Abstract
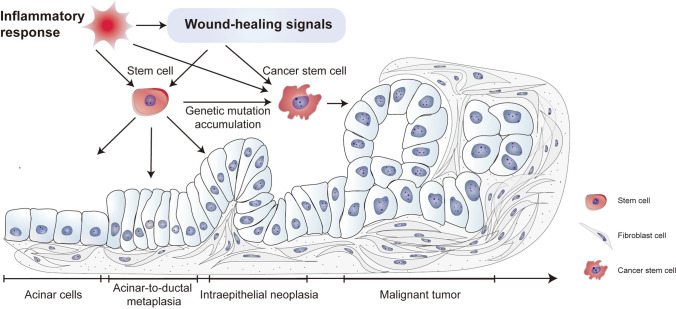
Cancer stem cells (CSCs) play an important role in cancer development. Based on advancements in CSC research, we propose a monophyletic model of cancer. This model is based on the idea that CSCs are stem cells with disordered differentiation whose original purpose was to repair damaged tissues. Inflammatory responses and damage repair signals are crucial for the creation and maintenance of CSCs. Normal quiescent stem cells are activated by environmental stimulation, such as an inflammatory response, and undergo cell division and differentiation. In the initial stage of cancer development, stem cell differentiation leads to heteromorphism due to the accumulation of gene mutations, resulting in the development of metaplasia or precancerosis. In the second stage, accumulated mutations induce poor differentiation and lead to cancer development. The monophyletic model illustrates the evolution, biological behavior, and hallmarks of CSCs, proposes a concise understanding of the origin of cancer, and may encourage a novel therapeutic approach.
Graphical Abstract
Keywords: Cancer stem cells, Inflammatory response, Tumor origin, Differentiation, Tissue repair signal
Introduction
According to the 2022 cancer statistics, cancer is the second leading cause of death after heart disease, comprising 21% of all deaths [1]. Even though genomics, transcriptomics, epigenomics, proteomics, and metabolomics have been employed to identify diverse hallmarks of cancer, targeted therapeutic strategies for cancer have achieved little efficacy [2, 3]. Understanding cancer remains a substantial challenge. For a long time, cancer was thought to be a “wound” that never heals, since many mesenchymal stem cells (MSCs) are recruited to the tumor microenvironment in a manner similar to the repair behaviors observed in damaged tissues [4, 5]. Current studies suggest that cancers arise from normal cells that accumulate oncogenic mutations. As only stem cells exhibit a lifelong capability for division and self-renewal, cancer stem cells (CSCs), which develop from stem cells, are considered the origin of cancer. CSCs exhibit striking similarities to normal stem cells in terms of differentiation, long-term proliferation (self-renewal), drug resistance, and anti-apoptosis [6, 7]. CSCs have been identified in tumors of the liver, pancreas, breast, brain, lung, and ovary. On the other hand, the classical CSCs model has faced a series of confusions and controversies regarding CSCs’ heterogenous origin, cell proportion, uncertain cell markers, and the genomic and phenotypic differences in different CSCs [8–10]. Experts attending The Year 2011 Working Conference on CSCs have suggested that a more accurate conceptual and practical framework of CSCs is important for their elimination [8]. The mechanisms underlying CSC tumorigenesis remain unclear [11]; there is therefore an urgent need for a new CSC theory to improve the understanding of CSC evolution, biology, identification, and to guide the development of effective therapeutic targets.
Recently, Liu reported the dualistic origin of human tumors, suggesting that tumors could arise from blastomeres generated from fertilized eggs, and stem cells generated from reprogrammed somatic cells [12]. This dualistic origin model attempted to explain the malignant characteristics of tumors, and its author claimed its superiority over other tumor origin models. However, this model ignored the dynamic changes that occur in the tumor histopathologic type. Cancers that undergo such changes include not only those that develop from benign tumors, but also those that convert to another histopathologic type after chemotherapy.
In this review, based on the classical CSC model, we propose a monophyletic origin of cancer. This monophyletic model suggests that CSCs are stem cells that lose control of differentiation. Stem cells, including totipotent stem cells, multipotent stem cells, and unipotent stem cells, accumulate crucial mutations that lead to disordered differentiation. Genetic alterations determine the degree of differentiation and whether a tumor is benign or malignant. Next, the model highlights that the primary cause of cancer progression is driven by CSCs. The division and differentiation of CSCs are dominated by tissue repair signals or inflammatory factors. Poorly differentiated cancer cells cannot repair damaged tissues. Thus, upstream signals continuously promote CSCs division and tumor proliferation. A high degree of tumor aggressiveness may correlate with a high-grade atypia in cancer cells. This theory applies to tumors in different tissues and of different pathological types.
CSCs Originate from Normal Stem Cells (NSCs)
The development of a fertilized egg into adulthood is a complex process. In the first few rounds of embryonic cell division, 2.4 mutations may occur for every generation of cell division [13]. Subsequently, chemical, radial, and inflammatory environmental factors cause DNA damage and tissue injuries [14, 15]. The risk of DNA damage is inherent during cell division and differentiation. Thus, genetic mutations may occur during tissue repair. Current research suggests that oncogenesis requires 3 to 7 crucial mutations to help cancer cells evade cell cycle checkpoints and apoptosis, and gain other malignant biological behaviors [5, 15, 16]. The mutational landscape indicates that normal tissues can usually carry “driver” mutations in cancer genes for decades, the burden of which increases with age [17]. However, only stem cells can self-renew and differentiate to repair damaged tissues throughout their lifespan. By accumulating plenty of oncogenic mutations, NSCs can transform into CSCs [6, 18]. Studies have shown that stem cells and progenitor cells in normal tissues are susceptible to carcinogenic transformation [19]. Tissues such as the intestinal epithelium, airway epithelium, liver, and pancreas are renowned for the strong regenerative ability of their resident stem cells and have a high incidence of cancer [1, 20, 21]. However, few cancers occur in peripheral nervous and myocardium tissue, which may be attributed to a lack of or weak stemness of resident stem cells in these tissues [21, 22].
