Abstract
Double-stranded RNA (dsRNA) induces gene-specific silencing in organisms from fungi to animals, a phenomenon known as RNA interference (RNAi). RNAi represents an evolutionarily conserved system to protect against aberrant expression of genes and a powerful tool for gene manipulation. Despite reports that RNAi can be induced in vertebrates, severe sequence-non-specific effects of long dsRNA have been documented in various systems. It has recently been shown in cultured mammalian cells that small interfering RNAs (siRNAs) of 21–23 nt can mediate RNAi but bypass the non-specific response induced by longer dsRNAs. However, the effectiveness of siRNAs has not been demonstrated in living vertebrates. In addition, the mechanism of siRNA suppression of gene expression in vertebrate cells remains to be elucidated. Here we show that synthetic 21 nt siRNAs can specifically inhibit the expression of exogenously introduced as well as endogenous genes in the embryos of Xenopus laevis. siRNAs significantly reduced the steady-state amount of both the mRNA and protein of the cognate gene target. Moreover, co-injection of siRNA with the target RNA transcript specifically suppressed the activity of the latter. Taken together, our findings establish siRNA-mediated post-transcriptional suppression of gene expression in Xenopus embryos.
INTRODUCTION
Double-stranded RNA (dsRNA) induces gene-specific silencing in living organisms ranging from fungi to animals, a phenomenon known as RNA interference (RNAi) (1–3). RNAi was observed first in the nematode Caenorhabditis elegans (4), and subsequently in insects and certain parasitic protozoa (5,6), as well as in oocytes and preimplantation embryos of mice (7). RNAi is closely related to the phenomena of co-suppression or post-transcriptional gene silencing in plants (8) and quelling in the fungus Neurospora (9). These processes represent an evolutionarily conserved system to protect against viral infection and the genomic instability caused by transposable elements and repetitive sequences. In addition, RNAi is a powerful tool for reverse genetic analysis and functional genomics (10,11).
In vitro biochemical analysis of RNAi in a lysate of Drosophila embryos or cells demonstrated that RNAi is mediated by 21–23 nt siRNAs cleaved from longer dsRNAs by an RNase III-like activity (12–15). The siRNAs function as guide RNAs for a sequence-specific multicomponent nuclease that catalyzes the degradation of mRNA transcripts, thereby silencing the expression of cognate genes (16,17). The finding that target mRNA is cleaved in 21–23 nt intervals lends further support to this model of siRNA action (16). siRNAs of similar size have also been found in plants and animals undergoing RNAi (18,19).
Despite reports that specific RNAi can be induced in zebrafish and Xenopus, as well as oocytes and preimplantation embryos, embryonic stem cells and some cultured cell lines of mice (7,20–27), severe sequence-non-specific effects of long dsRNA have been documented in various vertebrate systems (27–30). Plausibly, the non-specific activities may be mediated through the well-characterized dsRNA-dependent protein kinase PKR, which shuts down translation by phosphorylating initiation factor eIF2α and induces apoptosis (31,32). In addition, dsRNA can activate 2′,5′-adenosine synthetase (33), leading to non-specific mRNA cleavage by 2′,5′-adenosine-activated RNase L. In an attempt to resolve the problem of non-specificity, it has recently been found that 21–23 nt siRNAs which mediate RNAi in cultured mammalian cells can bypass the non-specific response induced by longer dsRNAs (34–36). This new finding paves the way for the use of siRNA in vertebrates. However, the utility of siRNAs in living vertebrates has not been demonstrated. In addition, the mechanism through which siRNAs inhibit gene expression is still incompletely understood. Studies in different biological systems remain necessary to address basic questions as to whether siRNA-mediated RNAi occurs transcriptionally or post-transcriptionally in all vertebrates.
Xenopus laevis is a major model organism for cellular and developmental biology research (37). Thus, Xenopus has led the way in establishing the mechanisms of early embryonic development, morphogenesis and organogenesis. One major limitation of this system is the lack of genetic analysis. It has not yet been possible to genetically knockout particular genes in Xenopus nor to work with a large number of defined mutants. Major approaches to block gene function in Xenopus rely on antisense RNAs and dominant-negative proteins. However, it is understood that antisense and dominant-negative experiments may sometimes yield non-specific results that are unexpected and even unexplainable (37). Hence, a new method for specific loss-of-function analysis in Xenopus is desirable. To develop such a method based on siRNA-mediated suppression of gene expression and to shed light on the underlying mechanisms, in this study we have investigated whether chemically synthesized siRNAs of 21 nt in length might inhibit gene expression in Xenopus embryos in a sequence-dependent fashion. siRNAs targeting either exogenous or endogenous genes were tested. Furthermore, co-injection experiments were performed to evaluate whether siRNAs might act directly on the co-injected RNA target. Our work provides the first evidence that siRNAs mediate post-transcriptional suppression of homologous gene expression in Xenopus embryos.
