Abstract
Homing endonucleases are enzymes that catalyze the highly sequence-specific cleavage of DNA. We have developed an in vivo selection in Escherichia coli that links cell survival with homing endonuclease-mediated DNA cleavage activity and sequence specificity. Using this selection, wild-type and mutant variants of three homing endonucleases were characterized without requiring protein purification and in vitro analysis. This selection system may facilitate the study of sequence-specific DNA cleaving enzymes, and selections based on this work may enable the evolution of homing endonucleases with novel activities or specificities.
INTRODUCTION
Homing endonucleases are a recently characterized class of sequence-specific double-stranded DNA cleaving enzymes that are involved in the process of inserting mobile genetic elements into genomic DNA. Unlike restriction endonucleases, which typically operate on palindromic DNA sequences 4–8 bp in length, homing endonucleases recognize very long and frequently non-palindromic sequences ~12–40 bp in length (1–3). Despite the unusual sequence specificity of homing endonucleases and their resulting utility as highly specific DNA cleavage agents, our understanding of these enzymes remains relatively limited. The few X-ray crystal or NMR solution structures of homing endonucleases solved thus far (reviewed in 1) reveal a diverse set of base-specific hydrogen bonds, polar interactions and van der Waals contacts between protein and DNA, all of which may contribute to the specificity of these enzymes.
Mechanistic studies of DNA cleavage by homing endonucleases have primarily adopted one of two strategies. The substrate DNA sequence can be mutated and assayed in vitro to identify the bases required for substrate cleavage by the wild-type enzyme, or the homing endonuclease can be subjected to site-directed mutagenesis and the resulting mutant enzymes assayed to identify catalytically important residues. Both attempts require the cloning, expression and purification of one or more endonucleases, the synthesis of one or more DNA substrates and the analysis of in vitro cleavage reactions. Further complicating the in vitro characterization of these enzymes, the overexpression of some homing endonucleases in common expression systems has been reported to induce cell lysis (2), limiting the yields of active protein.
Since the availability of purified homing endonucleases and their mutants is a significant bottleneck for the rapid characterization of this class of proteins, we sought to develop a general activity assay for homing endonucleases that does not require overexpression and purification of the protein of interest. While several approaches have been reported that link DNA cleavage to an observable signal in vitro (4–8), very few general strategies exist to assay enzyme-catalyzed sequence-specific DNA cleavage in living cells (9). Our strategy is to link the catalytic activity of a homing endonuclease to the survival of an Escherichia coli cell containing the homing endonuclease gene. Such a selection system would circumvent the need for protein purification and in vitro assays but require making endonucleolytic activity, normally detrimental to a cell, a necessity for cell survival. In addition to facilitating the rapid characterization of wild-type and mutant homing endonucleases, our findings may enable the evolution in vivo of novel homing endonucleases with altered specificities or activities.
MATERIALS AND METHODS
Plasmid construction
To construct plasmid pSupE-nuclease, a cassette containing a supE suppressor tRNA under control of the lpp promoter and rrnC terminator was amplified by PCR from plasmid pACsupE (10) and subcloned into the large NotI–KpnI fragment of pBluescript II SK(+)-Nco [which is an A823G mutant of pBluescript II SK(+) containing a NcoI site] to provide pSupE. The genes encoding the homing endonucleases I-SceI, I-ScaI and PI-SceI were amplified by PCR from plasmids pSCM525 (11), pET11-p28bi2 (12) and pHisVDE (13), respectively, and subcloned into the large NcoI–NotI fragment of pSupE under the control of the constitutive lac promoter to afford the pSupE-nuclease plasmids. PCR primers used to amplify genes encoding the supE expression cassette and the I-SceI, I-ScaI and PI-SceI homing endonucleases were as follows: 5′-TATGCATAACGCGGCCGCCCCGAGGGCACCTGTCCTAC-3′ and 5′-TATCTGGGTACCGCATGCACCATTCCTTGCGG-3′ for the supE cassette; 5′-AGCTCCATGGCAATGAAAAACATCAAAAAAAACCAGG-3′ and 5′-TATCAAATGCGGCCGCTTATTATTTCAGGAAAGTTTCGGAGG-3′ for I-SceI, 5′-AGCTCCATGGAATATACCATGCTGATTAAAAG-3′ and 5′-TATCAAATGCGGCCGCTTATTACAGATAGTTGCCCAG-3′ for I-ScaI, and 5′-AGCTCCATGGGATCCGCATGCTTTGCCAAG-3′ and 5′-TATCAAATGCGGCCGCTCATCAGCAATTATGGACGACAACC-3′ for PI-SceI.
