Abstract
To meet the new challenge of generating the draft sequences of mammalian genomes, we describe the development of a novel high throughput 96-well method for the purification of plasmid DNA template using size-fractionated, acid-washed glass beads. Unlike most previously described approaches, the current method has been designed and optimized to facilitate the direct binding of alcohol-precipitated plasmid DNA to glass beads from alkaline lysed bacterial cells containing the insoluble cellular aggregate material. Eliminating the tedious step of separating the cleared lysate significantly simplifies the method and improves throughput and reliability. During a 4 month period of 96-capillary DNA sequencing of the Rattus norvegicus genome at the Baylor College of Medicine Human Genome Sequencing Center, the average success rate and read length derived from >1 800 000 plasmid DNA templates prepared by the direct lysis/glass bead method were 82.2% and 516 bases, respectively. The cost of this direct lysis/glass bead method in September 2001 was ∼10 cents per clone, which is a significant cost saving in high throughput genomic sequencing efforts.
INTRODUCTION
Rapid and efficient methods for nucleic acid purifications are essential for many molecular biology applications. Several research groups have described methods for the purification of plasmid DNA that produce templates of sufficient quality for DNA sequence applications (1–12). The introduction of capillary electrophoresis DNA sequencers, however, has put further demands on template purity when compared to automated slab gel systems. For high throughput DNA sequencing of whole mammalian genomes, purification methods must also be designed to meet the challenges of platform flexibility, increasing scalability and decreasing costs, while maintaining high quality. At the Baylor College of Medicine Human Genome Sequencing Center (BCM-HGSC), the effort required to generate a draft sequence of the Rattus norvegicus genome in a 2 year period (13) required a departure from the M13 DNA sequencing approach and development of a higher throughput, paired end plasmid DNA sequence strategy.
The majority of plasmid preparation protocols rely on the ‘alkaline lysis’ method, which uses a narrow pH range (12.0–12.5) to selectively denature linear, but not covalently closed circular DNA (1). The alkaline lysate is rapidly neutralized to form an insoluble aggregate consisting of genomic DNA, protein–detergent complexes and high molecular weight RNA. In the original method the cleared lysate is then carefully recovered by centrifugation and ethanol precipitated for isolation of crude plasmid DNA. Alternative methods, such as the boiling method (2) and microwave preparation (11), have been used to isolate plasmid DNA; however, the alkaline lysis method represents the most robust and stable platform for further development of high throughput purification methods.
To improve the purity of plasmid DNA, several innovations have been coupled to the step of processing the cleared lysate. These methods include agarose gel electrophoresis extraction, column chromatography, cesium chloride gradient centrifugation, and selective adsorption using solid phase supports. Of these strategies, the latter has the advantages of simplicity and parallel processing via multiplexing of samples. The required centrifugation step in preparing the cleared lysates, however, significantly hinders automation by robotics. A filtration or clearing plate (9) or selective binding conditions to carboxylated magnetic particles (12) have been used to remove the insoluble aggregate material from the soluble plasmid DNA; however, these clearing steps have the disadvantage of increasing costs in materials and efforts, which decrease the cycle time of the method.
Glass particles, powder and beads have proven useful for purifying nucleic acids. An early technique to isolate DNA from agarose gels involved the use of the chaotropic salts sodium iodide (14) and sodium perchlorate (15,16) to facilitate binding of the DNA to common silicate glass, flint glass and borosilicate glass (glass fiber filter). Plasmid DNA initially isolated as a cleared lysate has also been purified by binding to glass fiber filter powder in the presence of sodium perchlorate (3). Recently, Engelstein et al. described the purification of plasmid DNA under high sodium chloride and 10% polyethylene glycol (PEG) conditions with silica particles (9). All of these methods have the common element of high salt solutions, in which the adsorption of plasmid DNA onto the glass substrate occurs most likely by a mechanism similar to adsorption chromatography.
In the method presented in this article we utilize a direct lysis protocol, in which the lysate containing the insoluble aggregate is directly added to a 96-well filter plate containing binding solution and glass beads. Unlike the high salt methods, we have found that alcohol precipitates of plasmid DNA bind efficiently to glass substrates and can occur in the presence of the insoluble cellular aggregate lysate. The methods developed here, which allow compatibility of the direct lysis solution with plasmid DNA binding to glass beads, lends itself to a simplified high throughput method and has the flexibility for complete automation on standard robotic platforms. The cost of each plasmid DNA prepared by the glass bead method is currently ∼10 cents, which affords significant advantages in high throughput sequencing of mammalian genomes. Considering genome center discounts, this represents a 4-fold saving in reagents and materials and an ∼10-fold saving at market prices.