MATERIALS AND METHODS
Plasmids
Plasmid pSPluc+NF was from Promega. To generate Xenopus expression vector pCS2-Luc for the GL3 version of luciferase (GenBank accession no. U47123), a 1.7 kb BglII–EcoRI fragment was released from pSPluc+NF and subcloned into pCS2 (a gift from Dr Dave Turner, University of Michigan, MI; 38) predigested with BamHI and EcoRI.
RNAs
Twenty-one nucleotide RNAs were chemically synthesized using 2′-O-silylated ribonucleoside 3′-O-phosphoramidites and thymidine phosphoramidite (GENSET Oligos, France). RNA oligos were purified by PAGE. The siRNA sequences targeting the GL2 and GL3 versions of luciferase have been described (34). The siRNA sequences targeting Xenopus cyclins B1 and B2 were as follows: B1A5, 5′-CGU ACA GCC UUG GGA GAC ATT; B1A3, 5′-UGU CUC CCA AGG CUG UAC GTT; B2A5, 5′-GUU AAG ACU AAG CAU GUA CTT; B2A3, 5′-GUA CAU GCU UAG UCU UAA CTT. Cyclin B1 siRNA was derived from nucleotides 199–217 (GenBank accession no. J03166). Cyclin B2 siRNA was from nucleotides 252–270 (GenBank accession no. J03167). Both targeting sequences are in the coding regions. Sequences of the mock siRNA (targeting irrelevant human but not Xenopus sequences) were 5′-UUC AAA UUG AAC UCC UCC ATT (MOCK5) and 5′-UGG AGG AGU UCA AUU UGA ATT (MOCK3).
RNA oligos were reconstituted to 200 µM in DEPC-treated water and stored in aliquots at –80°C. For annealing of siRNAs, 40 µM single strands were incubated in annealing buffer (100 mM potassium acetate, 30 mM HEPES–KOH, pH 7.4, and 2 mM MgCl2) for 1 min at 90°C, followed by 1 h at 37°C. Annealed siRNAs were stored in 1 µl aliquots at –80°C.
For the production of long dsRNA, equimolar amounts of sense and antisense RNAs in annealing buffer were annealed at 68°C for 20 min and 37°C for 2 h. Annealed long dsRNAs were stored in 1 µl aliquots at –80°C. The quality of dsRNA was analyzed on native agarose gels using dsDNAs and single-stranded RNAs as controls. Xenopus 14-3-3ɛ cDNA was generated by PCR and was used as template for the preparation of mock dsRNA.
Luciferase-coding RNA was synthesized via in vitro transcription driven by SP6 RNA polymerase (MEGAscript; Ambion). Linearized pSPluc+NF was used as template. RNA was capped with reagents from Ambion, purified using a RNeasy Mini kit from Qiagen and stored in 1 µl aliquots at –80°C.
Xenopus embryos
Eggs were collected from X.laevis females (Xenopus Express, Cape Town, South Africa), which had been injected with 500–700 U human chorionic gonadotrophin (Sigma) 12 h before egg collection. Eggs were fertilized in vitro with minced testis. Embryos were staged as described (37).
Luciferase assay
DNA (150 pg) or RNA (2 ng) samples (5 nl) with or without siRNA (10–20 µM) were microinjected into both blastomeres of 2-cell stage embryos. Injected embryos were collected at different stages. For each stage eight embryos in four groups were picked out randomly and washed twice with phosphate-buffered saline (PBS). Each group was lysed in 50 µl of 1× passive lysis buffer (Promega) and 20 µl of the lysate was assayed for luciferase activity as recommended by the manufacturer.
Western blotting
siRNAs (20–40 µM in 5 nl) were injected into both blastomeres of 2-cell stage embryos. Eight embryos in four groups were collected at the 4-, 8- and 32-cell stages. Embryos were washed once with PBS and each group was homogenized in 100 µl of whole cell extraction buffer (25 mM Tris–HCl, pH 7.5, 70 mM KCl, 1 mM EDTA, 20% glycerol, 5 mM dithiothreitol, 1 µg/µl leupeptin and 1 µg/µl pepstatin). The lysate (20 µl) was loaded onto a 12% SDS–PAGE gel and transferred to a PVDF membrane (Millipore). Rabbit antiserum raised against Xenopus cyclin B1 (39) was kindly provided by Dr Ulrich Strausfeld (University of Konstanz, Germany). This primary antibody was used at 1:1000 dilution to detect endogenous cyclin B1. Mouse monoclonal antibody to α-tubulin (clone B-5-1-2) was from Sigma. Standard immunostaining was carried out using the ECL enhanced chemiluminescence technique (Amersham Pharmacia).
