Abstract
Detection of cellular senescence is important quality analytics of cell therapy products, including mesenchymal stromal cells (MSCs). However, its detection is critically limited by the lack of specific markers and the destructive assays used to read out these markers. Here, we establish a rapid, live-cell assay for detecting senescent cells in heterogeneous mesenchymal stromal cell (MSC) cultures. We report that the T2 relaxation time measured by microscale Magnetic Resonance Relaxometry, which is related to intracellular iron accumulation, correlates strongly with senescence markers in MSC cultures under diverse conditions, including different passages and donors, size-sorted MSCs by inertial spiral microfluidic device, and drug-induced senescence. In addition, the live-cell and non-destructive method presented here has general applicability to other cells and tissues and can critically advance our understanding of cellular senescence.
Keywords: senescence, mesenchymal stromal cells, magnetic resonance relaxometry, live-cell assay
Graphical Abstract
Significance Statement.
Detection of senescent MSCs is important quality analytics for MSC-based cell therapy. The results of this study establish that the detection of senescent cells in heterogeneous mesenchymal stromal cell (MSC) cultures is possible by a live-cell and non-destructive method called magnetic resonance relaxometry.
Introduction
Cellular senescence is a cell state that leads to cell cycle exit accompanied by genetic, metabolic, and morphological changes of cells due to aging and other external or internal conditions.1 Cellular senescence was described nearly 60 years ago based on the observation that cells in culture have a finite ability to divide.2 Senescent cells are also found in tissues in vivo and are substantially implicated in many important pathologies. Cellular senescence is broadly defined as a complex cell fate that can be induced during normal development, physiological aging (longer time scales), or by intra- and extra-cellular stress (acute responses). Cellular senescence can be important for preventing cancer initiation and tumor progression.3-6 On the other hand, the accumulation of senescent cells in the body can have harmful effects, especially in age-related diseases like neurodegeneration, cardiovascular disease, osteoarthritis, renal dysfunction, non-alcoholic fatty liver disease, type 2 diabetes, and cancer.4,7,8 The expression of genes involved in DNA replication, DNA repair, and cell cycle is often downregulated in senescent cells.9 In addition, senescent cells display a senescence-associated secretory phenotype (SASP), which causes an alteration in the tissue microenvironment, including local or systemic inflammation and disruption of normal tissue structure, leading to resistance to immune clearance of these pathological cells.4,7,10 Thus, the detection and elimination of cells with senescent characteristics have become a target for medical intervention.3
Detecting and quantifying senescent cells is especially critical in cell therapy bioproduction.11 For example, mesenchymal stromal cells (MSCs) have clinical applications for indications in ischemic, inflammatory, and degenerative disorders as well as for regenerative medicine.10 Yet, senescent MSCs impose significant challenges in their clinical applications.11,12 Despite the clinical potential of MSCs, proliferation arrest is observed in MSC populations, along with morphological and phenotype alterations, after prolonged culture or expansion in vitro.11,13-15 Senescent MSCs exhibit enlarged and flat cell morphologies with stiffened nuclei and granular cytoplasm.4,11,15,16 Senescent MSCs have decreased multi-lineage differentiation potential11,14,15,17 and altered secretory and immunomodulatory functions, resulting in reduced therapeutic value.4,18,19 Notably, MSCs isolated from older donors show a higher level of senescence than younger donors,20 while significant variation in senescence levels in MSCs is observed among younger donors as well. Therefore, it is crucial to determine the senescent state of MSCs and other cell-based products as part of quality control in therapeutic cell production.
A critical bottleneck is the lack of quantitative tools that agnostically detect senescent MSCs in heterogeneous cell populations. Phenotypic diversity of senescent cells and a lack of robust biomarkers have hampered progress in both our understanding of cellular senescence and therapeutic applications.21,22 The standard method for detecting senescent MSCs is histochemical staining of acidic lysosomal β‐galactosidase (β-gal).4,23 The staining procedure requires stable pH (~6), cell fixation, and a long incubation period (>12 h). Alternatively, a modified colony-forming unit assay (CFU-f) can detect cellular senescence by estimating the proliferative potential of the MSCs after multiple passages.15,24,25 Real-time polymerase chain reaction (RT-qPCR) can also detect senescence by measuring expression levels of senescence-associated markers p16, p21, p53.4,7,26 However, these assays require lengthy and laborious procedures and are destructive end-point assays, which are not adequate for quality control of MSCs during cell manufacturing.27 Therefore, a rapid detection method that allows real-time quantification of senescent MSCs is critical for the quality control of cell therapeutics.
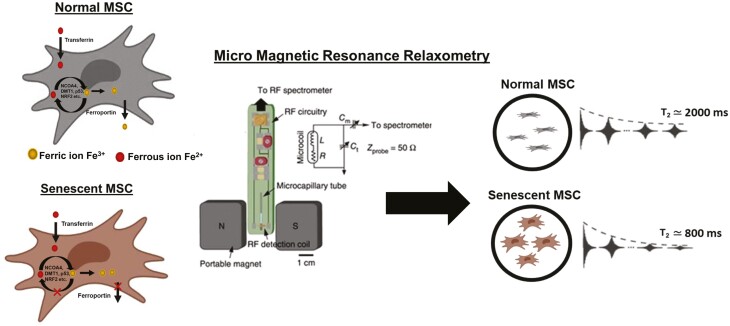
Significant evidence indicates that iron homeostasis and its dysfunction correlate with aging and related pathologies.28-32 Cellular iron is present in Fe2+ (reactive, labile, or “free” iron, diamagnetic) or Fe3+ (paramagnetic, often bound with iron storage protein ferritin) forms (Fig. 1). In iron-homeostatic cells, uptake, balancing, and recycling of Fe2+/Fe3+ are maintained by various iron transport system proteins (eg, transferrin (Tf), TfR1, ferroportin, NCOA4, and DMT1) and regulators (eg, p53, NRF2),33 involving many different molecular pathways.34 Notably, growing evidence indicates that an increase in intracellular iron may serve as a “natural marker” for cellular senescence. Senescent cells accumulate iron (ferritin-bound Fe3+) up to ~30-fold by adjusting their iron homeostasis proteins35 and inhibiting ferritinophagy36 (autophagy of Fe3+-bound ferritin) that recycles stored iron back to Fe2+. This sequestration of iron (Fe3+) in ferritin prevents ferroptosis,37 a unique cell death pathway requiring labile iron (Fe2+) released from ferritinophagy.38 Therefore, directly quantifying intracellular iron (Fe3+) in live cells could provide a measure for detecting cellular senescence in real-time during bioproduction.
Figure 1.
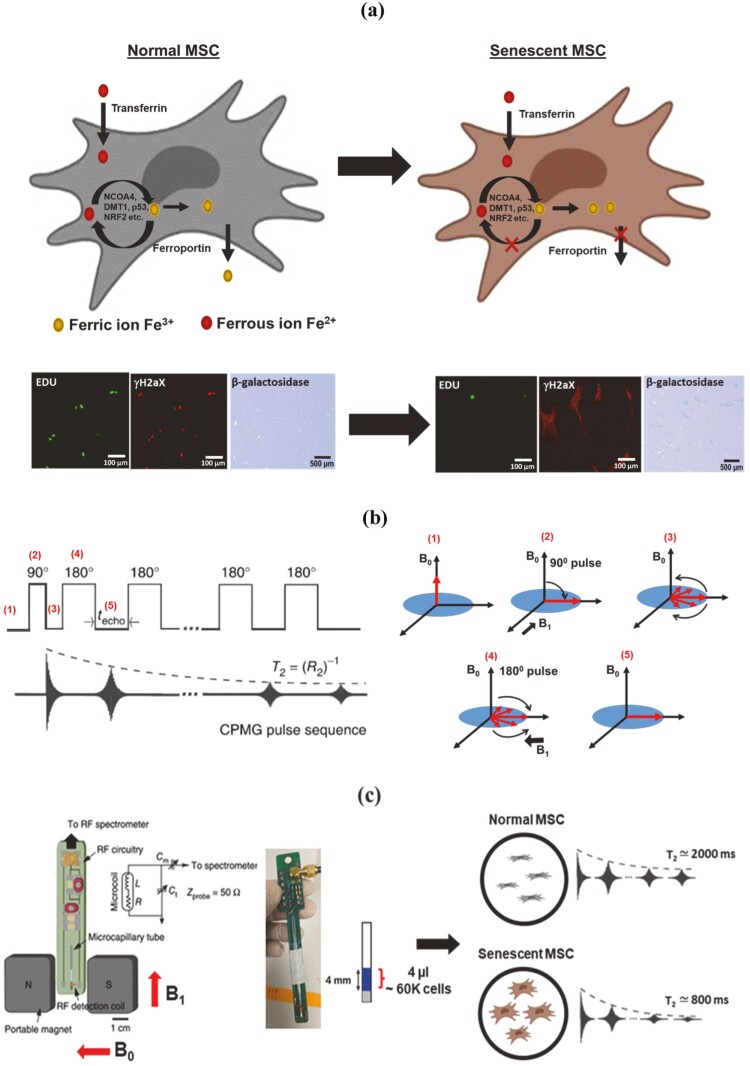
Live-cell and rapid detection of senescent MSCs using magnetic resonance relaxometry. (a) Normal cells maintain iron homeostasis, mediated by numerous iron transporters and iron-binding proteins. Senescent cells are correlated with the accumulation of paramagnetic Fe3+ (an inactive form of iron). Normal cells and senescent cells analyzed by standard assays are shown in the below images. The cells are stained by the senescence marker (γ-H2aX foci, red fluorescence), the proliferation marker, 5-ethynyl-2ʹ-deoxyuridine (Edu, green fluorescence), and β-galactosidase staining (blue fluorescence). (b) The CPMG (Carr-Purcell-Meiboom-Gill) pulse sequence for measuring the T2 values, which works efficiently in inhomogeneous magnetic fields produced by the permanent magnet of µMRR. A train of radiofrequency pulses is applied to the proton nuclei at the resonance frequency of 21.65 MHz with the inter echo time interval techo. It is repeated for thousands of echoes until an equilibrium is reached. This decaying peak height of successive echoes over time is called transverse relaxation time T2. (c) MRR system consists of a permanent magnet that provides a strong magnetic field (B0). A home-built radiofrequency (RF) detection probe is connected to an RF spectrometer. The MSC sample (young/proliferating/senescent MSCs) in the microcapillary tube is placed in the RF detection coil for T2 measurements. The microcapillary tube contains the 4 µL of MSC sample (blue) in the 4 mm detection range of the RF detection coil. The typical cell number required is 60 000 MSCs within the detection volume. The grey color is the crystoseal to seal the microcapillary tube. The right side shows the 1H spin-spin relaxation time T2 of normal and senescent MSCs, demonstrating decreased T2 in senescent MSCs compared to normal MSCs. Mesenchymal stem cell figures are adapted from the icon library of “Cell Types” by BioRender.com.
