Abstract
We report the synthesis of a novel trithiol-capped oligodeoxyribonucleotide and gold nanoparticle conjugates prepared from it. These DNA–gold nanoparticle conjugates exhibit substantially higher stability than analogs prepared from monothiol and cyclic disulfide-capped oligodeoxyribonucleotides, but comparable hybridization properties. A quantitative analysis of their stability under a range of conditions is provided. Significantly, this novel trithiol oligodeoxyribonucleotide can be used to stabilize particles >30 nm in diameter, which are essential for many diagnostic applications.
INTRODUCTION
Biopolymer patterned solid substrates and modified nanoparticles play important roles in biological diagnostics and nanotechnology. Among these biopolymers, DNA is of particular interest because of the biological information contained within its base sequence and its inherent programmability (1). In recent years, DNA–nanoparticle conjugates have been used to build a variety of two- and three-dimensional materials with novel optical, electrical and catalytic properties (2–9); these materials are being exploited in the development of several biodetection schemes (10–15).
There are many strategies available for preparing DNA–nanoparticle conjugates, including direct adsorption of alkylthiol- or disulfide-terminated oligonucleotides on a metal nanoparticle surface (2), covalent binding of oligonucleotides to a pre-activated nanoparticle surface (14) and adsorption of biotinylated oligonucleotides on a particle surface coated with avidin (3,16). So far, the methods employing oligonucleotides functionalized on the 5′ or 3′ end with alkylthiol are the most common ones. This is primarily due to the ease of synthesizing these modified oligonucleotides and the strong affinity of the thiol group for noble metal and chalcogenide surfaces. Such methods have been used for preparing stable oligonucleotide conjugates with Au (2), Au-coated Ag (17) and ZnS-coated CdSe (6) nanoparticles.
Recently, we developed a series of colorimetric DNA detection methods that utilize oligodeoxyribonucleotide-functionalized 16 nm diameter Au nanoparticles as probes (e.g. Probe I in Scheme 1A). These methods show great promise because of their very high sensitivity and selectivity for designated DNA targets (10,11,13). Although such nanoparticle probes have been demonstrated to be very stable under most DNA detection conditions, we found that larger Au nanoparticles (>30 nm) modified with the monothiol-terminated oligonucleotides slowly decompose at elevated temperatures (40∼90°C) and in high salt concentration buffers (0.3∼1.0 M NaCl). Moreover, when any size Au nanoparticle, modified with monothiol oligonucleotides such as 1 (Scheme 1A), are used in biological buffer systems containing dithiothreitol (DTT) or mercaptoethanol, the nanoparticle probes become inactive after prolonged exposure to the buffer, especially at elevated temperatures (40∼95°C) (18,19). Through fluorophore-labeling studies it has been shown that mercaptoethanol and DTT will displace the alkylthiol-capped oligonucleotides from the Au nanoparticle surface (19).
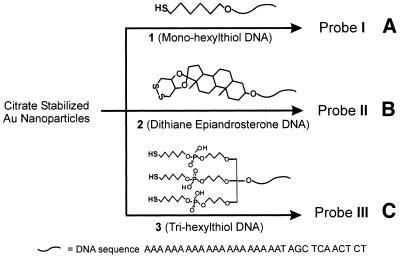
Scheme 1. Preparation schemes of oligonucleotide–Au nanoparticle conjugates with different anchor groups.
To increase the stability of the DNA–nanoparticle conjugates, we have previously developed a cyclic dithiane-epiandrosterone anchor group capped oligonucleotides such as 2 (Scheme 1B), which leads to significantly more stable DNA–Au conjugates (18). However, these relatively robust conjugates cannot survive prolonged cycling between room temperature and 95°C in the presence of 0.01 mM DTT. For some diagnostic applications (e.g. assays with duplex DNA), it would be desirable to use DNA–nanoparticle conjugates that retain their activity under these conditions. It is well known that polydentate ligands form much more stable metal–ligand complexes than do related monodentate ligands. In this paper, we take advantage of this chelating effect by designing and synthesizing oligonucleotides with three thiol-binding groups such as 3 (Scheme 1C), which can be strongly chemisorbed onto nanoparticle surfaces. Significantly, we show that the stabilities of Au nanoparticles (16, 50 and 100 nm) modified with these novel trithiol-capped oligonucleotides are substantially higher than comparable nanoparticle probes prepared from either the monothiol- or dithiane-functionalized oligonucleotides.