RT–PCR
Embryos were collected as above. Total RNA was isolated using Trizol reagent (Gibco BRL). Synthesis of first strand cDNA was primed by oligo(dT) and random hexamers. Primers for Xenopus cyclin B1 were 5′-TGT CAA GGA CAT TTA TGC TT and 5′-GGC TAA ACT ATC CTG TTC AG (360 bp fragment). Primers for Xenopus histone H4 were 5′-CGG GAT AAC ATT CAG GGT ATC ACT and 5′-ATC CAT GGC GGT AAC TGT CTT CCT (188 bp fragment). All RT–PCR amplifications were performed in the linear range.
RESULTS AND DISCUSSION
Specific inhibition of heterologous gene expression in Xenopus embryos by siRNA
Previous work in zebrafish and Xenopus embryos has suggested that RNAi may work in vertebrates (7,20–27). On the other hand, dsRNAs of >30 bp in length are also known to induce sequence-non-specific effects plausibly mediated through the activation of 2′,5′-adenosine synthetase and dsRNA-dependent protein kinase PKR (31–33). Thus, long dsRNAs induce non-specific degradation of all RNAs (28,29), leading to various gross defects in dsRNA-injected zebrafish and Xenopus embryos (27–29). The non-specific effects of long dsRNA hamper its further use for functional studies in model vertebrates such as zebrafish and Xenopus, as well as in differentiated somatic tissues in mice. Recent findings that 21–23 nt siRNAs can mediate RNAi in cultured mammalian cells without triggering the non-specific pathway leading to mRNA degradation and inhibition of protein synthesis (34–36) have paved the way for broader use of siRNAs. However, it remains to be seen whether siRNAs are as effective in living vertebrates as in cultured vertebrate cells. As a first step towards the application of siRNAs in Xenopus, we first examined whether siRNAs might inhibit expression of an exogenously introduced reporter gene during early embryogenesis.
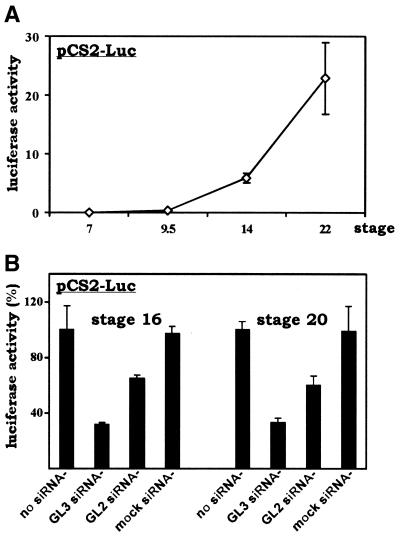
We microinjected pCS2-Luc, a Xenopus expression vector for the GL3 version of firefly luciferase, into 2-cell stage Xenopus embryos and assessed temporal expression of the luciferase gene at different developmental stages. Xenopus early embryos undergo synchronous divisions up to the 12th cell cycle. Cleavage cycles 2–12 are rapid and consist of alternating S and M phases only. Then the division cycle slows down and becomes asynchronous, zygotic transcription begins and gap phases (G1 and G2) are established. This change, occurring at stages 8 and 9, is known as the mid-blastula transition (MBT) (37). As expected, the luciferase reporter gene was not expressed before the MBT because the embryos were transcriptionally silent, but post-MBT expression of luciferase was robust and persistent, with a >20-fold increase from stage 9.5 to stage 22 (Fig. 1A). To examine the effects of siRNAs, we co-injected pCS2-Luc separately with buffer alone (no siRNA), with siRNA targeting the GL3 version of luciferase (GL3 siRNA), with siRNA designed against GL2 luciferase (GL2 siRNA) and with mock siRNA (Fig. 1B). Notably, the GL3 and GL2 versions of the luciferase gene share 95% identity in DNA sequence. In fact only 3 of 21 nt are different between the GL3 and GL2 siRNAs. The mock siRNA is against an irrelevant human cDNA. We observed that co-injection of GL3 siRNA led to >60% inhibition of expression of GL3 luciferase (Fig. 1B). In sharp contrast, the luciferase activity recovered from embryos co-injected with mock siRNA did not change noticeably if compared to the level in embryos that received no siRNA. The fact that mock siRNA could not inhibit luciferase expression verified the specificity of siRNA action. Interestingly, the GL2 siRNA, which is identical to GL3 siRNA except for 3 nt, also inhibited expression of the homologous GL3 luciferase, although the inhibition of GL3 luciferase by GL2 siRNA (∼25% inhibition) was significantly weaker than by GL3 siRNA (>60% inhibition). In the same setting, GL2 siRNA can specifically inhibit expression of GL2 luciferase (data not shown). Notably, similar patterns of inhibition by the GL2 and GL3 siRNAs were seen at both stages 16 and 20, suggesting that siRNA acts persistently, at least during neurulation.