MATERIALS AND METHODS
Reagents and supplies
The reagents necessary to perform the glass bead preparation method are readily available as standard components, such as glucose, Tris, EDTA, NaOH, IGEPAL CA-630 (formerly known as Nonidet P-40), potassium acetate, glacial acetic acid and 2-propanol (EM Science, Gibbstown, NJ) or as pre-made solutions I, II and III (catalog nos VW8869-1, VW8870-1 and VW8871-1, respectively; EM Science). Pure (100%) ethanol was purchased from Aaper (Shelbyville, KY). Solutions I–III are composed of, respectively: 51 mM glucose, 26 mM Tris–HCl, pH 8.0, and 51 mM EDTA; 0.9 N NaOH and 0.125% IGEPAL CA-630; and 387 mM potassium acetate, 15% glacial acetic acid. RNase A (Life Technologies, Rockville, MD) was added fresh to solution I (final concentration 0.35 mg/ml) prior to the lysis step. Glass beads were purchased as 212–150 µm size-fractionated, acid-washed glass beads from Sigma (St Louis, MO) or EM Science or as <106 µm size-fractionated, acid-washed glass beads. The 96-well filter used to retain the glass beads during the plasmid binding and washing steps was a Millipore 1.2 µm hydrophilic Durapore PVDF plate (Bedford, MA). For comparison, a 96-well Millipore 1.2 µm glass fiber type C and ground Whatman GF/C filter paper were used for 2-propanol binding experiments. Ground GF/C powder was prepared by grinding GF/C paper using a mortar and pestle, size fractionating the powder by three sedimentations and drying at 65°C for 2 h. Sedimentation was performed by resuspending the ground GF/C in 2-propanol, allowing the glass fiber to settle for 5 min and decanting the liquid suspension. To simplify the addition of a predetermined amount of glass beads to the Millipore filter, a predrilled 96-well plexiglass plate was constructed to reproducibly deliver ∼80 mg of 212–150 µm size-fractionated glass beads to the filter in a high throughput manner.
DNA purification method
Whole genome shotgun or BAC clone shotgun libraries from a Brown Norway strain of R.norvegicus were constructed using the ‘double adapter’ library protocol (17) and transformed into Epicurian Coli XL-10 Gold Ultracompetent cells (Stratagene, La Jolla, CA). Individual colonies were picked from square 24 × 24 cm plates either manually or using a Q-PIX robot (Genetix, Hampshire, UK) to inoculate 400 µl of Terrific Broth cultures containing carbenicillin (final concentration 60 µg/ml) in 96-well Beckman blocks. The cultures were grown for 17 h at 37°C with shaking at 300 r.p.m. The cells were recovered by centrifugation at 3000 r.p.m. for 15 min, followed by decanting the media. Blocks were inverted on absorbent towels, sealed with aluminum foil and stored at –20°C for further use.
To purify plasmid DNA, frozen cells were thawed for 30 min and then processed by adding 45 µl of solution I followed by shaking using an orbital shaker (Lab Line Instruments, Melrose Park, IL) at full speed for 10 min to facilitate cell lysis. Here, the cells appeared cloudy and had a somewhat ‘fuzzy’ appearance. The additions of solutions I–III were performed using a Hydra96 pipettor (Robbins Scientific, Sunnyvale, CA). An aliquot of 45 µl of solution II was added to the lysed cells and the mixture shaken for 1 min and allowed to stand for 2 min. Finally, 45 µl of solution III was added to neutralize the reaction, and the lysed cells were shaken for 2 min.