The transcriptional profile of cancer cells has many similarities with that of stem cells. The activation of stem cell signals, such as WNT, musashi and NOTCH, strongly contributes to cancer heterogeneity, progression, metastasis and therapy resistance [23–25]. NSCs have been considered the most likely source of CSCs. The earliest evidence for this hypothesis was that only hematopoietic stem cells (HSCs) with specific gene mutations could transform into hematological cancers. These HSCs have been used in targeted therapy for the treatment of hematological cancer patients [26, 27]. On the other hand, cancer initiation in young people or during childhood may occur because of key mutations that were inherited or acquired during the embryonic period. For example, patients with congenital heart disease have genetic variants that may increase the risk of cancer [28].
CSCs have been reported in many solid tumors, including breast cancer, colon cancer, glioblastoma, and pancreatic cancer. They play crucial roles in tumorigenesis, tumor growth, chemoresistance, metastasis, and recurrence [29–31]. In monophyletic model, because CSCs develop from NSCs, there are many similarities between CSCs and NSCs in terms of their cellular characteristics, such as their self-renewal ability and apoptosis inhibition programs [32]. Studies have identified various CSC markers, used in the isolation of CSCs, or for diagnostic, prognostic, and therapeutic purposes [33, 34]. However, there is no single specific marker that can be used to distinguish CSCs from cancer cells. Researchers usually use several markers or a combination of surface and intracellular markers to confirm the identity of CSCs. Here, we summarize the most prominent markers of CSCs, resident stem cells, and human embryonic stem cells (hESCs) in several high-incidence cancers, based on the 2022 cancer statistics (Table 1) [33–78].
Table 1.
The markers of CSCs, NSCs, and hESC
| Tissues | CSCs markers | Resident normal stem cells markers | References | hESC markers (33) |
|---|---|---|---|---|
| Breast | ALDH1, CD29, CD44, CD133, CD201, EpCAM, PODXL1, SSEA3, SSEA4, TRA-1–60, TRA-1–81, Tspan8 | Axin2, CD10, CD1d, CD24, CD29, CD49f, CD49b, CD61, CD90, CD133, CK5, CK8, CK14, CK18, CK19, c-Kit, Lrp5, Lgr5, Lgr6, EpCAM, Procr, sca1, Myh11 | (33–37) |
ABCG2, CD133, CD90, CD326, CD24, CD49f, CD146, CD10, CD117, CD26, CD29, CD9, CD166, Cripto1, Notch2, PODXL1, SSEA1, SSEA3, SSEA4, TRA-1–60, TRA-1–81 |
| Prostate | ABCG2, ALDH1, CD29, CD44, CD49f, CD133, CD151, CD166, Sca1, TRA-1–60 | CD29, CD49f, CD133, Sca1, Trop2, Lgr5 | (38–42) | |
| Lung | ABCG2, ALDH1, CD44, CD56, CD133, CD166, Cripto1, Notch2, Notch3, PODXL1, SSEA1, Sox2 | ABCA3, ASCL1, α-SMA, CD49f, CD271, CXCR4, GRP, KRT5, KRT8, KRT17, KRT19, LAMP3, NCAD, PDPN, ROBO, SCGB1A1, SCGB3A2, SFTPC, SMMHC, TP63, TM4SF1 | (33, 43–45) | |
| Colorectal | ALDH1, CD26, CD29, CD44, CD133, CD166, Cripto1, EpCAM, Lgr5 | Lgr5 | (33, 46–49) | |
| Melanoma of skin | ABCB5, ALDH1, CD20, CD133, CD271, Nanog, Oct3, Oct4 | CD29, CD46, CD49f, CD71, DLL1, DSG3, FRMD4A, LRIG1, MCSP | (50–56) | |
| Renal | ALDH1, CD24, CD44, CD105, CD133, CXCR4, SSEA1 | CD13, CD29, CD44, CD73, CD90, CD105, CD133, CD146, CD224, CK7, c-kit, NR3C2, Pax2, Pax8, SSEA4 | (57–62) | |
| Leukemia | ALDH1, CD9, CD25, CD26, CD32, CD33, CD34, CD44, CD47, CD93, CD96, CD97, CD99, CD103, CD123, CLL1, IL1RAP, PODXL1, TIM3 | ABCG2, CD133, CD26, CD34, CD44, CD45RA, CD49f, CD90, c-Kit, PODXL1, Sca1 | (33, 63–70) | |
| Pancreas | ABCG2, ALDH1, CD133, CD9, CD24, CD44, CD49f, c-Met, CXCR1,2, CXCR4, EpCAM, GLRX3, Notch2, Nestin, Notch3, Oct3,4, PODXL1, Tspan8 | Unsure markers: Cytokeratins, PDX1, Glb1 | (33,71–78) |
It is worth noting that most CSC markers are derived from markers present in hESCs. More than half of the CSC markers in prostate, breast, lung, and colorectal cancers are consistent with hESC markers. Existing drugs targeting CSC markers in solid cancers have shown poor efficacy in clinical trials [6, 19]. Another study reported that there were no statistical differences in the expression of several stemness markers between pancreatic cancer tissues and normal pancreas tissues. The expression of CSC markers is not related to tumor grade or differentiation grade [79]. Many pathways involved in NSC self-renewal or stemness maintenance promote the proliferation and invasion of CSCs [7]. This indicates that CSCs transform from NSCs in various tissues. CSC markers may be heterogeneous in cancers of different tissues. However, the specific markers of most normal adult stem cells in different tissues have yet to be identified [80]. Most NSC markers are derived from hESC markers. Thus, the true identity of CSCs developed from NSCs requires further exploration.