Plasmid pBar2 was constructed by ligating the ClaI–SphI fragment from pYsupA38B2 (10) containing araC and a two amber codon variant of barnase (Bar2) under the control of the pBAD promoter into the large ClaI–SphI fragment of pACYC184 to provide pACYC-Bar2. In addition, a synthetic cassette 5′-GCATGCCTGAGATCTTCGGATCA-3′ was ligated into the large SphI–HincII fragment of pACYC-TAG2 in order to create a BglII site. The recognition sequences of the enzymes I-SceI, I-ScaI and PI-SceI were inserted at this BglII site using the oligos 5′-GATCACACTGTCACATTGAGGTGCACTAGTTATTACCAGT-3′ and 5′-GATCACTGGTAATAACTAGTGCACCTCAATGTGACAT-3′ (recognition sequence of I-ScaI underlined; 5′-GATCATTACGCTAGGGATAACAGGGTAATATAACGT-3′ and 5′-GATCACGTTATATTACCCTGTTATCCCTAGCGTAAT-3′ (recognition site of I-SceI underlined); or 5′-GATCATCTGACGCCATTATCTATGTCGGGTGCGGAGAAAGAGGTAATGAAATGGCAGAAGTCTTGATGGAT-3′ and 5′-GATCATCCATCAAGACTTCTGCCATTTCATTACCTCTTTCTCCGCACCCGACATAGATAATGGCGTCAGAT-3′ (recognition site of PI-SceI underlined). Inserts containing multiple copies of the cleavage sites were obtained by self-ligation of the synthetic cassettes and gel purification of double-stranded fragments of the desired lengths. For I-SceI selections, pBar variants containing a dimer or a tetramer of the recognition site were used. For I-ScaI selections a trimer of the recognition site was used, and for PI-SceI selections a pBar variant containing a single copy of the recognition site was used. Variants of pBar containing more than four copies of any nuclease recognition sequence proved to be unstable in E.coli over many cell divisions. The relevant portions of all constructed plasmids were verified by automated DNA sequencing.
Selections
Selection strains were constructed by transformation of E.coli strain DH10B (Gibco BRL) with the appropriate variant of pBar2. Transformants were grown in 2xYT in the presence of chloramphenicol (40 µg/ml) and glucose (0.5%), and electrocompetent cells of the selection strains were prepared following standard procedures (14). For selections, typically 40 µl of competent cells were transformed with 10–100 ng of the appropriate variant of pSupE-nuclease. After electroporation, cells were immediately recovered in 2xYT + glucose (0.5%) and shaken at 37°C for 15–20 min. In order to estimate the total number of transformants, an aliquot of cells was plated on 2xYT + glucose (0.5%) + carbenicillin (125 µg/ml). A second aliquot of cells was washed with 2xYT and plated on 2xYT + arabinose (0.5%) + carbenicillin (125 µg/ml). All plates were incubated at 37°C for 8–18 h until colonies were clearly visible.
In vivo activity assays
Different pSupE-nuclease variants were adjusted to approximately equal concentrations (determined by gel densitometry and quantitation of the number of transformants under non-selective conditions), and selections were carried out as described above. Colonies surviving on glucose were used as an internal standard defined as 100% survival. For PI-SceI, the signal-to-background ratio was improved by an additional incubation at 37°C for 1 h in 2xYT + 125 µg/ml carbenicillin + 0.5% arabinose prior to plating. For the enzyme I-ScaI, the optimal signal-to-background ratio was observed by pre-incubating the transformants in 2xYT + 125 µg/ml carbenicillin + 0.5% glucose for 6 h at 37°C prior to plating.
RESULTS
Development of a conditionally toxic homing endonuclease substrate
Non-native DNA cleavage is typically detrimental to living cells. In order to transform DNA cleavage into an event necessary for cell survival, we took advantage of the ability of homing endonucleases to transform circular plasmid DNA into linear products. Since linear DNA does not replicate efficiently in E.coli and is rapidly degraded by the endogenous RecBCD nuclease (15), we hypothesized that endonuclease-catalyzed cleavage of a plasmid encoding a toxic protein could rescue the ability of cells to survive under suitably controlled growth conditions.