Figure 1.
siRNA suppression of heterologous gene expression in Xenopus embryos. (A) Temporal expression of the luciferase gene during early embryogenesis. Plasmid pCS2-Luc (150 pg in 5 nl), an expression vector for the GL3 version of luciferase, was microinjected into both blastomeres of 2-cell stage embryos. Injected embryos were collected at different stages, lysed and assayed for luciferase activity. All values represent the means of four groups of embryos and error bars indicate standard deviation from the mean. Experiments were repeated twice with similar results. (B) Gene-specific inhibition of luciferase expression by siRNA. Luciferase expression plasmid pCS2-Luc (150 pg) and the indicated siRNAs (20 µM) were co-injected into embryos as in (A). The injection volume was 5 nl. Embryos were collected at stages 16 and 20. The average luciferase activity recovered from control embryos without siRNA was set as 100%. GL3 siRNA, siRNA targeting GL3 luciferase; GL2 siRNA, siRNA targeting the GL2 version of luciferase; mock siRNA, siRNA targeting irrelevant human but not Xenopus sequences. Similar results were obtained using 10 µM siRNA (data not shown).
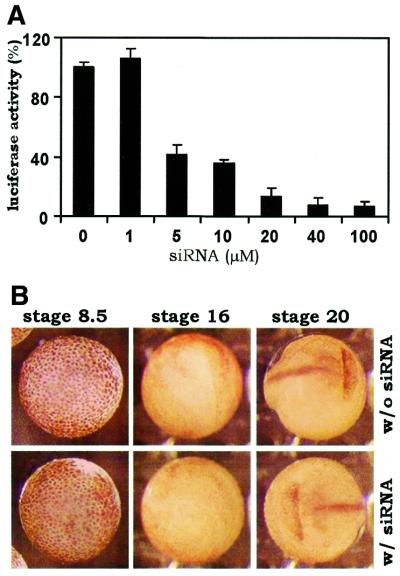
We also determined the concentration dependence of siRNA-mediated inhibition of gene expression. In contrast to the siRNA effect in nematodes, where as few as two molecules/cell are sufficient to induce repression (4), at least 5 µM GL3 siRNA was needed to mediate a significant inhibition of luciferase gene expression in Xenopus embryos (Fig. 2A). The dose-dependent effect of siRNA in Xenopus is generally consistent with previous findings on RNAi in mouse oocytes (26).
Figure 2.
Dose-dependent and specific inhibition of gene expression by siRNA. (A) Concentration-dependent effect of siRNA. PCS2-Luc (150 pg) was co-injected with the indicated concentrations of GL3 siRNA. The injection volume was 5 nl. (B) Representative examples of uninjected and GL3 siRNA-injected embryos at stages 8.5, 16 and 20. siRNA (20 µM in 5 nl) was injected at the 2-cell stage. At least 40 embryos were compared.
We noted that GL2 siRNA failed to inhibit expression of GL3 luciferase in four lines of cultured mammalian cells (34). We do not understand fully why GL2 siRNA behaves differently in the two systems. Nevertheless, the significantly weaker inhibition of GL3 luciferase expression mediated by GL2 siRNA in Xenopus embryos implies that siRNA inhibition is strictly sequence dependent, with 15% divergence in sequence making a dramatic difference. This is in agreement with results previously shown in other systems (19,40–42).
siRNA inhibition of gene expression in Xenopus embryos (Fig. 1B), albeit substantial and significant, is not as complete as in nematode and insect cells (4–6). These data are in line with published results obtained from cultured mammalian cells (34–36). Our findings reinforce the possibility that siRNAs may act through different mechanisms in vertebrates.
Specificity of action remains a major concern for siRNA-mediated inhibition of gene expression in Xenopus embryos. Previous studies from several groups have documented the induction of non-specific gross defects in zebrafish and Xenopus embryos by long dsRNA (25,27–29). In particular, it has been demonstrated that embryos injected with dsRNAs directed against lacZ or green fluorescent protein also display severe non-specific defects as early as 12 and 24 h post-fertilization (25,28,29). With this in mind, we compared the morphology of uninjected and GL3 siRNA-injected Xenopus embryos during the whole process of embryogenesis. We could not find any defects or abnormalities in the injected embryos. Neither could we discern any difference between the uninjected and injected groups at any developmental stage (representative embryos are shown in Fig. 2B). In particular, we performed semi-quantitative RT–PCR to compare the relative amounts of four mRNA transcripts (for cyclin B1, cyclin B2, CDC20/Fizzy/p55CDC and CDH1/Fizzy-related/HCT1) in the uninjected and injected embryos, but we failed to detect any change (data not shown). Thus, it is very unlikely that GL3 siRNA can induce non-specific degradation of other mRNAs. These results lend further support to the specificity of siRNA inhibition of gene expression.