CSCs Play a Leading Role in Cancer Development and Treatment Resistance
Chronic inflammation leads to the destruction of tissues and the activation of wound-healing signals. Prostaglandin E2 (PGE2) is a well-known signaling molecule whose release is induced by inflammation. PGE2 plays a vital role in stem cell differentiation, angiogenesis, and tissue regeneration in the heart, liver, intestine, kidney, and many other organs [81, 82]. PGE2 has been shown to have potent cancer-promoting effects [83, 84]. Notably, a study verified that PGE2 enhances the stemness of colon and gastric CSCs by activating many signaling pathways and promoting cancer metastasis [84, 85]. Other molecules, such as transforming growth factor beta (TGF-β) and yes-associated protein (YAP), have also been reported to regulate wound healing, cancer proliferation, and stemness maintenance [86–88]. YAP is essential for cancer initiation in many solid tumors and may be a potential therapeutic target [89]. Dysregulated Hippo pathway and YAP/TAZ–TEAD activity are reported to be related to tissue regeneration, wound healing, and CSC maintaining [90, 91]. Therefore, the pathways or molecules activated by inflammation during the wound healing process may trigger CSC proliferation and differentiation.
The original purpose of CSCs was to repair injured tissues. However, CSCs become various immature cell types, that is, cancer cells with high heterogeneity. Moreover, undifferentiated cells cannot restore the function of damaged tissues. Thus, the wound-healing signals promoting the proliferation and self-renewal of CSCs are continuously activated and result in a “wound that never heals” [92]. Alternatively, recruited MSCs differentiate into cancer-associated fibroblasts, which remodel the stroma and microenvironment, result in hypoxia, and accelerate angiogenesis and tumor metastasis [93].
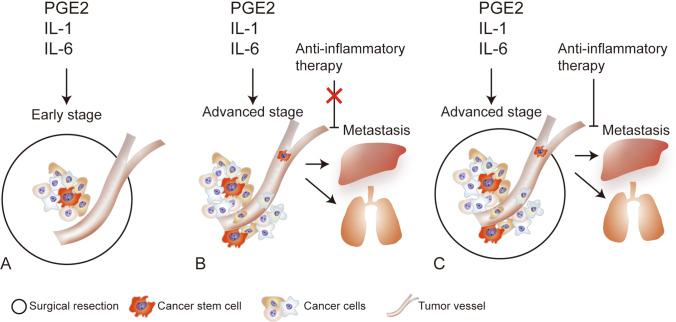
Since CSCs are derived from NSCs, undifferentiated cancer cells are homologous to normal cells to some extent. Even though immature cells are highly heterogeneous, it is difficult for the immune system to detect them. In recent years, next-generation sequencing, multi-omics, and single-cell sequencing technologies have developed rapidly, but very few specific molecular markers for CSCs have been identified [94, 95]. Immunotherapy, including neoantigen-based treatment and chimeric antigen receptor-T cell therapy, has shown limited effectiveness against solid tumors [96, 97]. Chemoresistance is a common characteristic of CSCs [98]. Currently, surgical resection remains an effective treatment for many solid tumors. It is believed that only surgery can completely remove a tumor, including CSCs, in the early stages of cancer (Fig. 1A) [99]. The 5-year relative survival rate associated with many tumors in the localized stage is higher than that associated with tumors in more advanced stages [1]. Many advanced-stage tumors have a poor prognosis, despite R0 resection [100–103]. Under some circumstances, surgical resection may increase the risk of cancer metastasis and progression, owing to systematic inflammation activation, ischemia/reperfusion injury, and immunosuppression [104]. Inflammatory factors, such as TGF-β, interleukin (IL)-1, IL-6, PGE2, and nuclear factor kappa B, have been confirmed to play an important role in cancer metastasis [105–107]. IL-6 has been shown to promote distant metastasis in cancers of the liver, lung, and breast, as well as in many other solid malignancies [108–110]. Tocilizumab, an inhibitor of the IL-6 receptor (IL-6R), suppresses the metastasis and progression of cancer when tested in cell lines and mouse models [111–114]. However, a large clinical randomized trial showed that IL-6R inhibitors are not effective against many cancers [115].
Fig. 1.
The treatment hypothesis based on monophyletic model. (A) Only surgery can completely remove a tumor, including CSCs, in the early stages of cancer. (B) Inflammatory factors, such as interleukin (IL)-1, IL-6, and PGE2, play an important role in cancer metastasis. Inflammatory inhibitors cannot prevent metastasis and achieve poor clinical efficacy when the primary tumor is not resected. (C) Inhibitors of inflammatory factors play an important role in preventing recurrence and progression after surgical resection
Circulating tumor cells (CTCs) are disseminated into peripheral blood and may lead to distant organ metastasis. Studies have reported that CTCs expressing CSC markers are associated with tumor metastasis and progression [116–118]. We hypothesized that tumors in the advanced stage disseminate CSCs, which initiates cancer metastasis. Inflammatory factors, such as IL-6, might be important accelerators of tumor formation and growth. Since many inflammatory factors are released from primary tumors, IL-6R inhibitors cannot prevent the metastasis induced by IL-6 and achieve poor clinical efficacy when the primary tumor is not resected (Fig. 1B). Occasionally, IL-6R inhibitors may hinder tumor progression. A small source of water is insufficient to extinguish large fires but may prevent reignition; similarly, inhibitors of inflammatory factors may play an important role in preventing recurrence and progression after surgical resection (Fig. 1C).