MATERIALS AND METHODS
Materials
Aqueous solutions of 50 and 100 nm diameter Au nanoparticles were purchased from British BioCell International, UK and smaller gold nanoparticles, 16 nm (15.7 1.6 nm), were prepared by citrate reduction of HAuCl4 via literature procedures (20). Nanopure water (resistance >18.1 MΩ) from a Barnstead NANOpure water system (Dubuque, USA) was used for all experiments. 5′-Thiol modifier C6 S-S phosphoramidite, trebler phosphoramidite and other reagents required for solid phase oligonucleotide synthesis were purchased from Glen Research (Sterling, USA).
General methods
Solid phase oligodeoxyribonucleotide synthesis was carried out on a Milligene Expedite DNA synthesizer. The oligonucletides were purified with high-performance liquid chromatography (HPLC) performed on a Hewlett-Packard (HP) series 1100 HPLC system. A Varian DYNAMAX C18 column (250 × 10.0 mm) was used for reverse phase (RP)–HPLC purification with 0.03 M triethylammonium acetate (TEAA) pH 7 and a 1%/min gradient of 95% CH3CN/5% 0.03 M TEAA at a flow rate of 3 ml/min, while monitoring the UV signal of DNA at 254 nm. Matrix-assisted laser desorption/ionization-time of flight mass-spectrometry (MALDI-TOF MS) was carried out with a PE Biosystems Voyager system 6050, calibrated by standard polythymine oligonucleotides. A transmission electron microscope (TEM; a Hitachi 8100 model) was used to determine the size and the morphology of Au nanoparticles. Fluorescence measurements were carried out with a Fluorolog-3 system (Instruments S.A. Inc., USA). UV-Vis measurements and melting analyses were performed using a HP 8453 diode array spectrophotometer equipped with a HP 89090a Peltier temperature controller.
Syntheses of single-stranded oligonucleotides with 5′-hexylthiol, 5′-dithiane epiandrosterone and 5′-trihexylthiol functionalities
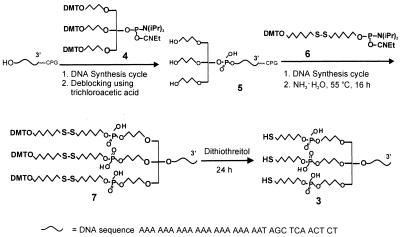
Oligodeoxyribonucleotide 1, with a hexylthiol headgroup at 5′ end, and oligodeoxyribonucleotide 2 [RP–HPLC retention time (tR) = 34.0 min; MALDI-TOF MS, 10323.48; calculated, 10329.76], with a cyclic dithiane-epiandrosterone headgroup at 5′ end, were synthesized as described previously (18,20). The synthesis of trithiol oligodeoxyribonucleotide 3 began with a coupling reaction between the –OH group at the 5′ end of an oligonucleotide derivative on 1000 Å controlled pore glass (CPG) beads and the phosphoramidite 4, ‘trebler’, under conditions for solid phase DNA synthesis (1 µmol scale, 15 min coupling time, >95% yield) (Scheme 2). The trebler moiety, which possesses three dimethoxyltrityl (DMT) protected hydroxyl groups, is used for subsequent thiol functionalization (21). Following removal of the three DMT groups with trichloroacetic acid in methylene chloride, the resulting triol 5 was further coupled with three equivalents of hexyldisulfide phosphoramidite 6 to form compound 7, which has three hexyldisulfide functionalities on the 5′ end of the olignucleotide. Note that we found a 15 min coupling time for this step, which is typically used for synthesizing monothiol oligonucleotides using the same hexyldisulfide phosphoramidite 6, failed to give high coupling yields. Therefore, an extended 120 min coupling time was used, which consisted of eight normal 15 min cycles without capping steps. With this procedure we obtained a ∼60% coupling yield. Compound 7 was cleaved from the CPG support with NH3·H2O (30%) by incubation CPG in 1 ml ammonia at 55°C for 16 h. After RP–HPLC purification (tR = 71.5 min), compound 7 was converted to the trithiol oligonucleotide 3 by reaction with 0.1 M DTT in 0.1 M phosphate buffer solution (pH 8) for 24 h. The purity of the as-prepared oligonucleotide 3 was further analyzed by RP–HPLC and MALDI-TOF MS (RP–HPLC tR = 29.9 min; MALDI-TOF MS, 10803.94; calculated, 10805.82), which show its purity to be >98%. This preparatory procedure typically yields 60∼100 nmol of 3 (1 µmol synthesis scale, 32 base oligonucleotide strand; see Scheme 2).