Specific inhibition of endogenous gene expression in Xenopus embryos by siRNA
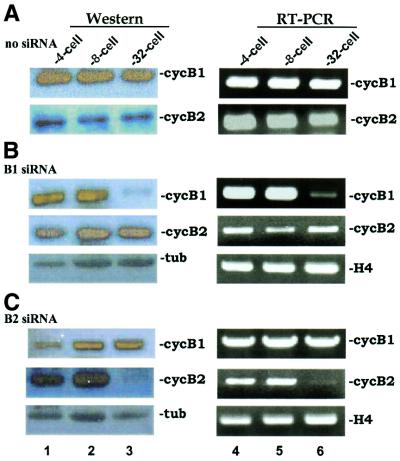
Above we have shown that siRNAs can specifically suppress expression of an exogenously introduced reporter gene. Suppression occurs after the MBT and is persistent until at least stage 20. Next we queried siRNA inhibition of endogenous genes. We designed two siRNAs against Xenopus cyclins B1 and B2. It is well known that cyclins oscillate during the early embryonic cell cycles (43). In particular, the cyclin B1 and B2 proteins are destroyed in mitosis through ubiquitin-dependent proteolysis catalyzed by the anaphase-promoting complex and are resynthesized from long-lasting maternal mRNA in the next S phase (43). When we microinjected siRNA against cyclin B1 (B1 siRNA) into 2-cell stage embryos, the steady-state amount of cyclin B1 protein dropped dramatically at the 32-cell stage after two successive rounds of division (compare Fig. 3B with A). As a control, the levels of α-tubulin and of cyclin B2 were not reduced in B1 siRNA-injected embryos. In contrast, the amount of cyclin B1 in embryos injected with siRNA against cyclin B2 (B2 siRNA), which was shown to specifically inhibit expression of cyclin B2 (Fig. 3C), was comparable to that in mock-injected embryos without siRNA (compare Fig. 3C with A), suggesting that inhibition mediated by siRNA has sequence specificity. The reduction in cyclins B1 and B2 is unlikely to be due to oscillations in cyclin production because early embryonic cell cycles in Xenopus are synchronized (37) and the control mock-injected embryos collected at the same time points expressed cyclin B1 and cyclin B2 abundantly (Fig. 3A).
Figure 3.
siRNA suppression of endogenous gene expression in Xenopus embryos. Both blastomeres of 2-cell stage embryos were mock injected (A), injected with siRNAs (20 µM in 5 nl) targeting Xenopus cyclin B1 (B) or injected with siRNA against cyclin B2 (C). Western blotting (left) and RT–PCR (right) were performed using the indicated antibodies and primers. Injection of 40 µM siRNAs yielded similar results (data not shown). B1 siRNA, cyclin B1-targeting siRNA; B2 siRNA, cyclin B2-targeting siRNA; cycB1, cyclin B1; cycB2, cyclin B2; tub, α-tubulin; H4, histone H4.
Since Xenopus embryos are transcriptionally silent before the MBT (37), inhibition of cyclin B1 expression by B1 siRNA likely occurs post-transcriptionally through inactivation of mRNA. To shed additional light on this, we performed semi-quantitative RT–PCR to check for cyclin B1 transcript. Results from this analysis were generally consistent with data from western blotting (Fig. 3). Again, the injection of B1 siRNA specifically reduced cyclin B1 mRNA in 32-cell stage embryos, but not histone H4 or cyclin B2 transcripts (Fig. 3B). The amounts of cyclin B1 mRNA in mock-injected (Fig. 3A) and B2 siRNA-injected (Fig. 3C) embryos were constant. We also noted that the injection of B1 or B2 siRNA broke synchronization of the cell division beyond the 32-cell stage (data not shown), so it is difficult to compare the mock-injected embryos continuously. Nevertheless, siRNA is effective for specific inhibition of an endogenous gene in the early cell cycles before onset of the MBT during Xenopus embryogenesis. Our data are in concordance with the model that siRNA induces gene-specific degradation of mRNA transcript.
Post-transcriptional suppression of gene expression in Xenopus embryos by siRNA
RNAi occurs both transcriptionally and post-transcriptionally in plants (8,15,44). However, the mechanisms of siRNA suppression of gene expression in vertebrate embryos and cells remain elusive. Our findings that injection of siRNA into Xenopus embryos led to a gene-specific reduction in mRNA transcript in a transcriptionally silent system prompt us to test more directly post-transcriptional suppression of gene expression by siRNA. Towards this end, we evaluated the influence of siRNA on the activity of an in vitro transcribed reporter RNA injected into Xenopus embryos.