Prior to transfer of the entire lysate solution to a 96-well filter, 205 µl of 2-propanol was dispensed into the glass bead filter plate using a Q-fill2 pipettor (Genetix). It should be noted that transfer of the insoluble cell aggregate, which has a similar appearance to that of a standard alkaline lysate, to the glass bead filter does not significantly affect the flow or overall quality of the plasmid preparation. After a static 5 min wait (i.e. no mixing) to facilitate binding of the 2-propanol-precipitated plasmid DNA to the glass beads, vacuum pressure was applied using a Millipore vacuum manifold to remove the cell lysate solution. The alcohol precipitate appears to selectively bind plasmid DNA over residual genomic DNA or high molecular weight RNA by agarose gel electrophoresis. The glass bead filters were then washed eight times with 200 µl of 80% aqueous ethanol by vacuum filtration. To semi-automate the washing step, a Q-fill2 pipettor was modified by mounting a Millipore vacuum manifold to the tray holder, which under continuous vacuum allowed the wells to dry before addition of the next wash solution. In this semi-automated manner four 96-well culture boxes can be processed as a batch from cell pellets to 80% ethanol washes in 30–35 min. Filter plates were then dried using a SpeedVac under heat, resuspended in 60 µl of 10 mM Tris–HCl, pH 8.0, 0.2 mM EDTA containing 0.16 mg/ml RNase A, allowed to stand for 1 min and then eluted by vacuum filtration into a 96-well collection plate. RNase is required to degrade low molecular weight RNA, which is not selectively aggregated after neutralization but binds to the glass beads following 2-propanol precipitation. In our experience, recovery of the plasmid elution volume is nearly quantitative by vacuum filtration. DNA templates stored for up to 14 days at 4°C produced similar high quality results to those templates sequenced immediately (x = 3840 reactions).
DNA sequencing and analysis
DNA templates were sequenced with BigDye Terminators v.3 (Applied Biosystems, Foster City, CA) using two miniTrak robots (CCS Packard, Torrance, CA) into 384-well cycle plates. The ‘1/8 reaction’ master mixes contained 1.0 µl BigDye v.3 diluted in 1.5 µl of 5× buffer, 3.5 µl of HPLC water and 1.0 µl of 4.0 pmol of either sequencing primer (5′-GTAAAACGACGGCCAGT) or reverse sequencing primer (5′-CAGGGAAACAGCTATAC). Following addition of 7.0 µl of the master mix, 4.0 µl of plasmid DNA from two 96-well plasmid collection plates were aliquoted in duplicate to pair both forward and reverse sequencing reactions into one 384-well plate. Sequencing plates were sealed using a PlateLoc plate sealer (Velocity11, Palo Alto, CA). Cycle sequencing was performed using either PTC-200 or PTC-225 thermal cyclers (MJ Research, Watertown, MA) using thermocycling conditions of 96°C for 30 s, 50°C for 20 s and 60°C for 4 min for 35 cycles. Sequencing reactions were then purified by ethanol precipitation (26 µl of 88.5% ethanol, 115 mM sodium acetate, pH 5.2), resuspended in 20 µl of HPLC water and analyzed using an ABI 3700 DNA sequencer.
RESULTS AND DISCUSSION
Initially, a number of parameters were investigated for the development of the glass bead preparation method, including bacterial growth conditions; different lysis reagents, solutions and detergents, washing conditions; a variety of glass materials; and different 96-well filter formats (data not shown). We determined three critical parameters that had a significant effect on performance of the direct lysis–glass bead preparation method and the recovery and quality of plasmid DNA. First, we found that an initial osmotic lysis step using glucose in solution I dramatically improved the flow of the direct cellular lysate through the filter plate. Using this reagent, the vacuum filtration behaved in a more consistent manner, which improved the quality and reproducibility of the primary sequencing data. Secondly, our experiments showed that irrespective of the type of glass material used, 2-propanol precipitation resulted in a higher recovery of plasmid DNA from the direct lysate compared to NaCl/PEG binding (see below). Compared to ethanol precipitation, we found that 2-propanol precipitation gave slightly higher plasmid yields and a slightly better flow performance (data not shown). Finally, we observed that the flow rate of the alcohol lysate through the filter plate was inversely proportional to the yield of plasmid binding to the glass beads. A flow rate of 0.5–1.0 ml/min typically gave optimal results and flow rates >3.0 ml/min gave lower yields of plasmid DNA. To optimize this parameter in a production setting, a number of 96-well filter plates were examined with respect to flow rate resistance. We found that the Millipore 1.2 µm hydrophilic Durapore PVDF plate gave sufficient flow resistance to provide the optimal flow rate at approximately –10 inches Hg vacuum pressure (typical house vacuum).
