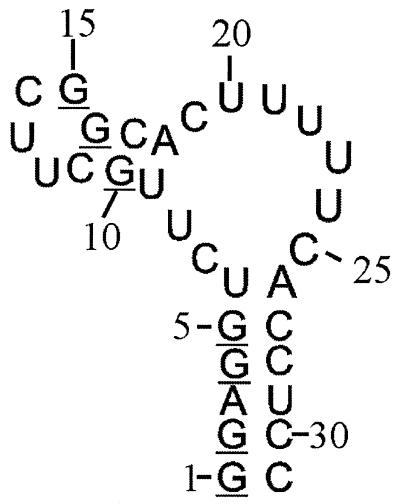Abstract
An effective in vitro enzymatic synthesis is described for the production of nucleoside triphosphates (NTPs) which are stereo-specifically deuterated on the H5′′ position with high selectivity (>98%), and which can have a variety of different labels (13C, 15N, 2H) in other positions. The NTPs can subsequently be employed in the enzymatic synthesis of RNAs using T7 polymerase from a DNA template. The stereo-specific deuteration of the H5′′ immediately provides the stereo-specific assignment of H5′ resonances in NMR spectra, giving access to important structural parameters. Stereo-chemical H-exchange was used to convert commercially available 1,2,3,4,5,6,6-2H-1,2,3,4,5,6-13C-d-glucose (d7-13C6-d-glucose) into [1,2,3,4,5,6(R)-2H-1,2,3,4,5,6-13C]-d-glucose (d6-13C6-d-glucose). [1′,3′,4′,5′′-2H-1′,2′,3′,4′,5′-13C]GTP (d4-13C5-GTP) was then produced from d6-13C6-d-glucose and guanine base via in vitro enzymatic synthesis employing enzymes from the pentose-phosphate, nucleotide biosynthesis and salvage pathways. The overall yield was ∼60 mg NTP per 1 g glucose, comparable with the yield of NTPs isolated from Escherichia coli grown on enriched media. The d4-13C5-GTP, together with in vitro synthesised d5-UTP, d5-CTP and non-labelled ATP, were used in the synthesis of a 31 nt RNA derived from the primer binding site of hepatitis B virus genomic RNA. (13C,1H) hetero-nuclear multiple-quantum spectra of the specifically deuterated sample and of a non-deuterated uniformly 13C/15N-labelled sample demonstrates the reduced spectral crowding and line width narrowing compared with 13C-labelled non-deuterated RNA.
INTRODUCTION
Structure determination by NMR of RNA molecules is intrinsically difficult because of the strong resonance overlap. This overlap is most severe in the H2′ to H5′/H5″ region of 1D and 2D 1H-NMR spectra, but is also evident in hetero-nuclear NMR correlation spectra, since many 13C/15N/31P resonances group in rather narrow regions. Thanks to uniform 13C/15N labelling, at present a size limit of structure determination by NMR of RNA molecules is reached of ∼50 nt (1). For larger RNA molecules, resonance overlap will further increase, compounded by the increased line broadening, which in turn will reduce the efficiency of hetero-nuclear NMR experiments (2). As a result, assignment of many proton resonances becomes more and more ambiguous. It may then become difficult, if not impossible, to derive a sufficient number of accurate distances from Nuclear Overhauser Effect (NOE) data to calculate a reliable NMR structure. To extend this size limit spectral simplification is necessary.
The special nature of the resonance overlap in RNAs, i.e. grouping of resonances in particular spectral regions (2,3), suggests that selective labelling methods are needed, rather than uniform labelling. It has been suggested on theoretical grounds that via a combination of selective deuteration and selective 13C/15N labelling, it should be possible to study RNAs with a size well over 100 nt (2,3). Substitution of hydrogens for deuterium both removes resonances from NMR spectra and reduces dipole–dipole induced relaxation, resulting in spectral simplification and improved efficiency of hetero-nuclear NMR experiments. It has been shown that deuteration of DNA and RNA oligonucleotides decreases the transverse relaxation rates of the remaining protons and results in decreased line widths and improves spectral resolution of those proton resonances (4–8).
Synthesis of specifically (deuterium) labelled RNA has been carried out successfully using phosphoramidite chemistry (9–11). This chemical synthesis has the advantage that non-uniform labelling schemes can easily be applied. However, in general, large isotopically labelled RNA oligonucleotides are prepared using labelled nucleoside triphosphates (NTPs) transcribed by T7 RNA polymerase directed by a DNA template (12–14). Isolation of the required NTPs from Escherichia coli grown on minimal medium composed of 90% 2H2O and sodium deuterioacetate, leads to uniform 2H labelling of ∼90% (4,5). The H5 and H8 protons can then be reintroduced via chemical exchange. The disadvantage of this biosynthetic approach is that sugar protons cannot be reintroduced.
A versatile alternative route towards selective labelling of larger RNAs is the in vitro synthesis of NTPs using enzymes from the pentose-phosphate, nucleotide biosynthesis and salvage pathways, originally proposed by Tolbert and co-workers (6,15,16). With this method specifically deuterated NTPs for labelled RNA can be synthesised in high yield from glucose. This synthetic scheme provides many different labelling patterns useful for RNA NMR studies by the combination of a variety of commercially available isotopically labelled variants of glucose with distinctly isotope labelled bases. In addition, this method provides a choice between fully deuterated and selectively deuterated ribose moieties. For example, protonation of both C1′ and C2′ (3,6) or only C2′ (17) have been reported. For such RNAs, the H3′, H4′, H5′ and H5′′ resonances are removed from the crowded H2′–H5′/H5′′ spectral region, which takes away the overlap of the (H2′,C2′) and (H3′,C3′) cross-peaks in (1H,13C) correlation experiments. The H2′ then become more easily assigned and the important sequential H2′–H6/8 NOE contacts become accessible. The sugar-pucker can still be determined, e.g. from JH1′H2′-couplings or via long-range JH2′C-couplings (18,19). In addition, removal of the H4′ takes away the dipolar interaction between H4′ and C4′, so that coherence transfer from C4′ to 31P becomes more efficient (2,17) and, thus, determination of the JC4′P3′- and JC4′P5′-couplings more accurate. The JC4′P3′-coupling together with JC2′P3′- and/or JH2H′3′-couplings unambiguously establish the ɛ- torsion angle (2). The JC4′P5′-coupling unambiguously establishes the β-torsion angle when it is trans; otherwise either the JH5′P5′- or JH5′′P5′-coupling is needed in addition (2).
