Abstract
The cannabinoids cannabidiol (CBD) and delta-9-tetrahydrocannabinol (THC) undergo extensive oxidative metabolism in the liver. Although cytochromes P450 form the primary, pharmacologically active, hydroxylated metabolites of CBD and THC, less is known about the enzymes that generate the major in vivo circulating metabolites of CBD and THC, 7-carboxy-CBD and 11-carboxy-THC, respectively. The purpose of this study was to elucidate the enzymes involved in forming these metabolites. Cofactor dependence experiments with human liver subcellular fractions revealed that 7-carboxy-CBD and 11-carboxy-THC formation is largely dependent on cytosolic NAD+-dependent enzymes, with lesser contributions from NADPH-dependent microsomal enzymes. Experiments with chemical inhibitors provided evidence that 7-carboxy-CBD formation is mainly dependent on aldehyde dehydrogenases and 11-carboxy-THC formation is mediated also in part by aldehyde oxidase. This study is the first to demonstrate the involvement of cytosolic drug-metabolizing enzymes in generating major in vivo metabolites of CBD and THC and addresses a knowledge gap in cannabinoid metabolism.
Keywords: cannabidiol, tetrahydrocannabinol, cannabinoid, drug metabolism
Cannabidiol (CBD) and delta-9-tetrahydrocannabinol (THC) (Figure 1) are two pharmacologically active natural products derived from cannabis. Though the chemical structures differ by only a single functional group, the pharmacology of these compounds differs significantly. THC is a partial agonist of cannabinoid receptors 1 and 2 (CB1 and CB2) located predominantly in the central nervous system and in immune tissues, respectively.1,2 The psychoactive effects of THC are mediated through binding to CB1.1 In contrast, CBD has low binding affinity for CB1 and CB2 and may decrease the activity of THC through negative allosteric modulation of CB1.1,3 CBD lacks the euphorigenic properties of THC and is commonly used as an anticonvulsant and anxiolytic agent. In 2018, CBD was approved by the U.S. Food and Drug Administration (FDA) to treat Lennox-Gastaut and Dravet syndromes, two rare and severe forms of epilepsy.4 Though the exact mechanism of action is unknown, CBD is not thought to exhibit anticonvulsant activity by interacting with cannabinoid receptors;4 rather, it may have a multimodal mechanism through binding to transient receptor potential vanilloid-1 (TRPV1), G protein-coupled receptor-55 (GPR55), and equilibrative nucleoside transporter 1 (ENT-1) in the brain.5
Figure 1.
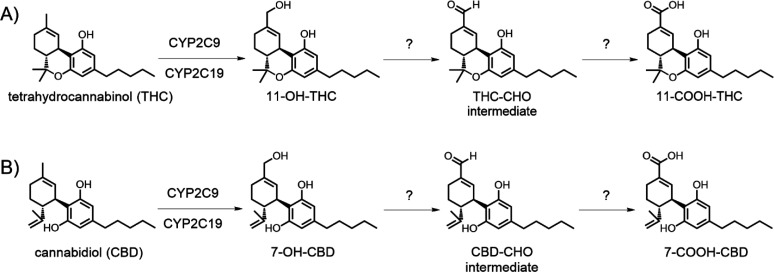
Proposed metabolic pathway for formation of 11-COOH-THC and 7-COOH-CBD. (A) THC is metabolized to the active metabolite 11-OH-THC by P450 enzymes. Subsequent oxidative metabolism generates the major circulating metabolite 11-COOH-THC through the putative multistep mechanism shown. (B) CBD similarly is metabolized by CYP2C9 and CYP2C19 to the pharmacologically active metabolite 7-OH-CBD, followed by oxidative metabolism to form 7-COOH-CBD.
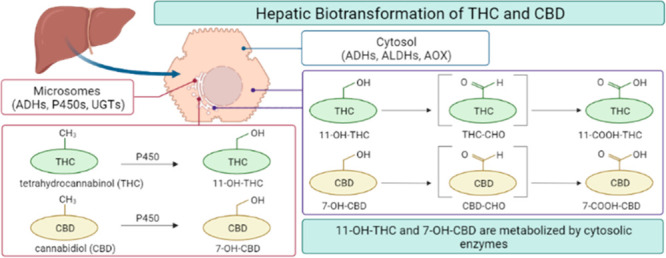
Both CBD and THC undergo oxidation and glucuronidation by cytochrome P450 (P450) and UDP-glucuronosyltransferase (UGT) enzymes, respectively, in the liver and gut prior to urinary and hepatoiliary excretion (Figure 1).4,6,7 The major circulating metabolites of CBD and THC in vivo are 7-carboxy-CBD (7-COOH-CBD) and 11-nor-9-carboxy-tetrahydrocannabinol (abbreviated as 11-carboxy-THC or 11-COOH-THC) formed from the pharmacologically active metabolites 7-hydroxy-CBD (7-OH-CBD) and 11-hydroxy-THC (11-OH-THC), respectively. Glucuronide metabolites of these carboxylic acids make up the majority of drug-related material recovered in urine.8−10 The plasma area under the concentration versus time curve (AUC) of 7-COOH-CBD is up to 40-fold higher than that of CBD following repeated administration of CBD to children.4,8 A similar AUC ratio of 11-COOH-THC to THC was measured following single-dose oral administration of THC to healthy adult volunteers,11 and 11-COOH-THC has been proposed as a biomarker for level of cannabis use.12−14 Additionally, the major glucuronide metabolite of 11-COOH-THC has been shown to inhibit multiple major P450 enzymes, including CYP2B6, CYP2C9, and CYP2D6 in vitro.(15) Factors affecting downstream 7-COOH-CBD and 11-COOH-THC formation may therefore impact the systemic exposure of the active metabolites of CBD and THC and hence the pharmacological effects of CBD and THC.
