Abstract
We describe here the development of a new and simple single-tube multiplex Pyrosequencing assay. Genomic DNA or cDNA was employed to PCR amplify region(s) using biotinylated and normal primer(s). Subsequent to capture of PCR products on streptavidin-coated beads, single-stranded DNA separation and hybridization of multiple sequencing primers, Pyrosequencing was performed. The obtained pyrogram resulted in a unique pattern in which the intensity of the signal determined the number of incorporated nucleotide(s). Here, we demonstrate the use of this multiplex Pyrosequencing for single nucleotide polymorphisms genotyping and microbial typing.
INTRODUCTION
Development of tools for the determination of variations within and between genomes has had profound effects on the studies of biological systems in the last decades. Several principles have been described thus far. These principles rely on the hybridization of oligonucleotides at either the upstream region of the variation or hybridization on the top of the sequence carrying the variation. Hybridization at the upstream region requires a DNA polymerase for reading through the variable region such as DNA sequencing techniques (1–4), while hybridization on the variable region may require ligase or nucleases (5–9). DNA sequencing has been the most accurate and informative technique despite its limited use for diagnostics. Limitation has been due mainly to the complexity of using these techniques. A user-friendly version of DNA sequencing, called Pyrosequencing (2), was described recently. Pyrosequencing is a real-time and non-electrophoretic technique, which dispenses with the need for labeled nucleotides and labeled primers. This technique employs enzymatic reactions, catalyzed by ATP sulfurylase and luciferase, to monitor the inorganic pyrophosphate released during nucleotide incorporation. Unreacted nucleotides are degraded by the enzyme apyrase, allowing the iterative addition of nucleotides (2). This technique generates quantitative signals and has already been used for different biological applications (10,11). Here, we report on the development of a multiplex Pyrosequencing assay, which was applied for genotyping of single nucleotide polymorphisms (SNP) and hepatitis C virus (HCV). This accurate method has important implications in typing and has the potential to be used for diagnosis in clinical settings.
MATERIALS AND METHODS
Samples and oligonucleotides
Genomic DNA from Drosophila melanogaster and 77 sera from HCV-positive veterans were obtained from Stanford Veteran Hospital. The oligonucleotides Dm2494F (5′-TTGCTACCGGAATGGATG) and BDm2494R (5′-B-GAACCAGTCATTAATTATGGGC), BGP25F (5′-GCCTTGGTGATTCACATTTGAATG), BGP23F (5′-CAATATCTTCAGGACAACCAACC), GP23R (5′-CTTTAACAGCCTGCACTATGGTG), GP25R (5′-CTGATTGATTGAATGCGGCGATG) were used for PCR amplification from 10 ng of genomic DNA. Sequencing primers SNP-1 (5′-AATACGAAATATTATATTTTATCTATCGGG), SNP-2 (5′-AAAGTGGATTTGCAACTAATATCTAATGCT), GP25SNP167 (5′-CTAGACGTTGAAAT) and GP23SNP69 (5′-GATTCCCATCTCAAA) were used for SNP analyses. The oligonucleotides HCV-PCR-OUTF (5′-CCCTGTGAGGAACTWCTGTCTTCACGC), HCV-PCR-OUTR (5′-GCTCATGRTGCACG-GTCTACGAGACCT), HCV-PCR-INF (5′-TCTAGCCATGGCGTTAGTAYGAGTGT), BHCV-PCR-INR (Biotin-5′-CACTCGCAAGCACCCTATCAGGCAGT), HCV-SEQF1 (5′-GGAACCGGTGAGTACACCGGAAT), HCV-SEQF2 (5′-GACYGGGTCCTTTCTTGGA) and HCV-SEQF3 (5′-ATTTGGGCGTGCCCCCGC) were used for HCV typing. All oligonucleotides were synthesized and HPLC purified by MWG Biotech (High Points, NC).