CSCs are Essentially Stem Cells with Disordered Differentiation
The concept of CSCs has been established for decades and has been verified by many researchers. If CSCs transform from NSCs and share many critical pathways with NSCs, what are the differences between them? Why do drugs that target CSC markers have poor clinical benefits? In our proposed monophyletic origin model, CSCs are derived from NSCs in different tissues from where the CSCs are located and have almost the same cellular characteristics as NSCs, including surface markers and intercellular pathways. The main biological feature that distinguishes CSCs from NSCs is that CSCs either differentiate into disordered cell types that do not restore the function of damaged tissues or fail to differentiate into mature tissues at all; for example, CSCs in hematopoietic cancers are blocked at a specific differentiation stage [119–121]. According to histopathological classification, the level of differentiation has been used to distinguish benign tumors from malignant ones, characterize the malignant behaviors of cancers, and predict prognosis [122, 123]. Epigenetics has been reported to play a vital role in the regulation of differentiation [124, 125]. Differentiation therapy has proven to be clinically useful for the induction of cancer cell differentiation in leukemia [126]. Therefore, signaling pathways that trigger differentiation failure may be an effective therapeutic target in the termination of CSC activation.
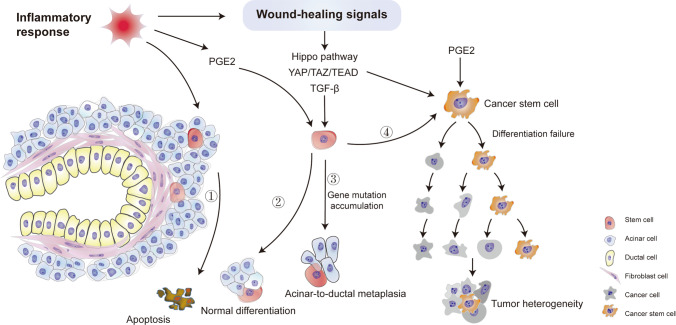
In our proposed model, the development of CSCs from NSCs is a long, continuous process. The most common cancers, such as gastric cancer, which develops from chronic atrophic gastritis; liver cancer induced by viral hepatitis or alcohol; cervical cancer caused by human papilloma virus; and pancreatic cancer resulting from chronic pancreatitis (CP), have been attributed to inflammation and immunoreaction [127–130]. The triggers of cancer are diverse, but chronic inflammation is the most common cause of tumor initiation [14]; pancreatic cancer has the poorest survival of all cancers and has a typical transformation process from CP to oncogenesis. Many studies have reported the key role of CSCs in pancreatic cancer, meanwhile, the pancreas is renowned for its capacity for regeneration [1, 78]. Based on the theory of monophyletic origin, we have proposed that CP progresses to pancreatic cancer; this will provide a better understanding of its progression and is suitable for other tumors (Fig. 2).
Fig. 2.
Progression model for pancreatic cancer based on the monophyletic model. Inflammatory responses in the pancreas induce cell apoptosis (①). PGE2 and wound-healing signals are activated to repair the injured tissues through NSC division and differentiation (②). NSCs dedifferentiate and adopt a novel status termed acinar-ductal metaplasia because of the gene mutation accumulation (③). NSCs become malignant CSCs (④). PGE2 and wound-healing signals maintain the division and proliferation of CSCs and lead to tumors
In the first stage of CP, alcohol, smoking, and other factors lead to acinar cell injury and inflammatory responses in the pancreas [131]. To repair damaged tissues and restore secretion by acinar cells, quiescent NSCs are activated, start the self-renewal process, and differentiate into acinar cells (Fig. 2). However, DNA damage and gene mutations, such as the Kras mutation that occurs during the repair process, may result in serious DNA damage-repair disorders in NSCs [132, 133]. At this stage, NSCs dedifferentiate and adopt a novel status termed acinar-to-ductal metaplasia (ADM) after the expression of genes regulating acinar lineage–specific transcription factors is inhibited (Fig. 2). However, the secretory functions of acinar cells are maintained in ADM [134, 135]. Metaplasia has also been shown to occur in the esophagus, stomach, lung airway, cervix, and mammary gland, and metaplastic lesions are considered precancerous lesions [132, 136]. Meanwhile, MSCs may be recruited to construct tumor-associated fibroblasts, which promote tumor microenvironment formation and tumor growth [137]. Tumor stromal remodeling induces a hypoxic environment and aggravates tumor progression [138].
If the environmental stress on the pancreas is not alleviated, the tissue repair process progresses to the second stage; NSCs transdifferentiate into a pathological morphology, termed pancreatic intraepithelial neoplasia (PanIN). In this state, some digestive enzyme secretion may be maintained. The degree of dysplasia determines the extent of PanIN lesions, that is, the number of cells with impaired secretory function. Disease progression is halted if the environmental stress is eliminated. However, because of ineffective treatment, many PanIN patients progress to the third stage, in which the NSCs become malignant CSCs (Fig. 2). This example supports the theory that CSCs are stem cells that have lost their ability to differentiate into mature tissues.