While the P450 enzymes such as CYP2C9, CYP2C19, and CYP3A4 are responsible for CBD and THC monohydroxylation,16−19 the enzyme(s) involved in generating 7-COOH-CBD and 11-COOH-THC remain unclear. Metabolism of alcohols such as 7-OH-CBD and 11-OH-THC by drug-metabolizing enzymes often occurs through multiple reaction steps, which may yield reactive aldehyde metabolites in the process.20 Common biotransformation pathways for primary hydroxylated metabolites involve conversion to aldehydes and carboxylic acids by cytochrome P450 enzymes, alcohol dehydrogenases (ADH), aldehyde dehydrogenases (ALDH), aldo-keto reductases, and aldehyde oxidase (AOX).21−23 Although all of these non-P450 enzymes are important in drug clearance, they may often be overlooked in preclinical drug metabolism studies.24
ADHs are NAD+-dependent enzymes that reversibly oxidize alcohols into aldehydes and ketones,21,22,25 including generation of the toxic ethanol metabolite acetaldehyde. There are over 20 human ADH enzymes organized into five classes based on amino acid sequence, activity, and tissue expression.23 ADH enzymes are variably expressed in the liver, GI tract, and other tissues.26,27 The competitive, nonspecific ADH inhibitor 4-methylpyrazole is used as an antidote for methanol and ethylene glycol poisoning.26
ALDHs are a superfamily of enzymes that reversibly catalyze the NAD(P)+-dependent oxidation of aldehydes into carboxylic acids.22 Mammalian ALDH enzymes have been organized into four different classes based on subcellular location and affinity toward the ethanol metabolite acetaldehyde.22 ALDHs are ubiquitously expressed within the mitochondria and cytosol of cells not only in the liver but also in the lungs, digestive tract, skin, kidneys, pancreas, and endocrine tissues, among others.22,27 In addition to their role in retinoic acid synthesis, ALDH enzymes are important for detoxication of reactive aldehyde metabolites formed during lipid peroxidation, as well as metabolites of cyclophosphamide, ifosphamide, and abacavir, among others.21,22,25
AOX is another cytosolic enzyme that catalyzes oxidation of aldehydes into carboxylic acids, as well as the oxidation of some nitrogen-containing aromatic ring systems.22 Like ALDH, AOX also synthesizes retinoic acid from retinaldehyde.28
CYP3A has been hypothesized to play a role in 7-COOH-CBD generation; however, direct evidence has not been reported in the literature, and little is known about the biotransformation of 7-OH-CBD to 7-COOH-CBD.16,29 Though clinical data show that 7-COOH-CBD is the most abundant metabolite, preliminary in vitro data from human liver microsomes (HLM) demonstrated relatively low turnover to 7-COOH-CBD when 7-OH-CBD was used as a substrate.18 Similar observations have been made in in vitro experiments examining the microsomal metabolism of THC to 11-OH-THC and 11-COOH-THC.17 Scaling the in vitro P450-mediated formation clearance of 11-COOH-THC from 11-OH-THC (4.6 L/h) to in vivo results in over 5-fold under-prediction of in vivo formation clearance of 11-COOH-THC (25.4 L/h),12,30 thus further suggesting that non-P450 enzymes play a role. While prior work has investigated 11-OH-THC and 7-OH-CBD oxidative metabolism in vitro, these studies have focused on microsomal P450 enzyme involvement, and metabolite formation by non-NADPH-dependent enzymes was not examined.7,16−18
In the present study, we performed reaction phenotyping experiments with human liver S9 (HLS9) and cytosol (HLC) to investigate whether NADPH- and NAD+-dependent enzymes form the major carboxylic acid metabolites of CBD and THC. Based on our findings, we conclude that 7-COOH-CBD and 11-COOH-THC may be generated through a multistep process catalyzed mainly by cytosolic dehydrogenases. This work demonstrates the importance of enzymes that were previously not known to be involved in cannabinoid clearance and addresses a knowledge gap in the literature related to the metabolism pathways of CBD and THC.
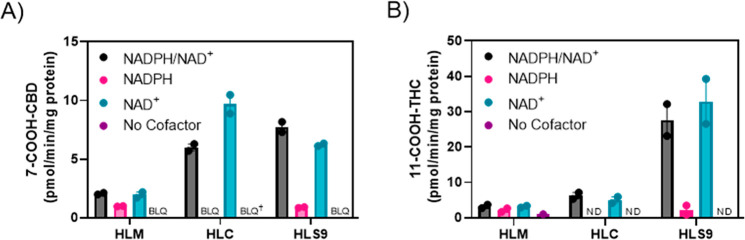
To determine the roles of NAD+- vs NADPH-dependent enzymes in 7-COOH-CBD formation, 7-COOH-CBD formation from 7-OH-CBD was measured in HLS9 containing both microsomal and cytosolic enzymes, and in HLM and HLC alone in the presence of the different cofactors (Figure 2). Metabolite generation was measured by LC-MS/MS (representative LC-MS/MS chromatograms are in the Supporting Information, Figure S1). Metabolite formation rates were determined to be linear with respect to time and protein concentration (Supporting Information, Figure S2). 7-COOH-CBD was formed in HLS9 and HLC largely by NAD+-dependent enzymes, whereas formation in incubations with HLS9 containing NADPH alone was negligible (Figure 2A). 7-COOH-CBD formation in HLM was lower compared to that in HLS9 and HLC. Given the comparable rates of metabolite formation observed with HLS9 and HLC, these findings indicate that 7-COOH-CBD formation is driven largely by NAD+-dependent cytosolic enzymes, with a minor contribution from NADPH-dependent enzymes. The results from initial experiments comparing rates of 7-OH-CBD depletion and 7-COOH-CBD formation in HLS9 at higher protein concentrations with different cofactors are shown in the Supporting Information, Tables S1 and S2, and Figure S3.
