Abstract
We have developed a dual reporter screen in Escherichia coli for identifying variants of the Flp site-specific recombinase that have acquired reactivity at an altered target site (mFRT). In one reporter, the lacZα gene segment is flanked by mFRTs in direct orientation. In the other, the red fluorescence protein (RFP) gene is flanked by the native FRTs. Hence, the color of a colony on an X-gal indicator plate indicates the recombination potential of the variant Flp protein expressed in it: blue if no recombination or only FRT recombination occurs, red if only mFRT recombination occurs and white if both FRT and mFRT recombinations occur. The scheme was validated by identification and in vivo characterization of Flp variants that show either relaxed specificity (active on FRT and mFRT) or moderately shifted specificity toward mFRT. We find that alteration of Lys-82 to Met, Thr, Arg or His enables the corresponding Flp variants to recombine FRT sites as well as altered FRT sites containing a substitution of G-C by C-G at position 1 of the Flp binding element (mFRT11). In contrast, wild-type Flp has no detectable activity on mFRT11. When Lys-82 is replaced by Tyr, the resulting Flp variant shows a small but reproducible preference for mFRT11 over FRT. However, this preference for mFRT11 is nearly lost when Tyr-82 is substituted by Phe.
INTRODUCTION
Site-specific recombinase Flp from Saccharomyces cerevisiae is a member of the integrase/tyrosine family (1). When bound to a pair of its target DNA sites (Flp recombination targets or FRTs), a Flp tetramer catalyzes two temporally separated strand exchange steps to mediate a recombination event. Each FRT site consists of two 13 bp Flp binding elements arranged in inverted orientation and separated by an 8 bp spacer (or strand exchange segment). Depending on the relative location and orientation of the recombination targets, the recombination outcome can be integration, excision, inversion or translocation of DNA. Recombination reactions mediated by Flp or by the related phage P1 recombinase Cre have been applied in a variety of organisms including prokaryotes, yeasts, plants, flies and mammals (2–4). One limitation to the use of Flp or Cre in genomic engineering stems from the strict target specificity of these proteins. In order to carry out a directed recombination reaction, the target site must be engineered into the genomic locale of interest. The power of site-specific recombination as a genetic engineering tool could be greatly expanded if one could pre-select a genomic site that closely resembles the normal recombination site, and coax the recombinase to acquire this new target specificity. Furthermore, availability of several recombinase variants with non-overlapping specificities would permit multiple gene manipulations to be carried out within a single cell without the impediment of undesirable DNA rearrangements resulting from cross reactivity. These considerations have provided the motivation for our efforts to investigate and alter DNA recognition by Flp.
Strategies for directed evolution aided by functional selection provide a powerful means for engineering nucleic acid and protein enzymes with desired properties (5,6). In applying these principles to generate altered DNA-binding specificities in Flp, we preferred to use a scheme that can identify events that not only result in the gain of ‘new’ DNA recognition but also in the simultaneous loss of ‘old’ DNA recognition. Otherwise the outcome is likely to be a pool of recombinase variants in which the vast majority have a ‘relaxed-specificity’ phenotype, and only a small minority have acquired a true shift in specificity. Although phage/surface display systems and challenge phage assays provide excellent tools for selection of novel DNA-binding properties in proteins (7,8), they do not readily offer a simultaneous ‘selection for and selection against’ strategy. Nor do they directly report on whether a selected protein is biochemically functional or not (9). Consequently, we developed a convenient assay for identification of recombinase variants that recognize and recombine new target sites in preference to the wild-type site.
We describe here a colony color based dual reporter screening system in Escherichia coli that may be employed in conjunction with mutagenesis or mutagenesis combined with gene shuffling to obtain novel target specificities in site-specific recombination. We have applied this experimental design to the Flp recombinase using an altered FRT site (mFRT11) that harbors a G-C to C-G substitution at position 1 of the Flp binding element. We have identified Flp variants that show either relaxed specificity towards FRT as well as mFRT11 or show shifted target specificity towards mFRT11. The amino acid substitutions in the variant Flp proteins map to position 82, which is a lysine in wild-type Flp. The crystal structure of a Flp–Holliday junction complex shows Lys-82 to be in contact with the G-C base pair at the first position of FRT (10) that has been mutated in mFRT11.
MATERIALS AND METHODS
E.coli strains
In all experiments E.coli strain DH10B (Invitrogen) was used [F–, mcrA, (mrr-hsdRMS-mcrBC), ø80dlacZM15, lacX74, deoR, recA1, endA1, ara139, D(ara, leu)7697, galU, galK, λ–, rpsL, nupG].
Oligonucleotides
Oligodeoxyribonucleotides were purchased from Integrated DNA Technologies, Inc. (Coralville, IA). The forward primers used for PCR amplifying lacZα or RFP reporters with flanking FRT, mFRT11 and FRTw2 (FRT with an altered spacer sequence), respectively, were: d(CCTTCCGCATGCGAAGTTCCTATACTTTCTAGAGAATAGGAACTTCCGTTGGC-CGATTCATTAATGCAGCTGGCACGACAGG), d(CCTTCCGCATGCGAAGTTCCTATAGTTTCTAGACTATAGGAACTTCCGTTGGCCGATTCATTAATGCAGCTGGCACGACAGG) and d(CCTTCCGCATGCGAAGTTCCTATACTATCTACAGAATAGGAACTTCCGTTGGCCGATTCATTAATGCAGCTGGCACGACAGG). The corresponding reverse primers were: d(CCTTCCAAGCTTGAAGTTCCTATTCTCTAGAAAGTATAGGAACTTCGGTGTTGGCGGGTGTCGGGGCTGGCTTAACTATGCG), d(CCTTCCAAGCTTGAAGTTCCTATAGTCTAGAAACTATAGGAACTTCGGTGTTGGCGGGTGTCGGGGCTGGCTTAACTATGCG), and d(CC- TTCCAAGCTTGAAGTTCCTATTCTGTAGATAGTATAGGAACTTCGGTGTTGGCGGGTGTCGGGGCTGGCTTAACTATGCG). Forward primers anneal ∼120 bp upstream of Plac in pUC18; reverse primers anneal ∼60 bp downstream of the NdeI site in pUC18.