We previously reported microscale-Magnetic Resonance Relaxometry (µMRR) as an efficient malaria diagnostic.39 The increased magnetic susceptibility of paramagnetic Fe3+ in the hemozoin crystals of infected red blood cells (RBCs) caused faster transverse relaxation of protons (T2) than the diamagnetic Fe2+ state in uninfected/healthy RBCs.39,40 In addition, µMRR-based phenotyping of the oxidative stress response in diabetes mellitus patients’ blood was reported as a potential alternative to the conventional Hb-A1c test.41 Others also reported the high-sensitivity detection of tumor cells,42-44 bacteria,45 and tuberculosis46 using a similar µMRR device with immunomagnetic labeling of molecular and cellular targets. Here, we show that µMRR is a non-destructive, label-free, and rapid method for detecting senescence in heterogeneous MSC cultures across various conditions. Thus, µMRR represents a robust methodology for quality control for cell therapy manufacturing, opening the door for broader adaptation of this method for detecting cellular senescence in other cells and tissues.
Materials and Methods
MSC Culture
Bone marrow-derived MSCs from 7 different donors as described in Table 2 were purchased from Lonza Pte. Ltd. and Rooster Bio Inc. The cells were expanded in tissue culture plate (TCP) at an initial cell density of 1500 cells/cm2 in low glucose Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% GlutaMAX, and 1% Penicillin/Streptomycin (Thermo Fisher Scientific, Singapore) at 37 °C in 5% CO2 atmosphere. The medium was changed every 2 days, and the cells were harvested at 80% confluency for further experiments or subcultures. During the cells harvesting, the cells were incubated with 0.25% Trypsin-EDTA (Thermo Fisher Scientific, Singapore) for 3 min. The cell number was calculated using a disposable hemocytometer (INCYTO, Korea) with the trypan blue exclusion method. MSCs in passages 3 (P3) to 6 (P6) were used for the experiments unless otherwise stated. MSC senescence induction was carried out by treatment with culture media containing 10 or 20 ng/mL of TGF-β1 for 3 days. For doxorubicin (DOX) treatment, cells were seeded at 1500 cells/cm2 and incubated with 1 µM doxorubicin (Sigma) for 24 h at 37 °C. Subsequently, the medium was removed, and a fresh medium was added to the cells to incubate for 24 h before analysis.
Table 2.
Description of donors.
| Name | Tissue type | Source | Age | Gender |
|---|---|---|---|---|
| Donor 1 | Bone marrow | Human MSC | 20 Y | Male |
| Donor 2 | Bone marrow | Human MSC | 22 Y | Male |
| Donor 3 | Bone marrow | Human MSC | 25 Y | Male |
| Donor 4 | Bone marrow | Human MSC | 19 Y | Female |
| Donor 5 | Bone marrow | Human MSC | 24 Y | Male |
| Donor 6 | Bone marrow | Human MSC | 36 Y | Male |
| Donor 7 | Bone marrow | Human MSC | 26 Y | Male |
MSC Sorting With Inertial Spiral Microchannel Device
The inertial spiral microchannel device was designed and fabricated in the same way as previously described.47 The device has 8 loops with a radius decreasing from 12 mm to 4 mm and a trapezoidal cross-section with 580 µm width, 85 µm inner height, and 133 µm outer height. It has one inlet for the introduction of the cell suspension to be sorted and 2 outlets for the collection of sorted cells. The design was carved on a micro-milled aluminum mold (Whits Technologies Inc., Singapore), and the device was cast from polydimethylsiloxane (PDMS) with a ratio of 10:1 base and curing agent mixture (Sylgard 184, Dow Corning Inc., USA).
Before sorting, MSCs were resuspended in culture media at 1-2 million cells/mL and loaded into syringes (Thermo Fisher Scientific, Singapore) that were connected to Tygon tubing (Spectra Teknik Pte. Ltd., Singapore). The tubing was inserted into the inlet of the device, and 2 separate tubings were inserted at the outlets for the collection of sorted cells. A syringe pump (PHD2000, Harvard Apparatus Inc., USA) was used to control the flow rate of cell suspension in the device. Two serial stages of sorting were performed to separate MSCs into 3 populations of different sizes. The first sorting was carried out at 3.5 mL/min, and the cells collected at the inner outlet were the largest subpopulation (22-26 µm). The cells collected at the outer outlet were then subjected to the second sorting at 1.5 mL/min, and the cells collected at the inner and outer outlets were medium size subpopulations (15-22 µm) and smallest subpopulations (11-15 µm), respectively. The unsorted MSCs were used as a control in this experiment. Real-time visualization of the separation within the inertial spiral microchannel device was achieved by an inverted microscope (IX71, Olympus Co., Japan) equipped with a high-speed CCD camera (Phantom v9, Vision Research Inc., USA).
MRR Measurement
µMRR instrument consists of a portable, permanent magnet (Metrolab Instruments, Plan-les-Ouates, Switzerland) with B0 = 0.5T and a bench-top type NMR console (Kea Magritek, Wellington, New Zealand). 1H MRR measurements were performed at the resonance frequency of 21.65 MHz inside the magnet. A single resonance proton MRR probe with a detection micro-coil of 1550-μm inner diameter was used for accommodating the MRR samples into the microcapillary tubes (o.d.: 1500 μm, i.d.: 950 μm) (22-260-950, Fisherbrand, Waltham, MA, USA). In the MRR probe, the electronic parts and coil were mounted on the single printed circuit board (Fig. 1c).39 All experiments were performed at 26.3 °C inside the magnet maintained by a temperature controller (RS component, UK).
For all MRR measurements, the normalized concentration (60K cells in 4 µL volume) of MSCs has been used unless otherwise stated. One milliliter culture of MSCs (3 × 105 cells/mL) was spun down at 300g for 5 min, and the supernatant was aspirated. The pellet is suspended in 20 µL of PBS and the 4 µL of this prepared sample was filled at a 4 mm length of the micro-capillary tube. The micro-capillary tube was sealed with critoseal (Leica Microsystems) and mounted into the coil for MRR measurements (Supplementary Fig. S1). Proton transverse relaxation times (T2) were measured by the standard Carr-Purcell-Meiboom-Gill (CPMG) pulse program48,49 (Fig. 1b). We maintained the transmitter power output at 12.5 mW for a single 90° pulse of pulse length 6 μs for all the T2 measurements. The CPMG train of pulses with inter echo time of 200 μs with 4000 echoes was used for all experiments. A recycle delay of 2 s, which is sufficient to allow all the spins to return to thermal equilibrium, was used. Twenty-four scans were performed for all experiments for signal averaging.
Real-Time Polymerase Chain Reaction (RT-qPCR) Analysis
Total RNA was extracted with the RNeasy Mini Kit (Qiagen, USA) following the manufacturer’s protocol. The concentration of RNA was determined using a NanoDrop UV-vis Spectrophotometer (NanoDrop Technologies, USA). Then, reverse transcription reaction was carried out with 200 ng total RNA using iScript cDNA synthesis kit (Biorad Laboratories, USA). Real-time PCR was performed using SYBR Green system with primers listed in Table 1. ABI 7500 real-time PCR system (Applied Biosystem, USA) was used to perform real-time PCR at 95 °C for 10 min and 40 cycles of amplification which encompasses denaturation step at 95 °C for 15 s and an extension step at 60 °C for 1 min. The gene expression level was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and calculated using the 2−ΔΔCt formula with reference to the respective control group. The data presented were representative of 3 technical replicates for each condition.
Table 1.
The primers for real-time PCR.
| Primer | Forward sequences | Reverse sequences |
|---|---|---|
| GAPDH | ATGGTGAAGGTGGAGTGAA | AATGAAGGGGTCATTGATGG |
| P16 | CACCGAATAGTTACGGTCGG | GCACGGGTCGGGTGAGAGTG |
| P21 | TCTTGTACCCTTGTGCCTCG | AGAAGATCAGCCGGCGTTTG |
Conditioned Media and ELISA
Size-sorted MSCs were seeded in T75 flasks for overnight adherence before media removal the next day. The cells were washed with PBS, and cultured in serum-free media for 48 h at 37 °C, with a 5% CO2 concentration. The media was then collected and centrifuged at 4500g for 15 min to remove cell debris. The supernatant was collected and concentrated to 500 µL with an Amicon Ultra 15 filter (3 kDa cut-off membrane) and stored at −80 °C before analysis. Cell number in each condition was determined to normalize the concentration values of the analytes of interest (MCP-1, IL-6, IL-8, and TGF-β1) obtained using Luminex-based multiplex assays (R&D Systems) according to manufacturer’s instructions. All samples were analyzed in technical duplicates and performed for 2 different donors. The data were obtained with a MAGPIX reader (Millipore), and concentrations were derived from measured mean fluorescence intensities (MFI) using fitted standard curves using 5-parameter logistic regression (SSL5) using the Milliplex Analyst software (Millipore).
In Vitro Multi-Lineage Differentiation Assay and Quantification
MSCs were seeded in 24-well plates for overnight adherence before media removal and reincubated with specific differentiation media (STEMCELL Technologies) for osteogenesis and adipogenesis, following the manufacturer’s protocol. Osteogenesis induction was confirmed with Alizarin Red S staining (ScienCell) for calcium deposits, while adipogenic differentiation was determined with Oil Red O staining (Sigma) for the detection of lipid droplets. For chondrogenesis, MSCs were pelleted at 1 × 106 cells/pellet in a 15 mL tube at 300g, 5 min. Chondrogenic media (STEMCELL Technologies) was added to this pellet, and media was changed every alternate day for 3 weeks before glycosaminoglycan staining with Safranin O.
The multi-lineage differentiation index (ie, adipogenesis index, osteogenesis index, and chondrogenesis index) was computed as the product of the relative stained area and the relative red color intensity of the stained area in an image.50 The positive staining areas were extracted by the adaptive threshold algorithm51 developed using MATLAB software (MathWorks, USA). The relative stained area was calculated with reference to the image area. The relative red color intensity was calculated with reference to the maximum intensity in the RGB image.
Senescence Assay and Quantification
MSCs were seeded on a 6-well plate at an initial cell seeding density of 1500 cells/cm2 and cultured for 3-5 days. Senescence β-galactosidase staining kit (Sigma Aldrich) was used to identify the senescent cells according to the manufacturer’s protocol. Briefly, the cells were fixed at room temperature for 10 min and incubated in a staining solution containing β-gal at 37 ˚C overnight. Cells stained positive with blue precipitate as a result of β-gal substrate cleavage were indicative of senescent cells.