Figure 2.
Cofactor dependence of 7-COOH-CBD and 11-COOH-THC formation. Pooled HLM, HLC, and HLS9 were incubated with 1 μM 7-OH-CBD (A) and 1 μM 11-OH-THC (B) in the presence of NADPH (1 mM), NAD+ (2 mM), their combination, or no cofactor. Bars represent the mean of two independent experiments, and points show the mean values in individual experiments performed as triplicates. ND = not detected; BLQ = below limit of quantitation (determined by mean rate of formation for 7-COOH-CBD). The limit of quantitation (LOQ) for 7-COOH-CBD was 0.69 pmol/min/mg protein. †For this condition, the mean rate of formation consisted of one result above the LOQ and one result below the LOQ (trial 1 rate = 0.98 pmol/min/mg protein, trial 2 rate = 0.53 pmol/min/mg protein, mean rate = 0.76 pmol/min/mg protein).
11-COOH-THC formation from 11-OH-THC was similarly studied in HLS9, HLM, and HLC in the presence and absence of the relevant cofactors (Figure 2B) and measured using LC-MS/MS under linear conditions for metabolite generation. (See Supporting Information, Figure S4 for representative chromatograms and Figure S5 for protein–time linearity data.) Formation of 11-COOH-THC was highest in HLS9 and also driven largely by the presence of NAD+. Since 11-COOH-THC formation was higher in HLS9 than in cytosol (HLC), the data suggest that both microsomal NAD+-dependent ADHs and cytosolic NAD+-dependent ALDHs and/or AOX contribute to 11-COOH-THC formation from 11-OH-THC.
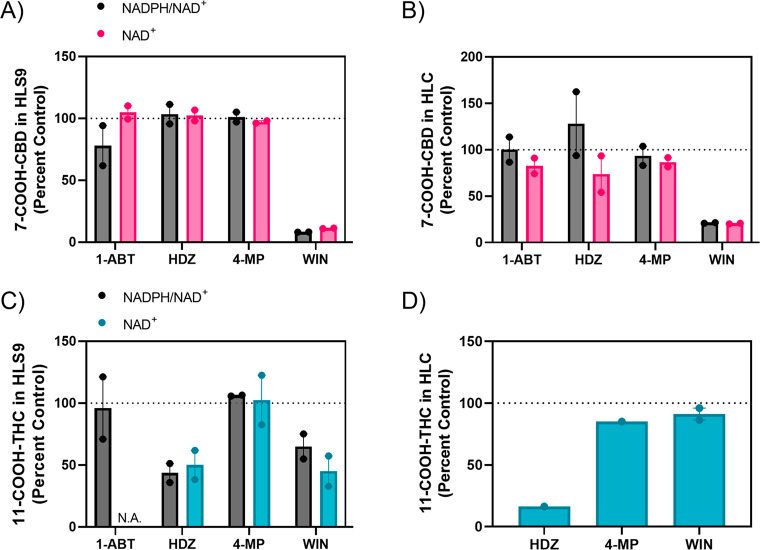
To identify the enzymes involved in 7-COOH-CBD and 11-COOH-THC formation, chemical inhibitors were used in incubations with HLS9 and HLC (Figure 3). The ALDH inhibitor WIN18,446 (WIN) reduced 7-COOH-CBD formation in HLS9 and HLC by 89% and 80%, respectively, in reactions supplemented with NAD+ (Figures 3A and 3B). In addition, the nonspecific P450 inhibitor 1-aminobenzotriazole (1-ABT) reduced NADPH-dependent 7-COOH-CBD formation by an average of 22% in HLS9. Hydralazine appeared to reduce NAD+-dependent 7-COOH-CBD formation in HLC; however, this finding was not replicated in incubations with both cofactors in HLC or in HLS9. Inhibition of ADH by 4-methylpyrazole did not appear to affect 7-COOH-CBD formation in either subcellular fraction. The absolute values for rates of 7-COOH-CBD formation are shown in the Supporting Information, Figure S6.
Figure 3.
Cannabinoid metabolite formation in the presence of selected chemical inhibitors. Pooled HLS9 and HLC were incubated with DMSO vehicle (0.5% v/v), 1-aminobenzotriazole (1-ABT, P450 inhibitor, 500 μM), hydralazine (HDZ, AOX inhibitor, 25 μM), 4-methylpyrazole (4-MP, ADH inhibitor, 250 μM), or WIN18,466 (WIN, ALDH inhibitor, 250 μM) with NADPH (1 mM) and NAD+ (2 mM) or NAD+ alone. Panels A and B show 7-COOH-CBD formation in HLS9 and HLC, respectively. Panels C and D show 11-COOH-THC formation in HLS9 and HLC, respectively. For panel C, experiments with 1-ABT were performed only with NADPH and NAD+ together. For panel D, experiments were performed with HLC in the presence and absence of NAD+ only. Bars represent the mean of two independent experiments, and the points show the range of the independent experiments performed in triplicate and normalized to matching control incubations without inhibitors. Gray bars = reactions performed with NADPH and NAD+, pink bars = reactions performed with 7-OH-CBD and NAD+, blue bars = reactions performed with 11-OH-THC and NAD+. N.A. = not applicable (experiment not performed).