Thus, although this in vitro synthesis method as described above is quite versatile, a drawback is that both H5′ and H5′′ are also removed. The H5′ (and H5′′) spins provide important structural parameters. Information from J-coupling involving either H5′ or H5′′ spins is required to determine the γ-torsion angle; for example, the JH5′C3′-coupling directly provides information on the γ-torsion angle (2,18). Also, as pointed out above, either H5′ or H5′′ is needed to unambiguously determine the β-torsion angle, e.g. together the JC4′P- and JH5′P-couplings unambiguously determine the β-torsion angle (2). In addition, the H5′–H2′ intra-sugar NOE contacts can provide information on sugar pucker, while intra-residue H5′–H6/8 NOEs give information on the γ-torsion angle, and most importantly H5′–H2′ sequential sugar NOE contacts strongly constrain the backbone (2). Note that in each of these examples it is assumed that the H5′ and H5′′ resonances could be stereo-specifically assigned.
To resolve this drawback, we have devised a synthetic method, which stereo-specifically reintroduces the H5′. This new approach is still based on the in vitro synthesis of NTPs via the pentose-phosphate and nucleotide biosynthesis and salvage pathways pathway, but employs a stereo-specifically labelled glucose as the starting product. This makes it possible to stereo-specifically deuterate the C5′ on the H5′′ position with a very high degree of selectivity. Here, we describe and demonstrate this isotope enrichment method. To illustrate the method, a 31 nt RNA molecule corresponding to the primer binding site (PBS; Fig. 1) of the pre-genomic RNA from hepatitis B virus (HBV) was synthesised with 13C and deuterium labels in the ribose ring at specific positions. Guanosine triphosphate (GTP) was synthesised with 13C labels in the ribose ring, deuterium labels at the 1′, 3′, 4′ and 5′′ positions and protons at the 2′ and 5′ positions, and incorporated into the PBS; cytidine triphosphate (CTP) and uridine triphosphate (UTP) were prepared with deuterium labels on the 1′, 3′, 4′, 5′ and 5′′ positions and a proton at the 2′ position in the sugar moiety, and also incorporated into the PBS.
Figure 1.
Secondary structure of the PBS of hepatitis B virus, synthesised to illustrate the stereo-specific labelling method. In one sample the G residues have 13C labels in the ribose ring, deuterium labels at 1′, 3′ and 4′ positions and stereo-specifically at the 5′′ position, the C and U residues have deuterium labels at the 1′, 3′, 4′, 5′ and 5′′ positions, and the A residues are fully protonated and not enriched in 13C or 15N. The stereo-specifically 5′′-deuterated G residues are underlined. In a second sample all residues were uniformly 13C/15N-labelled.
MATERIALS AND METHODS
Chemicals were purchased from Sigma (Tyreso, Sweden). [1,2,3,4,5,6,6-2H7-1,2,3,4,5,6-13C6]-d-glucose and [1,2,3,4,5,6,6-2H7]-d-glucose were purchased from Martek Corporation (Columbia, MD). The sodium salt of 3-phosphoglycerate was prepared from the barium salt of 3-phosphoglycerate by exchanging the barium for sodium with AGW50-X8 strong cation-exchanger, hydrogen form (200–400 mesh) obtained from BioRad (Sundbyberg, Sweden) (20). All enzymes were purchased from Sigma Chemical except for T7 polymerase (21), phosphoribosylpyro-phosphate synthetase (PRPP synthetase) (22,23), adenine phosphoribosyltransferase (24), CTP synthetase (25,26), xantine–guanine phosphoribosyltransferase (27,28) and uracil phosphoribosyltransferase (29,30), which were purified from over-expressing strains using a general purification scheme as described previously (6,15).
Synthesis of (6S)-1H-1,2,3,4,5,6-2H-1,2,3,4,5,6-13C-d-glucose by stereo-specific hydrogen exchange (9) (Fig. 2)
In view of the cost of 1,2,3,4,5,6,6-2H-1,2,3,4,5,6-13C-d-glucose (1), the synthetic sequence was first carried out with unlabelled d-glucose. This provided reference material for the synthesis with labelled material. Due to the extensive 2H and 13C labelling, routine 1H and 13C NMR experiments could not be used to fully characterise the products obtained from 1. Therefore, reactions with labelled material were monitored by TLC using unlabelled reference material, and labelled products were characterised by high resolution mass spectrometry.
1,2,3,4,5,6,6-2H-1,2,3,4,5,6-13C-1,2,3,4,6-penta-O-acetyl-d-glucopyranose (2)
A suspension of labelled d-glucopyranose (1, 4.00 g, 20.7 mmol) in a mixture of acetic anhydride (32 ml) and pyridine (36 ml) was stirred for 3 h. The solution was then concentrated and the residue was co-evaporated twice with toluene. The residue was dissolved in CHCl3 and washed three times with H2SO4 (3%, aq.), water, NaHCO3 (aq., sat.) and brine, and then dried (Na2SO4) and concentrated. The oily residue was crystallised from EtOH (95%) in two crops and the mother liquor was concentrated and purified by flash column chromatography (SiO2, heptane:ethyl acetate 1:1) to give 2 (8.04 g, 19.9 mmol, 96%). Rf 0.36 (SiO2, heptane:ethyl acetate 1:1). Fast atom bombardment mass spectroscopy (FAB MS): calculated 426.1693 [M + Na]+, found 426.1690.