Five to 15 random areas (except for Fig. 3b) were captured at the bright field by a color camera for quantifying the β-galactosidase staining-based senescence index (β-gal Sn index) using MATLAB image processing. The areas of cells stained positive were green/blue in color and reflected as the areas lacking red color. Thus, the positive staining areas were extracted by the adaptive threshold algorithm51 processing at the red channel of RGB images. β-gal Sn index was computed as the total sum of the complement of red intensity (mean of the red intensity of the image minus red intensity) in all selected areas. The final β-gal Sn index was normalized by the confluency of cells.
Figure 3.
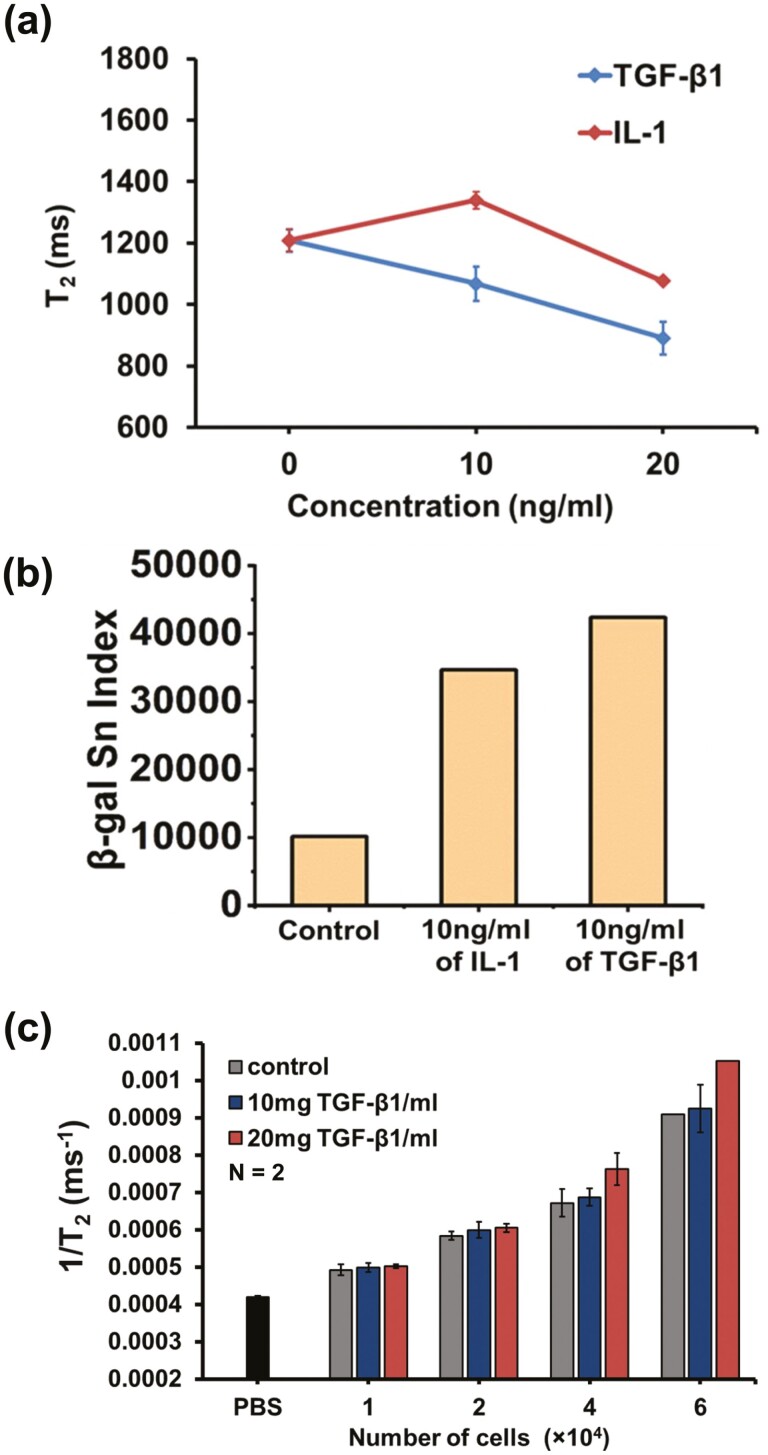
MRR detection of induced senescence by treatment of cytokines. (a) Average T2 values (n = 2) of MSCs (donor D4 as in Fig. 2, Passage 6) control and treated with TGB-β1 (blue) and IL-1 (red) at different concentrations (10 and 20 mg/mL). (b) Quantification of senescence index from β-galactosidase staining images of MSCs control, 10 ng/mL of IL-1 treated MSCs, and 10 ng/mL of TGF-β1 treated MSCs (one image for each sample group). The image of each sample contains more than 200 cells. (c) Limit of detection of MRR assay for senescent MSCs. Average 1/T2 values (n = 2) of MSCs with increasing cell numbers in microcapillary tubes of MRR for control and TGF-β1 (10 and 20 mg/mL) treated MSCs of the same donor at passage 6. The 1/T2 value (n = 2) of PBS in the microcapillary tube is shown as a black bar at the bottom of figure (b).
Fe3+ Staining and Quantification of MSCs
A reversible fluorescent Fe3+ sensor (RPE)52 was used to stain the intracellular iron (Fe3+) of MSCs. A stock solution of RPE (1 mM in acetonitrile) was diluted to a concentration of 20 µM in PBS for Fe3+ staining. 1 × 105 suspended cells were washed twice in PBS and incubated with PBS containing RPE (20 µM) at 37 °C for 30 min. After incubation, the cells were washed twice and suspended in PBS for flow cytometry measurement and microscope imaging within 10 min.
Immunofluorescence and Confocal Microscopy
For the detection of SA-βgal together with another senescence marker (γ-H2AX foci)53 and the proliferation marker, 5-ethynyl-2ʹ-deoxyuridine (EdU) in the cells, we performed the protocol as previously mentioned.54 Briefly, the cells were incubated with 10 µM EdU (Click-iT EdU Alexa Fluor 488 Imaging Kit, Invitrogen) for 24 h before fixation with 10% neutral buffered formalin (Sigma) and β-gal staining (Sigma-Aldrich) using components from the staining kit according to the manufacturer’s instructions (#9860, Cell Signaling Technology). Subsequently, EdU labeling was carried out following the manufacturer’s protocol (Invitrogen).
For immunostaining, cells were washed in PBS and incubated with a blocking buffer (PBS containing 0.5% bovine serum albumin) for 1 h before incubation with a mouse anti-phospho-Histone H2A.X (Ser139) antibody (05-636, Millipore) in blocking buffer for overnight at 4 °C. Cells were then incubated with Rhodamine Red-conjugated secondary antibody (Jackson Immuno Research Laboratories) for 1 h, washed thrice with PBS, and counterstained with NucBlue (Hoechst 33342)-containing mounting solution (Invitrogen). Images were acquired with an FV1200 Confocal Microscope using a 20× objective lens (Olympus). β-gal-stained cells were also visualized under phase-contrast microscopy.
Statistical Analysis
Statistical analyses were performed using Microsoft Excel and Origin Pro (Fig. 7). All data were presented as the mean ± SD. Groups were compared using the 2-tailed t-test. A P value < .05 was considered statistically significant.
Figure 7.
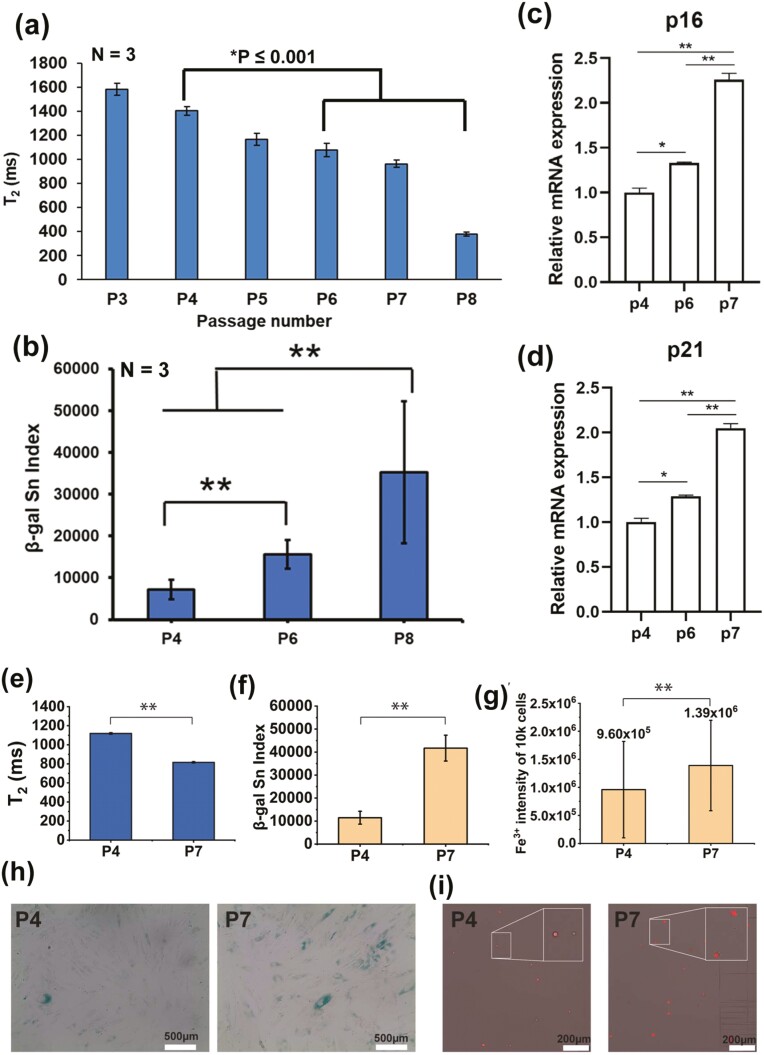
MRR analysis shows serial passage leads to an increase in the proportion of senescent MSCs and its intracellular iron (Fe3+) correlates senescent level. (a) Average T2 values (n = 3) of MSCs (donor D4), which are passaged serially from 3 to 8. Statistical significance is determined by a 2-tailed t-test. (b) Quantification of senescence of β-galactosidase stained MSCs from the same donor for passage P4, P6, and P8. Statistical significance is determined by a 2-tailed t-test, **indicates P < .001. Ten to 15 random areas were captured at the bright field by a color camera for quantifying the β-galactosidase staining-based senescence index (β-gal Sn index) using MATLAB image processing. (c and d) Relative mRNA expression values of P16 and P21 for MSCs from the same donor for passage P6 and P7 compared to passage P4 as a control. The experiment was performed in technical triplicates. An unpaired 2-tailed t-test was performed between selected pairs to determine the statistical significance. *Represents P ≤ 0.05, **represents P ≤ .01. (e) MRR measurements of donor D6 for the passages P4 and P7 (f and g) senescent and Fe3+ quantification of donor D6 (P4 and P7) respectively. (h) Representative images of β-galactosidase staining of MSCs for passages P4 and P7 where the cytoplasm of senescent MSCs was stained as blue. Those images (images number n = 5 for both 2 biological replicates; cell total number>1K was used to quantify the β-gal Sn index in (f). (i) Representative images of Fe3+ staining of MSCs, the more senescent P7 cells showing the brighter intensity compared with the cells from earlier passage P4. Intracellular iron (Fe3+) level of 10K Fe3+ staining of MSCs was measured by flow cytometry. Statistical significance is determined by a 2-tailed t-test, **indicates P < .001.