In contrast to the observations with 7-COOH-CBD formation, hydralazine reduced 11-COOH-THC formation in HLS9 by approximately 50% and in HLC by over 80% for NAD+-supplemented incubations (Figures 3C and 3D), suggesting a potential role of AOX in one of the steps in 11-COOH-THC formation from 11-OH-THC. WIN18,446 reduced 11-COOH-THC formation in HLS9 by 45% but had a minimal effect in HLC. Inhibition of P450 enzymes and ADH had little effect on 11-COOH-THC formation.
To further examine the role of ALDH enzymes in forming 7-COOH-CBD, the concentration resulting in 50% inhibition (IC50) by the ALDH inhibitor WIN18,446 was determined in HLC (Figure 4). WIN was a potent inhibitor of 7-COOH-CBD formation, with an estimated IC50 of 285 ± 17 nM. This IC50 is consistent with the value measured for WIN18,446 inhibition of retinoic acid formation by recombinant purified ALDH1A1 and ALDH1A3 and suggests that ALDH1A1 may play a role in 7-COOH-CBD formation,31 as ALDH1A3 is not found in HLS9.28
Figure 4.
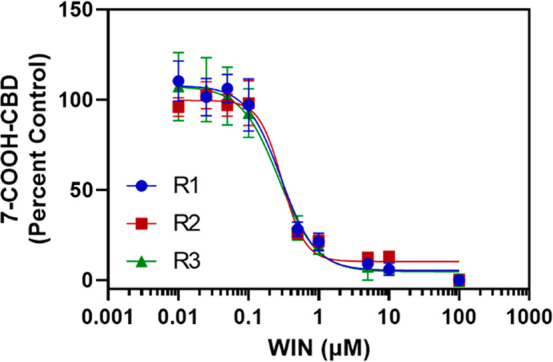
IC50of WIN18,446 for inhibition of 7-COOH-CBD formation in HLC. 7-OH-CBD (1 μM) was incubated with HLC (0.5 mg/mL) and NAD+ (2 mM) and the ALDH inhibitor WIN 18,446 (WIN, 0.01–100 μM) for 30 min. The estimated IC50 of WIN (285 ± 17 nM) was calculated by fitting a nonlinear dose–response inhibition model in GraphPad Prism to the data. Points represent the mean ± standard deviation of three replicates from a single experiment, and each curve was calculated from one of three independent experiments (R1–R3).
Given the potential role of AOX in 11-COOH-THC formation, the IC50 of the AOX inhibitor hydralazine was also determined in HLS9 (Figure 5). Hydralazine was found to strongly inhibit 11-COOH-THC generation, with an IC50 of 53 nM. This value is consistent with previously determined hydralazine IC50 toward AOX.28 It should be noted that hydralazine is a time-dependent inhibitor of AOX, and calculated IC50 values should be considered in the context of the incubation time used in the experiment. WIN18,446 was found to weakly inhibit 11-COOH-THC formation, with an estimated IC50 > 60 μM (Figure 6), suggesting ALDH1As and ALDH2 are not involved in 11-COOH-THC formation.
Figure 5.
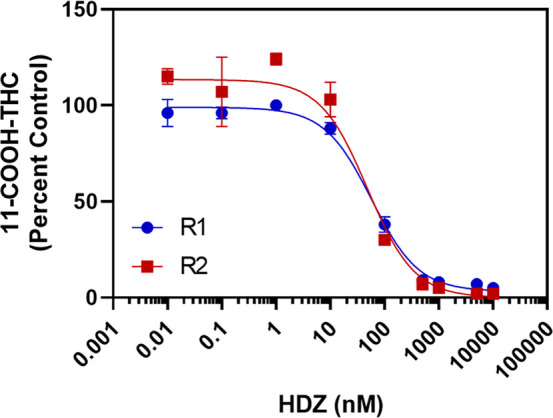
IC50of hydralazine for inhibition of 11-COOH-THC formation in HLS9. 11-OH-THC (3 μM) was incubated with HLS9 (0.1 mg/mL) and NAD+ (2 mM) and the AOX inhibitor hydralazine (HDZ, 0.01–10,000 nM) for 20 min. The estimated IC50 of HDZ (53 nM) was calculated by fitting a nonlinear dose–response inhibition model in GraphPad Prism to the data. Points represent the mean ± standard deviation of three replicates from a single experiment, and curves shown were calculated from two independent experiments (R1 and R2) completed across two different days.
Figure 6.
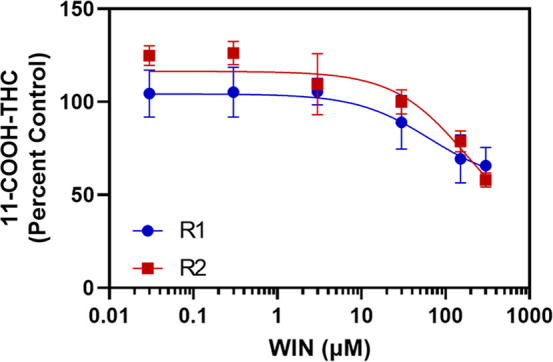
IC50of WIN18,446 for inhibition of 11-COOH-THC formation in HLS9. 11-OH-THC (3 μM) was incubated with HLS9 (0.1 mg/mL) and NAD+ (2 mM) with the ALDH inhibitor WIN (0.03–300 μM) for 10 min. The estimated IC50 of WIN (>60 μM) was calculated by fitting a nonlinear dose–response inhibition model to the data in GraphPad Prism. Points represent the mean ± standard deviation of three replicates from a single experiment, and curves were calculated from two independent experiments (R1 and R2) completed across two different days.