Pentachlorophenyl 1,2,3,4,5,6,6-2H-1,2,3,4,5,6-13C-2,3,4,6-tetra-O-acetyl-β-d-glucopyranoside (4)
HBr (33%) in acetic acid (30 ml) was added to a suspension of 2 (8.00 g, 19.8 mmol) in a mixture of acetic acid and acetic anhydride (2:1, 18 ml) at 0°C during 20 min. The resulting solution was stirred at room temperature (rt) for 90 min and was then partitioned between CHCl3 and water. The organic phase was washed three times with NaHCO3 (aq., sat.) and brine, and then dried (Na2SO4) and concentrated, yielding crude 3. Sodium (0.50 g, 22 mmol) and pentachlorophenol (5.82 g, 21.9 mmol) were added to EtOH (100 ml). After stirring for 1 h the solution was concentrated and the residue was dissolved in acetone (40 ml). Crude bromide 3 was added to this solution and heated at reflux for 5 h. After cooling to ambient temperature and filtration through celite, the solvent was removed and the residue was dissolved in CHCl3. The resulting solution was washed with water, dried (Na2SO4) and concentrated. Recrystallisation from EtOH (95%) gave 4 (7.01 g, 11.5 mmol, 58%). Rf 0.51 (SiO2, heptane:ethyl acetate 4:1). FAB MS: calculated 629.9959 [M + Na]+, found 629.9963.
1,2,3,4,5,6,6-2H-1,2,3,4,5,6-13C-1,6-anhydro-2,3,4-tri-O-benzoyl-d-glucopyranose (5)
A mixture of 4 (7.00 g, 11.5 mmol) and Amberlite IRA 400(OH) (20 ml) was stirred at 55°C for 2 h, then filtered and concentrated. The residue was dissolved in pyridine (145 ml) and benzoyl chloride (10.0 g, 71.5 mmol) was added, after which the solution was heated at 65–70°C for 7 h. After concentration, the oily crude product was dissolved in CHCl3 and washed three times with H2SO4 (3%, aq.), water, NaHCO3 (aq., sat.) and brine, and then dried (Na2SO4) and concentrated. Flash column chromatography (SiO2, heptane: ethyl acetate 4:1 → 3:1) gave 5 (2.23 g, 4.63 mmol, 40%) and perbenzoylated methyl d-glucoside (4.18 g, 6.70 mmol, 58%). Compound 5 had Rf 0.15 (SiO2, heptane:ethyl acetate 4:1). FAB MS: calculated 510.1846 [M + Na]+, found 510.1853.
(6S)-1,2,3,4,5,6-2H-1,2,3,4,5,6-13C-1,6-anhydro-2,3,4-tri-O-benzoyl-6-bromo-d-glucopyranose (6)
Compound 5 (2.10 g, 4.31 mmol) was dissolved in hot CCl4 (50 ml), and Br2 (2.70 g, 16.9 mmol) was added. After refluxing under high-intensity UV light for 10 h the mixture was diluted with CHCl3 (20 ml), washed with sodium bisulfite (aq., 10%), NaHCO3 (aq., sat.) and brine, then dried (Na2SO4) and concentrated. Flash column chromatography (SiO2, heptane:ethyl acetate 7:3) of the residue gave 6 (1.10 g) and unreacted 5 (915 mg). The procedure was repeated with recovered 5 to give additional 6 (in total of 1.91 g, 3.38 mmol, 78%). Rf 0.29 (SiO2, heptane:ethyl acetate 4:1). FAB MS: calculated 587.0889 [M + Na]+, found 587.0895.
(6R)-1,2,3,4,5,6-2H-1,2,3,4,5,6-13C-1,6-anhydro-2,3,4-tri-O-benzoyl-d-glucopyranose (7)
Compound 6 (1.91 g, 3.38 mmol) was dissolved in toluene (60 ml) after which AIBN (5 mg) and Bu3SnH (1.47 g, 5.07 mmol) were added. After heating at reflux for 40 min, the solvent was concentrated and the residue was purified by flash column chromatography (SiO2, heptane: ethyl acetate 7:3 → CHCl3:MeOH 10:1), yielding 7 (1.45 g, 2.98 mmol, 88%). Rf 0.15 (SiO2, heptane:ethyl acetate 4:1). FAB MS: calculated 509.1784 [M + Na]+, found 509.1783.
(6R)-1,2,3,4,5,6-2H-1,2,3,4,5,6-13C-1,2,3,4,6-penta-O-acetyl-d-glucopyranose (8)
A suspension of 7 (1.43 g, 2.94 mmol) in MeOH (35 ml) and methanolic NaOMe (2 M, 0.1 ml) was heated at reflux for 1 h. The solution was then cooled and neutralised with Amberlite IR120(H). The solvent was evaporated and the residue was dissolved in water, washed with toluene, treated with charcoal (1.2 g) and filtered through celite. The solvents were evaporated again and the residue was dissolved in a mixture of acetic anhydride (12 ml) and H2SO4 (0.2 ml) pre-cooled to 0°C. Stirring was continued for 3 h at this temperature and then H2O (70 ml) was added. The solution was extracted twice with CHCl3 and the combined organic phases were washed three times with NaHCO3 (aq., sat.) and then with brine. Concentration gave 8 (711 mg, 1.77 mmol, 60%), which was used directly in the next step. Compound 8 had Rf 0.29 (SiO2, heptane:ethyl acetate 1:1). FAB MS: calculated 425.1632 [M + Na]+, found 425.1638.
6(S)-1H-(6R)-1,2,3,4,5,6-2H-1,2,3,4,5,6-13C-d-glucose (9)
Compound 8 (691 mg, 1.72 mmol) was suspended in MeOH (40 ml) and methanolic NaOMe (2 M, 0.1 ml) was added. After stirring for 1 h, the solution was neutralised with Amberlite IR120(H), filtered and concentrated. The oily residue was crystallised from EtOH (95%) to give 9 (314 mg, 1.63 mmol, 95%). Rf 0.13 (SiO2, CH2Cl2:methanol 4:1). FAB MS: calculated 215.1103 [M + Na]+, found 215.1113.