The Relationship Between Monophyletic Origin Theory and the Hallmarks of Cancer
Researchers have identified various characteristics of cancer cells over the past few decades; Hanahan et al. summarized six hallmarks of cancer to establish a logical framework for to categorize tumors and provide a systematic understanding of tumor biological features [139]. Although the six hallmarks have been increased to eight and combined with additional characteristics, the central idea implicit in their theory should be noted: as normal cells develop into a malignant state, they acquire hallmark capabilities [3, 140]. According to the monophyletic origin theory, CSCs that initiate different cancers are transformed from resident NSCs. Thus, rather than acquiring the hallmark capabilities after malignant transformation, CSCs exhibited them before transformed from NSCs. Moreover, the hallmarks of cancer can be easily explained by the theory of monophyletic origin. The eight hallmarks of cancer include sustaining proliferative signaling, evading growth suppressors, resisting cell death, enabling replicative immortality, inducing vasculature, activating invasion and metastasis, reprogramming cellular metabolism, and avoiding immune destruction [140]. According to the monophyletic theory, the division and differentiation of CSCs is a mechanism for repairing injured tissues. Proliferative signaling is sustained by tissue-repair signals or inflammatory factors, because the cells produced by CSCs cannot restore the functions of damaged tissues, or because it is not possible to eliminate inflammatory factors, respectively. Hanahan hypothesized that inflammatory stroma may contribute to the generation and maintenance of CSCs [140]. The hypoxic environment induced by MSCs recruited to the tumor stroma facilitates the metastasis of cancer cells. Inflammatory cells have also been reported to promote cancer cell invasion by activating epithelial-mesenchymal transition [141].
Another hallmark of cancer is immune escape. Most studies have shown that the activity of immune cells in the tumor microenvironment is inhibited. However, immune cells and inflammatory responses may also promote tumorigenesis, metastasis, and progression [14, 142, 143]. Rather than killing tumor cells, immune cells present in the tumor microenvironment may be a manifestation of the inflammatory response [144]. These results indicate that inflammatory responses are the key triggers of cancer hallmarks.
In addition, mounting evidence suggests that the mechanisms orchestrating normal embryogenesis are strikingly similar to those associated with CSCs [145]. For example, telomerase activity in CSCs is consistent with that in NSCs and plays a crucial role in sustaining cell division and proliferation in cancer. Telomerase is considered a potent target for cancer therapy [146]. CSCs can switch between the quiescent and active states to escape chemotherapy [147]. Other features of CSCs, such as self-renewal, anti-apoptosis, and replicative immortality, were also confirmed to be exhibited by NSCs.
The most recently reported hallmarks of cancer indicate that cancer cells exhibit phenotypic plasticity. Cancer cells exhibit a complex, heterogeneous, and transformable pathogenesis that involves dedifferentiation, blocked differentiation, and transdifferentiation [3]. The monophyletic theory proposes that the cancer phenotype is determined by the degree of differentiation deficiency in CSCs. The failure to differentiate leads to diverse immature cancer cell phenotypes. Accumulating evidence suggests that many solid tumors are hierarchically organized. This may be because a single tumor evolves from distinct subpopulations of CSCs [148, 149]. Single-cell sequencing technologies have greatly contributed to our understanding of the origin and diversity of cancer [150].
The Superiority of the Monophyletic Model in Comparison with the Classical CSC Model
The classical CSC model has been established for decades based on the discovery that only a fraction of the population of cancer cells has the ability to self-renew, extensively proliferate, and form tumors, in both in vivo and in vitro experiments [151, 152]. The transformation of NSCs induced by genetic and epigenetic alterations may be the primary source of CSCs [7, 153–155]. CSCs have considerable biological similarities with NSCs and play a critical role in the initiation, metastasis, propagation, and therapy resistance of cancer [19]. The classical CSCs model relies on hypothetical origins of CSCs including NSCs, dedifferentiated mature cells, and induced pluripotent cancer cells [156, 157]. There are various controversies surrounding the CSCs model among cancer researchers; however, our monophyletic model scientifically summarizes the aberrant differentiation of cancer cells lead by CSCs, the dynamic evolution of CSCs from NSCs, and the driving role of wound-healing signals in cancer progression.
The monophyletic origin model considers that CSCs which originate from NSCs at different differentiation stages and from different resident tissues, have different abilities regarding multi-lineage differentiation. The degree of differentiation and the cellular activity of stem cells determines the biological aggressiveness and the intra-tumor heterogeneity of CSCs. For example, a mature teratoma can produce a variety of cell types similar to an organoid system, due to the potent differentiation capacity of embryonic stem cells. On the other hand, an immature teratoma dynamically transformed from a mature teratoma has very poor differentiation, and highly malignant biology [158, 159]. The monophyletic model presents the dynamic evolution process from normal tissue, to pre-malignant lesions and finally, malignant cancer. Evidence shows that, as the stemness, proliferative ability, and number of stem cells diminishes with age, the regeneration capacity of tissues reduces, such as of the skin and intestines [160, 161]. Tsang et al. found that younger breast cancer patients have a higher expression of the stem cell marker ALDH1 [162]. Studies indicated that cancers in young people have a more aggressive biological features, and lead to a higher mortality and poorer prognosis compared with cancers in older people [162–165]. These may provide a better understanding of the stemness of NSCs that CSCs transformed from decides the malignancy of the tumor.
The dynamic evolution in the monophyletic model is determined by the retained differentiative capacity of CSCs. The purpose of CSC division and proliferation is to repair damaged tissue; however, gene mutations bring about aberrant differentiation or de-differentiation, and lead to ineffective tissue repair. In cancerous tissue, CSCs may obtain mutations inducing transformation between CSCs and non-CSCs, thus resulting in a complex heterogeneous tumor. Furthermore, the monophyletic origin model proposes triggers of cancer progression including inflammatory response and wound-healing signals, such as PGE2, YAP, NOTCH, and WNT signals, which are necessary signals in normal tissue repair [161]. These signals also play critical roles in oncogenesis and cancer progression by controlling CSC activity. Strong correlations between the mechanisms of cancer growth and wound repair have been proven [166]. The resident stem cells performing wound repair have very similar transcriptomes to CSCs [167]. The monophyletic origin model provides a more comprehensive understanding of CSCs compared to the classical CSC model.