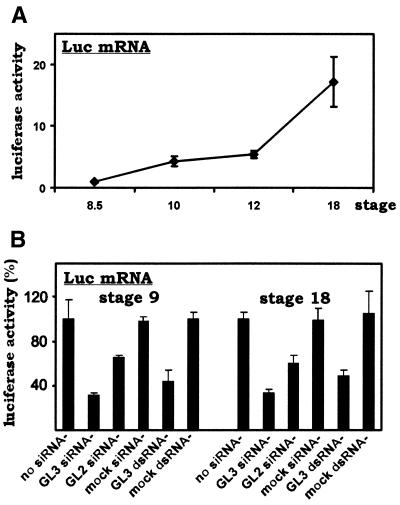
First, we microinjected an in vitro transcribed RNA coding for GL3 luciferase (Luc mRNA) into 2-cell stage embryos and determined the temporal expression pattern of luciferase during embryogenesis (Fig. 4A). We noted that luciferase activity was high at stages 9 and 18. Then we set out to co-inject siRNA with the reporter RNA in order to determine whether the siRNA acts at the transcriptional or post-transcriptional level. We reasoned that the siRNA should inhibit the activities of the reporter RNA and reporter DNA to the same degree if it acts directly on the RNA target solely at a post-transcriptional step. The siRNA should not affect the activity of the reporter RNA if its action is solely transcriptional. Finally, a partial inhibitory effect would be seen if the siRNA acts both transcriptionally and post-transcriptionally.
Figure 4.
Post-transcriptional suppression of gene expression in Xenopus embryos by siRNA and long dsRNA. (A) Temporal expression of luciferase-coding RNA during early embryogenesis. In vitro transcribed RNA encoding GL3 luciferase (Luc mRNA, 2 ng) was microinjected into one blastomere of 2-cell stage embryos. Injected embryos were collected at different stages, lysed and assayed for luciferase activity. (B) Gene-specific inhibition of luciferase expression by siRNA and long dsRNA. Luciferase-coding RNA (Luc mRNA, 2 ng) and the indicated siRNAs (20 µM) or dsRNAs (0.2 µg/µl) were microinjected into embryos as in (A). The injection volume was 5 nl. Embryos were collected at stages 9 and 18. The average luciferase activity recovered from control embryos without siRNA or dsRNA was set as 100%. GL3 siRNA, siRNA targeting GL3 luciferase; GL2 siRNA, siRNA targeting the GL2 version of luciferase; mock siRNA, siRNA targeting irrelevant human but not Xenopus sequences; GL3 dsRNA, 1.7 kb dsRNA corresponding to full-length GL3 luciferase-coding sequences; mock dsRNA, 1 kb dsRNA corresponding to an irrelevant gene. All values represent the means of four groups of embryos and error bars indicate standard deviation from the mean. Experiments were repeated twice with similar results.
Indeed, we observed the same degree of siRNA inhibitory effect on a reporter RNA (i.e. Luc mRNA; Fig. 4B) if compared to its inhibition of a reporter DNA (i.e. pCS2-Luc; Fig. 1B). Thus, at both stages 9 and 18 GL3 siRNA conferred 50–70% inhibition of the luciferase activity ascribed to the Luc mRNA. Notably, the phenotypes induced by GL2 siRNA and mock siRNA in the reporter RNA experiment (Fig. 4B) are also similar to the phenotypes induced in the reporter DNA experiments (Fig. 1B). Hence, while we cannot completely rule out the possibility of transcriptional suppression by siRNA, our data support post-transcriptional interference as the major mechanism for siRNA inhibition of gene expression.
We also observed that a long dsRNA targeting GL3 luciferase (GL3 dsRNA) can specifically inhibit the activity of reporter RNA at both stages 9 and 18 (Fig. 4B). The inhibitory effect of this 1.7 kb GL3 dsRNA is comparable to that mediated by GL3 siRNA. In this setting, a mock dsRNA of 1.0 kb did not induce non-specific inhibition of reporter RNA activity. Thus, long dsRNAs are capable of mediating specific suppression of homologous gene expression in Xenopus embryos, at least under certain circumstances. These findings are compatible with the recent documentation of specific RNAi in some cultured mammalian cells (23,24,26). However, we did notice that non-specific gross defects and abnormalities were found at significantly higher frequencies in Xenopus embryos injected with long dsRNAs than in uninjected or siRNA-injected counterparts (data not shown). Further investigations are required to fully document the phenotypes induced by long dsRNAs in Xenopus.
In this study we provide the first evidence that siRNAs are effective for sequence-specific suppression of heterologous and endogenous gene expression in Xenopus embryos (Figs 1–3). We also show that this suppression occurs post-transcriptionally through elimination of target mRNA (Figs 3 and 4). Considered together with recent reports that demonstrate siRNA inhibition of gene expression in cultured mammalian cells (34–36), our findings may prove useful for the further development of a new and powerful tool for blocking gene function in model vertebrates and, eventually, in humans. With further elucidation of the underlying mechanisms and further optimization of their uses, siRNAs may represent a new category of nucleic acid therapeutics for the treatment of human diseases.