Nucleotide synthesis
Enzymatic reactions were modified from Tolbert and co-workers (16). A brief, though complete, description is given below. All reactions were monitored by HPLC: Vydac 302IC4.6 column, detection at 260 nm. Mobile phase: A = NaH2PO4/Na2HPO4 (1:1 molar ratio) 25 mM in water, adjusted to pH 2.8 with acetic acid; B = NaH2PO4/Na2HPO4 (1:1 molar ratio) 125 mM in water, adjusted to pH 2.9 with acetic acid. Gradient: 0% B for 2 min, then linear form 0–100% B in 17 min, hold 100% for 2 min and return to 0% B in 0.1 min. Potassium phosphate buffer (50 mM) was used in reactions containing PRPP synthetase because the enzyme is inactivated in solutions with low phosphate concentration. The pH of the reactions was monitored periodically to maintain the pH between 7.0 and 7.9. Carbencillin (100 µg/ml) was added to prevent bacterial growth in the reactions. NTPs from the enzymatic reactions were purified by using boranate affinity chromatography on affigel 601 (BioRad) to remove the majority of salts and proteins in order to prepare the nucleotides for transcription (12,20).
Preparation of [1′,3′,4′,5′,-2H4-1′,2′,3′,4′,5′-13C5]GTP (d4-GTP)
Sodium 3-phosphoglycerate (4.5 mmol), guanine (0.75 mmol, 120 mg), α-ketoglutaric acid (3.0 mmol, 435 mg) and NH4Cl (4 mmol, 286 mg) were placed into a round bottom flask. This was dissolved in 60 ml of 50 mM potassium phosphate buffer pH 7.5, with 10 mM MgCl2 and 20 mM dithiothreitol (DTT). The pH was brought to 7.5 with 1 M NaOH. ATP (33 µmol, 18 mg), NADP+ (10 µmol, 3 mg) and [1,2,3,4,5,6,6 2H7-1,2,3,4,5,6-13C6]glucose (0.46 mmol, 90 mg) were added to the mixture. The reaction was started by adding 150 U phosphoglyceratemutase, 36 U enolase, 60 U pyruvate kinase, 36 U myokinase, 36 U l-glutamic dehydrogenase, 45 U hexokinase, 7 U glucose-6-phosphate dehydrogenase, 1.5 U 6-phosphogluconic dehydrogenase, 75 U phosphoriboisomerase, 1.5 U PRPP synthetase, 3 U XG PRT and 1.5 U guanylate kinase. After 4 days, an additional 1.5 mmol of Na-3-phosphoglycerate was added to the reaction together with 60 U pyruvate kinase, 40 U enolase and 40 U myokinase, and the reaction was stopped after 10 days. Boranate purification of the reaction and quantification with UV indicated that 65% of the glucose had been converted into isotopically labelled GTP, ɛ253 = 13 700 cm–1mol–1.
Preparation of [1′,3′,4′,5′,5′′-2H5]UTP (d5-UTP)
Typically, into a round-bottom flask sodium-3-phosphoglycerate (3 mmol), α ketoglutaric acid, (2.0 mmol, 290 mg), NH4Cl (2.8 mmol, 150 mg) and uracil (0.4 mmol, 45 mg) were placed. These were dissolved in 40 ml of a solution containing 10 mM MgCl2, 20 mM DTT and 50 mM potassium phosphate buffer, and the pH of the solution was adjusted to 7.5 with 1 M NaOH. ATP (5 µmol), NADP+ (13 µmol, 11 mg) and [1,2,3,4,5,6,62H7]glucose (0.4 mmol, 75 mg) were added to the mixture. The reaction was started by adding 150 U phosphoglycerate mutase, 50 U enolase, 75 U pyruvate kinase, 35 U myokinase, 25 U l-glutamic dehydrogenase, 50 U hexokinase, 10 U glucose-6-phosphate dehydrogenase, 1.5 U 6-phosphogluconic dehydrogenase, 100 U phosphoriboisomerase, 1 U PRPP synthetase, 3.75 U uracil phosphoribosyl transferase and 2 U nucleoside monophosphate kinase. At day 3, 5 and 7, 1.5 mmol sodium-3-phosphoglycerate, 10 µmol ATP, 50 U phosphoglycerate mutase, 25 U enolase, 25 U myokinase and 25 U pyruvate kinase were added, and after 11 days the reaction was frozen to stop it and purified by boranate chromatography. The yield was 85%, ɛ260 = 10 000 cm–1mol–1.
Preparation of [1′,3′,4′, 5′,5′′-2H5]CTP (d5-CTP) from d5-UTP
d5-UTP (0.1 mmol), 10 mmol NH4Cl and 0.50 mmol sodium-3-phosphoglycerate in 200 ml of a solution containing 5 mM MgCl2 and 1 mM DTT pH 7.5 were placed into a 500 ml three neck flask. To start the reaction 100 U phosphoglycerate mutase, 50 U enolase, 50 U pyruvate kinase, 50 U myokinase, 3 U CTP synthetase and 50 µmol ATP were added. The reaction was monitored with HPLC. The reaction was stopped after 48 h and purified by boronate chromatography. The yield of the reaction was 90% as determined by UV absorbance. CTP was quantified by absorbance at 259 and 280 nm: ATP ɛ259 = 15 400, ɛ280 = 1911, CTP ɛ259 = 7204, ɛ280 = 6905.
In vitro transcription
PBS RNA (5′-GGAGGUCUUGCUUCGGCACUUUUUCACCUCC-3′) was synthesised by in vitro transcription (28–30) with T7 RNA polymerase (16) using unlabelled NTPs from Pharmacia (Stockholm, Sweden), uniform 13C/15N-labelled NTPs from Silantes (Munchen, Germany) and the different isotopically labelled NTPs produced in this paper. Transcription conditions in general were 40 mM Tris–HCl pH 8.0, 5 mM DTT, 1 mM spermidine, 400 nM DNA template, 5 mM GMP, 0.01% Triton X-100, 80 mg/ml PEG 8000, 0.1 mg/ml T7 RNA polymerase. Uniform 13C/15N-labelled transcription had 1 mM 13C/15N-NTPs each and 30 mM MgCl2. Deuterated transcription had 2 mM 2H-NTPs and 25 mM MgCl2. The RNA was purified by 20% polyacrylamide gel electrophoresis, 8 M urea, electro-eluted, ethanol precipitated, desalted and dissolved in 100 mM NaCl, 0.1 mM EDTA, pH 6.2. The final concentration of the uniform 13C/15N-labelled sample was 0.3 mM and of the deuterated sample was 0.03 mM.