The forward and reverse primers used for randomizing the codon 82 in Flp were: d(CGATATTGTCAACAAATCACTCCAGTTTAAATACNNNACGCAAAAAGCAACAATTCTGGAAGCC), and d(GGCTTCCAGAATTGTTGCTTTTTGCGTNNNGTATTTAAACTGGAGTGATTTGTTGACAATATCG).
Construction of reporters
Two reporters for screening recombination by blue/white colony color, pBU1 and pBU11, respectively, were constructed by cloning lacZα flanked by either FRT or mFRT11 in direct orientation into the vector pBAD24 (Ampr, ColE1 origin) (11). In the first step of construction, pUC18 was digested with SacI and HindIII, treated with Klenow polymerase and circularized by self ligation. This step eliminated all restriction sites in the multiple cloning sites except for EcoRI. The lacZα gene from the resulting plasmid was PCR-amplified using oligonucleotides with flanking FRT or mFRT11 and then cloned into pBAD24 digested with SphI and HindIII to obtain pBU1 or pBU11, respectively.
The two reporters for red/white screening were constructed by cloning the red fluorescence protein (RFP) gene flanked by either FRTw2 or mFRT11 in direct orientation into the vector pBAD33 (Cmr, p15a origin) (11), and were named p33Rdw2 and p33Rdm11, respectively. First, the coding region of the RFP gene was PCR amplified from plasmid pDsRed1-N1 (Stratagene, La Jolla, CA) and cloned into pUC18 digested with EcoRI and NdeI. Then the RFP region was PCR amplified using primers harboring FRTw2 or mFRT11 and cloned into pBAD33 digested with SphI and HindIII to obtain p33Rdw2 or p33Rdm11, respectively.
The expression vector for Flp
In the Flp expression plasmid p4B-Flp, the FLP gene was placed under the control of the arabinose-inducible PBAD promoter (11). Initially, a DNA fragment spanning the araC-rrnB region from pBAD33 was inserted into pBBR1MCS-2 (Kmr, R1 origin) (purchased from the Netherlands Culture Collection of Bacteria, Utrecht, Netherlands) (12) digested with NsiI and Bsu36I. The mob gene present on the resulting plasmid was inactivated by digesting the plasmid with SfiI and BspHI, treating with Klenow polymerase and re-circularizing the plasmid by self-ligation to obtain the expression vector p4B. The wild-type or variant FLP genes were placed into p4B between the SacI and HindIII sites to obtain p4B-Flp or p4B-mFlp. In these plasmids, Flp or Flp variant expression can be rapidly induced by l-arabinose. The gene can also be rapidly repressed when l-arabinose is removed from the medium (11). It should be noted that the wild-type Flp used here was Flpe, a more thermostable enzyme than native Flp (13). Flpe contains P2S, L33S, Y108N and S294P substitutions. The Flp variants described here also retain these amino acid changes. The rationale for starting with Flpe rather than Flp in attempts to evolve new target specificities was that the former is a more robust protein at 37°C, the optimum growth temperature for E.coli.
Mutagenesis of Flp
Mutagenesis of Flp was carried out using error-prone PCR (13) to yield a mutation frequency of 1–6 nt per FLP gene. The mutagenized pool of FLP genes was cloned into the expression vector described above, and assayed in vivo in E.coli for recombination activity on native or mutant FRTs.
To randomize codon 82 in the FLP gene, site-specific mutagenesis was performed using complementary PCR primers in which the nucleotides specifying codon 82 were degenerate (the primer sequences are listed above). The plasmid p4B-Flp was amplified using the mutagenic primers and Pfu polymerase. The PCR reaction and subsequent procedures were carried out according to the recommendations of Stratagene.
In vivo recombination assays
The in vivo dual reporter recombination assays were carried out as follows. Competent cells harboring pBU11 and p33Rdw2 were transformed with p4B-mFlp (either as individual variants or a mutagenised pool). LB medium [10 g/l NaCl (Sigma), 10 g/l tryptone peptone (Difco) and 5 g/l yeast extract (Difco)] was added to the cells and Flp was expressed by the addition of l-arabinose to a final concentration of 0.1% for 2.5 h at 37°C. Then cells were plated on LB-plates (LB plus bacto agar; Difco) supplemented with 100 mg/l ampicillin, 30 mg/l chloramphenicol, 25 mg/l kanamycin and 100–200 mg/l X-gal. Plates were incubated at 37°C for 36–48 h, by which time the red color of the colonies was readily visible under UV light.
When retesting candidate Flp variants of interest for target preference, the host strain contained only the lacZα reporter: either pBU1 or pBU11. The protocols of transient expression of Flp or Flp variants were the same as described above, except that transformed cells were plated on LB plates supplemented with ampicillin, kanamycin and X-gal. Colonies were scored for their color (blue or white) after 24 h of growth of the transformants.