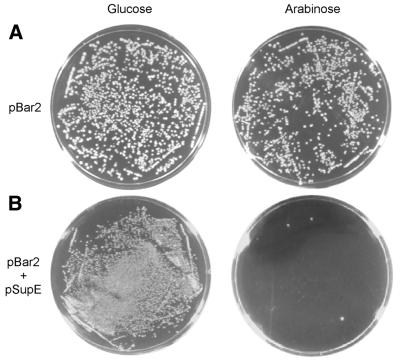
The first requirement of implementing this strategy is ‘caging’ the toxicity of a toxic gene such that it is not lethal to cells until a homing endonuclease has had the opportunity to catalyze the toxic plasmid’s cleavage. To effect this caging, we used a mutant form of barnase, the highly toxic RNase from Bacillus amyloliquefaciens (16–20), in which two non-essential residues (Gln2 and Asp44) had been mutated to amber (TAG) stop codons (10). The amber-mutated barnase gene (Bar2) was placed under the control of the PBAD promoter (21), allowing barnase expression to be induced with arabinose and repressed with glucose. Efforts to cage the toxicity of wild-type barnase simply by repressing its expression using glucose were unsuccessful, suggesting that the low level of barnase expression even under PBAD repression conditions is lethal to E.coli. Plasmids containing Bar2 were introduced into E.coli strain DH10B, which has minimal ability to suppress amber nonsense codons. The resulting cells were viable in the presence of glucose as well as in the presence of arabinose, indicating that the caging strategy successfully abrogates the toxicity of barnase (see Fig. 2A).
Figure 2.
(A) Cells harboring the pBar2 plasmid show similar viability on glucose and arabinose, indicating that the toxicity of barnase has been successfully caged by the two amber codons. Identical numbers of transformants were plated on arabinose and glucose plates. (B) Transforming pSupE into cells harboring pBar2 results in cell death upon induction of barnase expression with arabinose (right) but survival in the presence of glucose (left). Identical numbers of transformants were plated on arabinose and glucose plates.
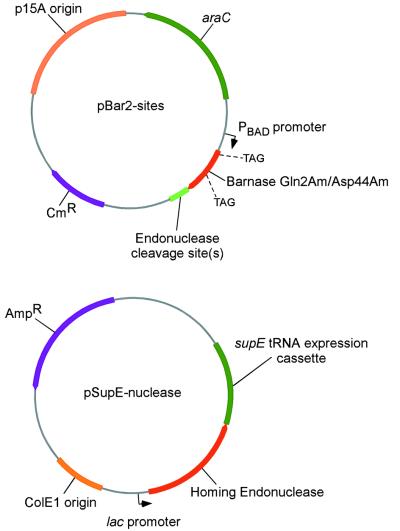
An expression cassette encoding the efficient amber suppressor tRNA supE (22) was introduced into a separate plasmid, designated pSupE, containing a compatible origin of replication (Fig. 1B). These amber suppressor tRNA expression plasmids were transformed into competent cells harboring pBar2 plasmids and plated on growth media supplemented with carbenicillin (to ensure the presence of pSupE) and containing either glucose or arabinose, but lacking chloramphenicol. We hypothesized that even in the absence of chloramphenicol (which normally ensures maintenance of the pBar2 plasmid) the 10–20 copies of pBar2 per cell at the time of transformation would be sufficiently toxic in the presence of the amber suppressor tRNA to be lethal. Indeed, essentially no growth of cells harboring pBar2 and pSupE was observed on arabinose (Fig. 2B). In contrast, cells harboring pBar2 and pSupE were viable when grown in the presence of glucose (Fig. 2B).
Figure 1.
The selection system for homing endonuclease activity is based on the two compatible plasmids pBar2-sites and pSupE-nuclease. The former plasmid contains nuclease cleavage sites of interest and places expression of an amber nonsense mutated barnase gene under control of the arabinose-induced and glucose-repressed PBAD promoter. The latter plasmid expresses the homing endonuclease enzyme together with an amber suppressor tRNA.
These results demonstrate that the amber suppression of plasmids expressing nonsense-mutated barnase genes is lethal to E.coli cells even in the absence of selective pressure to maintain the barnase-encoding plasmids. Our findings suggest, therefore, that the rate of barnase-induced cell death is faster than the rate of spontaneous loss of all copies of the pBar2 plasmid.