NMR experiments
NMR data were collected on a Bruker DRX 600 spectrometer with a Bruker z-gradient TXI cryo probe at a temperature of 296 K. Two-dimensional hetero-nuclear multiple-quantum (2D HMQC) spectra were acquired with a spectral width of 9057 Hz in the proton dimension and 3396 Hz in the carbon dimension. The spectra were recorded with 512 complex data points in the t2 dimension (proton) and 256 points in the t1 dimension (carbon). The spectra of the uniformly labelled sample were recorded in 90% H2O/10% D2O with 16 scans per t1 increment, and of the deuterated sample in 100% D2O with 192 scans per t1 increment.
RESULTS AND DISCUSSION
Preparation of specifically deuterated NTPs
The synthesis of the specifically deuterated NTPs is similar to previously published procedures (16,17). Glucose is converted to ribose-5-phosphate and subsequently attached to a base in a single coupled enzymatic reaction.
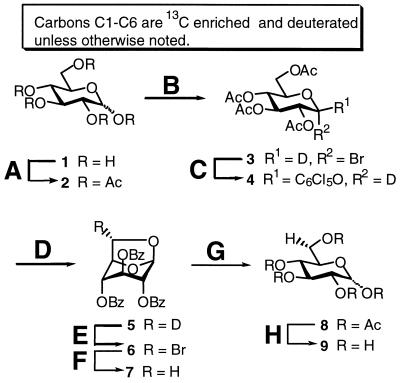
Specific labelling at the C5′ position was achieved by stereo-specific hydrogen exchange of the D6(S) to a proton in perdeuterated glucose. The 6(S) proton of glucose becomes the H5′ of ribose during the enzymatic NTP synthesis. Exchange of D6(S) in perdeuterated uniformly 13C-labelled glucose (d7-13C6-glucose) to a proton yields 6(S)-1H-1,2,3,4,5,6(R)-2H-13C6-d-glucose (d6-13C6-glucose; 9) (31). Synthesis of 9 was performed essentially as described for (6R)-6-2H-d-glucose (Fig. 2) (31). In brief, anomeric bromide 3, which was obtained from per-acetylated glucose 2, was substituted with sodium pentachlorophenoxide to give 4. Treatment of 4 with methanolic sodium methoxide afforded 1,6-anhydro glucose 5, but was also accompanied by formation of substantial amounts (32%) of methyl β-d-glucoside. A different procedure was therefore investigated in an attempt to avoid this problem (32). When unlabelled glucoside 4 was treated with Amberlite IRA400(OH) in methanol, the desired 1,6-anhydroglucose was formed as the only product in good yield (82%). Surprisingly, when this was repeated using labelled glucose, the yield dropped to 40%, and methyl β-d-glucoside was formed as main product (58%). The methyl glucoside could, however, be converted to 2 by refluxing in HBr in acetic acid. Photobromination of 5 gave 6, which was reduced to 7 with tri-n-butyltinhydride. Removal of the benzoyl groups in methanolic sodium methoxide, followed by acid catalysed opening of the 1,6-anhydro-pyranoside, gave per-acetylated glucose 8. Finally, deacetylation and crystallisation afforded 9 (d6-13C6-glucose) in 9% overall yield from 1.
Figure 2.
Reagents and conditions (yields): (A) Ac2O, pyridine, rt (96%); (B) 33% HBr in AcOH, AcOH/Ac2O, 0°C, rt; (C) C6Cl5ONa, acetone, reflux (58% from 2); (D) Amberlite IRA400 (OH), MeOH, 55°C, then BzCl, pyridine, 65°C (40%); (E) Br2, CCl4, hυ, reflux (78%); (F) Bu3SnH, AIBN, toluene, reflux (88%); (G) NaOMe, MeOH, reflux, then Ac2O, H2SO4, 0°C (60%); (H) NaOMe, MeOH, rt (95%).
Subsequent application of d6-13C6-glucose in the GTP forming reaction yielded a GTP with 13C labels in the whole ribose and a deuterium at the 5′′ position and a proton at the 5′ position. The enzymatic reactions were carried out in H2O to also obtain a proton at the C2′ position. Indeed, a GTP was produced with protons in the H2′ and H5′ positions (d4-GTP). Surprisingly, in 20% of the synthesised GTP a proton was detected at the C1′. Hydrogen exchange at the C1′ can occur when the proton on C2 of glucose-6-phosphate is exchanged. This is because the pentose-phosphate pathway oxidatively removes the C1 of glucose, making the C2 of glucose into the C1 of ribose-5-phosphate. The exchange at the C2 of glucose can be accomplished during enzymatic synthesis by glucose-6-phosphate isomerase (G6P isomerase). G6P isomerase catalyses the isomerisation of a C1 aldose (glucose-6-phosphate) into a C2 ketose (fructose-6-phophate) through an enediolate intermediate (6). However, we never deliberately added G6P isomerase to our synthesis of d4-GTP. For the moment we conjecture that the enzymes we used for the synthesis were contaminated with small amounts of G6P isomerase as we have observed this H-exchange in some of our other syntheses (33). The overall yield of the (d4-GTP) from d7-13C6-glucose is 6%. This yield might seem low; however, it is comparable with the yield obtained from isolating NTPs from E.coli grown on isotope-enriched medium (∼60 mg NTPs/g glucose). However, one cannot obtain specific labelling patterns with the latter method.
NTPs (d5-UTP and d5-CTP) with a proton on C2′ and deuterons at the rest of the positions of the sugar ring were prepared from [1,2,3,4,5,6,6 2H7]glucose (d7-glucose) in water. The atom at the C2′ position in nucleotides comes from exchange with the solvent (6,34). Half of the purified d5-UTP was converted to d5-CTP with CTP transferase. The yields were 85 and 90% for d5-UTP and d5-CTP, respectively, as determined by UV absorbance (UTP ɛ260 = 10 000 cm–1mol–1, CTP ɛ259 = 7204, ɛ280 = 6905). The enrichment level of the ribose is high with >98% deuteration at positions H1′, H3′, H4′, H5′ and H5′′. The deuteration at the H2′ is <5%, as estimated from peak volumes.