Results
MRR Detection of Senescent MSCs and Its Correlation With Standard Assays
Senescent cells in MSC cultures often diminish their therapeutic efficacy in preclinical and clinical trials. We sought to measure proliferation and senescence using µMRR by comparison with conventional assays to correlate cell state directly with the T2 values. The CPMG (Carr-Purcell-Meiboom-Gill) pulse sequence (Fig. 1b) is used for measuring the T2 values, which works efficiently in inhomogeneous magnetic fields produced by a permanent magnet of µMRR. The µMRR system (Fig. 1c) consists of a 0.5T permanent portable magnet and a radio frequency (RF) detection probe connected to a spectrometer. For MRR experiments, a small-sized sample (4 µL sample volume, which fits into the detection range of RF detection coil) is required, which is loaded into the microcapillary tube with minimal processing. T2 values of the samples having 6 × 104 cells are measured within 5 min (Supplementary Fig. S1).
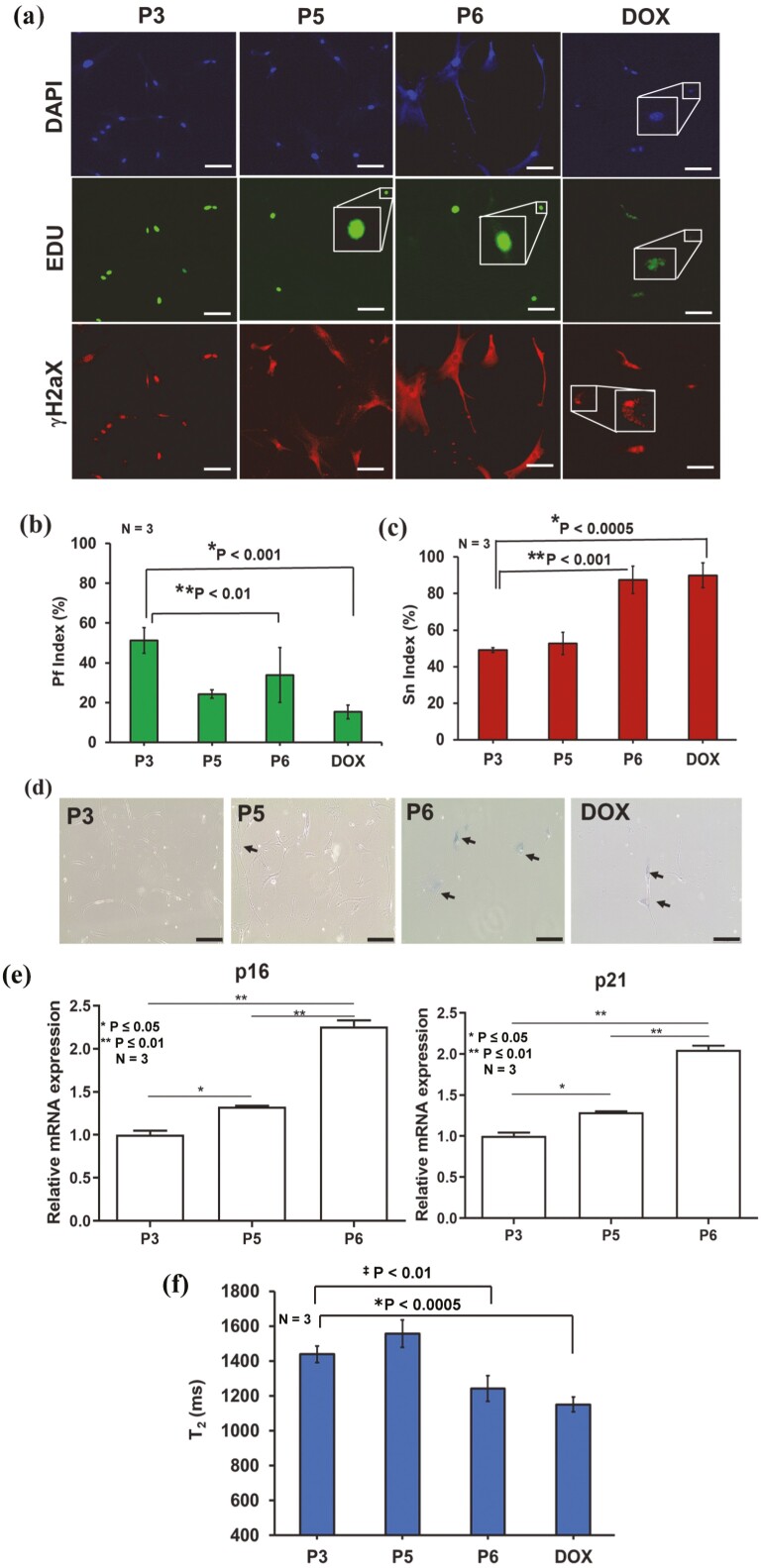
For comparison between conventional senescence assays and µMRR, we cultured MSCs in vitro from the same donor to passage 3 (P3), passage 5 (P5), and passage 6 (P6). As a positive control for senescence cells, we also treated cells from P3 for 24 h with 1 µM of doxorubicin (DOX), a chemotherapy agent that produces DNA damage and reactive oxygen species known to induce senescence in tumor cells.55 To measure DNA synthesis, EdU (5-ethynyl-2ʹ-deoxyuridine) was used as an indicator of active cell proliferation (shown in green). In contrast, senescent cells are known to accumulate γ-H2AX foci (indicated in red). A proliferation (Pf) index and senescence (Sn) index were calculated for all passages and DOX-treated MSCs from these images (see Methods and Materials) (Fig. 2a) and plotted respectively (Fig. 2b, 2c). Based on the Pf/Sn index graphs, more proliferating cells (EdU) were present in P3. Pf gradually decreased with the later passages (P5 and P6) and was lowest in DOX-treated cells. The Sn index showed the opposite trend, where more senescent-like cells (γ-H2aX foci) were observed in P6 and DOX-treated cells than in P3 and P5. Moreover, we observed a higher proportion of β-gal-stained cells in later passages (P6) (Fig. 2d). Similarly, qPCR of the senescence markers p16 and p21 showed higher expression in P6 (Fig. 2e). Thus, senescent MSCs were clearly visible at later passages and in response to DOX treatment.
Figure 2.
Correlating MRR with standard assays. (a) Immunofluorescent images of MSCs (donor D4) that are culture-expanded to passages P3, P5, P6, and doxorubicin treated MSCs from P3. The cells are stained by the senescence marker (γ-H2aX foci, red fluorescence) and the proliferation marker, 5-ethynyl-2ʹ-deoxyuridine (Edu, green fluorescence). The cell nuclei are counterstained with NucBlue (Hoechst 33342), which emits blue fluorescence (shown as DAPI) when bound to DNA. The experiment was performed with 3 technical replicates and more than 100 cells in each replicate. The scale bars are 100 µm. (b and c) The proliferation (Pf) index (%) and senescent (Sn) index (%) are calculated by counting the number of green and red fluorescent stains relative to the number of blue fluorescent stains in all images. A total of 10 images from each of 3 technical replicates of staining experiments in MSCs of P3, P5, P6, and DOX treated were used for the calculation of Pf index (%) and Sn index (%), which are plotted in (b) and (c) respectively. Statistical analyses of P3 with DOX and P6 (n = 3) are done by a 2-tailed t-test. For (b) the P values are *P < .001 and ‡P < .01. For (c) the P values are *P < .0005 and ‡P < .001. (d) β-galactosidase staining of MSCs for passages P3, P5, and P6 where the cytoplasm of senescent MSCs was stained blue (black arrows). The scale bars are 200 µm. (e) Relative mRNA expression values of senescence-associated markers p16 and p21 for MSCs from a single donor at different passages (P), namely P3, P5, and P6. The data shown are representative of 3 technical replicates for each passage. An unpaired 2-tailed t-test was performed between selected pairs to determine the statistical significance. *Represents P ≤ .05, **Represents P ≤ .01. (f) Average T2 results (n = 3) of P3, P5, P6 and DOX treated. Statistical analysis of P3 with DOX and P6 (n = 3) is done by a 2-tailed t-test, and the P values are *P < .0005 and ‡P < .01.
We next compared these data with T2 measurements by performing parallel µMRR experiments using a normalized cell number of 6 × 104 in 4 µL volume in the microcapillary tube. MSCs were added to the micro-capillary tube and placed inside the radio frequency (RF) detection coil of the µMRR machine. We demonstrate that the T2 measurements correlated well with results from conventional end-point assays. Specifically, we observed a significant decrease in T2 levels, an indicator of increased iron accumulation, at later passages (P6) and after DOX treatment (Fig. 2f). Thus, MRR T2 measurements correlate well with known biochemical senescence markers.
MRR Detection of Induced Senescence by Treatment of Cytokines
Cytokines such as transforming growth factor-β1 (TGFβ1) and interleukin-1 (IL-1) induce senescence in MSCs.56 TGFβ1 is a TGF family member that controls numerous cellular functions, including proliferation and cell death.57,58 IL-1 is a pro-inflammatory cytokine whose expression is associated with SASP.59 We next treated MSCs (same donor as in Fig. 2, passage 6) with different concentrations (10 ng and 20 ng/mL) of TGFβ1 and IL-1 for 3 days and performed measurements using µMRR. T2 values of TGFβ1 and IL-1 treated cells decreased with increasing cytokine concentrations (Fig. 3a), predicting that TGFβ1 and IL-1 induce MSC senescence in culture. Quantification of β-galactosidase staining shows higher levels of senescent cells in TGF-β1 compared to IL-1 treated cells (see Fig. 3b), in accordance with T2 values.
Since MSCs donors typically show variability in proliferative ability, it is important to find the limit of detection (LOD) of the µMRR assay in terms of the cell number. Thus, we analyzed the LOD by titirating the number of cells to 4 × 104, 2 × 104, and 1 × 104 cells for µMRR detection as shown in Fig. 3c; Supplementary Fig. S2. MSCs (same donor as Fig. 2, P6) were treated with TGFβ1 at different concentrations (10 and 20 mg/mL) to induce senescence, and batches of 1, 2, and 4 (×104 cells) in 4 µL detection volume of TGFβ1 treated and untreated MSCs were collected after 3 days for µMRR detection. When there is no iron or no paramagnetic component in the sample (eg, PBS buffer), the T2 value of the sample is essentially the same as the natural T2 relaxation time of pure water, which is typically above 2000 ms. However, depending on the amount of paramagnetic component in the sample, this T2 relaxation process is disrupted, and therefore, relaxation occurs much more quickly, reducing the T2 time (Supplementary Fig. S3). The 1/T2 value of PBS buffer was also compared with 1/T2 value of MSCs (treated and untreated) as a control since PBS has abundant water protons and no iron redox stages are there to reduce the T2 relaxation of water protons. This analysis shows a significant difference in T2 value of TGFβ1 treated MSCs when compared to the T2 value of untreated MSCs and PBS at cell numbers at or larger than 4 × 104 cells. This establishes the cell number necessary for a reliable µMRR quantification of senescent cells in live cultures, and we used 6 × 104 cells per assay in all of our experiments.