Nucleic acid extraction and PCR
Genomic DNA from D.melanogaster was extracted according to the standard procedures. For SNP genotyping, 10 ng of total genomic DNA was used for PCR amplification using Dm2494F and BDm2494R. For HCV genotyping, RNA was extracted from 100 µl of patient sera using Ambion’s Totally RNA isolation kit (www.ambion.com). cDNA was synthesized using the kit SuperScript™ pre-amplification system from Life Technologies (www.lifetech.com). First strand cDNA synthesis employed an RNA/primer mixture containing 5 µl RNA and 1 µl 0.5 µg/µl random primer, which was incubated at 70°C for 10 min and then placed on ice for at least 1 min. A reaction mixture containing 2 µl 10× PCR buffer (200 mM Tris–HCl pH 8.4, 500 mM KCl), 2 µl 25 mM MgCl2, 10 mM dNTP mix and 0.1 M dithiothreitol, was added to each RNA/primer mixture, mixed gently, collected by brief centrifugation and then incubated at 42°C for 5 min. After addition of 200 U of SuperScript II RT, the tube was incubated at 40°C for 50 min. The reaction was terminated at 70°C for 15 min and then chilled on ice. The reactions were collected by brief centrifugation. One microliter of RNase H was added to each tube and incubated for 20 min at 37°C. Outer PCR was performed on 1 µl of cDNA using HCV-PCR-OUTF and HCV-PCR-OUTR to amplify a 270-nt-long fragment from 5′ untranslated region (5′ UTR) (12). The outer PCR was diluted by 500 000 times and 1 µl of that was used as a template for inner PCR using primers HCV-PCR-INF and HCV-PCR-INR.
Template preparation for Pyrosequencing
The biotinylated PCR products were immobilized onto streptavidin-coated super paramagnetic Dynabeads™ M280-streptavidin (Dynal A.S., Oslo, Norway). Single-stranded DNA template was obtained by discarding the supernatant after incubation of the immobilized PCR product in 0.10 M NaOH for 3 min. For multiplex Pyrosequencing, 5 pmol of each of the sequencing primer for SNP analysis or HCV genotyping were added to the immobilized strand(s) and hybridization was performed as described previously (13).
Multiplex Pyrosequencing
Primed DNA templates were placed in a microtiter plate and Pyrosequencing™ substrate and enzymes were dispensed using the fully automated microtiter plate-based PSQ™ Pyrosequencing machine (www.pyrosequencing.com). For multiplex SNP genotyping two sequencing primers were hybridized either to a single or two DNA template(s). For HCV genotyping, three primers were hybridized to a single template. The progress of sequencing was followed in real time using Pyrosequencing SNP software (www.pyrosequencing.com). Randomly, 14 of the HCV PCR products and the template containing SNPs used in the multiplex were analyzed by standard Pyrosequencing.
RESULTS
Principle of the multiplex Pyrosequencing
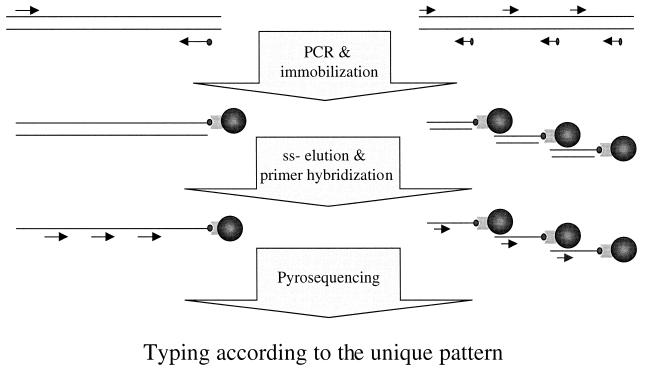
Multiplex Pyrosequencing enables simultaneous analyses of multiple target DNA. PCR was employed to amplify target DNA template(s) containing more than one variable region on genomic DNA. Biotinylated-target DNA template(s) was captured onto streptavidin-coated beads. Subsequently, single-stranded DNA was eluted and multiple primers, complementary to the DNA sequences before the variations, were hybridized to the template and Pyrosequencing was performed for sequence determination (Fig. 1). The obtained pyrogram resulted a unique pattern in which the signal intensity determined the number of incorporated nucleotides. As the reference sequences were known, the sequence of variable region could be determined.