Scheme 2. Synthesis of single-stranded oligonucleotide with a 5′-trihexylthiol functional group.
Preparation of thiolated olignucleotide–Au nanoparticle conjugates
Au nanoparticles (16, 50 and 100 nm)/trithiolated oligonucleotide conjugates were prepared by the procedure described previously for binding monothiolated oligonucleotides to Au nanoparticles (20). For comparison purposes, Probe I, Probe II and Probe III were prepared with the appropriate thiolated oligonucleotides under the same conditions (Scheme 1).
Stability tests of oligonucleotide–Au nanoparticle conjugates
Colloids of each conjugate (using 16 nm Au particles, 0.9 ml, 2 nM) were dispersed in 0.3 M NaCl and 10 mM phosphate buffer. DTT (0.1 ml, 100 mM) was added to each colloid at 40°C, which eventually resulted in nanoparticle aggregation. As the reaction proceeded, a gradual color change from red to blue to black was observed. The optical changes were monitored by UV-Vis spectroscopy and used as a diagnostic indicator to study probe stability.
Melting properties of hybrids formed with 5′-thiolated oligonucleotide functionalized Au nanoparticle probes
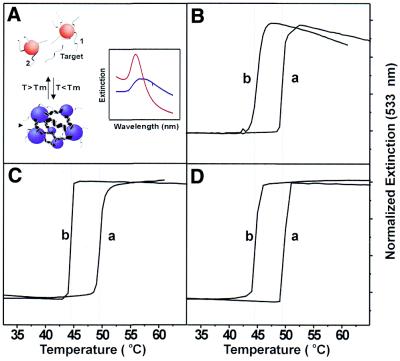
The hybridization properties of the nanoparticle probes were studied in 0.3 M NaCl and 10 mM phosphate buffer (pH 7) as previously described (20). Two sets of nanoparticle probes were used, one set bearing the sequence 5′-X-A10-ATC CTT ATC AAT ATT-3′ and the other set bearing 5′-X-A10-TAA CAA TAA TCC CTC-3′, where X designates one of the three different types of thiol groups as shown in Scheme 1. A 30mer target 5′-GAG GGA TTA TTG TTA AAT ATT GAT AAG GAT-3′, (a), or a 30mer single-base mismatched target 5′-GAG GGA TGA TTG TTA AAT ATT GAT AAG GAT-3′, (b), was used to link together both sets of nanoparticle probes to form DNA linked nanoparticle assemblies (see Fig. 2A). The melting experiment was performed on a HP 8453 diode array spectrophotometer equipped with a HP 89090a Peltier temperature controller. The temperature ramping range was from 20 to 70°C with an interval of 0.5°C and 1 min holding time for each temperature point.
Figure 2.
The melting properties of 50 nm Type-I, Type-II and Type-III Probes. (A) Scheme describing the hybridization between two sets of probes and a 30mer complementary target to form nanoparticle aggregates; the inset shows the melting of aggregates resulting in a sharp transition in extinction at 533 nm. Melting curves for Type-I Probes (B), Type-II Probes (C) and Type-III Probes (D). Note that in each case (B–D) both a perfectly complementary target (a) and single base mismatched one (b) were studied (see Materials and Methods for base sequences).