Potential Therapeutic Targets Based on the Monophyletic Model
Based on the classical CSCs model, researchers mainly focused on the discovery of biomarkers for the identification of and targeted therapies against CSCs. However, no specific biomarkers could be used to screen CSCs accurately until now. The drugs targeting CSCs have shown unsatisfactory results [19]. The monophyletic model proposes that aberrant differentiation is the root cause of cancer. Clarifying the molecular mechanisms of cell differentiation is crucial in facilitating novel cancer therapies [168]. Differentiation therapy has been proven to be an effective treatment method against some types of cancer [88, 169]. In addition, according to the monophyletic model, anti-inflammatory response signals and anti-wound-healing signals are pivotal to treatment success and recurrence prevention. Many biomarkers of CSCs play an important role in the inflammatory response. The signaling pathways regulating the inflammatory response and wound repair are primary factors of CSC activity and cancer progression [104–107]. However, it is important to explore and define the appropriate stage for intervention with anti-inflammatory and anti-wound repair therapy, such as post operation.
Conclusion
Cancer remains a global threat to human health. Currently, no drugs or therapeutic methods can cure cancer. However, significant progress has been made in the development of targeted therapies, immunotherapy, biological therapy, chemoradiotherapy, and surgical treatment [116, 170, 171]. CSCs are considered tumor-initiating cells because they exhibit self-renewal, treatment resistance, metastasis, and tumor formation capabilities [11]. The monophyletic origin model of cancer provides a concise account of how NSCs can transform into CSCs and how certain trigger factors can promote and sustain proliferation, invasion, and chemoresistance. In our view, the biological behaviors of cancer cells should be described as dynamic, chaotic, and self-preserving, rather than aggressive; cancer cells do not intend to destroy organs. In the past few decades, eliminating cancer cells and CSCs has been the main therapeutic principle behind anti-cancer strategies. It may be time to view the biological features of cancer in a different light and pay more attention to the redifferentiation of CSCs and the factors driving CSC differentiation.
Acknowledgements
We would like to thank Editage (www.editage.cn) for English language editing.
Author Contribution
Q. L., and T. Q. developed the proposal and drafted the manuscript. P. Y., and P. L., contributed to the literature search. All the authors have reviewed and approved the manuscript.
Funding
This work was supported by the Key Science and Technology Research Project of Henan Province (212102310151).
Data Availability
Not available.
Declarations
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Competing Interests
The authors declare no competing financial interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel RL, et al. Cancer statistics, 2022. CA: A Cancer Journal for Clinicians. 2022;72(1):7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Ushijima T, Clark SJ, Tan P. Mapping genomic and epigenomic evolution in cancer ecosystems. Science. 2021;373(6562):1474–1479. doi: 10.1126/science.abh1645. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D. Hallmarks of cancer: New dimensions. Cancer discovery. 2022;12(1):31–46. doi: 10.1158/2159-8290.CD-21-1059. [DOI] [PubMed] [Google Scholar]
- 4.Fu X, et al. Mesenchymal stem cell migration and tissue repair. Cells. 2019;8(8):784. doi: 10.3390/cells8080784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yin W, et al. Cancer and stem cells. Experimental Biology and Medicine. 2021;246(16):1791–1801. doi: 10.1177/15353702211005390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke MF. Clinical and therapeutic implications of cancer stem cells. The New England Journal of Medicine. 2019;380(23):2237–2245. doi: 10.1056/NEJMra1804280. [DOI] [PubMed] [Google Scholar]
- 7.Reya T, et al. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 8.Valent P, et al. Cancer stem cell definitions and terminology: The devil is in the details. Nature Reviews Cancer. 2012;12(11):767–775. doi: 10.1038/nrc3368. [DOI] [PubMed] [Google Scholar]
- 9.Antoniou A, et al. Cancer stem cells, a fuzzy evolving concept: A cell population or a cell property. Cell Cycle. 2013;12(24):3743–3748. doi: 10.4161/cc.27305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen LV, et al. Cancer stem cells: An evolving concept. Nature Reviews Cancer. 2012;12(2):133–143. doi: 10.1038/nrc3184. [DOI] [PubMed] [Google Scholar]
- 11.Bisht S, et al. Cancer stem cells: From an insight into the basics to recent advances and therapeutic targeting. Stem Cells International. 2022;2022:9653244. doi: 10.1155/2022/9653244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J. The dualistic origin of human tumors. Seminars in Cancer Biology. 2018;53:1–16. doi: 10.1016/j.semcancer.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park S, et al. Clonal dynamics in early human embryogenesis inferred from somatic mutation. Nature. 2021;597(7876):393–397. doi: 10.1038/s41586-021-03786-8. [DOI] [PubMed] [Google Scholar]
- 14.Greten FR, Grivennikov SI. Inflammation and cancer: Triggers, mechanisms, and consequences. Immunity. 2019;51(1):27–41. doi: 10.1016/j.immuni.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Basu AK. DNA damage, mutagenesis and cancer. International Journal of Molecular Sciences. 2018;19(4):970. doi: 10.3390/ijms19040970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams GH, Stoeber K. The cell cycle and cancer. The Journal of Pathology. 2012;226(2):352–364. doi: 10.1002/path.3022. [DOI] [PubMed] [Google Scholar]
- 17.Moore L, et al. The mutational landscape of normal human endometrial epithelium. Nature. 2020;580(7805):640–646. doi: 10.1038/s41586-020-2214-z. [DOI] [PubMed] [Google Scholar]
- 18.Coates PJ, Lorimore SA, Wright EG. Cell and tissue responses to genotoxic stress. The Journal of Pathology. 2005;205(2):221–235. doi: 10.1002/path.1701. [DOI] [PubMed] [Google Scholar]