Figure 1.
Schematic presentation of the principle of multiplex Pyrosequencing on a single template (left) and on multiple templates (right). PCR was performed with one of the primers biotinylated. Subsequent to immobilization of the product on streptavidin-coated magnetic beads and single-stranded elution, several sequencing primers were hybridized to the template(s) and Pyrosequencing was performed. A unique pattern was obtained, determining the exact sequence of different nucleotide positions.
Multiplex Pyrosequencing for SNP genotyping
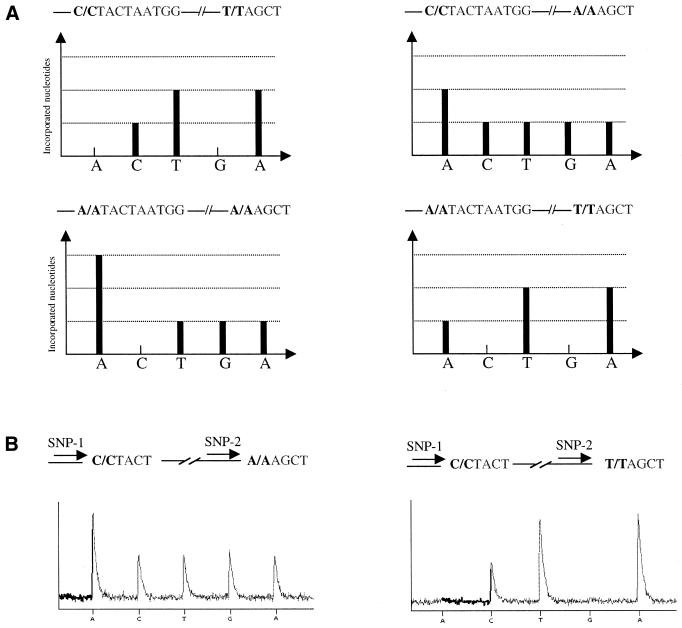
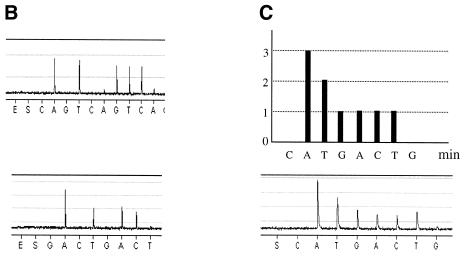
As described earlier (2), Pyrosequencing takes advantage of enzymatic reactions culminating in the production of photons to determine the nucleotide sequence of DNA. The intensity of signals obtained in a pyrogram correlate with the number of nucleotides incorporated by DNA polymerase. For multiplex SNP Pyrosequencing, one single template containing two SNPs (Fig. 2) or two templates each containing a polymorphism (Fig. 3) were PCR amplified. Two sequencing primers were simultaneously hybridized to the obtained PCR products and Pyrosequencing was performed. As the genomic DNA was from an inbred organism, four different patterns could be expected (Fig. 2A). The pyrogram obtained from SNP analyses demonstrates either the C:C and A:A or C:C and T:T sequences (Fig. 2B). Multiplex Pyrosequencing on multiple templates was performed on PCR products generated by multiplex PCR. Figure 3A demonstrates gel electrophoresis analysis on a duplex PCR. Standard Pyrosequencing using just one primer (Fig. 3B) and multiplex Pyrosequencing using both sequencing primers were performed (Fig. 3C).
Figure 2.
Schematic demonstration of the expected patterns obtained from multiplex analyses of two SNPs residing on a DNA template (A). Raw data in the pyrogram demonstrate two of the four possible expected patterns (B). The correct sequences are shown above each pyrogram and the order of dispensation is demonstrated below the pyrograms.
Figure 3.