The results of this study demonstrate that formation of 7-COOH-CBD and 11-COOH-THC is mainly NAD+-dependent in cytosolic and HLS9 hepatocellular fractions, with lesser contributions from NADPH-dependent enzymes. Formation of both 7-COOH-CBD and 11-COOH-THC was lower in reactions containing only the microsomal fraction regardless of the cofactor(s) added. The interpretation of reaction phenotyping experiments performed with chemical inhibitors shown in Figure 3 may be limited by the multistep and possibly multienzyme formation of 7-COOH-CBD and 11-COOH-THC from the respective alcohol metabolites and the potential involvement of an unstable aldehyde intermediate. For these reasons, it is difficult to determine which step of metabolite formation may be affected by the inhibitors used in this study, and whether one or both steps are inhibited. For example, it is possible that AOX supports one of the steps in 11-COOH-THC formation but not both and hence 11-COOH-THC formation cannot be observed in the absence of a cofactor. However, the contribution of AOX to one of the steps becomes apparent when NAD+ is added, allowing the two-step process for formation of 11-COOH-THC.
The difference in enzyme selectivity for some of the chemical inhibitors is another limitation for identifying the specific enzymes involved in 7-COOH-CBD and 11-COOH-THC formation. For example, 4-methylpyrazole is known to inhibit not only ADH but also CYP2E1.32 4-Methylpyrazole is a nonspecific inhibitor of multiple ADH isoenzymes, with preference for class I enzymes (i.e., ADH1A, -1B, and -1C).26,33 The class II enzyme ADH4 and class III enzyme ADH5 are far less sensitive to 4-methylpyrazole inhibition.23,33 Since each of these ADH enzymes is hepatically expressed, the lack of inhibition by 4-methylpyrazole is a potential limitation with the inhibition experiments performed in this study to evaluate the role of ADHs.26 In addition, WIN18,446 broadly inhibits multiple ALDH enzymes, including ALDH1A1, ALDH1A2, and ALDH1A3, as well as ALDH2.31,34 However, as ALDH2 is a mitochondrial enzyme, it is unlikely to contribute to 7-COOH-CBD or 11-COOH-THC formation observed in HLS9 or HLC. Similarly, ALDH1A2 and ALDH1A3 are not detected in HLS9,28 while ALDH1A1 expression is high in the human liver, and hence ALDH1A2 and ALDH1A3 are unlikely to be the enzymes inhibited by WIN18,446 in human liver fractions here.
For CBD, ALDH enzymes appear to be most important in forming 7-COOH-CBD, with minor contributions by P450 enzymes. WIN18,446 was a potent inhibitor of 7-COOH-CBD formation in HLC. The IC50 of WIN18,446 for 7-COOH-CBD formation in HLC was approximately 285 nM, supporting the involvement of ALDH1A1. This IC50 is similar to the values determined for WIN18,446 inhibiting retinaldehyde metabolism by recombinant ALDH1A1 (102 nM) and ALDH1A3 (187 nM).31 P450 enzymes may catalyze formation of an aldehyde intermediate, and ALDH may oxidize this intermediate to 7-COOH-CBD. Note that while P450s may be involved in this process, they do not appear to be required; in fact, formation of 7-COOH-CBD was higher in HLC and HLS9 compared to that in microsomes alone. Further, 7-COOH-CBD generation was higher in HLC incubations supplemented with NAD+ alone than when both cofactors (NADPH and NAD+) were added (Figure 1A). This finding suggests possible involvement of a NAD(P)H-dependent cytosolic enzyme, such as an aldo-keto reductase, capable of reducing the aldehyde intermediate back into an alcohol.23,35
Interestingly, while ALDH (likely ALDH1A1 inhibited by WIN18,446) was found to be important for forming 7-COOH-CBD, AOX (inhibited by hydralazine) appears more important for 11-COOH-THC formation. Hydralazine markedly decreased 11-COOH-THC formation in HLC. Inhibition of P450s had virtually no effect on forming 11-COOH-THC in HLS9. The inhibition of 11-COOH-THC formation by hydralazine in HLS9 (IC50 = 53 nM) appears more potent compared to the IC50 value reported in recombinant AOX for retinoic acid formation (420 nM),28 likely due to the longer incubation times employed in this study and inactivation of AOX by hydralazine.
Aldehyde intermediates such as the proposed metabolites in Figure 1 are electrophilic and reactive. Prior evidence shows that aldehyde intermediates may be capable of forming reactive oxygen species and binding to intracellular thiols and amines, leading to toxicity.20−22 Multiple approved drugs, such as cyclophosphamide and derivatives felbamate, sunitinib, pazopanib, and abacavir form toxic aldehyde metabolites that may contribute to adverse effects.21 Further work is needed to confirm the presence of an aldehyde intermediate in forming the cannabinoid carboxylic acid metabolites.
Notably, CBD and THC inhibit different UGT and P450 enzymes and can contribute to enzyme-mediated drug interactions;15,19,36,37 however, inhibition of ALDH, ADH, and AOX by these compounds (or their metabolites) has not been studied. The findings presented here may have clinical implications for patients using concomitant ALDH, ADH, or AOX substrates or inhibitors with cannabis. This may include xenobiotics (e.g., ethanol, chemotherapeutic agents, acrolein, or benzaldehyde) or endogenous compounds such as retinaldehyde or lipid peroxidation products, among others. Additional investigation of the inhibitory properties for these cannabinoids is therefore warranted.
Collectively, this study demonstrates that NAD+-dependent non-P450 enzymes play a critical role in cannabinoid metabolism. To our knowledge, this is the first report to demonstrate the involvement of non-P450 enzymes in the formation of 7-COOH-CBD and 11-COOH-THC. Further work is needed to determine the specific enzyme contributions to individual steps of 7-COOH-CBD and 11-COOH-THC formation, i.e., conversion to the aldehyde intermediate and subsequent oxidation to the respective carboxylic acid.