Effects of deuteration
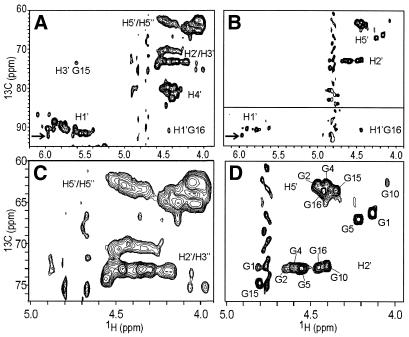
The d4-13C5-GTP, together with the in vitro synthesised d5-UTP, d5-CTP and non-labelled ATP were used in the T7-polymerase directed synthesis of the 31 nt RNA derived from the PBS of HBV genomic RNA as described in the Material and Methods. Also, a fully protonated and uniformly 13C/15N-labelled PBS was synthesised. To illustrate the effect of the labelling scheme we have recorded HMQC spectra of the fully protonated and uniformly 13C/15N-labelled PBS (Fig. 3A and C), and of the selectively deuterated PBS (Fig. 3B and D). Comparison of Figure 3A and C with B and D shows that spectral crowding is greatly reduced by the deuteration. While in the uniformly labelled PBS the H2′ and H3′ resonances overlap in the same spectral region (Fig. 3C), in the G-labelled sample the individual H2′ resonances of the seven G residues are clearly distinguishable (Fig. 3D). The H2′ resonances can even be assigned (see labels) via comparison of their chemical shifts with the known H2′ chemical shift of the G residues in the UUCG loop and its closing base pair and with the H2′ shifts of the 5′-G1-G2 sequence. The H3′ of the G residue from the UUCG loop has a rather unusual resonance position, 5.6 p.p.m., due to the syn-χ-angle of this residue (35), and is indeed present in Figure 3A. The absence of this peak in Figure 3B confirms that the deuteration scheme functioned, i.e. no 3′ protons on the G residues.
Figure 3.
Spectral simplification by deuteration demonstrated by the HMQC spectra of the 31 nt PBS RNA shown in Figure 1. (A) Uniformly 13C/15N-labelled PBS. (B) PBS with 1′,2′,3′,4′,5′-13C5-1′,3′,4′,5′′-2H4-Gs, 1′,3′,4′,5′,5′′-2H5-Us, 1′,3′,4′,5′,5′′-2H5-Cs and unlabelled As. The C1′–H1′ cross-peaks (bottom part, below horizontal line) are displayed with deeper contour levels, so that they appear with approximately the same intensities as the C2′–H2′ and C5′–H5′ cross-peaks in the upper half of the spectrum. The C1′–H1′ peak heights are ∼20% of the C2′–H2′ and C5′–H5′ peaks. (C) Expanded region of spectrum (A). (D) Expanded region of (B). The cross-peaks in (D) are labelled with their assignment (see text); some other peak labels are described in text. The arrows in (A) and (B) point to the G16 H1′–C1′ cross-peak used in Figure 4.
Most importantly, the individual H5′ can easily be distinguished in the G-labelled sample (Fig. 3D), whereas they still overlap with the H5′′ protons in the spectrum of the uniformly labelled sample (Fig. 3C). As for the H2′ they can be assigned via chemical shift comparison.
The labelling is stereo-selective to a very high degree, i.e. no H5′′ could be detected in the spectra. We estimate from the noise level in the HMQC spectrum compared to the H5′ peak heights that the stereo-selectivity is better than 98%. This is important, because in this way stereo-specific assignment of H5′ versus H5′′ does not have to be based on intensity comparisons. More importantly, stereo-specific assignment of H5′ and H5′′ protons makes the extraction of important structural parameters possible. For example, it is required for the unambiguous determination of the torsion angles γ and β from J-couplings. It also provides many useful intra-residue, sequential and long-range NOE contacts (2).
The H1′ protons resonate in a separate spectral region and usually do not overlap with other resonances in HMQC spectra. However, as is visible from Figure 3A, with 31 resonances, quite some overlap is present between the individual H1′ resonances of the uniformly labelled PBS. The residue-specific labelling resolves the overlap (Fig. 3B). Immediately, six (out of seven) resonances can be identified, including a H1′ proton with a rather unusual chemical shift (at 97/4.2 p.p.m.), which can be attributed to G16. The peak immediately confirms the formation of the UUCG loop.
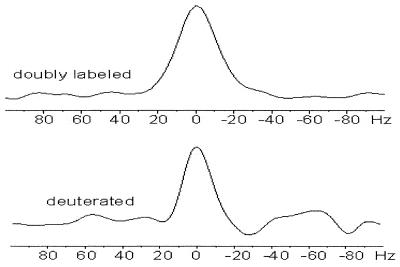
The acquired line narrowing due to the specific deuteration can be appreciated from the 1H-1D traces shown in Figure 4. Here, 1H-1D traces of a single H1′/C1′ cross-peak (6.0/92.2 p.p.m.) in the HMQC spectrum of the deuterated and of the uniformly labelled RNA (Fig. 3, arrows) are compared. The resonance from the deuterated RNA has a line width of ∼18 Hz, compared with ∼25 Hz in the uniformly labelled RNA (part of it is caused by the JH1′H2′-couplings, max. 8 Hz). The line narrowing (of ∼8 Hz) can be attributed to the removal of part of the 1H–1H dipolar interactions. For a molecule of this size the 1H–1H dipolar interactions can be roughly estimated to contribute ∼10 Hz to the line width of the sugar protons (2,3,17). Even narrower lines are expected for the U and C residues in the deuterated sample, since here the 13C-1H dipolar interaction is also removed (also ∼10 Hz). The line narrowing translates into improved resolution, which further benefits assignment. It also improves coherence transfer efficiencies in hetero-nuclear experiments, for example in HCP- and HCN-correlation experiments (2,3,17).
Figure 4.