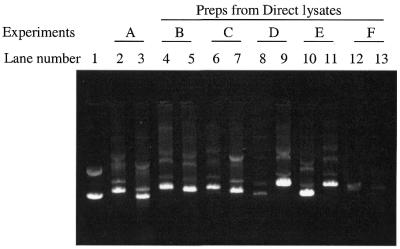
The binding efficiency of the direct lysis protocol was further characterized by examining different glass materials, including different sized acid-washed glass beads (common silicate glass) and intact and ground glass fiber filters type C (GF/C) (borosilicate glass) (Fig. 1). To normalize the different glass materials, the amounts of glass beads and ground GF/C were measured using a fixed volume plexiglass dispenser (see Materials and Methods). We found that the direct lysis method coupled with 2-propanol precipitation gave high quality plasmid DNA in good yield using all the glass supports examined in Figure 1 (lanes 4–11). Typically, significantly lower yields of plasmid DNA were recovered when binding of the direct lysate was facilitated in a 1.25 M NaCl, 10% PEG solution (final concentration) (lanes 12 and 13). Performance of the direct lysis/2-propanol preparation method using the different glass materials, however, differed with respect to performance and recovery of reproducible quantities of plasmid DNA. In general, the 1.2 µm GF/C filters gave greater flow resistance during filtering of the direct lysate and the subsequent washing steps. Furthermore, the ground GF/C material gave variable recovery of plasmid DNA volumes eluted (lanes 8 and 9). For routine usage we found that 212–150 µm size-fractionated, acid-washed glass beads gave the most reliable results regarding plasmid DNA yield and protocol performance. The average plasmid yield was 21 ± 12 µg/ml of culture (x = 192).
Figure 1.
Comparison of different glass supports and precipitation solutions for rat whole genome shotgun clones: lane 1, 500 ng pGEM plasmid template control; lanes 2 and 3, experiment A, ethanol precipitation of standard cleared alkaline lysates; lanes 4–11, 2-propanol precipitation: lanes 4 and 5, experiment B, using 212–150 µm acid-washed glass beads; lanes 6 and 7, experiment C, using <106 µm size-fractionated, acid-washed glass beads; lanes 8 and 9, experiment D, using a ground glass fiber filter (Whatman GF/C); lanes 10 and 11, experiment E, using a Millipore 1.2 µm glass fiber type C plate; lanes 12–13, experiment F, precipitation by 1.25 M NaCl, 10% PEG solution using 212–150 µm acid-washed glass beads. For each experiment 8.3% of the eluted product (5 µl) was loaded for the different glass support and precipitation solution variables prepared from 0.4 ml of bacterial cultures.
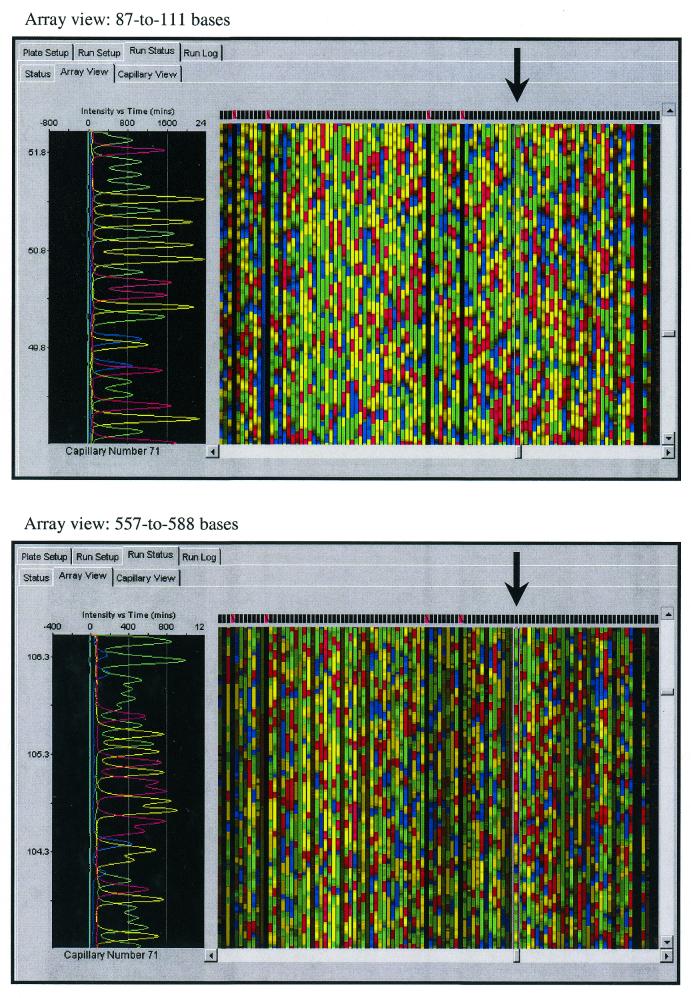
The quality of the glass bead template preparation method was further characterized for DNA sequencing by examining the primary data and the Phred (18) base quality data. As shown in the array view from the ABI 96-capillary DNA sequencer (Fig. 2), the glass bead preparation method showed good signal and low background intensities in the primary DNA sequencing data generated from diluted ‘1/8’ BigDye v.3 terminator chemistry reactions. Both the early (87–111 bases) and late (557–588 bases) images show good baseline resolution of the fluorescent-labeled DNA fragments, which taken together are parameters that are characteristic of high quality plasmid templates. Since the introduction into the rat BCM-HGSC production–sequencing pipeline in May 2001, >1 800 000 plasmid DNA templates have been prepared using this direct lysis/glass bead preparation method (Table 1). The average success rate over this 4 month period was 82.2%, which was calculated as the percentage of sequencing reactions yielding a minimum of 100 Phred bases with quality values ≥20 (Q20). The average read lengths for successful sequencing reactions were calculated by counting the total number of Q20 bases and dividing by the number of sequencing reads. The average read length for the entire data set was 516 bases, with the latter 2 months yielding even better success rates and read lengths. Overall, these data show good quality and reproducibility of plasmid DNA templates prepared using the direct lysis method for high throughput capillary DNA sequencing of the rat genome.