MRR Detects an Increase in Senescence of Large MSCs Enriched by Spiral Microfluidic Device
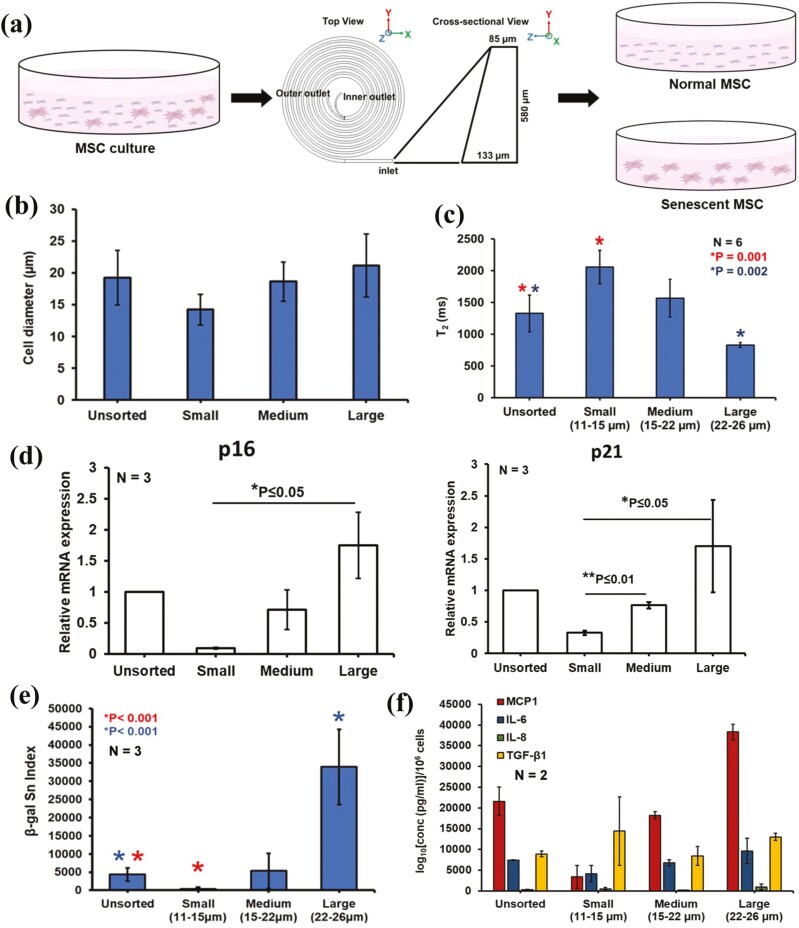
Previously, we and others identified cell size as a marker for distinguishing MSC phenotypes.60,61 Smaller MSCs are generally multipotent MSCs, whereas larger MSCs tend to display limited growth and differentiate more prominently toward the bone lineage. We found that large MSCs sorted by inertial microfluidic devices displayed senescent phenotypes with limited growth potential and longer doubling times compared to smaller MSCs47 (Fig. 4a; Supplementary Fig. S4). Using the same microfluidic device, we sorted MSCs into 3 size groups: Unsorted, small (11-15 µm), medium (15-22 µm), and large (22-26 µm) (Fig. 4b). Analysis of the sorted MSCs showed decreasing T2 value from small to large cells (Fig. 4c), indicating water proton nuclear relaxation, which is ~2000 ms in pure water, is significantly reduced due to the paramagnetic iron (Fe3+) impurity in senescent MSCs. In addition, expression of senescence-associated markers p16 and p21 (measured by RT-qPCR) were elevated in the Large cells compared to medium and small groups (Fig. 4d). We also observed a high β-gal Sn Index in large and unsorted group cells (Fig. 4e; Supplementary Fig. S5), in line with the lower T2 values measured by µMRR. These results support previous reports of Fe3+ storage in senescent cells,35 because faster T2 relaxation is due to increased paramagnetic Fe3+ content in the cells. We also measured the secretion of molecules associated with the senescence-associated secretory phenotype (SASP) for the unsorted and size-sorted MSC subpopulations (small, medium, and large) (Fig. 3f). Using Luminex assays, we found higher expression of key secreted markers associated with SASP such as MCP-1 (significantly higher expression), IL-6, IL-8,62,63 and TGFβ156 in larger MSCs,50 consistent with the decrease in T2 values.
Figure 4.
MRR assay shows large MSCs are more senescent based on size sorting using a spiral microfluidic device. (a) Size separation of young/proliferating and senescent MSCs using a spiral microfluidic device from MSC cultures. The MSC culture is pumped into the microfluidic device and sorted at various speeds to collect different-sized MSCs at the outlets. Mesenchymal stem cell figures are adapted from the icon library of “Cell Types” by BioRender.com. (b) Cell diameter (µm) of sorted MSCs using spiral microfluidic sorting device. (c) Average T2 values (n = 6) of unsorted and size-sorted MSCs from 2 different donors (D3 and D4 passage 3) using a spiral microfluidic device. Large cells (22-26 µm) are the size-sorted MSCs collected in the first round of sorting, and medium (15-22 µm) and small (11-15 µm) cells are collected from the 2nd round of sorting. Statistical analysis of unsorted and small cells (n = 6, *P = .001) and unsorted & large cells (n = 6, *P = .002) are done by unpaired 2-tailed t-test. (d) The mRNA expression level of unsorted and size-sorted MSCs showing the overexpression of senescent markers P16 and P21 in LargeMSCs. The data shown are representative of 3 technical replicates for each group. An unpaired 2-tailed t-test was performed between selected pairs to determine the statistical significance. *Represents P ≤ .05, **Represents P ≤ 0.01. (e) Quantification of senescence index from the β-galactosidase images of unsorted and size-sorted MSCs. Five to 10 random areas were captured at the bright field by a color camera for quantifying the β-galactosidase staining-based senescence index (β-gal Sn index) using MATLAB image processing. Biological replicates N = 3. The images of each sample contain more than 500 cells. (f) Secretome profiling of factors associated with senescence, secreted from unsorted and size-sorted MSC subpopulations in 2 donors. Each sample was performed in technical duplicates. Concentrations of the secreted analytes normalized per million cells were plotted.
Correlation of MRR Measurements with Population Doubling Times (PDT)
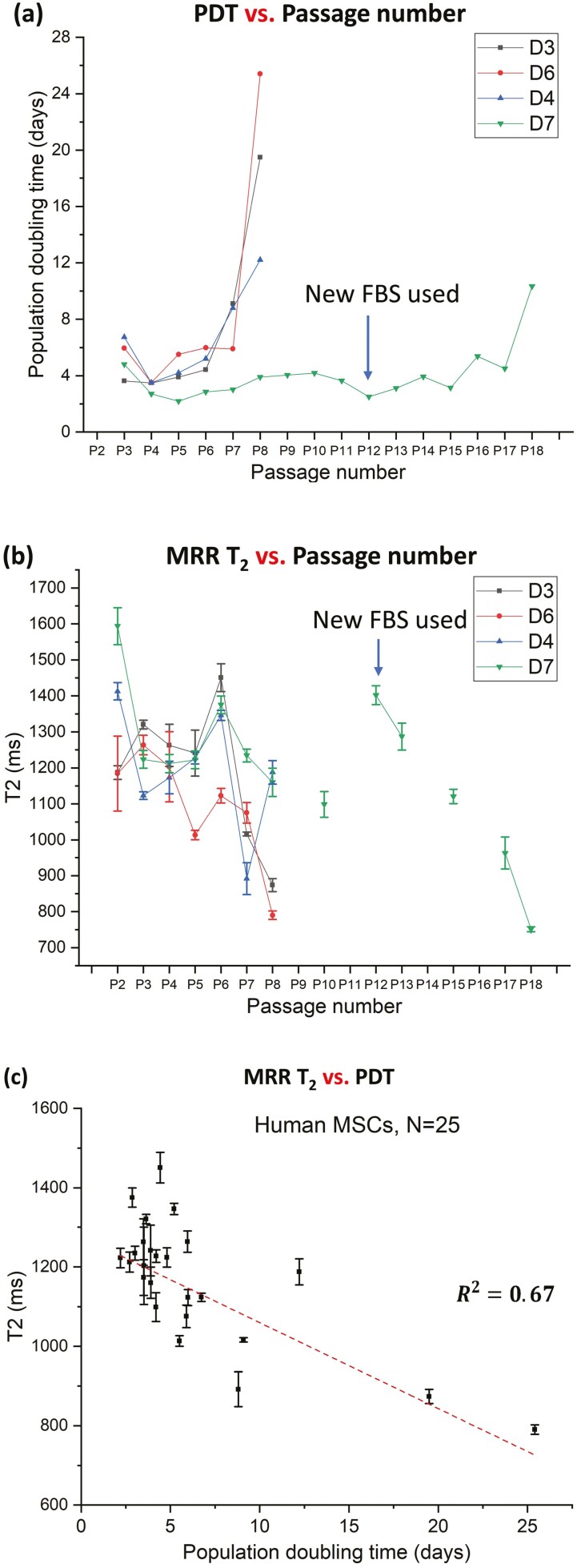
To understand the correlation of MRR measurements (T2 values) with the population doubling times (PDT), 4 different donors (D3, D4, D6, and D7, see Table 2) were cultured in serial passages from P2 to P18. The PDTs were measured for each passage for different donors and plotted in Fig. 5a. The T2 values of the respective passages of each donor cells are shown in Fig. 5b. For early passages (up to P6), the T2 values are rather fluctuating, along with PDT (some passages grow faster, while others do not). However, once the PDT is significantly increased (a clear sign of senescence), the T2 values drop below 1000 ms. Cells from a donor (D7) are especially of high quality and could maintain a good growth profile to passage 17. However, when this cell eventually turned senescent, one can observe that T2 value dropped below 900 ms, reflecting a severe Fe3+ accumulation. The correlation graph of PDTs against T2 values (Fig. 5c) shows a significant correlation (R2 = 0.67). When T2 values are less than 1000 ms, the human MSCs show significant senescence and will not proliferate well (PDT > 8 days). Therefore, µMRR assay has sufficient sensitivity to detect senescent MSCs in culture for the quality control purpose of MSCs.