Separation of Flp expression plasmids from the reporter plasmids
In order to determine the sequence of a Flp variant of interest, it had to be separated from the reporter plasmids harbored by the E.coli cells in which recombination was assayed. First, a mixture of all three plasmids was isolated from the chosen colony. Then 1 µl of the plasmid DNA was diluted 1000-fold to a final concentration of ∼0.3 ng/µl. An aliquot of 1 µl of the diluted plasmid was transformed into 20 µl of competent plasmid-free DH10B (equivalent to ∼107 log-phase cells processed for induction of competence), and transformants were selected on LB plates supplemented with kanamycin (50 mg/l). They were replica plated on LB-Amp and LB-Cm plates to identify those that had lost the reporter plasmids (Amp and Cm sensitive). Following isolation, the structural integrity of the p4B-mFlp plasmids was ascertained by restriction enzyme digestion. In routine assays using this protocol, it is not necessary to do the secondary screen on LB-Amp and LB-Cm plates. In our hands, plasmid preparations from Kan-resistant colonies contained p4B-mFlp free of the other plasmids at a frequency of one-half or better.
RESULTS AND DISCUSSION
Several factors were taken into consideration in designing the experimental protocols for obtaining recombination positive variants of Flp that have acquired new DNA target recognition. They are briefly discussed below.
Choice of the Flp binding element in the altered FRT site
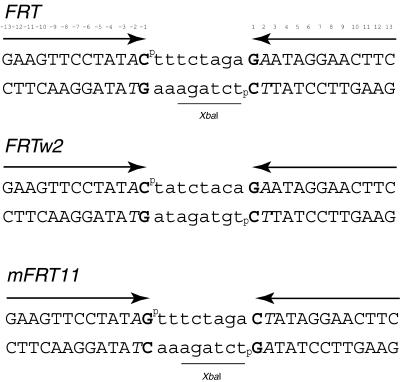
Senecoff et al. (14) carried out a detailed mutational analysis of each position of the 13 bp Flp binding element in the FRT site. To create an altered Flp target (mFRT11), we introduced a transversion mutation at position 1 of the native FRT, from G-C to C-G (Fig. 1). We also introduced a symmetrized position 2 in mFRT11 by placing a T-A base pair in both the Flp binding elements. In FRT, this position is asymmetric, being A-T in one binding element and T-A in the other. According to Senecoff et al. (14), it takes an Flp excess of >100-fold to get detectable recombination on this mutant FRT. By designing a site that was essentially a ‘non-substrate’ for wild-type Flp, we expected to virtually eliminate ‘background recombination’ and thus readily detect even those Flp variants that had acquired only modest reactivity on mFRT11. At this time we cannot rule out the possibility that the symmetrization of position 2 also contributes to the cognitive attributes of mFRT11. However, in experiments not reported here we have tested the first position C-G mutation in the context of the native FRT site (containing A-T and T-A, respectively, at the second position of the two Flp binding elements). This altered site behaved like mFRT11 in that it was not a recombination substrate for wild-type Flp but yielded recombination when tested against two of the Flp variants discussed later [Flp(K82Y) and Flp(K82M)].
Figure 1.
The Flp recombination target sites and its variants. The sequences of the two ‘wild-type’ FRT sites (FRT and FRTw2) and the mutant site mFRT11 used in this study are shown. The Flp binding elements, denoted by the head-to-head arrows, are separated by the 8 bp spacer shown in lowercase letters. The phosphates at the strand exchange points are indicated by ‘p’. Position 1 in both Flp binding elements of mFRT11 is changed from the G-C bp in FRT to the C-G bp (indicated in bold). Note that position 2 in the Flp binding elements of FRT (shown in italics) is asymmetric: an T-A base pair in the left binding element and a A-T base pair in the right one. In mFRT11, this position has been symmetrized: a T-A base pair in both binding elements. The XbaI site in the native spacer is underlined.
Use of distinct spacer sequences in the FRT and mFRT sites
Previous studies have shown that a modified FRT site, severely debilitated in recombination due to a mutation at one of the positions in the Flp binding element, can be partially rescued by a wild-type FRT site in an intermolecular recombination reaction, provided both FRTs have identical spacers (15–17). Since the recombination reaction is carried out by four Flp monomers, two monomers bound to each substrate, the rescue can be accounted for by protein cooperativity within the recombination complex. In the dual reporter assay, we wanted to avoid even rare intermolecular recombination events between native and mutant FRT sites (see below). This reaction would fuse the two reporter plasmids and create a pair of hybrid FRT–mFRT sites at the recombination junctions. Recombination events between a hybrid site and native or mutant FRTs in the fusion plasmid could randomly eliminate either the lacZα or the RFP gene, and thus give misleading results.
Whereas the sequence of the spacer in FRT per se is not critical in Flp recombination (although high G/C content can reduce recombination efficiency; 18), perfect homology between partner spacers is absolutely essential for the reaction (16,19). We could thus avoid the fusion reaction between the reporter plasmids by having two non-homologous spacers in wild-type and mutant FRTs. After comparing the efficiencies of a set of sequence-altered spacers to the native spacers (5′-TTTCTAGA-3′) in a wild-type Flp/FRT reaction in vivo, we identified 5′-TATCTACA-3′ as the optimal alternative spacer under the Flp expression conditions employed in our assay. Hence the FRT sites flanking the RFP gene were constructed with the 5′-TATCTACA-3′ spacer (referred to as FRTw2) and the mFRT11 sites flanking lacZα with the 5′-TTTCTAGA-3′ spacer (Fig. 1). The base pair changes in the FRTw2 spacer do disrupt polypyrimidine tracts that are believed to be important for optimal efficiency of recombination (18). We have verified that the outcomes from our assays are not biased in any fashion by the alternative spacer sequence. When tested against FRT or FRTw2 in separate assays, concordant results were obtained with Flp or a Flp variant of interest.
