determinants of RNA-binding specificity of the heterogeneous nuclear ribonucleoprotein C proteins. J. Biol. Chem., 269, 23074–23078. [PubMed] [Google Scholar]
- 28.Oubridge C., Ito,N., Evans,P.R., Teo,C.H. and Nagai,K. (1994) Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature, 372, 432–438. [DOI] [PubMed] [Google Scholar]
- 29.Allain F.H., Bouvet,P., Dieckmann,T. and Feigon,J. (2000) Molecular basis of sequence-specific recognition of pre-ribosomal RNA by nucleolin. EMBO J., 19, 6870–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conte M.R., Grune,T., Ghuman,J., Kelly,G., Ladas,A., Matthews,S. and Curry,S. (2000) Structure of tandem RNA recognition motifs from polypyrimidine tract binding protein reveals novel features of the RRM fold. EMBO J., 19, 3132–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crowder S.M., Kanaar,R., Rio,D.C. and Alber,T. (1999) Absence of interdomain contacts in the crystal structure of the RNA recognition motifs of Sex-lethal. Proc. Natl Acad. Sci. USA, 96, 4892–4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deo R.C., Bonanno,J.B., Sonenberg,N. and Burley,S.K. (1999) Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell, 98, 835–845. [DOI] [PubMed] [Google Scholar]
- 33.Handa N., Nureki,O., Kurimoto,K., Kim,I., Sakamoto,H., Shimura,Y., Muto,Y. and Yokoyama,S. (1999) Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature, 398, 579–585. [DOI] [PubMed] [Google Scholar]
- 34.Wang X. and Tanaka Hall,T.M. (2001) Structural basis for recognition of AU-rich element RNA by the HuD protein. Nature Struct. Biol., 8, 141–145. [DOI] [PubMed] [Google Scholar]
- 35.Shamoo Y., Krueger,U., Rice,L.M., Williams,K.R. and Steitz,T.A. (1997) Crystal structure of the two RNA binding domains of human hnRNP A1 at 1.75 Å resolution. Nature Struct. Biol., 4, 215–222. [DOI] [PubMed] [Google Scholar]
- 36.Xu R.M., Jokhan,L., Cheng,X., Mayeda,A. and Krainer,A.R. (1997) Crystal structure of human UP1, the domain of hnRNP A1 that contains two RNA-recognition motifs. Structure, 5, 559–570. [DOI] [PubMed] [Google Scholar]
- 37.Shamoo Y., Abdul-Manan,N. and Williams,K.R. (1995) Multiple RNA binding domains (RBDs) just don’t add up. Nucleic Acids Res., 23, 725–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allain F.H., Gubser,C.C., Howe,P.W., Nagai,K., Neuhaus,D. and Varani,G. (1996) Specificity of ribonucleoprotein interaction determined by RNA folding during complex formulation. Nature, 380, 646–650. [DOI] [PubMed] [Google Scholar]
- 39.Jokhan L., Dong,A.P., Mayeda,A., Krainer,A.R. and Xu,R.M. (1997) Crystallization and preliminary X-ray diffraction studies of UP1, the two-RRM domain of hnRNP A1. Acta Crystallogr. D, 53, 615–618. [DOI] [PubMed] [Google Scholar]
- 40.Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 41.Brunger A.T., Adams,P.D., Clore,G.M., DeLano,W.L., Gros,P., Grosse-Kunstleve,R.W., Jiang,J.S., Kuszewski,J., Nilges,M., Pannu,N.S. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 42.Sheldrick G.M. and Schneider,T.R. (1997) SHELXL: high-resolution refinement. Methods Enzymol., 277, 319–343. [PubMed] [Google Scholar]
- 43.Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 44.McRee D.E. (1999) XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol., 125, 156–165. [DOI] [PubMed] [Google Scholar]
- 45.Read R.J. (1986) Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. A, 42, 140–149. [Google Scholar]
- 46.Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK—a program to check the stereochmical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- 47.Cohen G.H. (1997) ALIGN: a program to superimpose protein coordinates, accounting for insertions and deletions. J. Appl. Crystallogr., 30, 1160–1161. [Google Scholar]
- 48.Burley S.K. and Petsko,G.A. (1985) Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science, 229, 23–28. [DOI] [PubMed] [Google Scholar]
- 49.Yamano A. and Teeter,M.M. (1994) Correlated disorder of the pure Pro22/Leu25 form of crambin at 150 K refined to 1.05-Å resolution. J. Biol. Chem., 269, 13956–13965. [PubMed] [Google Scholar]
- 50.Caspar D.L., Clarage,J., Salunke,D.M. and Clarage,M. (1988) Liquid-like movements in crystalline insulin. Nature, 332, 659–662. [DOI] [PubMed] [Google Scholar]
- 51.Clarage J.B., Clarage,M.S., Phillips,W.C., Sweet,R.M. and Caspar,D.L. (1992) Correlations of atomic movements in lysozyme crystals. Proteins, 12, 145–157. [DOI] [PubMed] [Google Scholar]
- 52.Dallaire F., Dupuis,S., Fiset,S. and Chabot,B. (2000) Heterogeneous nuclear ribonucleoprotein A1 and UP1 protect mammalian telomeric repeats and modulate telomere replication in vitro. J. Biol. Chem., 275, 14509–14516. [DOI] [PubMed] [Google Scholar]
- 53.Fiset S. and Chabot,B. (2001) hnRNP A1 may interact simultaneously with telomeric DNA and the human telomerase RNA in vitro. Nucleic Acids Res., 29, 2268–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee A.L., Kanaar,R., Rio,D.C. and Wemmer,D.E. (1994) Resonance assignments and solution structure of the second RNA-binding domain of sex-lethal determined by multidimensional heteronuclear magnetic resonance. Biochemistry, 33, 13775–13786. [DOI] [PubMed] [Google Scholar]