Linking homing endonuclease activity and specificity with cell survival
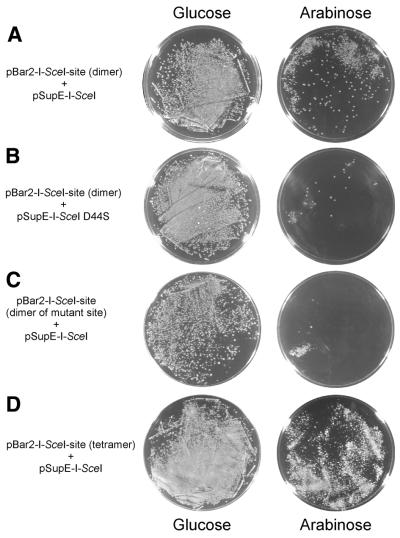
We used the homing endonuclease I-SceI to develop a link between homing endonuclease activity and the alleviation of pBar2-mediated toxicity. I-SceI is a monomeric 237 amino acid protein belonging to the LAGLIDADG family of homing endonucleases. This enzyme cleaves the 18 bp recognition sequence 5′-TAG GGA TAA // CAG GGT AAT-3′ leaving a 4 nt 3′ overhang (23). We ligated two repeats of this recognition sequence into the pBar2 plasmid affording pBar2-I-SceI-site. The gene encoding I-SceI was subcloned into pSupE behind a constitutive lac promoter resulting in pSupE-I-SceI. When competent cells harboring pBar2-I-SceI-site were transformed with pSupE-I-SceI under the conditions described above, we observed significant levels of survival (~25%) on arabinose (Fig. 3A). As a control, we mutated the critical active site aspartate in the P1 motif of I-SceI (2) from Asp44 to Ser. When introduced into cells containing pBar2-I-SceI-site, the pSupE-I-SceI-D44S mutant was unable to yield viable colonies on arabinose at a significant rate (<1%; Fig. 3B). As an additional control, we repeated the selection using a mutant I-SceI recognition site (5′-TAG GGA TAA CAa GGT AAT-3′) that is known not to be cleaved by I-SceI (23–25). Transformation of the selection strain containing this mutant recognition site with wild-type pSupE-I-SceI also resulted in very low levels of survival on arabinose (Fig. 3C). Taken together, these results demonstrate that the selection system described above successfully links cell survival with both homing endonuclease activity and DNA sequence specificity.
Figure 3.
(A) Transformation of cells harboring pBar2-I-SceI-site with pSupE-I-SceI results in significant cell survival upon induction with arabinose. (B) In contrast, the same selection strain transformed with pSupE-I-SceID44S encoding an inactive endonuclease results in very low survival rates on arabinose. (C) Repeating the assay in (A) with a pBar2-I-SceI-site variant in which one critical base of the I-SceI cleavage site has been mutated (see text) also results in non-viable cells. (D) Increasing the intracellular concentration of homing endonuclease substrate by using a variant of pBar2-I-SceI-site containing four copies of the I-SceI cleavage site results in a higher level of survival compared with the two-copy variant of pBar2-I-SceI-site shown in (A).
In addition to the selection strain harboring pBar2-I-SceI-site containing two copies per plasmid of the I-SceI cleavage site (Fig. 3A), we also generated and characterized a selection strain with a pBar2-I-SceI-site variant containing four copies per plasmid of the wild-type cleavage site. When transformed with pSupE-I-SceI, the four-copy variant reproducibly survived at an ~2-fold higher rate compared with the survival rate of the two-copy strain, consistent with the hypothesis that elevating the concentration of substrate DNA sites in vivo increases the efficiency of pBar2 cleavage (Fig. 3D). This result suggests that the stringency of the homing endonuclease selection can be modulated by varying the number of nuclease cleavage sites in the pBar2 plasmid. Variants of pBar2 containing more than four copies of endonuclease cleavage sites proved unstable when propagated in E.coli.
A sensitive in vivo activity assay for homing endonucleases
We next evaluated the suitability of the selection system described above as a general and semi-quantitative assay for homing endonucleases activity and site specificity. To test the generality of our strategy, we established selection systems similar to the I-SceI system described above for two additional homing endonucleases: PI-SceI and I-ScaI. Although these enzymes also belong to the LAGLIDADG endonuclease family, there is no appreciable sequence homology among the I-ScaI, I-SceI and PI-SceI proteins. Further, the lengths of their cleavage sites (16, 18 and ~30 bp for I-ScaI, I-SceI and PI-SceI, respectively) vary significantly and the DNA sequences cleaved by these enzymes are unrelated (12,13,23), suggesting that a selection system compatible with all three enzymes is likely to be applicable to homing endonucleases in general.