- 19.Lytle NK, Barber AG, Reya T. Stem cell fate in cancer growth, progression and therapy resistance. Nature Reviews Cancer. 2018;18(11):669–680. doi: 10.1038/s41568-018-0056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanpain C, Fuchs E. Stem cell plasticity. Plasticity of epithelial stem cells in tissue regeneration. Science. 2014;344(6189):1242281. doi: 10.1126/science.1242281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X. Stem cells in tissues, organoids, and cancers. Cellular and Molecular Life Sciences. 2019;76(20):4043–4070. doi: 10.1007/s00018-019-03199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De la Rosa MB, Kozik EM, Sakaguchi DS. Adult stem cell-based strategies for peripheral nerve regeneration. Advances in Experimental Medicine and Biology. 2018;1119:41–71. doi: 10.1007/5584_2018_254. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez ME, et al. EZH2 expands breast stem cells through activation of NOTCH1 signaling. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(8):3098–3103. doi: 10.1073/pnas.1308953111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fox RG, et al. Image-based detection and targeting of therapy resistance in pancreatic adenocarcinoma. Nature. 2016;534(7607):407–411. doi: 10.1038/nature17988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tammela T, et al. A Wnt-producing niche drives proliferative potential and progression in lung adenocarcinoma. Nature. 2017;545(7654):355–359. doi: 10.1038/nature22334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nature Medicine. 1997;3(7):730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 27.Yung Y, et al. Targeting abnormal hematopoietic stem cells in chronic myeloid leukemia and philadelphia chromosome-negative classical myeloproliferative neoplasms. International journal of molecular sciences. 2021;22(2):659. doi: 10.3390/ijms22020659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morton SU, et al. Association of damaging variants in genes with increased cancer risk among patients with congenital heart disease. JAMA Cardiology. 2021;6(4):457–462. doi: 10.1001/jamacardio.2020.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saygin C, et al. Targeting cancer stemness in the clinic: From hype to hope. Cell Stem Cell. 2019;24(1):25–40. doi: 10.1016/j.stem.2018.11.017. [DOI] [PubMed] [Google Scholar]
- 30.Lathia JD, et al. Cancer stem cells in glioblastoma. Genes & Development. 2015;29(12):1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee CJ, Dosch J, Simeone DM. Pancreatic cancer stem cells. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2008;26(17):2806–2812. doi: 10.1200/JCO.2008.16.6702. [DOI] [PubMed] [Google Scholar]
- 32.Signore M, Ricci-Vitiani L, De Maria R. Targeting apoptosis pathways in cancer stem cells. Cancer letters. 2013;332(2):374–382. doi: 10.1016/j.canlet.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 33.Walcher L, et al. Cancer stem cells-origins and biomarkers: Perspectives for targeted personalized therapies. Frontiers in Immunology. 2020;11:1280. doi: 10.3389/fimmu.2020.01280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim WT, Ryu CJ. Cancer stem cell surface markers on normal stem cells. BMB Reports. 2017;50(6):285–298. doi: 10.5483/BMBRep.2017.50.6.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheung SK, et al. Stage-specific embryonic antigen-3 (SSEA-3) and β3GalT5 are cancer specific and significant markers for breast cancer stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(4):960–965. doi: 10.1073/pnas.1522602113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bai X, et al. Cancer stem cell in breast cancer therapeutic resistance. Cancer Treatment Reviews. 2018;69:152–163. doi: 10.1016/j.ctrv.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 37.Zhu R, et al. TSPAN8 promotes cancer cell stemness via activation of sonic Hedgehog signaling. Nature Communications. 2019;10(1):2863. doi: 10.1038/s41467-019-10739-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taurin S, Alkhalifa H. Breast cancers, mammary stem cells, and cancer stem cells, characteristics, and hypotheses. Neoplasia. 2020;22(12):663–678. doi: 10.1016/j.neo.2020.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rajasekhar VK, et al. Tumour-initiating stem-like cells in human prostate cancer exhibit increased NF-κB signalling. Nature Communications. 2011;2:162. doi: 10.1038/ncomms1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li T, et al. ALDH1A1 is a marker for malignant prostate stem cells and predictor of prostate cancer patients' outcome. Laboratory Investigation. 2010;90(2):234–244. doi: 10.1038/labinvest.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tu SM, Lin SH. Prostate cancer stem cells. Clinical Genitourinary Cancer. 2012;10(2):69–76. doi: 10.1016/j.clgc.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoogland AM, et al. Validation of stem cell markers in clinical prostate cancer: α6-integrin is predictive for non-aggressive disease. The Prostate. 2014;74(5):488–496. doi: 10.1002/pros.22768. [DOI] [PubMed] [Google Scholar]
- 43.Li JJ, Shen MM. Prostate stem cells and cancer stem cells. Cold Spring Harbor Perspectives in Medicine. 2019;9(6):a030395. doi: 10.1101/cshperspect.a030395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schaal CM, et al. Regulation of Sox2 and stemness by nicotine and electronic-cigarettes in non-small cell lung cancer. Molecular Cancer. 2018;17(1):149. doi: 10.1186/s12943-018-0901-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maiuthed A, Chantarawong W, Chanvorachote P. Lung cancer stem cells and cancer stem cell-targeting natural compounds. Anticancer Research. 2018;38(7):3797–3809. doi: 10.21873/anticanres.12663. [DOI] [PubMed] [Google Scholar]
- 46.Parekh KR, et al. Stem cells and lung regeneration. American Journal of Physiology. Cell Physiology. 2020;319(4):C675–C693. doi: 10.1152/ajpcell.00036.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Munro MJ, et al. Cancer stem cells in colorectal cancer: A review. Journal of Clinical Pathology. 2018;71(2):110–116. doi: 10.1136/jclinpath-2017-204739. [DOI] [PubMed] [Google Scholar]