1D traces in 1H direction through the G16 H1′–C1′ cross-peak (at 6.0/92.2 p.p.m.) in the HMQC spectrum of Figure 3, showing the acquired line narrowing upon deuteration. The G16 H1′–C1′ resonance has half height-line widths of 25 Hz (top) and 18 Hz (bottom) in the non-deuterated (indicated as doubly labelled) and deuterated samples, respectively.
Finally, we note that, thanks to the deuteration and the use of cryoprobe technology, the HMQC spectra could be recorded with good signal-to-noise overnight (∼14 h) for the deuterated RNA, which had a concentration of only 30 µM.
CONCLUSIONS
We have described an effective enzymatic synthesis for the production of NTPs, which are stereo-selectively deuterated on the H5′′ position with high selectivity. The yield is ∼6% (60 mg NTPs/g glucose), comparable with the yield from NTPs isolated from E.coli bacteria grown on enriched media. The stereo-selectivity we estimate to be better than 98% from the HMQC spectra. The method employs the NTP synthesis route based on enzymes of the pentose-phosphate, nucleotide biosynthesis and salvage pathways. In this way a host of different specific labelling patterns can be incorporated into RNAs, including stereo-selectively H5′′ deuterated residues.
We have demonstrated the versatility of this method to produce specific labelling patterns via a 31 nt RNA sample. The RNA was synthesised with fully protonated adenine residues, with cytidine and uridine residues deuterated only on the ribose in the 1′, 3′, 4′ and 5′/5′′ positions, and with guanine residues with 13C labels in the ribose ring and deuterium labels on the 1′, 3′, 4′ and 5′′ positions.
This selective, non-uniform labelling can solve many of the spectral overlap problems. The deuteration reduces spectral crowding by removal of 1H resonances, while the resulting line narrowing further improves spectral resolution. Moreover, it improves the coherence transfer efficiency (e.g. from C4′ to 31P in HCP experiments), so that isotope-edited NMR experiments can be fully exploited to further reduce resonance overlap.
The possibility of producing NTPs via the enzymatic route has important additional advantages. The H5′ can now be stereo-specific assigned, so that the γ- and β-torsion angles can unambiguously be determined, while intra- and inter-residue NOEs involving H5′ become accessible, further improving structure definition.
It is expected that structure determination of larger RNA molecules (>50 nt) is possible when a rationally designed labelling scheme is applied. This requires non-uniform and stereo-selective labelling which is now available with the enzymatic synthesis of NTPs described above.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank J. Williamson for generously providing the over-producing strains for the enzymes needed in the synthesis described. J. J. Dunn is thanked for providing the strain for producing T7 RNA polymerase. This study was supported by grants from the Structural Biology Network of the Strategic Research in Sweden (SSF), Swedish National Research Council, Bioteknik Medel, Umeå University Sweden to S.W. and from an EMBO fellowship to J.C. to visit the laboratory of C. W. Hilbers (Nijmegen, The Netherlands).
REFERENCES
- 1.Kolk M.H., van der Graaf,M., Wijmenga,S.S., Pleij,C.W.A., Heus,H.A. and Hilbers,C.W. (1998) NMR structure of a classical pseudoknot: interplay of single- and double-stranded RNA. Science, 280, 234–238. [DOI] [PubMed] [Google Scholar]
- 2.van Buuren B.N.M. and Wijmenga,S.S. (1998) The use of NMR methods for conformational studies of nucleic acids. Prog. NMR Spec., 32, 287–387. [Google Scholar]
- 3.Cromsigt J., van Buuren,B., Schleucher,J. and Wijmenga,S. (2001) Resonance assignment and structure determination for RNA. Methods Enzymol., 338, 371–399. [DOI] [PubMed] [Google Scholar]
- 4.Nikonowicz E.P., Michnicka,M., Kalurachchi,K. and DeJong,E. (1997) Preparation and characterization of a uniformly 2H/15N-labeled RNA oligonucleotide for NMR studies. Nucleic Acids Res., 25, 1390–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nikonowicz E.P. (2001) Preparation and use of 2H-labeled RNA oligonucleotides in nuclear magnetic resonance studies. Methods Enzymol., 338, 320–341. [DOI] [PubMed] [Google Scholar]
- 6.Tolbert T.J. and Williamson,J.R. (1997) Preparation of specifically deuterated and 13C-labeled RNA for NMR studies using enzymatic synthesis. J. Am. Chem. Soc., 119, 12100–12108. [Google Scholar]
- 7.Nikonowicz E.P., Kalurachchi,K. and DeJong,E. (1997) Comparison of H5 and H8 relaxation rates of a 2H/13C/15N labeled RNA oligonucleotide with selective protonation at C5 and C8. FEBS Lett., 415, 109–113. [DOI] [PubMed] [Google Scholar]