Figure 2.
Snapshot images of ABI model 3700 capillary array views from two different chromatographic regions from a rat BAC clone skimmed project. The vertical chromatogram to the left is partially processed fluorescence data from capillary number 71 showing good baseline resolution of fluorescent bands near 600 bases. The signal reduction in the later region is the result of electrokinetic injection of sequencing reactions into capillaries.
Table 1. Summary statistics of the BCM-HGSC rat genome sequence effort using the glass bead preparation between 12 May and 15 September 2001.
| Month (2001) | No. of sequencing reactions loaded | Pass quality (%) | Read length (Q20 bases) |
|---|---|---|---|
| May | 738 256 | 80.2 | 503 |
| Junea | 455 441 | 79.8 | 491 |
| July | 684 342 | 82.4 | 506 |
| August | 1 306 970 | 83.5 | 534 |
| September | 444 236 | 84.2 | 529 |
| Total | 3 619 245 | 82.2 | 516 |
Reactions were loaded on 69 Applied Biosystems Model 3700 DNA sequencers, which included both WGS and BAC clone shotgun skim sequence reactions. The number of plasmid DNA templates required to generate the total number of paired end sequence reactions is half (∼1 810 000). DNA sequencing quality was determined using the base caller Phred (18) and the average success rates are reported as a percentage of sequencing reactions, which produced a minimum of 100 Q20 bases, and the average read lengths of successful reads are derived from the total number of Q20 bases divided by the number of sequencing reads.
aLower production numbers were the result of tropical storm Allison hitting Houston, Texas on 8 June 2001.
The development of a simplified and robust method for plasmid DNA templates described here meets the challenges for high throughput sequencing of whole mammalian genomes. Automation and increased well number per plasmid plate can achieve additional increases in throughput. Elimination of the cleared lysate step simplifies the method and streamlines the process from pelleted bacterial cells in growth boxes to highly purified plasmid DNA templates in a completely automated manner without interruption. Currently we are evaluating the performance of the direct lysis/glass bead preparation method on a commercial robotic platform at the BCM-HGSC. Moreover, our sequencing pipeline is currently configured to combine two 96-well template plates into one 384-well sequencing plate for paired end sequencing of plasmid DNA, which is potentially problematical in that it can mix different BAC clone projects. While a 384-well format is useful for DNA sequencing, we are investigating the feasibility of a 192-well template format. Besides the advantage of doubling throughput, the 192-well format has the additional benefit of simplifying the tracking of multiple BAC clone projects by single plate-to-plate transfers for paired end sequencing.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to a number of BCM-HGSC staff members, including George Weinstock, Erica Sodergren, Graham Scott, Paul Havlak, Lora Lewis, Stephanie Moore, Shereen Thomas, Carrie Burkett and Hailey Stanley, for their technical expertise in their respective roles in the BCM-HGSC DNA production–sequencing pipeline. This work was supported by grants 1-U54-HG02139 and 1-U54-HG02345 from the National Institutes of Health.