Experimental Procedures
A description of the chemicals and reagents used, experimental procedures, and LC-MS/MS instrumentation and analysis performed is provided in the Supporting Information.
Acknowledgments
LC-MS/MS analysis of CBD and CBD metabolites was conducted in part at the UNC Biomarker Mass Spectrometry Core Facility, which is supported by NIH NIEHS award number P30ES010126. The authors thank Peter Hans Cable and Dr. Zhenfa Zhang (University of North Carolina at Chapel Hill) for LC-MS/MS technical support.
Glossary
Abbreviations
- 11-COOH-THC
11-nor-9-carboxy-tetrahydrocannabinol, or 11-carboxy-tetrahydrocannabinol
- 1-ABT
1-aminobenzotriazole
- 4-MP
4-methylpyrazole
- 7-COOH-CBD
7-carboxy-cannabidiol
- ADH
alcohol dehydrogenase
- ALDH
aldehyde dehydrogenase
- AOX
aldehyde oxidase
- BLQ
below limit of quantitation
- CBD
cannabidiol
- HDZ
hydralazine
- LOQ
limit of quantitation
- NAD+
nicotinamide adenine dinucleotide
- NADP+
nicotinamide adenine dinucleotide phosphate
- NADPH
dihydronicotinamide adenine dinucleotide phosphate
- ND
not detected
- P450
cytochrome P450
- THC
delta-9-tetrahydrocannabinol
- WIN
WIN18,446, or N,N′-1,8-octanediylbis[2,2-dichloroacetamide]
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.3c00017.
Chemicals, reagents, and experimental procedures; Table S1, 7-OH-CBD depletion in HLS9 in the presence and absence of cofactors; Table S2, 7-COOH-CBD formation in HLS9 in the presence and absence of cofactors; Figure S1, representative LC-MS/MS chromatograms of cofactor dependence of 7-COOH-CBD formation; Figure S2, protein–time linearity of 7-COOH-CBD formation in HLC, HLM, and HLS9; Figure S3, 7-OH-CBD substrate depletion and 7-COOH-CBD formation in HLS9; Figure S4, representative LC-MS/MS chromatograms of cofactor dependence of 11-COOH-THC formation; Figure S5, protein–time linearity of 11-COOH-THC formation in HLC, HLM, and HLS9; and Figure S6, formation rates of cannabinoid metabolites in the presence of selected chemical inhibitors (PDF)
Author Contributions
J.L.B.: Conceptualization, methodology, validation, formal analysis, investigation, writing - original draft, writing - review and editing, visualization. A.K.A.: Conceptualization, methodology, validation, formal analysis, investigation, writing - original draft, writing - review and editing, visualization. N.I.: Conceptualization, writing - review and editing, supervision, funding acquisition. K.D.J.: Conceptualization, writing - review and editing, supervision, funding acquisition.
J.L.B. is supported by the National Institute of General Medical Sciences of the National Institutes of Health under award T32GM086330. A.K.A. is supported in part by the National Institute of General Medical Sciences of the National Institutes of Health under award T32GM007750. A.K.A. and N.I. are supported in part by the National Institute on Drug Abuse of the National Institutes of Health under award P01DA032507. N.I. is supported in part as the University of Washington School of Pharmacy Milo Gibaldi Endowed Chair of Pharmaceutics. K.D.J. is supported in part by the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM143044 and by the UNC Eshelman School of Pharmacy.
The research reported is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors declare no competing financial interest.
Supplementary Material
References
- Lucas C. J.; Galettis P.; Schneider J. The Pharmacokinetics and the Pharmacodynamics of Cannabinoids. Br. J. Clin. Pharmacol. 2018, 84, 2477–2482. 10.1111/bcp.13710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertwee R. G. The Diverse CB1 and CB2 Receptor Pharmacology of Three Plant Cannabinoids: Delta9-Tetrahydrocannabinol, Cannabidiol and Delta9-Tetrahydrocannabivarin. Br. J. Pharmacol. 2008, 153, 199–215. 10.1038/sj.bjp.0707442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laprairie R. B.; Bagher A. M.; Kelly M. E. M.; Denovan-Wright E. M. Cannabidiol Is a Negative Allosteric Modulator of the Cannabinoid CB1 Receptor. Br. J. Pharmacol. 2015, 172, 4790–4805. 10.1111/bph.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwich Biosciences . EPIDIOLEX® (cannabidiol) prescribing information, 2023. https://www.epidiolex.com/sites/default/files/pdfs/1120/EPX-03645-1120_EPIDIOLEX_(cannabidiol)_USPI.pdf.