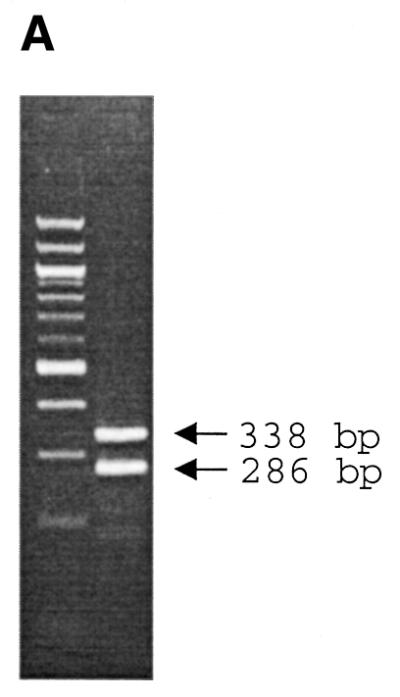
Pyrogram demonstrating raw data obtained from multiplex Pyrosequencing using multiple templates obtained from multiplex PCR. Gel electrophoresis separation resolves two templates obtained from multiplex PCR (A). The left-hand lane represents the DNA marker, which is a 100 bp DNA ladder. Raw pyrogram data from each DNA template using a single primer are demonstrated in (B) and using multiple primers are shown in (C). Theoretical pattern expected from each nucleotide addition is demonstrated in the top panel of (C). The order of nucleotide dispensation is demonstrated below the pyrograms.
Multiplex Pyrosequencing for HCV genotyping
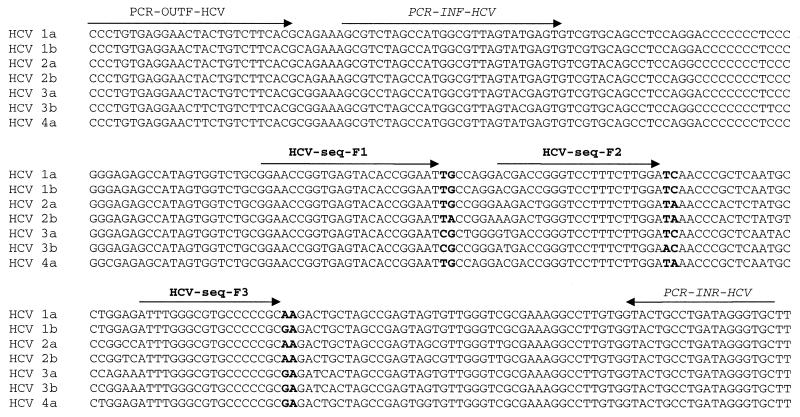
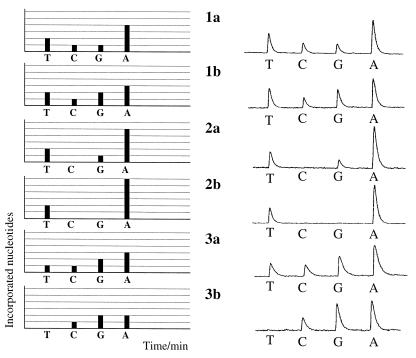
Alignment of the obtained data from standard Pyrosequencing showed that by just sequencing 6 nt residing in three regions of a 230-nt-long fragment (as highlighted in Fig. 4) it is possible to subtype different HCV in the population analyzed (Fig. 4). Three sequencing primers were hybridized to the amplified 230-nt-long fragment from HCV genome and Pyrosequencing was performed. A fingerprint was obtained in which the height of each peak reflects the sum of the number of incorporated nucleotides at the 3′ termini of the three primers. Figure 5, left, demonstrates the expected pattern of nucleotide incorporation during multiplex Pyrosequencing for six of the major subtypes of HCV and the raw data obtained from pyrogram is presented in Figure 5, right. Of the 77 American sera analyzed by Pyrosequencing, 35% had the 2T:1C:1G:4A pattern of incorporation, 29% had the 2T:1C:2G:3A pattern of incorporation, 21% had the 2T:0C:1G:5A pattern of incorporation, 4% had the 2T:0C:0G:6A pattern of incorporation, 1% had the 1T:1C:2G:3A pattern of incorporation and 10% had the 0T:1C:2G:2A pattern of incorporation. The data obtained were consistent with the data obtained from standard Pyrosequencing.