The dual reporter system for recombination assays
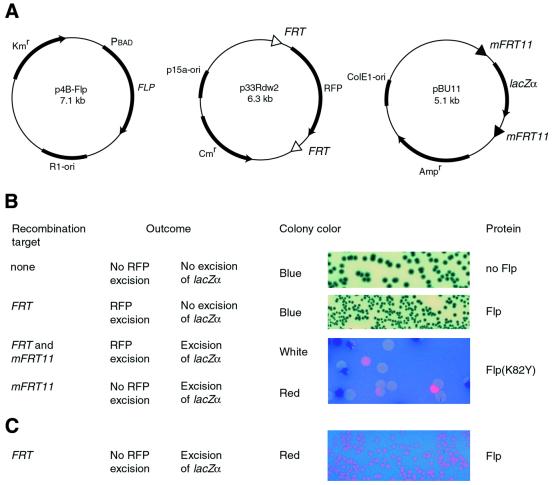
In order to challenge Flp variants with FRT and mFRT11 simultaneously, three mutually compatible plasmid constructs were employed (Fig. 2A). One was the Flp expression vector p4B-Flp (R1 replication origin, kanamycin resistance, and lowest copy number among the three) from which Flp could be rapidly and reversibly turned on from the arabinose-inducible promoter PBAD. The other two, p33Rdw2 and pBU11, harbored the FRTw2-RFP-FRTw2 and mFRT11-lacZα-mFRT11 cassettes, respectively. Plasmid pBU11 (Ampr) was based on the ColE1 replication origin and had a higher copy number than p33Rdw2 (Cmr), which harbored the p15a origin (11,12). This configuration of the reporter plasmids raises the stringency of the screen by demanding that a Flp variant with altered specificity prove itself by preferentially and quantitatively recombining the mFRT11 sites present at a higher copy number relative to the FRT sites.
Figure 2.
The dual reporter screening system. (A) The Flp expression plasmid and the recombination reporter plasmids used in the assay are schematically diagrammed at the top. The spacer of FRT was altered from its native sequence 5′-TTTCTAGA-3′ to 5′-TATCTACA-3′ to create FRTw2 (see Fig. 1) to prevent potential cross recombination between FRT and mFRT11. (B) The consequences of recombination (or lack thereof) in one or both reporters and the resultant colony color in presence of X-gal are indicated. Flp(K82Y) was obtained in this study and is described in detail in the text. Flp(K82Y) yields white colonies because it can act on mFRT11 and FRT to delete both lacZα and RFP and a rare red colony because of its slight preference for mFRT11 (complete excision of lacZα and incomplete excision of RFP). The few blue colonies indicate the incomplete deletion of lacZα. The plate was photographed under UV light and digitally processed using Adobe Photoshop software. (C) Wild-type Flp and the non-standard reporter configuration pBU1 (FRT-lacZα-FRT) and p33Rdm11 (mFRT11-RFP-mFRT11) were used in this assay. The observed phenotype, red colonies with rare blue colonies and a nearly complete absence whites colonies, is that predicted in the normal dual reporter assay (FRTw2-RFP-FRTw2; mFRT11-lacZα-mFRT11) for a Flp variant that has completely switched its target specificity from FRT to mFRT11. The photograph was taken under UV light.
In the dual reporter screen, the colony color distinguishes the following among the different recombination possibilities: recombination at either FRT or mFRT, recombination at both sites and recombination at neither site (Fig. 2B). Absence of recombination or recombination only between FRT sites would be seen as blue colony color on X-gal plates. The red color of RFP is normally masked by the blue color produced by β-galactosidase. However, lack of FRT recombination and the presence of RFP can be inferred by a red halo that surrounds a blue colony after several days. Recombination at the FRT sites only and loss of RFP would yield blue colonies that do not show the delayed appearance of the red halo. Recombination at both FRT and mFRT sites (indicating relaxed DNA specificity) would excise both RFP and lacZα reporters to give white colonies. Finally, recombination at mFRT only, due to altered target specificity of a Flp variant, would delete lacZα, and the resulting colony color would be red. Under the optimized copy number conditions set for the reporters, a Flp variant with relaxed specificity can give rise to white colonies interspersed with blue ones but can almost never yield a red colony. This is because incomplete excision of the lower copy RFP cassettes against complete excision of the higher copy lacZα cassettes is highly unlikely when the lacZα and the RFP reporters are equally efficient as recombination substrates.
Accuracy of the dual reporter assay in identifying recombination specificity
We first verified the robust properties of the dual reporter system by performing the colony color assay using wild-type Flp and a variation of the reporter plasmid configurations. The bacterial host harboring pBU1 (which has FRT sites flanking lacZα) and p33Rdm11 (which has mFRT11 sites flanking RFP gene) was transformed with p4B-Flp (Fig. 2C). Following expression of Flp, roughly 95% of the colonies were red (as expected for excision of the lacZα but not the RFP reporter), none were white (as would be the case for both lacZα and RFP reporter excision) and the rest (∼5%) were blue (Table 1). Thus, under the altered reporter design, wild-type Flp mimics a ‘perfect’ switch in specificity from mFRT11 to FRT. Conversely, in an assay with wild-type Flp and the standard reporters pBU11 (mFRT11-lacZα-mFRT11) and p33Rdw2 (FRTw2-RFP-FRTw2), the frequency of occurrence of white colonies was <0.01% (Table 1). No red halos were observed around the blue colonies even after several days of incubation. These findings are consistent with no (or a very limited) recognition of mFRT11 by Flp contrasted by its highly efficient recombination activity at the FRT sites. The combined results from the two assays demonstrate that the dual reporter system faithfully reflects the target specificity of wild-type Flp rather than any particular aspect of reporter plasmid configurations.