- Gray R. A.; Whalley B. J. The Proposed Mechanisms of Action of CBD in Epilepsy. Epileptic Disord. 2020, 22, 10–15. 10.1684/epd.2020.1135. [DOI] [PubMed] [Google Scholar]
- Mazur A.; Lichti C. F.; Prather P. L.; Zielinska A. K.; Bratton S. M.; Gallus-Zawada A.; Finel M.; Miller G. P.; Radomińska-Pandya A.; Moran J. H. Characterization of Human Hepatic and Extrahepatic UDP-Glucuronosyltransferase Enzymes Involved in the Metabolism of Classic Cannabinoids. Drug Metab. Dispos. 2009, 37, 1496–1504. 10.1124/dmd.109.026898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K.; Yamaori S.; Funahashi T.; Kimura T.; Yamamoto I. Cytochrome P450 Enzymes Involved in the Metabolism of Tetrahydrocannabinols and Cannabinol by Human Hepatic Microsomes. Life Sci. 2007, 80, 1415–1419. 10.1016/j.lfs.2006.12.032. [DOI] [PubMed] [Google Scholar]
- Ujváry I.; Hanuš L. Human Metabolites of Cannabidiol: A Review on Their Formation, Biological Activity, and Relevance in Therapy. Cannabis Cannabinoid Res. 2016, 1, 90–101. 10.1089/can.2015.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chayasirisobhon S. Mechanisms of Action and Pharmacokinetics of Cannabis. Perm. J. 2020, 25, 19.200. 10.7812/TPP/19.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P. L.; Moffat A. C. Identification in Human Urine of Delta 9-Tetrahydrocannabinol-11-Oic Acid Glucuronide: A Tetrahydrocannabinol Metabolite. J. Pharm. Pharmacol. 2011, 32, 445–448. 10.1111/j.2042-7158.1980.tb12966.x. [DOI] [PubMed] [Google Scholar]
- Sachse-Seeboth C.; Pfeil J.; Sehrt D.; Meineke I.; Tzvetkov M.; Bruns E.; Poser W.; Vormfelde S. V.; Brockmöller J. Interindividual Variation in the Pharmacokinetics of Delta9-Tetrahydrocannabinol as Related to Genetic Polymorphisms in CYP2C9. Clin. Pharmacol. Ther. 2009, 85, 273–276. 10.1038/clpt.2008.213. [DOI] [PubMed] [Google Scholar]
- Huang W.; Czuba L. C.; Manuzak J. A.; Martin J. N.; Hunt P. W.; Klatt N. R.; Isoherranen N. Objective Identification of Cannabis Use Levels in Clinical Populations Is Critical for Detecting Pharmacological Outcomes. Cannabis Cannabinoid Res. 2022, 7, 852–864. 10.1089/can.2021.0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabritius M.; Favrat B.; Chtioui H.; Battistella G.; Annoni J.-M.; Appenzeller M.; Dao K.; Fornari E.; Lauer E.; Mall J.-F.; Maeder P.; Mangin P.; Staub C.; Giroud C. THC-COOH Concentrations in Whole Blood: Are They Useful in Discriminating Occasional from Heavy Smokers?. Drug Test. Anal. 2014, 6, 155–163. 10.1002/dta.1581. [DOI] [PubMed] [Google Scholar]
- Hädener M.; Martin Fabritius M.; König S.; Giroud C.; Weinmann W. Assessing Cannabis Consumption Frequency: Is the Combined Use of Free and Glucuronidated THCCOOH Blood Levels of Diagnostic Utility?. Drug Test. Anal. 2017, 9, 1043–1051. 10.1002/dta.2114. [DOI] [PubMed] [Google Scholar]
- Nasrin S.; Watson C. J. W.; Perez-Paramo Y. X.; Lazarus P. Cannabinoid Metabolites as Inhibitors of Major Hepatic CYP450 Enzymes, with Implications for Cannabis-Drug Interactions. Drug Metab. Dispos. 2021, 49, 1070–1080. 10.1124/dmd.121.000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R.; Yamaori S.; Takeda S.; Yamamoto I.; Watanabe K. Identification of Cytochrome P450 Enzymes Responsible for Metabolism of Cannabidiol by Human Liver Microsomes. Life Sci. 2011, 89, 165–170. 10.1016/j.lfs.2011.05.018. [DOI] [PubMed] [Google Scholar]
- Patilea-Vrana G. I.; Anoshchenko O.; Unadkat J. D. Hepatic Enzymes Relevant to the Disposition of (−)-Δ9-Tetrahydrocannabinol (THC) and Its Psychoactive Metabolite, 11-OH-THC. Drug Metab. Dispos. 2019, 47, 249–256. 10.1124/dmd.118.085548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beers J. L.; Fu D.; Jackson K. D. Cytochrome P450-Catalyzed Metabolism of Cannabidiol to the Active Metabolite 7-Hydroxy-Cannabidiol. Drug Metab. Dispos. 2021, 49, 882–891. 10.1124/dmd.120.000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal S.; Paine M. F.; Unadkat J. D. Comprehensive Predictions of Cytochrome P450 (P450)-Mediated In Vivo Cannabinoid-Drug Interactions Based on Reversible and Time-Dependent P450 Inhibition in Human Liver Microsomes. Drug Metab. Dispos. 2022, 50, 351–360. 10.1124/dmd.121.000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalgutkar A. S. Liabilities Associated with the Formation of “Hard” Electrophiles in Reactive Metabolite Trapping Screens. Chem. Res. Toxicol. 2017, 30, 220–238. 10.1021/acs.chemrestox.6b00332. [DOI] [PubMed] [Google Scholar]
- O’Brien P. J.; Siraki A. G.; Shangari N. Aldehyde Sources, Metabolism, Molecular Toxicity Mechanisms, and Possible Effects on Human Health. Crit. Rev. Toxicol. 2005, 35, 609–662. 10.1080/10408440591002183. [DOI] [PubMed] [Google Scholar]
- Ahmed Laskar A.; Younus H. Aldehyde Toxicity and Metabolism: The Role of Aldehyde Dehydrogenases in Detoxification, Drug Resistance and Carcinogenesis. Drug Metab. Rev. 2019, 51, 42–64. 10.1080/03602532.2018.1555587. [DOI] [PubMed] [Google Scholar]