Acknowledgments
ACKNOWLEDGEMENTS
We thank C. M. Wong for technical assistance with the computer graphics, D. Turner and U. Strausfeld for gifts of reagents and Y. Peng, A. C. S. Chun, C. M. Wong and O. G. W. Wong for critical reading of the manuscript. D.-Y.J. is a Leukemia and Lymphoma Society Scholar. K.H.K. is an MSc project student. This work was supported in part by grants (to D.-Y.J.) from the University Research Committee of the University of Hong Kong.
REFERENCES
- 1.Sharp P.A. (2001) RNA interference—2001. Genes Dev., 15, 485–490. [DOI] [PubMed] [Google Scholar]
- 2.Hammond S.M., Caudy,A.A. and Hannon,G.J. (2001) Post-transcriptional gene silencing by double-stranded RNA. Nature Rev. Genet., 2, 110–119. [DOI] [PubMed] [Google Scholar]
- 3.Zamore P.D. (2001) RNA interference: listening to the sound of silence. Nature Struct. Biol., 8, 746–750. [DOI] [PubMed] [Google Scholar]
- 4.Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 5.Kennerdell J.R. and Carthew,R.W. (1998) Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell, 95, 1017–1026. [DOI] [PubMed] [Google Scholar]
- 6.Ngo H., Tschudi,C., Gull,K. and Ullu,E. (1998) Double-stranded RNA induces mRNA degradation in Trypanosoma brusei. Proc. Natl Acad. Sci. USA, 95, 14687–14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wianny F. and Zernicka-Goetz,M. (2000) Specific interference with gene function by double-stranded RNA in early mouse development. Nature Cell Biol., 2, 70–75. [DOI] [PubMed] [Google Scholar]
- 8.Waterhouse P.M., Graham,M.W. and Wang,M.B. (1998) Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl Acad. Sci. USA, 95, 13959–13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cogoni C., Irelan,J.T., Schumacher,M., Schmidhauser,T.J., Selker,E.U. and Macino,G. (1996) Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA–DNA interactions or DNA methylation. EMBO J., 15, 3153–3163. [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser A.G., Kamath,R.S., Zipperlen,P., Martinez-Campos,M., Sohrmann,M. and Ahringer,J. (2000) Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature, 408, 325–330. [DOI] [PubMed] [Google Scholar]
- 11.Gönczy P., Echeverri,G., Oegema,K., Coulson,A., Jones,S.J., Copley,R.R., Duperon,J., Oegema,J., Brehm,M., Cassin,E. et al. (2000) Functional genomic analysis of cell devision in C. elegans using RNAi of genes on chromosome III. Nature, 408, 331–336. [DOI] [PubMed] [Google Scholar]
- 12.Zamore P.D., Tuschl,T., Sharp,P.A. and Bartel,D.P. (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell, 101, 25–33. [DOI] [PubMed] [Google Scholar]
- 13.Yang D., Lu,H. and Erickson,J.W. (2000) Evidence that processed small dsRNAs may mediate sequence-specific mRNA degradation during RNAi in Drosophila embryos. Curr. Biol., 10, 1191–1200. [DOI] [PubMed] [Google Scholar]
- 14.Elbashir S.M., Lendeckel,W. and Tuschl,T. (2001) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev., 15, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernstein E., Caudy,A.A., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. [DOI] [PubMed] [Google Scholar]
- 16.Tuschl T., Zamore,P.D., Lehmann,R., Bartel,D.P. and Sharp,P.A. (1999) Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev., 13, 3191–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammond S.M., Bernstein,E., Beach,D. and Hannon,G.J. (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature, 404, 293–296. [DOI] [PubMed] [Google Scholar]
- 18.Hamilton A.J. and Baulcombe,D.C. (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science, 286, 950–952. [DOI] [PubMed] [Google Scholar]
- 19.Parrish S., Fleenor,J., Xu,S.Q., Mello,C. and Fire,A. (2000) Functional anatomy of a dsRNA trigger: differential requirement for the two trigger strands in RNA interference. Mol. Cell, 6, 1077–1087. [DOI] [PubMed] [Google Scholar]
- 20.Li Y.-X., Farrell,M.J., Liu,R., Mohanty,N. and Kirby,M.L. (2000) Double-stranded RNA injection produces null phenotypes in zebrafish. Dev. Biol., 217, 394–405. [DOI] [PubMed] [Google Scholar]