Figure 5.
MRR analysis correlates with the population doubling time (PDT). (a) Population doubling time of MSCs from 4 different donors (D3, D4, D6, and D7), measured at each passage while passaging from P2 to P18. PDT values were calculated by comparing the total cell number seeded with the total cell number harvested at the end of each passage. At the beginning of the passage 12, MSCs from D7 were cultured with a new batch of media, which may have partially delayed the onset of senescence until P18. (b) T2 values of MSCs from the same 4 different donors, measured on the harvested cells at the end of selected passage. (c) T2 values vs. population doubling time (PDT) showing T2 correlates well with PDT (R2 = 0.67).
MRR Measurements Correlate With the Chondrogenic Potential of MSCs
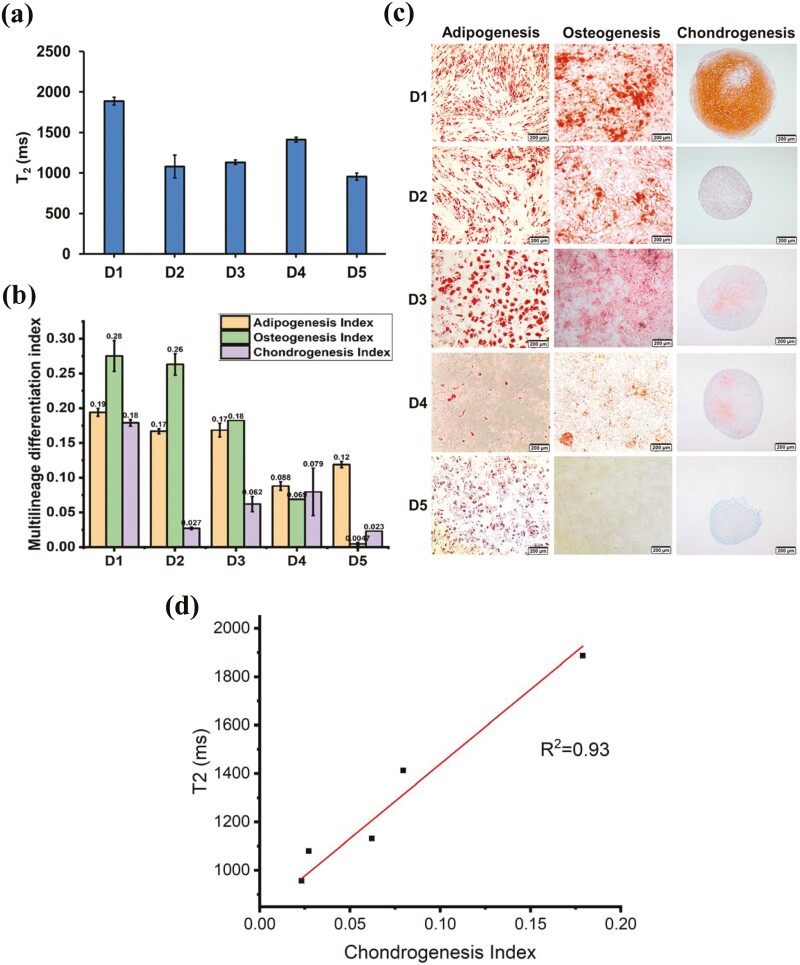
The most critical therapeutic quality attribute of MSCs is their ability to differentiate into cell lineages such as adipocytes, chondrocytes, and osteoblasts. MSCs isolated from different donors vary significantly in their multi-lineage differentiation potential, a critical and poorly understood limitation in achieving consistent quality control of MSCs for therapy. Here, we analyzed MSCs from 5 different donors (D1-D5; See Methods for description of donors and Table 2) cultured in vitro to P3. For µMRR analysis, we used a normalized cell number of 6 × 104 for each T2 experiment. Fig. 6a shows the T2 profile of MSCs from different donors. Multi-lineage differentiation potential of MSCs from the corresponding donors (D1-D5) was then measured by staining Alizarin red S, Oil Red O, and Safranin O for osteogenesis, adipogenesis, and chondrogenesis, respectively. The multi-lineage differentiation index was calculated (Fig. 6b, 6c), and T2 values of MSCs from each donor were correlated with differentiation potential. Chondrogenesis index correlated with the T2 value (Fig. 6d), where the highest T2 value was recorded for MSCs from D1 and the lowest T2 from D5. Hence, the lower T2 values corresponded to a decline in chondrogenic differentiation potential and reduced proliferation. Thus, decreased differentiation potential in MSCs from certain donors may be affected by the presence of higher proportions of senescent cells in the population, and this could also influence cell proliferation.
Figure 6.
MRR analysis measures altered multi-lineage differentiation in MSCs from different donors. (a) T2 values (n = 3) of MSCs from different donors D1 to D5 (passage 3). (b) Image-based quantification of multi-lineage differentiation potential. Differentiation index (Adipogenesis index, osteogenesis index, and chondrogenesis index) quantified from the images of respective MSC donors in passage 3. (c) Multi-lineage differentiation (adipogenesis, osteogenesis, and chondrogenesis) images of MSCs from different donors. (d) T2 vs. chondrogenesis index from different donors D1 to D5 showing T2 correlates well with chondrogenesis potential (R2 = 0.93).
MRR Measurements Show Serial Passage Leads to an Increase in the Proportion of Senescent MSCs
During in vitro culture of MSCs, cellular senescence tends to increase during serial passaging. Senescent cells can affect the proliferation of MSCs, differentiation potential, and hence the quality of cells for their clinical use. Therefore, it is important to identify senescent cells and potentially remove them from each passage of MSC culture.47 We analyzed MSCs (D4) from serial passages P3 to P8 under in vitro culture conditions. Using µMRR, T2 values of normalized concentrations of MSCs from each passage are shown in Fig. 7a. We observed the highest T2 value for MSCs in the earliest passage, whereas these levels progressively decreased at later passages. MSCs from the later passage, where the Senescence index was greater based on the intensity and number of cells displaying β-gal staining, showed the lowest T2 value (Fig. 7b; Supplementary Fig. S6). Similarly, RT-qPCR of the cell cycle inhibitors p16 and p21 showed a trend toward the highest expression in cells from late passages as compared to early passages (Fig. 7c, 7d). Together, our data demonstrate that the MRR T2 measurement is a non-destructive and label-free method for detecting the accumulation of senescent MSCs across a range of conditions and donors in live cultures.
MRR Measurements Correlate With Intracellular Iron (Fe3+) and Senescent Level of MSCs
Because recent evidence suggests that senescent cells show an accumulation of iron (Fe3+), we next investigated the correlation between T2 values and Fe3+ levels. Fe3+ was quantified by a reversible fluorescent Fe3+ sensor (RPE)52 at the different passages of MSCs (D6, P4 vs. P7) in parallel with µMRR measurements. As shown in Fig. 7e, MSCs at the P7 show a statistically significant decrease in T2 value (P < .001, 2-tailed t-test) compared to earlier passage P4, correlating with an increase of classical senescence markers (Fig. 7f, 7h). Fig. 7g; Supplementary Fig. S7 show that later MSC passages exhibit higher Fe3+ levels (ie, higher Fe3+ mean intensity of 10K cells) as quantified by the RPE using flow cytometry and also with the brighter fluorescent intensity of cells in the images (Fig. 7i). These results show the MRR measurement correlates with the Fe3+ accumulations in MSCs that also show a senescence phenotype.
Discussion
We have established a rapid and non-destructive method for quantifying senescent cells in vitro. Our approach requires a small number of cells (<105) and does not require any reagents or sample preparation steps. Direct Fe3+ quantification by µMRR provides well-correlated results with conventional end-point and time-intensive assays for senescence, such as RT-qPCR, Luminex assay, and β-galactosidase staining. Since paramagnetic Fe3+ (but not diamagnetic Fe2+) increases the magnetic susceptibility, which stimulates proton nuclear relaxation of water molecules in the cells, our measurement is specifically quantifying the Fe3+ content of cellular iron. Previously, intracellular Fe2+ (labile iron) was measured by colorimetric and other assays,64 yet reliable and quantitative detection of Fe2+ has generally been challenging,65,66 presumably due to the reactive nature of Fe2+. For Fe3+ quantification, ferritin is often used as a surrogate maker,67 which is a large (~480 kDa) protein complex that can accommodate up to 4500 atoms of Fe3+ iron. Thus, ferritin concentration may not correlate well with the Fe3+ amount stored in the cell. ICP-MS quantification68 of total iron concentration measurement is the current standard for iron quantification; however, this metric may not correctly reflect the stored Fe3+, which is more closely correlated to cellular senescence.
In contrast to the conventional iron quantification methods, µMRR is uniquely positioned to advance iron biology in general. µMRR allows the phenotyping of live cells, allowing downstream biological and functional measurements of other indicators on the same cells. This ability enables direct correlations between MRR T2 and other biochemical and functional measurements on the same cells. Thus, adapting µMRR is potentially transformative for the field of cell therapy production, especially given the lack of consensus cell surface markers specific for senescent cells.
In this study, the chondrogenic potential of MSCs from different donors also correlates well with MRR results, with the highest T2 value measured from MSCs having robust chondrocyte differentiation potential. This technology will be instrumental in screening MSCs for the highest growth and differentiation potential in a patient-specific manner in real-time. One can also adopt this new assay for optimizing the biomanufacturing of MSCs or other cell therapy products. For example, MRR could be used to monitor cell cultures in real-time in a way that would allow the removal of senescent cells using microfluidic sorting.47 Thus, our approach could overcome a major bottleneck to produce higher-grade MSCs and cell products for specific therapeutic modalities. In addition, recent data suggest the important roles of iron homeostasis in the maintenance of iPSCs69,70 and HSCs,71,72 raising the prospect that this technology can impact a broader class of therapeutic cells in the future. We envision µMRR as a rapid, live-cell quality screening tool for the detection of cellular senescence across a range of cell types and tissues. More broadly, the “iron-imaging” modality utilized here is already widely used in T1 and T2 weighted imaging using MRI.73-76 Therefore, any insights from MRR quantification of cellular senescence could readily be compared to in vivo pathology. Given the resolution of modern MRI approaches below 100 µm,77 non-invasive detection of senescent cells in live tissue may be feasible in the future.
Conclusion
Using intracellular iron (Fe3+) accumulation as a natural biomarker, we have developed a rapid and non-destructive assay for senescence in MSC cultures, which is well-correlated with all the conventional biochemical assays of cellular senescence. It is ideally suited to monitor and improve the quality of MSCs produced for cell therapy applications.
Supplementary Material
Contributor Information
Smitha Surendran Thamarath, Singapore-MIT Alliance for Research and Technology (SMART–Critical Analytics for Manufacturing of Personalized Medicine (CAMP) IRG 1 Create Way, Singapore, Singapore; Singapore-MIT Alliance for Research and Technology (SMART)—Anti-Microbial Resistance (AMR) IRG 1 Create Way, Innovation Wing, Singapore, Singapore.