Table 1. Relative recombination activity of Flp and Flp variants in the in vivo dual reporter assay.
| Colony color (%) | |||
|---|---|---|---|
| Blue | White | Red | |
| Flp* | 5 ± 3* | <0.01* | 95 ± 3* |
| Flp | >99.99 | <0.01 | <0.01 |
| Flp(K82M) | 18 ± 9 | 82 ± 9 | <0.01 |
| Flp(K82H) | 27 ± 6 | 73 ± 6 | <0.01 |
| Flp(K82Y) | 21 ± 3 | 53 ± 7 | 26 ± 5 |
| Flp(K82M) + Flp(K82H) + Flp(K82Y) | 19 ± 6 | 74 ± 8 | 7 ± 5 |
Recombination was assayed by colony color following 2.5 h of Flp or Flp variant expression in cells harboring the RFP and lacZα reporter plasmids. The asterisks in row 1 denote the non-standard reporter configurations in this particular assay, FRT sites flanking lacZα and mFRT11 sites flanking RFP. In all other assays, lacZα and RFP were flanked by mFRT11 sites and FRT sites, respectively. Each experiment was repeated three times with a particular batch of LB medium. With different batches of LB, we observed differences in the absolute level of recombination. However, relative recombination efficiencies on FRT versus mFRT11 were always consistent.
Flp variants with relaxed and shifted DNA specificity
In an initial test of the optimized dual reporter system using a pool of PCR-mutagenised FLP genes cloned in the expression vector p4B, we identified potential Flp variants with relaxed DNA specificity by the appearance of rare white colonies (indicating FRT × FRT and mFRT11 × mFRT11 recombinations) against a large background of blue colonies (at a frequency of ∼10–4). DNA sequencing of a subset of these variant Flp clones revealed, in a majority, a mutation in codon 82, changing the native Lys-82 to Met, Thr or Arg. The Lys-82 codon (AAG) can yield Met (ATG), Thr (ACG) and Arg (AGG) codons by single hits within it. The probability of multiple mutations in the same codon would have been quite rare under our mutagenesis protocol.
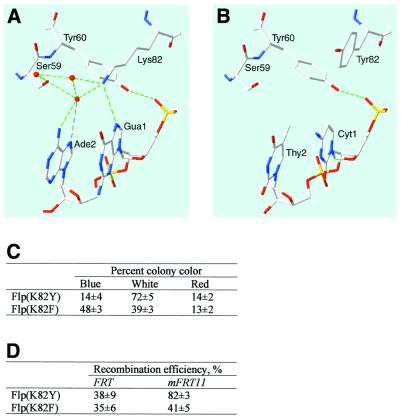
The structure of the Flp/DNA co-crystal (10) shows Lys-82 to be in direct contact with the N7 atom of guanine at position 1 of the native Flp binding element (see Fig. 5A). Since mFRT11 is mutated at this G, we subjected Lys-82 to directed mutagenesis and screened the codon-specific library using the dual reporter assay. Two additional variants, Flp(K82H) and Flp(K82Y), were identified among plasmids isolated from white colonies. Flp(K82Y) but not Flp(K82H) was also found in a few of the red colonies that arose at a much lower frequency than the whites. We tentatively assigned Flp(K82H) to the relaxed specificity group, and suspected that Flp(K82Y) has likely acquired a shift in specificity, albeit modest, towards mFRT11. We then tested Flp(K82M), Flp(K82H) and Flp(K82Y) individually in the dual reporter assay. Transient expression of Flp(K82M) led to the appearance of ∼80% white colonies (indicating FRT as well as mFRT11 recombinations), 20% blue colonies (denoting no recombination or only FRT recombination) and no red colonies (signifying the absence of preferred mFRT11 recombination over FRT recombination) (Table 1). The values for Flp(K82H) were 70% white colonies, 30% blue colonies and no red colonies (Table 1). In contrast, transient expression of Flp(K82Y) resulted in ∼55% white, 25% red and 20% blue colonies (Table 1).
Figure 5.
Comparison between Flp(K82Y) and Flp(K82F) in FRT and mFRT11 recombination. (A) The interaction between Lys-82 of wild-type Flp and the guanine at the first position of FRT is adapted from the Flp-DNA crystal structure, PDB code 1FLOA (10). Numbering of base pairs in FRT is the same as in Figure 1. The distance between the NZ-atom of Lys-82 and the N7-atom of Gua-5 is 3.21 Å. (B) Replacement of Lys-82 by Tyr, in the absence of changes in peptide conformations in its vicinity, would likely preclude direct interaction with DNA since the minimum distance between the aromatic ring of Tyr-82 and the C5 atom of cytosine in mFRT11 would be >4 Å. The program WHAT IF (20) was used to model Cyt-1 and Thy-2 in place of Gua-1 and Ade-2. The Swiss PDB Viewer 3.7 software was used to model Tyr-82 in place of Lys-82 (21). (C) The results of a dual reporter assay for Flp(K82Y) and Flp(K82F) carried out in parallel are shown. For details of the assay, see the legend to Table 1. (D) The reactivities of Flp(K82Y) and Flp(K82F) were compared by testing them in parallel against pBU1 (FRT) and pBU11 (mFRT11) as described in Table 2.
The three Lys-82 variants and Flp were also tested in separate, non-competitive assays against FRT or mFRT11 using pBU1 (FRT-lacZα-FRT) and pBU11 (mFRT11-lacZα-mFRT11) plasmids as recombination substrates. The utilization of target sites by the mutants was consistent with the expectations from the dual reporter assay (Table 2). In particular, Flp(K82Y) showed a roughly 2-fold preference for mFRT11.
Table 2. Relative recombination activity of Flp variants in non-competitive single reporter assays.
| |
Recombination efficiency (%) |
|
|---|---|---|
| FRT | mFRT11 | |
| Flp | 96 ± 2 | <0.01 |
| Flp(K82M) | 85 ± 12 | 83 ± 6 |
| Flp(K82H) | 67 ± 5 | 58 ± 7 |
| Flp(K82Y) | 53 ± 10 | 78 ± 5 |
Recombination efficiency is expressed as the ratio of white colonies to the sum of white and blue colonies after 2.5 h of Flp or Flp variant expression in the cells harboring the lacZα reporter pBU1 or pBU11. Experiments were repeated four times. Comments regarding different batches of LB (see legend to Table 1) also apply here.