REFERENCES
- 1.Birnboim H.C. and Doly,J. (1979) A rapid alkaline extraction procedure for screening plasmid DNA. Nucleic Acids Res., 7, 1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holmes D.S. and Quigley,M. (1981) A rapid boiling method for the preparation of bacterial plasmid. Anal. Biochem., 114, 193–197. [DOI] [PubMed] [Google Scholar]
- 3.Marko M.A., Chipperfield,R. and Birnboim,H.C. (1982) A procedure for the large-scale isolation of highly purified plasmid DNA using alkaline extraction and binding to glass powder. Anal. Biochem., 121, 382–387. [DOI] [PubMed] [Google Scholar]
- 4.Zhou C., Yang,Y. and Jong,A.Y. (1990) Mini-prep in ten minutes. Biotechniques, 8, 172–173. [PubMed] [Google Scholar]
- 5.Huang G.M., Wang,K., Kuo,C., Paeper,B. and Hood,L. (1994) A high-throughput plasmid DNA preparation method. Anal. Biochem., 223, 35–38. [DOI] [PubMed] [Google Scholar]
- 6.Hawkins T.L., O’Connor-Morin,T., Roy,A. and Santillan,C. (1994) DNA purification and isolation using a solid-phase. Nucleic Acids Res., 22, 4543–4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng W., Schummer,M., Cirisano,F.D., Baldwin,R.L., Karlan,B.Y. and Hood,L. (1996) High throughput plasmid mini preparations facilitated by micro-mixing. Nucleic Acids Res., 24, 5045–5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itoh M., Carninci,P., Nagaoka,S., Sasaki,N., Okazaki,Y., Ohsumi,T., Muramatsu,M. and Hayashizaki,Y. (1997) Simple and rapid preparation of plasmid template by a filtration method using microtiter filter plates. Nucleic Acids Res., 25, 1315–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engelstein M., Aldredge,T.J., Madan,D., Smith,J.H., Mao,J.-I., Smith,D.S. and Rice,P.W. (1998) An efficient, automatable template preparation for high throughput sequencing. Microbial Comp. Genomics, 3, 237–241. [DOI] [PubMed] [Google Scholar]
- 10.Itoh M., Kitsunai,T., Akiyama,J., Shibata,K., Izawa,M., Kawai,J., Tomaru,Y., Carninci,P., Shibata,Y., Ozawa,Y., Muramatsu,M., Okazaki,Y. and Hayashizaki,Y. (1999) Automated filtration-based high-throughput plasmid preparation system. Genome Res., 9, 463–470. [PMC free article] [PubMed] [Google Scholar]
- 11.Marra M.A., Kucaba,T.A., Hillier,L.W and Waterston,R.H. (1999) High-throughput plasmid DNA purification for 3 cents per sample. Nucleic Acids Res., 27, e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elkin C.J., Richardson,P.M., Fourcade,H.M., Hammon,N.M., Pollard,M.J., Predki,P.F. Glavina,T. and Hawkins,T.L. (2001) High-throughput plasmid purification for capillary sequencing. Genome Res., 11, 1269–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marshall E. (2001) Rat genome spurs an unusual partnership. Science, 291, 1872. [DOI] [PubMed] [Google Scholar]
- 14.Vogelstein B. and Gillespie,D. (1979) Preparative and analytical purification of DNA from agarose. Proc. Natl Acad. Sci. USA, 76, 615–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang R., Lis,J. and Wu,R. (1979) Elution of DNA from agarose gels after electrophoresis. Methods Enzymol., 68, 176–182. [DOI] [PubMed] [Google Scholar]
- 16.Chen C.W. and Thomas,C.A.,Jr (1980) Recovery of DNA segments from agarose gels. Anal. Biochem., 101, 339–341. [DOI] [PubMed] [Google Scholar]
- 17.Andersson B., Wentland,M.A., Ricafrente,J.Y., Liu,W. and Gibbs,R.A. (1996) A “double adaptor” method for improved shotgun library construction. Anal. Biochem., 236, 107–113. [DOI] [PubMed] [Google Scholar]
- 18.Ewing B., Hillier,L., Wendl,M.C. and Green,P. (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res., 8, 175–185. [DOI] [PubMed] [Google Scholar]