RESULTS AND DISCUSSION
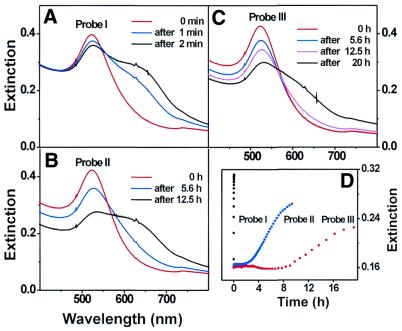
When DTT displaces the oligonucleotides from the Au nanoparticle surface, irreversible aggregation takes place. This results in a red shifting and broadening of the surface plasmon band at 524 nm for 16 nm diameter Au particles (Fig. 1A–C), which is well understood by Mie theory (22,23). The important observation is that the rate of aggregation is substantially slower for Probe III as compared with Probes I and II. Indeed, Probe I completely aggregated within 2 min in 10 mM DTT, 0.3 M NaCl and 10 mM phosphate buffer at 40°C, while Probe II began to aggregate after 2 h and is completely aggregated after 6 h (Fig. 1D). The solution containing Probe III maintained the optical properties of the dispersed nanoparticles for 10 h but began to gradually aggregate over the next 10 h as evidenced by a gradual red shifting and broadening of the surface plasmon band over the next 10 h. The aggregation reactions involving Probes I to III were further confirmed by TEM.
Figure 1.
Time evolution of UV-Vis spectra showing the stability of conjugates of 16 nm Au nanoparticle/oligonucleotide with different anchor groups in 10 mM DTT and 0.3 M NaCl solutions at 40°C. (A) Probe I (mono-hexylthiol), (B) Probe II (cyclic-disulfide), (C) Probe III (tri-hexylthiol), (D) extinction changes at 600 nm for Probe I, Probe II and Probe III during the reactions monitored in (A–C).
The surface coverages of oligonucletide I, II and III on the 16 nm Au nanoparticle surfaces were determined by a previously reported fluorophore-labeling method (19) (Table 1). The surface coverage values are in the range between 84 and 129 oligonucleotide strands per particle with Probe III exhibiting the highest coverage followed by II and then I. The higher surface coverage of III is thus attributed to the fact that trithiol groups bind to a gold nanoparticle surface more efficiently than the disulfide or monothiol moieties. All of these results strongly support the conclusion that trithiol oligonucleotide III exhibits a higher binding affinity for the Au nanoparticle surface than the monothiol or steroid disulfide-terminated oligonucleotides.
Table 1. Average surface coverages (strands per particle) for Oligo I, Oligo II and Oligo III on the 16 nm diameter Au nanoparticles, determined by a fluorophore labeling method.
| Oligo I | Oligo II | Oligo III | |
|---|---|---|---|
| Surface coverage (strands per particle) | 84 ± 5 | 94 ± 6 | 129 ± 8 |
It is important to note that the melting properties of the hybridization products derived from the three probes are qualitatively the same. When 50 nm Au particles (denoted as Type-I, Type-II and Type-III probes) are hybridized to complementary 30mer target strands, the melting temperature for all the three types of probes falls within the 48.8–49.4°C range (Fig. 2B–D). Note that the single base mismatched target, b, can be differentiated easily from the complementary target, a, due to the extremely sharp melting transitions associated with the nanoparticle probes (compare melting curves ‘a’ and ‘b’ in Fig. 2B–D). Indeed, the Tm values for aggregates formed from the complementary and mismatched target strands largely differ by 5°C, and the full width at half-maximum of the first derivative of the melting curves are in the 0.5–1.0°C range. These data show that although the oligonucleotide binding head groups dramatically affect the stability of the nanoparticle probes, they have very little effect on the hybridization and melting properties of DNA–Au conjugates.