- Parkinson A.; Ogilvie B. W.; Buckley D. B.; Kazmi F.; Parkinson O.. Biotransformation of Xenobiotics. In Casarett and Doull’s Toxicology: The Basic Science of Poisons, 9th ed.; Klaassen C. D., Ed.; McGraw-Hill Education: New York, NY, 2019. [Google Scholar]
- Argikar U. A.; Potter P. M.; Hutzler J. M.; Marathe P. H. Challenges and Opportunities with Non-CYP Enzymes Aldehyde Oxidase, Carboxylesterase, and UDP-Glucuronosyltransferase: Focus on Reaction Phenotyping and Prediction of Human Clearance. AAPS J. 2016, 18, 1391–1405. 10.1208/s12248-016-9962-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh J. S.; Reese M. J.; Thurmond L. M. The Metabolic Activation of Abacavir by Human Liver Cytosol and Expressed Human Alcohol Dehydrogenase Isozymes. Chem. Biol. Interact. 2002, 142, 135–154. 10.1016/S0009-2797(02)00059-5. [DOI] [PubMed] [Google Scholar]
- Di L.; Balesano A.; Jordan S.; Shi S. M. The Role of Alcohol Dehydrogenase in Drug Metabolism: Beyond Ethanol Oxidation. AAPS J. 2021, 23, 20. 10.1208/s12248-020-00536-y. [DOI] [PubMed] [Google Scholar]
- Uhlén M.; Fagerberg L.; Hallström B. M.; Lindskog C.; Oksvold P.; Mardinoglu A.; Sivertsson Å.; Kampf C.; Sjöstedt E.; Asplund A.; Olsson I.; Edlund K.; Lundberg E.; Navani S.; Szigyarto C. A.; Odeberg J.; Djureinovic D.; Takanen J. O.; Hober S.; Alm T.; Edqvist P. H.; Berling H.; Tegel H.; Mulder J.; Rockberg J.; Nilsson P.; Schwenk J. M.; Hamsten M.; von Feilitzen K.; Forsberg M.; Persson L.; Johansson F.; Zwahlen M.; von Heijne G.; Nielsen J.; Pontén F. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Zhong G.; Seaman C. J.; Paragas E. M.; Xi H.; Herpoldt K.-L.; King N. P.; Jones J. P.; Isoherranen N. Aldehyde Oxidase Contributes to All-Trans-Retinoic Acid Biosynthesis in Human Liver. Drug Metab. Dispos. 2021, 49, 202–211. 10.1124/dmd.120.000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison G.; Crockett J.; Blakey G.; Sommerville K. A Phase 1, Open-Label, Pharmacokinetic Trial to Investigate Possible Drug-Drug Interactions Between Clobazam, Stiripentol, or Valproate and Cannabidiol in Healthy Subjects. Clin. Pharmacol. Drug Dev. 2019, 8, 1009–1031. 10.1002/cpdd.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patilea-Vrana G. I.; Unadkat J. D. Quantifying Hepatic Enzyme Kinetics of (−)-Δ9-Tetrahydrocannabinol (THC) and Its Psychoactive Metabolite, 11-OH-THC, through In Vitro Modeling. Drug Metab. Dispos. 2019, 47, 743–752. 10.1124/dmd.119.086470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold S. L.; Kent T.; Hogarth C. A.; Schlatt S.; Prasad B.; Haenisch M.; Walsh T.; Muller C. H.; Griswold M. D.; Amory J. K.; Isoherranen N. Importance of ALDH1A Enzymes in Determining Human Testicular Retinoic Acid Concentrations. J. Lipid Res. 2015, 56, 342–357. 10.1194/jlr.M054718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirogi R.; Palacharla R. C.; Uthukam V.; Manoharan A.; Srikakolapu S. R.; Kalaikadhiban I.; Boggavarapu R. K.; Ponnamaneni R. K.; Ajjala D. R.; Bhyrapuneni G. Chemical Inhibitors of CYP450 Enzymes in Liver Microsomes: Combining Selectivity and Unbound Fractions to Guide Selection of Appropriate Concentration in Phenotyping Assays. Xenobiotica. 2015, 45, 95–106. 10.3109/00498254.2014.945196. [DOI] [PubMed] [Google Scholar]
- Lee S.-L.; Shih H.-T.; Chi Y.-C.; Li Y.-P.; Yin S.-J. Oxidation of Methanol, Ethylene Glycol, and Isopropanol with Human Alcohol Dehydrogenases and the Inhibition by Ethanol and 4-Methylpyrazole. Chem. Biol. Interact. 2011, 191, 26–31. 10.1016/j.cbi.2010.12.005. [DOI] [PubMed] [Google Scholar]
- Amory J. K.; Muller C. H.; Shimshoni J. A.; Isoherranen N.; Paik J.; Moreb J. S.; Amory D. W.; Evanoff R.; Goldstein A. S.; Griswold M. D. Suppression of Spermatogenesis by Bisdichloroacetyldiamines Is Mediated by Inhibition of Testicular Retinoic Acid Biosynthesis. J. Androl. 2011, 32, 111–119. 10.2164/jandrol.110.010751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosemond M. J. C.; Walsh J. S. Human Carbonyl Reduction Pathways and a Strategy for Their Study in Vitro. Drug Metab. Rev. 2004, 36, 335–361. 10.1081/DMR-120034154. [DOI] [PubMed] [Google Scholar]
- Gaston T. E.; Bebin E. M.; Cutter G. R.; Liu Y.; Szaflarski J. P. UAB CBD Program. Interactions between Cannabidiol and Commonly Used Antiepileptic Drugs. Epilepsia 2017, 58, 1586–1592. 10.1111/epi.13852. [DOI] [PubMed] [Google Scholar]
- Bansal S.; Maharao N.; Paine M. F.; Unadkat J. D. Predicting the Potential for Cannabinoids to Precipitate Pharmacokinetic Drug Interactions via Reversible Inhibition or Inactivation of Major Cytochromes P450. Drug Metab. Dispos. 2020, 48, 1008–1017. 10.1124/dmd.120.000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.