Figure 4.
Sequence alignment of seven different 5′ UTRs of HCV sequences. PCR-INR-HCV and PCR-INF-HCV demonstrate the hybridization positions of nested PCR primers. HCV-seq-F1, HCV-seq-F2 and HCV-seq-F3 show positions of the sequencing primers. Pyrosequencing of highlighted nucleotides result in a specific pattern of nucleotide incorporation.
Figure 5.
Theoretical pattern (left) and pyrogram (right) obtained from six different subtypes using Multiplex Pyrosequencing. The height of the y-axis is determined by the number of incorporated nucleotides and the x-axis is determined by time. Nucleotides were added according to the specified order at 1 min intervals. The added nucleotide is indicated below the pyrogram.
DISCUSSION
In this report, we describe the development of a multiplex Pyrosequencing assay, which is applied for multiple SNP analyses and sequence determination of variable regions of different HCV genome. There are several advantages with genotyping by Pyrosequencing compared with the available methods: (i) Pyrosequencing determines the exact sequence thereby providing the same accuracy as conventional sequencing methods; (ii) the technique dispenses with the need for labeled nucleotides, labeled primers and electrophoresis; and (iii) large number of samples can be analyzed in a short time. This technique has now been multiplexed which enables multiple analyses of one or several target DNA thereby decreasing the cost for PCR, template preparation and genotyping. Furthermore, the analysis time can be shortened allowing processing of a large number of samples. In addition, several peaks are obtained in a pyrogram of multiplex Pyrosequencing and their heights can be compared in order to confirm the number of incorporated nucleotides, thereby making the typing very accurate.
Here, we have focused on showing the feasibility of multiplexing in the context of Pyrosequencing. We showed that the pyrogram from multiplex data were consistent with the sequence of those templates as ascertained by standard Pyrosequencing and conventional DNA sequencing. For multiplex SNP analysis, the number of multiplexing can be increased when inbred material is used. For example, if three SNPs are analyzed from heterozygous DNA template, 27 different patterns can be expected while the number is decreased to eight possible patterns in inbred materials. Software is under development enabling a higher level of multiplexing and automatic scoring of different genotypes (www.pyrosequencing.com). For HCV genotyping, we used a population that was earlier analyzed by standard Pyrosequencing and we expected six different subtypes. However, for the samples not sequenced previously, the multiplex pyrogram of each was in agreement with several HCV subtypes. For example, the 2T:1C:1G:4A pattern is compatible with the sequence of HCV subtypes 1a, 1b, 1e, 8b and 9a. Nevertheless, in veteran people, 1a is the major subtype showing this pattern (14). For enhancing the discrimination power of Pyrosequencing for HCV subtyping one could increase the number of sequencing primers or the number of nucleotide dispensation in order to obtain a unique fingerprint. A particular 7-nt dispensation is expected to produce 27 different patterns on 62 5′ UTR HCV sequences belonging of 42 different subtypes. Although eight of these patterns would be consistent with sequences of more than one subtype, each of the remaining 19 would be consistent with only one or more sequence belonging to the same subtype (data not shown). The simpler 4-nt dispensation protocol may be adequate for typing organisms when genomes show less variability than HCV.
One inherent challenge with standard Pyrosequencing is proportionality of the signal intensity in a pyrogram when >5 nt in a row are incorporated. This problem is due mainly to the longer time needed for polymerization by DNA polymerase and rapid degradation of nucleotides by apyrase. However, in multiplex Pyrosequencing, more proportional signals are expected in a pyrogram because of the possibility of multiple independent nucleotide incorporations from different sequencing primers by different DNA polymerases.
Two fully automated machines using this system are now available that analyze 96 or 384 samples simultaneously, which enable analyses of 5000 samples per 8 h or 50 000 per day, respectively. Using the latter machine the cost for each analysis including PCR, template preparation and Pyrosequencing can be reduced to 20 cents per DNA fragment and the number of multiplexing further decreases the cost proportionally.