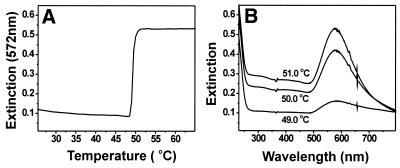
Also, note that 50 nm Type-I probes begin to decompose at elevated temperatures (>50°C), resulting in a decrease in the intensity of the surface plasmon band at 533 nm and showing a tailing-off in extinction during the end of the melting analysis (Fig. 2B). Previous studies in our group have shown that large Au nanoparticles (>30 nm) modified with monothiol oligonucleotides are not stable in high salt concentration solutions and at elevated temperatures. Significantly, we have found that the trithiol oligonucleotides can stabilize even 100 nm diameter Au particles, making them useful as probes in DNA diagnostic schemes. For example, Type-III probes formed from 100 nm Au particles exhibit sharp melting transitions in 0.3 M NaCl, 10 mM PBS solution after reacting with complementary target (Fig. 3). Indeed, a transition with no tailing-off in extinction is observed for such particles as was observed in the case of the 50 nm particles prepared from the monothiol (see Fig. 2B). Finally, the UV-Vis spectra of such probes measured as a function of temperature show no evidence of particle decomposition or aggregation, which would be reflected in the surface plasmon band broadening or irreversible changes in intensity.
Figure 3.
(A) The melting curve of 100 nm Type-III Probes. (B) The UV-Vis spectra at different temperatures during the melting analysis.
CONCLUSION
We have developed a synthetic route for making trithiol terminated oligonucleotides, which can be used to prepare extremely stable DNA–nanoparticle conjugates. Although the exact bonding of the trithiol headgroup on the gold nanoparticle surface is not properly understood, the method yields methodologically useful diagnostic probes. Specifically, the work has several significant practical implications. First, probe stability is essential for diagnostic applications. Until now, the larger particles (>30 nm) have exhibited varying degrees of stability that have restricted their use in assays such as those based on light scattering that rely on their reversible hybridization properties (13,24). Secondly, an increase in probe stability will increase the utility of such structures in more complex environments. Finally, this synthetic strategy can be extended readily to make oligonucleotides with multidentate binding groups that can be used to modify and stabilize other noble metal and metal chalcogenide surfaces (nanoparticles or planar structures). Efforts in this direction are underway.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
REFERENCES
- 1.Storhoff J.J. and Mirkin,C.A. (1999) Programmed materials synthesis with DNA. Chem. Rev., 99, 1849–1862. [DOI] [PubMed] [Google Scholar]
- 2.Mirkin C.A., Letsinger,R.L., Mucic,R.C. and Storhoff,J.J. (1996) A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature, 382, 607–609. [DOI] [PubMed] [Google Scholar]
- 3.Alivisatos A.P., Johnsson,K.P., Peng,X., Wilson,T.E., Loweth,C.J., Bruchez,M.P.,Jr and Schultz,P.G. (1996) Organization of nanocrystal molecules using DNA. Nature, 382, 609–611. [DOI] [PubMed] [Google Scholar]
- 4.Mucic R.C., Storhoff,J.J., Mirkin,C.A. and Letsinger,R.L. (1998) DNA-directed synthesis of binary nanoparticle network materials. J. Am. Chem. Soc., 120, 12674–12675. [Google Scholar]
- 5.Loweth C.J., Caldwell,W.B., Peng,X., Alivisatos,A.P. and Schultz,P.G. (1999) DNA-based assembly of gold nanocrystals. Angew. Chem. Int. Ed., 38, 1808–1812. [DOI] [PubMed] [Google Scholar]
- 6.Mitchell G.P., Mirkin,C.A. and Letsinger,R.L. (1999) Programmed assembly of DNA functionalized quantum dots. J. Am. Chem. Soc., 121, 8122–8123. [Google Scholar]
- 7.Taton T.A., Mucic,R.C., Mirkin,C.A. and Letsinger,R.L. (2000) The DNA-mediated formation of supermolecular mono- and multilayered nanoparticle structures. J. Am. Chem. Soc., 122, 6305–6306. [Google Scholar]