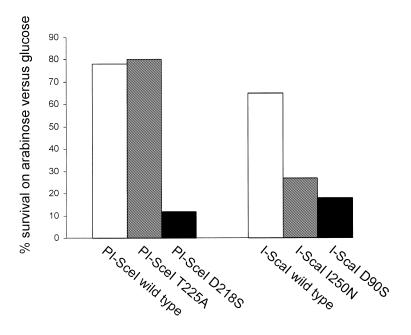
We generated wild-type pSupE-PI-SceI and pSupE-I-ScaI plasmids as well as variants encoding the catalytically inactive D218S (26) and D90S (27) mutants of PI-SceI and I-ScaI, respectively. Plasmids pBar2-PI-SceI-site and pBar2-I-ScaI-site containing the wild-type cleavage sites of these two homing endonucleases were also constructed. For both the PI-SceI and I-ScaI enzymes we observed high survival rates of cells containing the wild-type pSupE-nuclease plasmid and the matched pBar2-site wild-type cleavage site when grown in the presence of arabinose (Fig. 4). In contrast, cells expressing the inactive nuclease mutants survived on arabinose at a much lower rate (Fig. 4). To further evaluate the utility of this assay, we constructed mutant pSupE plasmids expressing mutant endonucleases with previously characterized activities. For I-ScaI we chose the I250N mutant that possesses activity too low to detect in vitro but which can be observed in vivo (27), while for PI-SceI we used the T225A mutant that exhibits slightly higher activity in vitro than wild-type PI-SceI (28). The signals generated by these mutant enzymes (the percentage of colonies surviving selection on arabinose versus on glucose) were compared with those generated by the wild-type enzymes (Fig. 4). The wild-type I-ScaI nuclease induces survival on arabinose at a 65% rate relative to survival on glucose. In contrast, the I250N I-ScaI mutant causes survival at a 27% rate, while cells expressing the inactive D90S mutant survive at an 18% rate. Among PI-SceI variants, the wild-type enzyme results in 78% survival under selection conditions, while cells expressing the T225A mutant survive at an 80% rate (not statistically distinguishable from the wild-type survival rate) and those expressing the inactive D218S mutant survive at a 12% rate. These results are consistent with the previously reported relative activities of these homing endonuclease variants and suggest that the selection system described above can serve as a general semi-quantitative in vivo assay for homing endonuclease activity.
Figure 4.
Quantitative analysis of the activities of six homing endonuclease variants of I-ScaI and PI-SceI. Cells harboring pBar2-PI-SceI-site (left three bars) or pBar2-I-ScaI-site (right three bars) were transformed with the pSupE-nuclease plasmids encoding the six nucleases listed and processed as described in the Materials and Methods. The percentage of surviving colonies on arabinose-containing media relative to the number of colonies arising from an identical number of transformants plated on glucose-containing media is shown for each nuclease. Values reflect the average of three independent trials and standard deviations were <15% of each value reported.
DISCUSSION
Here we describe an in vivo selection system in E.coli for homing endonuclease-catalyzed DNA cleavage. In this system, one plasmid contains a cleavage site of interest together with a caged toxic gene, while a second plasmid encodes the homing endonuclease to be studied and a suppressor tRNA that enables the functional expression of the toxic gene. In the absence of homing endonuclease activity, cells harboring both plasmids are largely not viable in media containing arabinose. The small amount of background growth observed under these conditions is likely due to the rare but detectable rejection of all, or nearly all, copies of the pBar2 plasmid by the cells in the absence of the plasmid maintenance marker (chloramphenicol) during recovery and selection. Consistent with this hypothesis, we have found that the majority of these background colonies are chloramphenicol sensitive (data not shown). Expression of an active homing endonuclease presumably leads to cleavage of its recognition site on the pBar2 plasmid, degradation of the linearized pBar2 DNA, and reduction of the pBar2 copy number to an extent that the resulting cells are viable in the presence of arabinose. The system was evaluated for three homing endonucleases that all belong to the LAGLIDADG family (I-ScaI, I-SceI and PI-SceI), and in each case an active enzyme–substrate combination was required for cell survival.