- 21.Oelgeschläger M., Larrain,J., Geissert,D. and De Robertis,E.M. (2000) The evolutionarily conserved BMP-binding protein Twisted gastrulation promotes BMP signalling. Nature, 405, 757–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang S., Tutton,S., Pierce,E. and Yoon,K. (2001) Specific double-stranded RNA interference in undifferentiated mouse embryonic stem cells. Mol. Cell. Biol., 21, 7807–7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Billy E., Brondani,V., Zhang,H., Müller,U. and Filipowicz,W. (2001) Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl Acad. Sci. USA, 98, 14428–14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paddison P.J., Caudy,A.A. and Hannon,G.J. (2002) Stable suppression of gene expression by RNAi in mammalian cells. Proc. Natl Acad. Sci. USA, 99, 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wargelius A., Ellingsen,S. and Fjose,A. (1999) Double-stranded RNA induces specific developmental defects in zebrafish embryos. Biochem. Biophys. Res. Commun., 263, 156–161. [DOI] [PubMed] [Google Scholar]
- 26.Svoboda P., Stein,P., Hayashi,H. and Schultz,R.M. (2000) Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development, 127, 4147–4156. [DOI] [PubMed] [Google Scholar]
- 27.Nakano H., Amemiya,S., Shiokawa,K. and Taira,M. (2000) RNA interference for the organizer-specific gene Xlim-1 in Xenopus embryos. Biochem. Biophys. Res. Commun., 274, 434–439. [DOI] [PubMed] [Google Scholar]
- 28.Oates A.C., Bruce,A.E.E. and Ho,R.K. (2000) Too much interference: injection of double-stranded RNA has non-specific effects in the zebrafish embryo. Dev. Biol., 224, 20–28. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Z., Cao,Y., Li,M. and Meng,A. (2001) Double-stranded RNA injection produces nonspecific defects in zebrafish. Dev. Biol., 229, 215–223. [DOI] [PubMed] [Google Scholar]
- 30.Caplen N.J., Fleenor,J., Fire,A. and Morgan,R.A. (2000) dsRNA-mediated gene silencing in cultured Drosophila cells: a tissue culture model for the analysis of RNA interference. Gene, 252, 95–105. [DOI] [PubMed] [Google Scholar]
- 31.Samuel C.E. (1993) The eIF-2α protein kinases, regulators of translation in eukaryotes from yeasts to humans. J. Biol. Chem., 268, 7603–7606. [PubMed] [Google Scholar]
- 32.Der S.D., Yang,Y.L., Weissmann,C. and Williams,B.R.G. (1997) A double-stranded RNA-activated protein kinase-dependent pathway mediating stress-induced apoptosis. Proc. Natl Acad. Sci. USA, 94, 3279–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghosh S.K., Kusari,J., Bandyopadhyay,S.K., Samanta,H., Kumar,R. and Sen,G.C. (1991) Cloning, sequencing and expression of two murine 2′-5′-oligoadenylate synthetases. Structure–function relationships. J. Biol. Chem., 266, 15293–15299. [PubMed] [Google Scholar]
- 34.Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 35.Caplen N.J., Parrish,S., Imani,F., Fire,A. and Morgan,R.A. (2001) Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl Acad. Sci. USA, 98, 9742–9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caplen N.J., Taylor,J.P., Statham,V.S., Tanaka,F., Fire,A. and Morgan,R.A. (2002) Rescue of polyglutamine-mediated cytotoxicity by double-stranded RNA-mediated RNA interference. Hum. Mol. Genet., 11, 175–184. [DOI] [PubMed] [Google Scholar]
- 37.Sive H.L., Grainger,R.M. and Harland,R.M. (2000) Early Development of Xenopus laevis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Turner D.L. and Weintraub,H. (1994) Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev., 8, 1434–1447. [DOI] [PubMed] [Google Scholar]
- 39.Findeisen M., El-Denary,M., Kapitza,T., Graf,R. and Strausfeld,U. (1999) Cyclin A-dependent kinase activity affects chromatin binding of ORC, Cdc6 and MCM in egg extracts of Xenopus laevis. Eur. J. Biochem., 264, 415–426. [DOI] [PubMed] [Google Scholar]
- 40.Boutla A., Delidakis,C., Livadaras,I., Tsagris,M. and Tabler,M. (2001) Short 5′-phosphorylated double-stranded RNAs induce RNA interference in Drosophila. Curr. Biol., 11, 1776–1780. [DOI] [PubMed] [Google Scholar]
- 41.Scadden A.D.J. and Smith,C.W.J. (2001) RNAi is antagonized by A→I hyper-editing. EMBO Rep., 2, 1107–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elbashir S.M., Martinez,J., Patkaniowska,A., Lendeckel,W. and Tuschl,T. (2001) Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J., 20, 6877–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morgan D.O. (1999) Regulation of the APC and the exit from mitosis. Nature Cell Biol., 1, E47–E53. [DOI] [PubMed] [Google Scholar]
- 44.Wassenegger M. (2000) RNA-directed DNA methylation. Plant Mol. Biol., 43, 203–220. [DOI] [PubMed] [Google Scholar]