Ching Ann Tee, Singapore-MIT Alliance for Research and Technology (SMART–Critical Analytics for Manufacturing of Personalized Medicine (CAMP) IRG 1 Create Way, Singapore, Singapore; NUS Tissue Engineering Program is NUS Tissue Engineering Program, Life Science Institute, National University of Singapore, Singapore, Singapore.
Shu Hui Neo, Singapore-MIT Alliance for Research and Technology (SMART–Critical Analytics for Manufacturing of Personalized Medicine (CAMP) IRG 1 Create Way, Singapore, Singapore.
Dahou Yang, Singapore-MIT Alliance for Research and Technology (SMART–Critical Analytics for Manufacturing of Personalized Medicine (CAMP) IRG 1 Create Way, Singapore, Singapore.
Rashidah Othman, Singapore-MIT Alliance for Research and Technology (SMART–Critical Analytics for Manufacturing of Personalized Medicine (CAMP) IRG 1 Create Way, Singapore, Singapore.
Laurie A Boyer, Singapore-MIT Alliance for Research and Technology (SMART–Critical Analytics for Manufacturing of Personalized Medicine (CAMP) IRG 1 Create Way, Singapore, Singapore; Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA, USA; Department of Biology, Massachusetts Institute of Technology, Cambridge, MA, USA.
Jongyoon Han, Singapore-MIT Alliance for Research and Technology (SMART–Critical Analytics for Manufacturing of Personalized Medicine (CAMP) IRG 1 Create Way, Singapore, Singapore; Singapore-MIT Alliance for Research and Technology (SMART)—Anti-Microbial Resistance (AMR) IRG 1 Create Way, Innovation Wing, Singapore, Singapore; Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA, USA; Department of Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge, MA, USA.
Funding
This research is supported by the National Research Foundation, Prime Minister’s Office, Singapore under its Campus for Research Excellence and Technological Enterprise (CREATE) programme, through Singapore-MIT Alliance for Research and Technology (SMART): Critical Analytics for Manufacturing Personalized-Medicine (CAMP) Inter-Disciplinary Research Group (IRG) as well as Anti-Microbial Resistance (AMR) IRG.
Conflict of Interest
J.H., L.A.B, S.S.T., and S.H.N. filed an intellectual property with a patent that is wholly owned and managed by MIT. J.H. also declared honoraria from TGS management for a seminar on an unrelated topic (desalination). The other authors declared no potential conflicts of interest.
Author Contributions
S.S.T., L.A.B., J.H.: designed the overall experimental plan. S.S.T.: carried out the MRR experiments with support from C.A.T, S.H.N. D.Y., C.A.T., S.H.N., R.O.: performed various biochemical assays. S.S.T., D.Y.: performed data analysis. The manuscript was written by S.S.T. All authors participated in editing and revising the manuscript.
Data Availability
The data that support the findings of this study are available at the Mandeley Data repository (DOI: 10.17632/83cchm9fjz.1), which can be previewed by accessing the link cited here.78
References
- 1. Kuilman T, Michaloglou C, Mooi WJ, Peeper DS.. The essence of senescence Thomas. Genes Dev. 2010;24:2463-2479. https://doi.org/ 10.1101/gad.1971610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hayflick L, Moorhead PS.. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585-621. https://doi.org/ 10.1016/0014-4827(61)90192-6 [DOI] [PubMed] [Google Scholar]
- 3. Van Deursen JM. The role of senescent cells in ageing. Nature. 2014;509:439-446. https://doi.org/ 10.1038/nature13193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Campisi J. Aging, cellular senescence, and cancer. Annu Rev Physiol. 2013;75:685-705. https://doi.org/ 10.1146/annurev-physiol-030212-183653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hanahan D, Weinberg RA.. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. [DOI] [PubMed] [Google Scholar]
- 6. Collado M, Blasco MA, Serrano M.. Cellular senescence in cancer and aging. Cell. 2007;130:223-233. https://doi.org/ 10.1016/j.cell.2007.07.003 [DOI] [PubMed] [Google Scholar]
- 7. Naylor RM, Baker DJ, van Deursen JM.. Senescent cells: a novel therapeutic target for aging and age- related diseases. Clin Pharmacol Ther. 2013;93:105-116. https://doi.org/ 10.1038/clpt.2012.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McHugh D, Gil J.. Senescence and aging: causes, consequences, and therapeutic avenues. J Cell Biol. 2018;217(1):65-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wagner W, Horn P, Castoldi M, et al. Replicative senescence of mesenchymal stem cells: a continuous and organized process. PLoS One. 2008;3:e2213. https://doi.org/ 10.1371/journal.pone.0002213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mareschi K, Ferrero I, Rustichelli D, et al. Expansion of mesenchymal stem cells isolated from pediatric and adult donor bone marrow. J Cell Biochem. 2006;97:744-754. https://doi.org/ 10.1002/jcb.20681 [DOI] [PubMed] [Google Scholar]
- 11. Bonab MM, Alimoghaddam K, Talebian F, et al. Aging of mesenchymal stem cell in vitro. BMC Cell Biol. 2006;7(14):1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Drela K, Stanaszek L, Nowakowski A, et al. Experimental strategies of mesenchymal stem cell propagation: adverse events and potential risk of functional changes. Stem Cells Int. 2019;2019:1-10. https://doi.org/ 10.1155/2019/7012692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rubin H. Promise and problems in relating cellular senescence in vitro to aging in vivo. Arch Gerontol Geriatr. 2002;34:275-286. https://doi.org/ 10.1016/s0167-4943(01)00221-7 [DOI] [PubMed] [Google Scholar]
- 14. Banfi A, Muraglia A, Dozin B, et al. Proliferation kinetics and differentiation potential of ex vivo expanded human bone marrow stromal cells: implications for their use in cell therapy. Exp Hematol. 2000;28:707-715. https://doi.org/ 10.1016/s0301-472x(00)00160-0 [DOI] [PubMed] [Google Scholar]
- 15. Schellenberg A, Lin Q, Schüler H, et al. Replicative senescence of mesenchymal stem cells causes DNA-methylation changes which correlate with repressive histone marks. Aging. 2011;3:873-888. https://doi.org/ 10.18632/aging.100391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li Y, Wu Q, Yujia W, et al. Senescence of mesenchymal stem cells (review). Int J Mol Med. 2017;39(4):775-782. [DOI] [PubMed] [Google Scholar]
- 17. Noer A, Boquest AC, Collas P.. Dynamics of adipogenic promoter DNA methylation during clonal culture of human adipose stem cells to senescence. BMC Cell Biol. 2007;8(18):1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liang H, Hou H, Yi W, et al. Increased expression of pigment epithelium-derived factor in aged mesenchymal stem cells impairs their therapeutic efficacy for attenuating myocardial infarction injury. Eur Heart J. 2013;34:1681-1690. https://doi.org/ 10.1093/eurheartj/ehr131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Campioni D, Rizzo R, Stignani M, et al. A decreased positivity for CD90 on human mesenchymal stromal cells (MSCs) is associated with a loss of immunosuppressive activity by MSCs. Cytometry B: Clin Cytom. 2009;76(3):225-230. [DOI] [PubMed] [Google Scholar]
- 20. Zhou S, Greenberger JS, Epperly MW, et al. Age-related intrinsic changes in human bone-marrow-derived mesenchymal stem cells and their differentiation to osteoblasts. Aging Cell. 2008;7:335-343. https://doi.org/ 10.1111/j.1474-9726.2008.00377.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang L, Pitcher LE, Prahalad V, Niedernhofer LJ, Robbins PD.. Recent advances in the discovery of senolytics. Mech Ageing Dev. 2021;200:111587. https://doi.org/ 10.1016/j.mad.2021.111587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mehdizadeh M, Aguilar M, Thorin E, et al. The role of cellular senescence in cardiac disease: basic biology and clinical relevance. Nat Rev Cardiol. 2022;19(4):250-264. 10.1038/s41569-021-00624-2 [DOI] [PubMed] [Google Scholar]
- 23. Dimri GP, Lee X, Basile G, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363-9367. https://doi.org/ 10.1073/pnas.92.20.9363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schellenberg A, Hemeda H, Wagner W.. Tracking of replicative senescence in mesenchymal stem cells by colony-forming unit frequency. Methods Mol Biol. 2013;976:143-154. https://doi.org/ 10.1007/978-1-62703-317-6_11 [DOI] [PubMed] [Google Scholar]
- 25. Penfornis P, Pochampally R.. Colony forming unit assays. In: Gnecchi M, ed. Mesenchymal Stem Cells. Methods in Molecular Biology. Vol 1416. Humana Press, Springer protocols; 2016:159-169. [DOI] [PubMed] [Google Scholar]
- 26. Cheng H, Qiu L, Ma J, et al. Replicative senescence of human bone marrow and umbilical cord derived mesenchymal stem cells and their differentiation to adipocytes and osteoblasts. Mol Biol Rep. 2011;38:5161-5168. https://doi.org/ 10.1007/s11033-010-0665-2 [DOI] [PubMed] [Google Scholar]
- 27. Debacq-Chainiaux F, Erusalimsky JD, Campisi J, Toussaint O.. Protocols to detect senescence-associated beta-galactosidase (SA-βgal) activity, a biomarker of senescent cells in culture and in vivo. Nat Protocols. 2009;4:1798-1806. https://doi.org/ 10.1038/nprot.2009.191 [DOI] [PubMed] [Google Scholar]
- 28. Fairweather-Tait SJ, Wawer AA, Gillings R, et al. Iron status in the elderly. Mech Ageing Dev. 2014;136–137:22-28. 10.1016/j.mad.2013.11.00 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu J, Jia Z, Knutson MD, Leeuwenburgh C.. Impaired iron status in aging research. Int J Mol Sci. 2012;13:2368-2386. https://doi.org/ 10.3390/ijms13022368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Puxeddu E, Comandini A, Cavalli F, et al. Iron laden macrophages in idiopathic pulmonary fibrosis: the telltale of occult alveolar hemorrhage? Pulm Pharmacol Ther. 2014;28:35-40. https://doi.org/ 10.1016/j.pupt.2013.12.002 [DOI] [PubMed] [Google Scholar]
- 31. Ogilvie-Harris DJ, Fornaiser VL.. Synovial iron deposition in osteoarthritis and rheumatoid arthritis. J Rheumatol. 1980;7(1):30-36. [PubMed] [Google Scholar]