- 8.Mbindyo J.K.N., Reiss,B.D., Martin,B.R., Keating,C.D., Natan,M.J., Mallouk,T.E. (2001) DNA-directed assembly of gold nanowires on complementary surfaces. Adv. Mater., 13, 249–254. [Google Scholar]
- 9.Willner I., Patolsky,F. and Wasserman,J. (2001) Photoelectrochemistry with controlled DNA-cross-linked CdS nanoparticle arrays. Angew. Chem. Int. Ed., 40, 1861–1864. [PubMed] [Google Scholar]
- 10.Elghanian R., Storhoff,J.J., Mucic,R.C., Letsinger,R.L. and Mirkin,C.A. (1997) Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science, 277, 1078–1080. [DOI] [PubMed] [Google Scholar]
- 11.Taton T.A., Mirkin,C.A. and Letsinger,R.L. (2000) Scanometric DNA array detection with nanoparticle probes. Science, 289, 1757–1760. [DOI] [PubMed] [Google Scholar]
- 12.Taylor J.R., Fang,M.M. and Nie,S. (2000) Probing specific sequences on single DNA molecules with bioconjugated fluorescent nanoparticles. Anal. Chem., 72, 1979–1986. [DOI] [PubMed] [Google Scholar]
- 13.Taton T.A., Lu,G. and Mirkin,C.A. (2001) Two-color labeling of oligonucleotide arrays via size-selective scattering of nanoparticle probes. J. Am. Chem. Soc., 123, 5164–5165. [DOI] [PubMed] [Google Scholar]
- 14.Pathak S., Choi,S.K., Arnheim,N. and Thompson,M.E. (2001) Hydroxylated quantum dots as luminescent probes for in situ hybridization. J. Am. Chem. Soc., 123, 4103–4104. [DOI] [PubMed] [Google Scholar]
- 15.He L., Musick,M.D., Nicewarner,S.R., Salinas,F.G., Benkovic,S.J., Natan,M.J. and Keating,C.D. (2000) Colloidal Au-enhanced surface plasmon resonance for ultrasensitive detection of DNA hybridization. J. Am. Chem. Soc., 122, 9071–9077. [Google Scholar]
- 16.Niemeyer C.M., Burger,W. and Peplies,J. (1998) Covalent DNA-Streptavidin conjugates as building blocks for novel biometallic nanostructures. Angew. Chem. Int. Ed., 37, 2265–2268. [DOI] [PubMed] [Google Scholar]
- 17.Cao Y.W., Jin,R. and Mirkin,C.A. (2001) DNA-modified core-shell Ag/Au nanoparticles. J. Am. Chem. Soc., 123, 7961–7962. [DOI] [PubMed] [Google Scholar]
- 18.Letsinger R.L., Elghanian,R., Viswanadham,G. and Mirkin,C.A. (2000) use of steroid cyclic disulfide anchor in constructing gold nanoparticle-oligonucleotide conjugates. Bioconjug. Chem., 11, 289–291. [DOI] [PubMed] [Google Scholar]
- 19.Demers L.M., Mirkin,C.A., Mucic,R.C., Reynolds,R.A.,III and Letsinger,R.L. (2000) Fluorescence-based method for determining the surface coverage and hybridization efficiency of thiol-capped oligonucleotides bound to gold thin films and nanoparticles. Anal. Chem., 72, 5535–5541. [DOI] [PubMed] [Google Scholar]
- 20.Storhoff J.J., Elghanian,R., Mucic,R.C., Mirkin,C.A. and Letsinger,R.L. (1998) One-pot colorimetric differentiation of polynucleotides with single base imperfections using gold nanoparticle probes. J. Am. Chem. Soc., 120, 1959–1964. [Google Scholar]
- 21.Shchepinov M.S., Udalova,I.A., Bridgman,A.J. and Southern,E.M. (1997) Oligonucleotide dendrimers: synthesis and use as polylabelled DNA probes. Nucleic Acids Res., 25, 4447–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Storhoff J.J., Lazarides,A.A., Mucic,R.C., Mirkin,C.A., Letsinger,R.L. and Schatz,G.C. (2000) What controls the optical properties of DNA-linked gold nanoparticle assemblies? J. Am. Chem. Soc., 122, 4640–4650. [DOI] [PubMed] [Google Scholar]
- 23.Lazarides A.A. and Schatz,G.C. (2000) DNA-linked metal nanosphere materials: structural basis for the optical properties. J. Phys. Chem. B, 104, 460–467. [Google Scholar]
- 24.Reynolds R.A. III, Mirkin,C.A. and Letsinger,R.L. (2000) Homogeneous nanoparticle- based quantitative colorimetric detection of oligonucleotides. J. Am. Chem. Soc., 122, 3795–3796. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.