The dual reporter assay correctly distinguishes between relaxed and altered specificities
As a further test for the specificity shift of Flp(K82Y), we mixed Flp(K82Y), Flp(K82M) and Flp(K82H) (cloned in the expression vector p4B) in equimolar amounts and transformed the host strain harboring the FRTw2-RFP-FRTw2 and mFRT11-lacZα-mFRT11 reporter cassettes with the plasmid mixture. The resultant colonies, following recombinase expression, showed the following color distribution: ∼20% blue, 70% white and 10% red (Table 1). We randomly picked a set of red and white colonies (10 of each color), isolated the Flp expression plasmid from each and sequenced them in their FLP coding region. Consistent with previous results, the red colonies (mFRT11 × mFRT11 recombination) contained only Flp(K82Y). On the other hand, the white colonies (FRT × FRT and mFRT11 × mFRT11 recombination) contained any one of the three Flp variants.
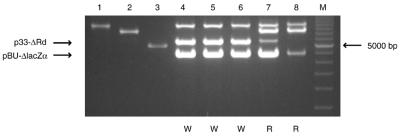
Plasmids isolated from the white and red colonies were analyzed by HindIII digestion to ascertain the parental or recombined configuration of the reporter cassettes. The analysis also included plasmids from a subset of the red colonies obtained in the positive control assay described above, in which Flp was expressed in the presence of p33Rdm11 (mFRT11-RFP-mFRT11) and pBU1 (FRTw2-lacZα-FRTw2) as reporter substrates (Fig. 2C). Results from representative samples are shown in Figure 3. The Flp expression plasmid (p4B-mFlp) was present unaltered, as judged by its size, in all the white and the red colonies analyzed (Fig. 3, compare lanes 4–8 with lane 1). As expected, the size of the RFP reporter (p33Rdm11; mFRT11) in the red colonies from the positive control assay matched the size of the parental form, indicating the absence of recombination between mFRT11 sites by wild-type Flp (Fig. 3, compare lanes 8 and 2). The RFP reporter (p33Rdw2; FRT) from the red colonies formed following Flp(K82Y) expression was present predominantly in the parental form; however, a minor fraction of a shortened version (p33-ΔRd) corresponding to RFP gene excision by FRT × FRT recombination was also detected (Fig. 3, compare lanes 7 and 8 with lane 2). This result is consistent with the fact that the discrimination by Flp against mFRT11 is much stronger than that by Flp(K82Y) against FRT. Note, though, that the lacZα reporter was shortened to pBU-ΔlacZα (consistent with the deletion of lacZα) in the red colonies produced by Flp (FRT recombination) as well as by Flp(K82Y) (mFRT11 recombination), and no parent plasmid could be detected in them (Fig. 3, compare lanes 7 and 8 with lane 3). This finding not only reflects the well-established exquisite specificity of Flp for FRT but also underscores its acquired activity on mFRT11 by virtue of the K82Y mutation. In all the white colonies, derived by the action of Flp(K82Y), Flp(K82M) or Flp(K82H), the lacZα and the RFP reporters were nearly quantitatively converted to the shortened forms pBU-ΔlacZα (mFRT11 × mFRT11 recombination) and p33-ΔRd (FRT × FRT recombination), respectively (Fig. 3, compare lanes 4–6 with 2 and 3).
Figure 3.
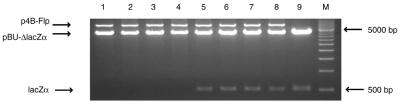
Restriction analysis of the plasmids isolated from white and red colonies obtained as a result of Flp or variant Flp expression. The control plasmids and plasmid mixtures isolated from the colonies were digested with HindIII, which cuts each plasmid only once. Lanes 1–3 provide the reference bands for the Flp expression plasmid and the parental forms of the reporter plasmids. Lanes 4–8 show the representative profiles from red or white colonies. For each recombinase/dual reporter configuration, at least 10 colonies of the indicated color were analyzed. For lanes 4–8, the expression vector harboring FLP or variant FLP and the two reporter plasmids (included in parentheses) are listed below. The pBU1 and pBU11 reporters contain the lacZα cassette and the p33Rdm11 and p33Rdw2 reporters contain the RFP cassette. The pBU1 and p33Rdw2 plasmids harbor FRT and FRTw2, respectively; pBU11 and p33Rdm11 harbor mFRT11. W and R refer to white and red colonies, respectively. Lane 1, p4B-Flp; lane 2, p33Rdw2; lane 3, pBU11; lane 4, p4B-Flp(K82M)+(pBU11, p33Rdw2); lane 5, p4B-Flp(K82H)+(pBU11, p33Rdw2); lane 6, p4B-Flp(K82Y)+(pBU11, p33Rdw2); lane 7, p4B-Flp(K82Y)+(pBU11, p33Rdw2); lane 8, p4B-Flp+(pBU1, p33Rdm11); lane M, 500 bp DNA marker ladder (Bio-Rad).