Our results suggest that this selection system can be used as a sensitive in vivo activity assay for studying combinations of double-strand cleaving homing endonucleases and cleavage sites of interest. A selection strain containing a pBar2 plasmid with a cleavage site of interest allows the semi-quantitative determination of the ability of wild-type or mutant homing endonucleases to cleave that site. Each assay is internally controlled by comparing survival under selection conditions (in the presence of arabinose) with survival in the absence of selective pressure (in the presence of glucose). This internal control normalizes the signal relative to the total number of transformants and corrects for differences in transformation efficiencies between experiments, although variable expression levels among different endonuclease mutants may also affect survival rates. The endpoints of the signal are conveniently calibrated by measuring the survival rates of wild-type and inactive mutants under selection conditions.
Traditionally, the effect of site-directed or random mutagenesis on the activity of homing endonucleases is determined by in vitro DNA cleavage using purified nucleases and subsequent gel electrophoresis of the resulting DNA fragments. The system described here circumvents laborious protein overexpression and purification and involves simple plasmid transformation and cell plating rather than in vitro cleavage assays. In addition, the ability of this selection to detect cleavage activity of the I250N mutant of I-ScaI—activity that was not detectable by in vitro assay but is known to exist in vivo (27)—suggests that this system may be able to detect low levels of activity difficult to observe using traditional in vitro endonuclease assay methods. Finally, in vivo selection allows enzyme activities to be assayed in the living cell under complex conditions that in some cases may be more relevant than artificial in vitro conditions. This selection system should, therefore, facilitate structure–function analyses of homing endonucleases and moreover may assist the study of other sequence-specific DNA cleavage agents capable of functioning in living cells.
The successful development of an in vivo selection system linking homing endonuclease activity and specificity with cell survival may also enable the evolution of homing endonucleases with altered cleavage specificities. Mutant endonucleases capable of cleaving DNA sequences of interest may be selected using libraries of pSupE-nuclease plasmids and pBar2 variants containing desired cleavage sites. Efforts to further develop this system to evolve novel homing endonucleases, which may require reducing background survival to a level comparable with the frequency of generating desired endonucleases in protein libraries, are in progress.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Bernard Dujon for providing plasmid pSCM525, Claude Monteilhet for plasmid pET11-p28bi2, Frauke Christ for plasmid pHisVDE and Dan Rosenbaum for providing plasmid pBluescript II SK(+)-Nco. M.G. is supported by the Emmy Noether Stipendium of the Deutsche Forschungsgemeinschaft. K.C. and I.S. were supported by Harvard College Research Program grants. This work was supported by NSF CAREER Award MCB-0094128 and Harvard University.
REFERENCES
- 1.Chevalier B.S. and Stoddard,B.L. (2001) Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res., 29, 3757–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jurica M.S. and Stoddard,B.L. (1999) Homing endonucleases: structure, function and evolution. Cell. Mol. Life Sci., 55, 1304–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belfort M. and Roberts,R.J. (1997) Homing endonucleases: keeping the house in order. Nucleic Acids Res., 25, 3379–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J.J., Geyer,R. and Tan,W. (2000) Using molecular beacons as a sensitive fluorescence assay for enzymatic cleavage of single-stranded DNA. Nucleic Acids Res., 28, e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McLaughlin L.W., Benseler,F., Graeser,E., Piel,N. and Scholtissek,S. (1987) Effects of functional group changes in the EcoRI recognition site on the cleavage reaction catalyzed by the endonuclease. Biochemistry, 26, 7238–7245. [DOI] [PubMed] [Google Scholar]
- 6.Waters T.R. and Connolly,B.A. (1992) Continuous spectrophotometric assay for restriction endonucleases using synthetic oligodeoxynucleotides and based on the hyperchromic effect. Anal. Biochem., 204, 204–209. [DOI] [PubMed] [Google Scholar]
- 7.Jeltsch A., Fritz,A., Alves,J., Wolfes,H. and Pingoud,A. (1993) A fast and accurate enzyme-linked immunosorbent assay for the determination of the DNA cleavage activity of restriction endonucleases. Anal. Biochem., 213, 234–240. [DOI] [PubMed] [Google Scholar]
- 8.Lee S.P. and Han,M.K. (1997) Fluorescence assays for DNA cleavage. Methods Enzymol., 278, 343–363. [DOI] [PubMed] [Google Scholar]
- 9.Seligman L.M., Stephens,K.M., Savage,J.H. and Monnat,R.J.,Jr (1997) Genetic analysis of the Chlamydomonas reinhardtii I-CreI mobile intron homing system in Escherichia coli. Genetics, 147, 1653–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu D.R. and Schultz,P.G. (1999) Progress toward an organism with an expanded genetic code. Proc. Natl Acad. Sci. USA, 96, 4780–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perrin A., Buckle,M. and Dujon,B. (1993) Asymmetrical recognition and activity of the I-SceI endonuclease on its site and on intron-exon junctions. EMBO J., 12, 2939–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monteilhet C., Dziadkowiec,D., Szczepanek,T. and Lazowska,J. (2000) Purification and characterization of the DNA cleavage and recognition site of I-ScaI mitochondrial group I intron encoded endonuclease produced in Escherichia coli. Nucleic Acids Res., 28, 1245–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wende W., Grindl,W., Christ,F., Pingoud,A. and Pingoud,V. (1996) Binding, bending and cleavage of DNA substrates by the homing endonuclease Pl-SceI. Nucleic Acids Res., 24, 4123–4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tabor S. and Struhl,K. (1989) Current Protocols in Molecular Biology. John Wiley and Sons, New York.