- 32. Morris CJ, Blake DR, Wainright AC, et al. Relationship between iron deposits and tissue damage in the synovium: an ultrastructural study. Ann Rheum Dis. 1986;45(1):21-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen X, Yu C, Kang R, et al. Iron metabolism in ferroptosis. Front Cell Dev Biol. 2020;8(590226):1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhou RP, Chen Y, Wei X, et al. Novel insights into ferroptosis: implications for age-related diseases. Theranostics. 2020;10:11976-11997. https://doi.org/ 10.7150/thno.50663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Masaldan S, Clatworthy SAS, Gamell C, et al. Iron accumulation in senescent cells is coupled with impaired ferritinophagy and inhibition of ferroptosis. Redox Biol. 2018;14:100-115. https://doi.org/ 10.1016/j.redox.2017.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mancias JD, Wang X, Gygi SP, Harper JW, Kimmelman AC.. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509:105-109. https://doi.org/ 10.1038/nature13148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060-1072. https://doi.org/ 10.1016/j.cell.2012.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gao M, Monian P, Pan Q, et al. Ferroptosis is an autophagic cell death process. Cell Res. 2016;26:1021-1032. https://doi.org/ 10.1038/cr.2016.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peng WK, Kong TF, Ng CS, et al. Micromagnetic resonance relaxometry for rapid label-free malaria diagnosis. Nat Med. 2014;20:1069-1073. https://doi.org/ 10.1038/nm.3622 [DOI] [PubMed] [Google Scholar]
- 40. Thamarath SS, Xiong A, Lin PH, et al. Enhancing the sensitivity of micro magnetic resonance relaxometry detection of low parasitemia Plasmodium falciparum in human blood. Sci Rep. 2019;9(1):1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Peng WK, Chen L, Boehm BO, et al. Molecular phenotyping of oxidative stress in diabetes mellitus with point-of-care NMR system. NPJ Aging Mech Dis. 2020;6(11):1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Castro CM, Ghazani AA, Chung J, et al. Miniaturized nuclear magnetic resonance platform for detection and profiling of circulating tumor cells. Lab Chip. 2014;14(1):14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee H, Sun E, Ham D, Weissleder R.. Chip-NMR biosensor for detection and molecular analysis of cells Hakho. Nat Med. 2008;14:869-874. https://doi.org/ 10.1038/nm.1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Haun JB, Devaraj NK, Hilderbrand SA, Lee H, Weissleder R.. Bioorthogonal chemistry amplifies nanoparticle binding and enhances the sensitivity of cell detection. Nat Nanotechnol. 2010;5:660-665. https://doi.org/ 10.1038/nnano.2010.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liong M, Hoang AN, Chung J, et al. Magnetic barcode assay for genetic detection of pathogens. Nat Commun. 2013;4(1752):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Issadore D, Min C, Liong M, et al. Miniature magnetic resonance system for point-of-care diagnostics. Lab Chip. 2011;11:2282-2287. https://doi.org/ 10.1039/c1lc20177h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yin L, Wu Y, Yang Z, et al. Microfluidic label-free selection of mesenchymal stem cell subpopulation during culture expansion extends the chondrogenic potential: in vitro. Lab Chip. 2018;18:878-889. https://doi.org/ 10.1039/c7lc01005b [DOI] [PubMed] [Google Scholar]
- 48. Carr HY, Purcell EM.. Effects of diffusion on free precession in nuclear magnetic resonance experiments. Phys Rev. 1954;94:630-638. https://doi.org/ 10.1103/physrev.94.630 [DOI] [Google Scholar]
- 49. Meiboom S, Gill D.. Modified spin-echo method for measuring nuclear relaxation times. Rev Sci Instrum. 1958;29:688-691. https://doi.org/ 10.1063/1.1716296 [DOI] [Google Scholar]
- 50. Yin L, Yang Z, Wu Y, et al. Label-free separation of mesenchymal stem cell subpopulations with distinct differentiation potencies and paracrine effects. Biomaterials. 2020;240:119881. https://doi.org/ 10.1016/j.biomaterials.2020.119881 [DOI] [PubMed] [Google Scholar]
- 51. Yang D, Subramanian G, Duan J, et al. A portable image-based cytometer for rapid malaria detection and quantification. PLoS One. 2017;12:e0179161. https://doi.org/ 10.1371/journal.pone.0179161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Y. Wei, Aydin Z., Zhang Y., Liu Z., Guo M.. A turn-on fluorescent sensor for imaging labile Fe(3+) in live neuronal cells at subcellular resolution. Chembiochem. 2012;13(11):1569–1573. [DOI] [PubMed] [Google Scholar]
- 53. Bernadotte A, Mikhelson VM, Spivak IM.. Markers of cellular senescence. Telomere shortening as a marker of cellular senescence. Aging. 2016;8:3-11. https://doi.org/ 10.18632/aging.100871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Itahana K, Itahana Y, Dimri GP.. Colorimetric detection of senescence-associated β galactosidase. Methods Mol Biol. 2013;965:143-156. 10.1007/978-1-62703-239-1_8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kozhukharova I, Zemelko V, Kovaleva Z, et al. Therapeutic doses of doxorubicin induce premature senescence of human mesenchymal stem cells derived from menstrual blood, bone marrow and adipose tissue. Int J Hematol. 2018;107:286-296. https://doi.org/ 10.1007/s12185-017-2346-6 [DOI] [PubMed] [Google Scholar]
- 56. Tominaga K, Suzuki HI.. TGF-β signaling in cellular senescence and aging-related pathology. Int J Mol Sci . 2019;20:5002. https://doi.org/ 10.3390/ijms20205002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wu J, Niu J, Li X, Wang X, Guo Z.. Zhang F. TGF-β1 induces senescence of bone marrow.pdf. BMC Dev Biol. 2014;14(21):1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ksia̧zek K. A comprehensive review on mesenchymal stem cell growth and senescence. Rejuvenation Res. 2009;12(2):105–116. [DOI] [PubMed] [Google Scholar]
- 59. Lunyak VV, Amaro-Ortiz A, Gaur M.. Mesenchymal stem cells secretory responses: senescence messaging secretome and immunomodulation perspective. Front Genet. 2017;8(19):1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Colter DC, Sekiya I, Prockop DJ.. Identification of a subpopulation of rapidly self-renewing and multipotential adult stem cells in colonies of human marrow stromal cells. Proc Natl Acad Sci USA. 2001;98:7841-7845. https://doi.org/ 10.1073/pnas.141221698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lee WC, Shi H, Poon Z, et al. Multivariate biophysical markers predictive of mesenchymal stromal cell multipotency. Proc Natl Acad Sci USA. 2014;111:E4409-E4418. https://doi.org/ 10.1073/pnas.1402306111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Suvakov S, Cubro H, White WM, et al. Targeting senescence improves angiogenic potential of adipose-derived mesenchymal stem cells in patients with preeclampsia. Biol Sex Differ. 2019;10:49. https://doi.org/ 10.1186/s13293-019-0263-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Soto-Gamez A, Demaria M.. Therapeutic interventions for aging: the case of cellular senescence. Drug Discov Today. 2017;22:786-795. https://doi.org/ 10.1016/j.drudis.2017.01.004 [DOI] [PubMed] [Google Scholar]
- 64. Hirayama T, Nagasawa H.. Chemical tools for detecting Fe ions. J Clin Biochem Nutr. 2017;60:39-48. https://doi.org/ 10.3164/jcbn.16-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tenopoulou M, Kurz T, Doulias PT, Galaris D, Brunk U T.. Does the calcein-AM method assay the total cellular “labile iron pool” or only a fraction of it? Biochem J. 2007;403:261-266. https://doi.org/ 10.1042/bj20061840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Abbasi U, Abbina S, Gill A, et al. A facile colorimetric method for the quantification of labile iron pool and total iron in cells and tissue specimens. Sci Rep. 2021;11(6008):1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mackenzie EL, Iwasaki K, Tsuji Y.. Intracellular iron transport and storage: from molecular mechanisms to health implications. Antioxid Redox Signal. 2008;10:997-1030. https://doi.org/ 10.1089/ars.2007.1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Denoyer D, Pearson HB, Clatworthy SAS, et al. Copper as a target for prostate cancer therapeutics: copper-ionophore pharmacology and altering systemic copper distribution. Oncotarget. 2016;7:37064-37080. https://doi.org/ 10.18632/oncotarget.9245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Han Z, Yu Y, Xu J, et al. Iron Homeostasis determines fate of human pluripotent stem cells via glycerophospholipids-epigenetic circuit. Stem Cells. 2019;37:489-503. https://doi.org/ 10.1002/stem.2967 [DOI] [PubMed] [Google Scholar]
- 70. Han Z, Xu Z, Chen L, et al. Iron overload inhibits self-renewal of human pluripotent stem cells via DNA damage and generation of reactive oxygen species. FEBS Open Bio. 2020;10:726-733. https://doi.org/ 10.1002/2211-5463.12811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Xin J, Xiaoyuan H, Xiaoli C, et al. Iron overload impairs normal hematopoietic stem and progenitor cells through reactive oxygen species and shortens survival in myelodysplastic syndrome mice. Haematologica. 2018;103(10):1627-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Muto Y, Nishiyama M, Nita A, Moroishi T, Nakayama KI.. Essential role of FBXL5-mediated cellular iron homeostasis in maintenance of hematopoietic stem cells. Nat Commun. 2017;8:16114. https://doi.org/ 10.1038/ncomms16114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Dusek P, Dezortova M, Wuerfel J.. Chapter 9-Imaging of iron. In: Bhatia KP, Schneider SA, eds. International Review of Neurobiology, Vol 110. Academic Press; 2013:195-239. [DOI] [PubMed] [Google Scholar]
- 74. Van Walderveen MAA, Lycklama à Nijeholt GJ, Adér HJ, et al. Hypointense lesions on T1-weighted spin-echo magnetic resonance imaging: relation to clinical characteristics in subgroups of patients with multiple sclerosis. Arch Neurol. 2001;58(1):76-81. [DOI] [PubMed] [Google Scholar]
- 75. Golay X, Johanna Silvennoinen M, Zhou J, et al. Measurement of tissue oxygen extraction ratios from venous blood T2: increased precision and validation of principle. Magn Reson Med. 2001;46(2):282-291. [DOI] [PubMed] [Google Scholar]
- 76. Parikh T, Drew SJ, Lee VS, et al. Focal liver lesion detection and characterization with diffusion-weighted MR imaging: comparison with standard breath-hold T2-weighted imaging. Radiology. 2008;246:812-822. https://doi.org/ 10.1148/radiol.2463070432 [DOI] [PubMed] [Google Scholar]
- 77. Stucht D, Danishad KA, Schulze P, et al. Highest resolution in vivo human brain MRI using prospective motion correction. PLoS One. 2015;10:e01339211-e01339217. https://doi.org/ 10.1371/journal.pone.0133921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.https://data.mendeley.com/datasets/83cchm9fjz/draft?a=75bb28c4-88c4-48be-9635-a54c2b27ec72
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available at the Mandeley Data repository (DOI: 10.17632/83cchm9fjz.1), which can be previewed by accessing the link cited here.78