Authentication of recombination specificities of Flp variants by single reporter assays
Once the dual reporter assay reveals a desired target specificity, subsequent tests of this phenotype are carried out using the single reporter assays (Table 2). To further verify that colony color is an authentic indicator of recombination preference, we also analyzed plasmid DNA from blue and white colonies obtained after expressing Flp(K82Y), Flp(K82M) or Flp(K82H) in cells harboring reporters with lacZα flanked by FRTs in one case and by mFRT11s in the other (pBU1 and pBU11, respectively). The results of XbaI plus HindIII digestion in Figure 4 were obtained with Flp(K82M); however, the other two Flp variants gave identical results (data not shown). The XbaI sites, present only in the reporter plasmid and not on the expression plasmid, are located in the middle of the spacer sequence of the FRT or mFRT11 sites. One of the XbaI sites is neighbored by a HindIII site, their separation being only 20 bp. Flp-mediated excision of lacZα would shorten the reporter precisely by the XbaI–XbaI fragment. As shown in Figure 4, this fragment was present in the reporter plasmid from the blue colonies (Fig. 4, lanes 5–8), whereas it was absent in the shorter plasmid version from the white colonies (Fig. 3, lanes 1–4). The main backbone fragment (XbaI–HindIII) was the same in the parental and recombined forms of the reporters (Fig. 4, lanes 1–8), indicating that there were no obvious unsuspected DNA rearrangements within them. The third product from the digestion of the reporter (a 20 bp XbaI–HindIII fragment) would have run past the bottom of the gel during electrophoresis. HindIII action would linearize the expression plasmid p4B-mFlp, which showed uniform size regardless of the colony color (Fig. 4, lanes 1–8). Even when this analysis was performed on a large number of pooled white or blue colonies, no new digestion fragments indicative of unexpected DNA rearrangements were seen (data not shown).
Figure 4.
Restriction analysis of plasmids isolated from white and blue colonies obtained by transient expression of p4B-Flp(K82M) in cells harboring the lacZα reporter pBU11. All plasmid mixtures and the control plasmid were digested with XbaI and HindIII. XbaI cuts the reporter plasmid within the spacer region. HindIII linearizes the expression plasmid and cuts the reporter plasmid immediately outside one of the two FRT sites. Lanes 1–4, plasmids purified from white colonies; lane 5–8, plasmids purified from blue colonies; lane 9, pBU11; lane M, 500 bp DNA marker ladder (Bio-Rad).
The sum of the results from Figures 3 and 4 attests to the reliability and accuracy of the colony color based dual reporter assay in identifying novel recombination specificities.
The mFRT11 preference of Flp(K82Y) is nearly lost in Flp(K82F)
The contact made by Lys-82 of Flp with the guanine at position 1 of the FRT, illustrated in Figure 5A, does not provide a simple explanation for how replacement of the Lys by Tyr could have provided dual recognition of FRT and mFRT11, with the latter being a slightly preferred target. Modeling Tyr and cytosine in place of Lys-82 in Flp and guanine at the first position of FRT11, respectively, indicate the closest approach of Tyr and the C5 atom of cytosine to be more than 4 Å apart (Fig. 5B). Tyr-82 would also be at the same distance from the N7-atom of guanine at the first position in native FRT (not shown). Thus, direct amino acid-to-base contact mediated by Tyr-82 is unlikely although water mediated contacts cannot be excluded. We also cannot rule out the possibility that K82Y substitution alters the local conformation of the peptide chain in its vicinity to mediate differential recognition of mFRT11 or FRT. Perhaps the phenolic ring of Tyr-82 may then promote hydrophobic interactions with the C5-C6 atoms of cytosine (mFRT11) and repulsion from the N7 atom of guanine (FRT). This more favorable adaptive conformational adjustment towards mFRT11 could account for the slight shift in the specificity of Flp(K82Y). In the absence of structural information, these ideas must be considered speculative.
To test whether the hydroxyl group of Tyr-82 is essential for shifted specificity, we constructed Flp(K82F) by directed mutagenesis and tested it in parallel with Flp(K82Y) in the double reporter assay. The relative proportions of blue, white and red colonies obtained for the two proteins are shown in Figure 5C. We also tested them in parallel against FRT and mFRT11 in the single reporter assays using pBU1 and pBU11 as substrate plasmids. The recombination efficiencies in this assay, estimated as the ratio of white colonies to the sum of blue and white colonies, are listed in Figure 5C. The data from the two types of assays are mutually consistent, and indicate that Flp(K82F) was more or less comparable to Flp(K82Y) in activity on FRT. However, it was ∼2-fold less active on mFRT11 than Flp(K82Y). These results suggest that both the aromatic ring and the phenolic hydroxyl of Tyr-82 are required for the small mFRT11 bias shown by Flp(K82Y).
Summary
We have described here a functional recombination assay in E.coli using a dual reporter strategy for detecting relaxed as well as switched target specificities in the Flp site-specific recombinase. We have discussed the various parameters that were considered and optimized in designing the screening system. Having demonstrated the basic utility of this strategy, it is now feasible to improve its efficacy by housing within the recombination cassettes reporter genes that can be selected for or against under appropriate conditions. The selection method would vastly improve the number of variant recombinases that can be sampled in a single assay.
Based on our present studies, the yield of relaxed specificity Flp variants from a combination of mutagenesis and the dual reporter screen is of the order of one in 104 for the particular altered FRT site tested. Candidate colonies indicated to harbor relaxed specificity in the first screening round do contain some false positives. In a typical experiment, out of approximately 20 colonies scored as potential relaxed specificity variants, less than half would turn out to be spurious. For our purposes, this is quite an acceptable level of false positives as they are easily weeded out by examining their plasmid configurations (as in Fig. 3) or, if necessary, with the help of a second screen utilizing two separate single reporters.