- 8.Agback P., Maltseva,T.V., Yamakage,S.I., Nilson,F.P.R., Foldesi,A. and Chattopadhyaya,J. (1994) The difference in the T-2 relaxation rates of the protons in the selectively deuterated and fully protonated sugar residues in a large oligo-DNA (‘NMR-window’) gives complementary structural information. Nucleic Acids Res., 22, 1404–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamakage S.I., Maltseva,T.V., Nilson,F.P., Foldesi,A. and Chattopadhyaya,J. (1993) Deuteration of sugar protons simplify NMR assignments and structure determination of large oligonucleotide by the 1H-NMR window approach. Nucleic Acids Res., 21, 5005–5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glemarec C., Kufel,J., Foldesi,A., Maltseva,T.V., Sandström,A., Kirsebom,L.A. and Chattopadhyaya,J. (1996) The NMR structure of 31mer RNA domain of Escherichia coli RNase P RNA using its non-uniformly deuterium labelled counterpart [the ‘NMR-window’ concept]. Nucleic Acids Res., 24, 2022–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foldesi A., Yamakage,S.I., Nilsson,F.P., Maltseva,T.V. and Chattopadhyaya,J. (1996) The use of non-uniform deuterium labelling [‘NMR-window’] to study the NMR structure of a 21mer RNA hairpin. Nucleic Acids Res., 24, 1187–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nikonowicz E.P., Sirr,A., Legault,P., Jucker,F.M., Baer,L.M. and Pardi,A. (1992) Preparation of 13C and 15N labelled RNAs for heteronuclear multi-dimensional NMR studies. Nucleic Acids Res., 20, 4507–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milligan J.F., Groebe,D.R., Witherell,G.W. and Uhlenbeck,O.C. (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res., 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milligan J.F. and Uhlenbeck,O.C. (1989) Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol., 180, 51–62. [DOI] [PubMed] [Google Scholar]
- 15.Tolbert T.J. and Williamson,J.R. (1996) Preparation of specifically deuterated RNA for NMR studies using a combination of chemical and enzymatics synthesis. J. Am. Chem. Soc., 118, 7929–7940. [Google Scholar]
- 16.Scott L.G., Tolbert,T.J. and Williamson,J.R. (2000) Preparation of specifically 2H- and 13C-labeled ribonucleotides. Methods Enzymol., 317, 18–38. [DOI] [PubMed] [Google Scholar]
- 17.Cromsigt J.A.M.T.C., Schleucher,J., Kidd-Ljunggren,K. and Wijmenga,S.S. (2000) Synthesis of specifically deuterated nucleotides for NMR studies on RNA. J. Biomol. Struct. Dyn., c11, 211–220. [DOI] [PubMed] [Google Scholar]
- 18.Ippel J.H., Wijmenga,S.S., de Jong,R., Heus,H.A., Hilbers,C.W., de Vroom,E., van der Marel,G.A. and van Boom,J.H. (1996) Heteronuclear scalar couplings in the bases and sugar rings of nucleic acids: their determination and application in assignment and conformational analysis. Magn. Reson. Chem., 34, S156–S176. [Google Scholar]
- 19.Hines J.V., Landry,S.M., Varani,G. and Tinoco,I.,Jr (1994) Carbon-proton scalar couplings in RNA: 3D heteronuclear and 2D isotope edited NMR of a 13C labelled extra stable hairpin. J. Am. Chem. Soc., 116, 5823–5831. [Google Scholar]
- 20.Batey R.T., Battiste,J.L. and Williamson,J.R. (1995) Preparation of isotopically enriched RNAs for heteronuclear NMR. Methods Enzymol., 261, 300–322. [DOI] [PubMed] [Google Scholar]
- 21.Davanloo P., Rosenberg,A., Dunn,J. and Studier,F.W. (1984) Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc. Natl Acad. Sci. USA, 81, 2035–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hove-Jensen B. (1985) Cloning and characterization of the prs gene encoding phosphoribosylpyrophosphatase synthetase of Escherichia coli. Mol. Gen. Genet., 201, 269–276. [DOI] [PubMed] [Google Scholar]
- 23.Switzer R.L. and Gibson,K.J. (1978) Phosphoribosylpyrophosphate synthetase (ribose-5-phosphate pyrophosphokinase) from Salmonella typhimurium. Methods Enzymol., 51, 3–11. [DOI] [PubMed] [Google Scholar]
- 24.Arnold W.J. and Kelley,W.N. (1978) Adenine phosphoryltransferase. Methods Enzymol., 51, 568–574. [DOI] [PubMed] [Google Scholar]
- 25.Anderson P.M. (1983) CTP synthetase from Eschericia coli: an improved purification procedure and characterization of hysteretic and enzyme concentration effects on kinetic properties. Biochemistry, 22, 3285–3292. [DOI] [PubMed] [Google Scholar]
- 26.Long C. and Koshland,D.E.,Jr (1978) Cytidine triphosphate synthetase. Methods Enzymol., 51, 79–83. [DOI] [PubMed] [Google Scholar]
- 27.Pratt D. and Subramani,S. (1983) Nucleotide sequence of the Eschericia coli xanthine-guanine phosphoribosyl transferase gene. Nucleic Acids Res., 11, 8817–8823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu S.W. and Milman,G. (1983) Purification and characterizaton of Eschericia coli guanine-xanthine phosphoribosyltransferase produced by a high efficiency expression plasmid utilizing a lambda PL promotor and CI857 temperature-sensitive respressor. J. Biol. Chem., 258, 7469–7475. [PubMed] [Google Scholar]
- 29.Rasmussen U.B., Mygind,B. and Nygaard,P. (1986) Purification and some properties of uracil phosphoribiosyl transferase from Escherichia coli K12. Biochim. Biophys. Acta, 881, 268–275. [DOI] [PubMed] [Google Scholar]
- 30.Andersen P.S., Smith,J.M. and Mygind,B. (1992) Characterization of the upp gene coding uracil phosphoribosyltransferase of Escherichia coli K12. Eur. J. Biochem., 204, 51–56. [DOI] [PubMed] [Google Scholar]
- 31.Ohrui H., Horiki,H., Kishi,H. and Meguro,H. (1983) Synthesis of (6R) and (6S)-d-glucose-6-2H through stereospecific photo-bromination of 1,6-anhydro-β-d-glucopyranose derivative. Agric. Biol. Chem., 47, 1101–1106. [Google Scholar]
- 32.Boons G.J., Isles,S. and Setälä,P. (1995) An improved procedure for the preparation of 1,6-anhydro sugars. Syn. Lett., 7, 755–756. [Google Scholar]
- 33.Flodell S., Cromsigt,J., Schleucher,J., Kidd-Ljunggren,K. and Wijmenga,S. (2002) Structure elucidation of the hepatitis B virus encapsidation signal by NMR on selectively labeled RNAs. J. Biomol. Struct. Dyn., 19, 1–10. [DOI] [PubMed] [Google Scholar]
- 34.Lienhard G.E. and Rose,I.A. (1964) The mechanism of action of 6-phosphogluconate dehydrogenase. Biochemistry, 3, 190–193. [DOI] [PubMed] [Google Scholar]
- 35.Cromsigt J.A., Hilbers,C.W. and Wijmenga,S.S. (2001) Prediction of proton chemical shifts in RNA. Their use in structure refinement and validation. J. Biomol. NMR, 21, 11–29. [DOI] [PubMed] [Google Scholar]