- 15.Kuzminov A. and Stahl,F.W. (1997) Stability of linear DNA in recA mutant Escherichia coli cells reflects ongoing chromosomal DNA degradation. J. Bacteriol., 179, 880–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Axe D.D., Foster,N.W. and Fersht,A.R. (1996) Active barnase variants with completely random hydrophobic cores. Proc. Natl Acad. Sci. USA, 93, 5590–5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin C., Richard,V., Salem,M., Hartley,R. and Mauguen,Y. (1999) Refinement and structural analysis of barnase at 1.5 Å resolution. Acta Crystallogr. D Biol. Crystallogr., 55, 386–398. [DOI] [PubMed] [Google Scholar]
- 18.Jucovic M. and Hartley,R.W. (1995) In vivo system for the detection of low level activity barnase mutants. Protein Eng., 8, 497–499. [DOI] [PubMed] [Google Scholar]
- 19.Hartley R.W. (1989) Barnase and barstar: two small proteins to fold and fit together. Trends Biochem. Sci., 14, 450–454. [DOI] [PubMed] [Google Scholar]
- 20.Hartley R.W. (1988) Barnase and barstar. Expression of its cloned inhibitor permits expression of a cloned ribonuclease. J. Mol. Biol., 202, 913–915. [DOI] [PubMed] [Google Scholar]
- 21.Guzman L.M., Belin,D., Carson,M.J. and Beckwith,J. (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol., 177, 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu D.R., Magliery,T.J. and Schultz,P.G. (1997) Characterization of an ‘orthogonal’ suppressor tRNA derived from E. coli tRNA2Gln. Chem. Biol., 4, 685–691. [DOI] [PubMed] [Google Scholar]
- 23.Monteilhet C., Perrin,A., Thierry,A., Colleaux,L. and Dujon,B. (1990) Purification and characterization of the in vitro activity of I-SceI, a novel and highly specific endonuclease encoded by a group I intron. Nucleic Acids Res., 18, 1407–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colleaux L., D’Auriol,L., Galibert,F. and Dujon,B. (1988) Recognition and cleavage site of the intron-encoded omega transposase. Proc. Natl Acad. Sci. USA, 85, 6022–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beylot B. and Spassky,A. (2001) Chemical probing shows that the intron-encoded endonuclease I-SceI distorts DNA through binding in monomeric form to its homing site. J. Biol. Chem., 276, 25243–25253. [DOI] [PubMed] [Google Scholar]
- 26.Christ F., Schoettler,S., Wende,W., Steuer,S., Pingoud,A. and Pingoud,V. (1999) The monomeric homing endonuclease PI-SceI has two catalytic centres for cleavage of the two strands of its DNA substrate. EMBO J., 18, 6908–6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szczepanek T., Jamoussi,K. and Lazowska,J. (2000) Critical base substitutions that affect the splicing and/or homing activities of the group I intron bi2 of yeast mitochondria. Mol. Gen. Genet., 264, 137–144. [DOI] [PubMed] [Google Scholar]
- 28.He Z., Crist,M., Yen,H., Duan,X., Quiocho,F.A. and Gimble,F.S. (1998) Amino acid residues in both the protein splicing and endonuclease domains of the PI-SceI intein mediate DNA binding. J. Biol. Chem., 273, 4607–4615. [DOI] [PubMed] [Google Scholar]