Using the screening system in its present configuration, we have identified the variant Flp(K82Y) that prefers the altered target site mFRT11 to the native site FRT. The virtual absence of mFRT11 preference upon placing Phe at position 82 in Flp suggests that both the aromatic ring and the phenolic hydroxyl group contribute to target discrimination. We also identified two other variants, Flp(K82M) and Flp(K82H), that show relaxed specificity, and are able to utilize FRT and mFRT11 with approximately equal efficiency. By comparison, wild-type Flp is virtually inactive on mFRT11 under the in vivo assay conditions but recombines FRT nearly quantitatively. Taken together, these results indicate that the amino acid at position 82 of Flp is particularly important in target site selectivity. Consistent with this notion, preliminary data suggest that the magnitude of specificity switch in Flp(K82Y) can be further enhanced by additional amino acid substitutions in Flp (Y.Voziyanov, F.Stewart and M.Jayaram, unpublished data). The dual reporter assay can be adapted to other recombinases for generating new DNA specificities. Expanding the range of site-specific recombinases by designing new DNA targets and evolving matching recombinase variants opens new options for biotechnology and genome engineering.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Youming Zhang, Phoebe Rice and Frank Buchholz for discussions. This work was supported in part by an Alexander von Humboldt Research Fellowship to Y.V., a grant from the Volkswagen Foundation, Program on Conditional Mutagenesis to A.F.S. and an NIH grant to M.J. Partial support was provided by the Robert F. Welch Foundation and by a Research Grant to M.J. from the University of Texas at Austin.
REFERENCES
- 1.Nunes-Duby S.E., Kwon,H.J., Tirumalai,R.S., Ellenberger,T. and Landy,A. (1998) Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res., 26, 391–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kilby N.J., Snaith,M.R. and Murray,J.A. (1993) Site-specific recombinases: tools for genome engineering. Trends Genet., 9, 413–421. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez C.I., Buchholz,F., Galloway,J., Sequerra,R., Kasper,J., Ayala,R., Stewart,A.F. and Dymecki,S.M. (2000) High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nature Genet., 25, 139–140. [DOI] [PubMed] [Google Scholar]
- 4.Mills A.A. and Bradley,A. (2001) From mouse to man: generating megabase chromosome rearrangements. Trends Genet., 17, 331–339. [DOI] [PubMed] [Google Scholar]
- 5.Tobin M.B., Gustafsson,C. and Huisman,G.W. (2000) Directed evolution: the ‘rational’ basis for ‘irrational’ design. Curr. Opin. Struct. Biol., 10, 421–427. [DOI] [PubMed] [Google Scholar]
- 6.Kurtzman A.L., Govindarajan,S., Vahle,K., Jones,J.T., Heinrichs,V. and Patten,P.A. (2001) Advances in directed protein evolution by recursive genetic recombination: applications to therapeutic proteins. Curr. Opin. Biotechnol., 12, 361–370. [DOI] [PubMed] [Google Scholar]
- 7.Grabherr R., Ernst,W., Oker-Blom,C. and Jones,I. (2001) Developments in the use of baculoviruses for the surface display of complex eukaryotic proteins. Trends Biotechnol., 19, 231–236. [DOI] [PubMed] [Google Scholar]
- 8.Sidhu S.S. (2000) Phage display in pharmaceutical biotechnology. Curr. Opin. Biotechnol., 11, 610–616. [DOI] [PubMed] [Google Scholar]
- 9.Cheng Q., Swalla,B.M., Beck,M., Alcaraz,R.,Jr, Gumport,R.I. and Gardner,J.F. (2000) Specificity determinants for bacteriophage Hong Kong 022 integrase: analysis of mutants with relaxed core-binding specificities. Mol. Microbiol., 36, 424–436. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y., Narendra,U., Iype,L.E., Cox,M.M. and Rice,P.A. (2000) Crystal structure of a Flp recombinase-Holliday junction complex: assembly of an active oligomer by helix swapping. Mol. Cell., 6, 885–897. [PubMed] [Google Scholar]
- 11.Guzman L.M., Belin,D., Carson,M.J. and Beckwith,J. (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol., 177, 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kovach M.E., Elzer,P.H., Hill,D.S., Robertson,G.T., Farris,M.A., Roop,R.M.,II and Peterson,K.M. (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene, 166, 175–176. [DOI] [PubMed] [Google Scholar]
- 13.Buchholz F., Angrand,P.O. and Stewart,A.F. (1998) Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat. Biotechnol., 16, 657–662. [DOI] [PubMed] [Google Scholar]
- 14.Senecoff J.F.P., Rossmeissl,P.J. and Cox,M.M. (1988) DNA recognition by FLP recombinase of the yeast 2 micron plasmid: a mutational analysis of the FLP binding site. J. Mol. Biol., 201, 405–421. [DOI] [PubMed] [Google Scholar]
- 15.Andrews B.J., McLeod,M., Broach,J. and Sadowski,P.D. (1986) Interaction of the FLP recombinase of the Saccharomyces cerevisiae 2 micron plasmid with mutated target sequences. Mol. Cell. Biol., 6, 2482–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prasad P.V., Horensky,D., Young,L.J. and Jayaram,M. (1986) Substrate recognition by the 2 micron circle site-specific recombinase: effect of mutations within the symmetry elements of the minimal substrate. Mol. Cell. Biol., 6, 4329–4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Govind N.S. and Jayaram,M. (1987) Rapid localization and characterization of random mutations within the 2 micron circle site-specific recombinase: a general strategy for analysis of protein function. Gene, 51, 31–41. [DOI] [PubMed] [Google Scholar]
- 18.Umlauf S.W. and Cox,M.M. (1988) The functional significance of DNA sequence structure in a site-specific genetic recombination reaction. EMBO J., 6, 1845–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Senecoff J.F., Bruckner,R.C. and Cox,M.M. (1985) The FLP recombinase of the yeast 2-micron plasmid: characterization of its recombination site. Proc. Natl Acad. Sci. USA, 82, 7270–7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vriend G. (1990) WHAT IF: A molecular modeling and drug design program. J. Mol. Graph., 8, 52–56. [DOI] [PubMed] [Google Scholar]
- 21.Guex N., Diemand,A. and Peitsch,M.C. (1999) Protein modelling for all. Trends Biochem. Sci., 24, 364–367. [DOI] [PubMed] [Google Scholar]