Acknowledgments
ACKNOWLEDGEMENTS
The authors from Stanford Genome Technology Center are supported from NIH grant 2P0I HG00205. E.E. is supported by a grant from the Iranian Blood Transfusion Center.
REFERENCES
- 1.Sanger F., Nicklen,S. and Coulson,A.R. (1977) DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA, 74, 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ronaghi M., Uhlen,M. and Nyren,P. (1998) A sequencing method based on real-time pyrophosphate. Science, 281, 363–365. [DOI] [PubMed] [Google Scholar]
- 3.Fu D.J., Broude,N.E., Koster,H., Smith,C.L. and Cantor,C.R. (1995) A DNA sequencing strategy that requires only five bases of known terminal sequence for priming. Proc. Natl Acad. Sci. USA, 24, 10162–10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Syvanen A.C., Sajantila,A. and Lukka,M. (1993) Identification of individuals by analysis of biallelic DNA markers, using PCR and solid-phase minisequencing. Am. J. Hum. Genet., 52, 46–59. [PMC free article] [PubMed] [Google Scholar]
- 5.Tyagi S., Bratu,D.P. and Kramer,F.R. (1998) Multicolor molecular beacons for allele discrimination. Nat. Biotechnol., 16, 49–53. [DOI] [PubMed] [Google Scholar]
- 6.Samiotaki K., Kwiatkowski,M., Patrik,J. and Landegren,U. (1994) Dual-color detection of DNA sequence variants by ligase-mediated analysis. Genomics, 20, 238–242. [DOI] [PubMed] [Google Scholar]
- 7.Day I.N.M. and Humpheries,S.E. (1994) Electrophoresis for genotyping: microtiter array diagonal gel electrophoresis on horizontal polyacrylamide gels, hydrolink, or agarose. Anal. Biochem., 29, 152–162. [DOI] [PubMed] [Google Scholar]
- 8.Nilsson M., Malmgren,H., Samiotaki,M., Kwiatkowski,M., Chowdhary,B.P. and Landegren,U. (1994) Padlock probes: circularizing oligonucleotides for localized DNA detection. Science, 265, 2085–2088. [DOI] [PubMed] [Google Scholar]
- 9.Marshall D.J., Heisler,L.M., Lyamichev,V., Murvine,C., Olive,D.M., Ehrlich,G.D., Neri,B.P. and de Arruda,M. (1997) Determination of hepatitis C virus genotypes in the United States by cleavase fragment length polymorphism analysis. J. Clin. Microbiol., 35, 3156–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ronaghi M. (2001) Pyrosequencing sheds light on DNA sequencing. Genome Res., 11, 3–11. [DOI] [PubMed] [Google Scholar]
- 11.Nordstrom T., Gharizadeh,B., Pourmand,N., Nyren,P. and Ronaghi,M. (2001) Method enabling fast partial sequencing of cDNA clones. Anal. Biochem., 15, 266–271. [DOI] [PubMed] [Google Scholar]
- 12.Han J.H., Shyamala,V., Richman,K.H., Brauer,M.J., Irvine,B., Urdea,M.S., Tekamp-Olson,P., Kuo,G., Choo,Q.L. and Houghton,M. (1991) Characterization of the terminal regions of hepatitis C viral RNA: identification of conserved sequences in the 5′ untranslated region and poly(A) tails at the 3′ end. Proc. Natl Acad. Sci. USA, 88, 1711–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ronaghi M., Nygren,M., Lundeberg,J. and Nyren,P. (1999) Analyses of secondary structures in DNA by pyrosequencing. Anal. Biochem., 267, 65–71. [DOI] [PubMed] [Google Scholar]
- 14.Cheung R.C. (2000) Epidemiology of hepatitis C virus infection in American veterans. Am. J. Gastroenterol., 95, 740–747. [DOI] [PubMed] [Google Scholar]