- 48.Wahab SMR, et al. The identifications and clinical implications of cancer stem cells in colorectal cancer. Clinical Colorectal Cancer. 2017;16(2):93–102. doi: 10.1016/j.clcc.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 49.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449(7165):1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 50.Yui S, et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5+ stem cell. Nature Medicine. 2012;18(4):618–623. doi: 10.1038/nm.2695. [DOI] [PubMed] [Google Scholar]
- 51.Parmiani G. Melanoma cancer stem cells: Markers and functions. Cancers. 2016;8(3):34. doi: 10.3390/cancers8030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones PH, Watt FM. Separation of human epidermal stem cells from transit amplifying cells on the basis of differences in integrin function and expression. Cell. 1993;73(4):713–724. doi: 10.1016/0092-8674(93)90251-K. [DOI] [PubMed] [Google Scholar]
- 53.Li A, Simmons PJ, Kaur P. Identification and isolation of candidate human keratinocyte stem cells based on cell surface phenotype. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(7):3902–3907. doi: 10.1073/pnas.95.7.3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tani H, Morris RJ, Kaur P. Enrichment for murine keratinocyte stem cells based on cell surface phenotype. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(20):10960–10965. doi: 10.1073/pnas.97.20.10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wan H, et al. Desmosomal proteins, including desmoglein 3, serve as novel negative markers for epidermal stem cell-containing population of keratinocytes. Journal of Cell Science. 2003;116(Pt 20):4239–4248. doi: 10.1242/jcs.00701. [DOI] [PubMed] [Google Scholar]
- 56.Legg J, et al. Role of melanoma chondroitin sulphate proteoglycan in patterning stem cells in human interfollicular epidermis. Development. 2003;130(24):6049–6063. doi: 10.1242/dev.00837. [DOI] [PubMed] [Google Scholar]
- 57.Tan DW, et al. Single-cell gene expression profiling reveals functional heterogeneity of undifferentiated human epidermal cells. Development. 2013;140(7):1433–1444. doi: 10.1242/dev.087551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saeednejad Zanjani L, et al. Expression of CD105 cancer stem cell marker in three subtypes of renal cell carcinoma. Cancer Biomarkers. 2018;21(4):821–837. doi: 10.3233/CBM-170755. [DOI] [PubMed] [Google Scholar]
- 59.Rasti A, et al. Reduced expression of CXCR4, a novel renal cancer stem cell marker, is associated with high-grade renal cell carcinoma. Journal of Cancer Research and Clinical Oncology. 2017;143(1):95–104. doi: 10.1007/s00432-016-2239-8. [DOI] [PubMed] [Google Scholar]
- 60.Fang P, et al. Targeting strategies for renal cancer stem cell therapy. Current Pharmaceutical Design. 2020;26(17):1964–1978. doi: 10.2174/1381612826666200318153106. [DOI] [PubMed] [Google Scholar]
- 61.Li X, et al. Human urine-derived stem cells protect against renal ischemia/reperfusion injury in a rat model via exosomal miR-146a-5p which targets IRAK1. Theranostics. 2020;10(21):9561–9578. doi: 10.7150/thno.42153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bharadwaj S, et al. Multipotential differentiation of human urine-derived stem cells: Potential for therapeutic applications in urology. Stem Cells. 2013;31(9):1840–1856. doi: 10.1002/stem.1424. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y, et al. Urine derived cells are a potential source for urological tissue reconstruction. The Journal of Urology. 2008;180(5):2226–2233. doi: 10.1016/j.juro.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 64.Karantanos T, Jones RJ. Acute myeloid leukemia stem cell heterogeneity and its clinical relevance. Advances in Experimental Medicine and Biology. 2019;1139:153–169. doi: 10.1007/978-3-030-14366-4_9. [DOI] [PubMed] [Google Scholar]
- 65.Saito Y, et al. Identification of therapeutic targets for quiescent, chemotherapy-resistant human leukemia stem cells. Science Translational Medicine. 2010;2(17):17ra9. doi: 10.1126/scitranslmed.3000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jin L, et al. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nature Medicine. 2006;12(10):1167–1174. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]
- 67.Majeti R, et al. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 2009;138(2):286–299. doi: 10.1016/j.cell.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Herrmann H, et al. Delineation of target expression profiles in CD34+/CD38- and CD34+/CD38+ stem and progenitor cells in AML and CML. Blood Advances. 2020;4(20):5118–5132. doi: 10.1182/bloodadvances.2020001742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kikushige Y, et al. TIM-3 is a promising target to selectively kill acute myeloid leukemia stem cells. Cell Stem Cell. 2010;7(6):708–717. doi: 10.1016/j.stem.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 70.Hu L, et al. Antioxidant N-acetyl-L-cysteine increases engraftment of human hematopoietic stem cells in immune-deficient mice. Blood. 2014;124(20):e45–e48. doi: 10.1182/blood-2014-03-559369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garg S, Madkaikar M, Ghosh K. Investigating cell surface markers on normal hematopoietic stem cells in three different niche conditions. International Journal of Stem Cells. 2013;6(2):129–133. doi: 10.15283/ijsc.2013.6.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ishiwata T, et al. Pancreatic cancer stem cells: Features and detection methods. Pathology Oncology Research. 2018;24(4):797–805. doi: 10.1007/s12253-018-0420-x. [DOI] [PubMed] [Google Scholar]
- 73.Hermann PC, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1(3):313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 74.Jo JH, et al. GLRX3, a novel cancer stem cell-related secretory biomarker of pancreatic ductal adenocarcinoma. BMC Cancer. 2021;21(1):1241. doi: 10.1186/s12885-021-08898-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Matsuda Y, Kure S, Ishiwata T. Nestin and other putative cancer stem cell markers in pancreatic cancer. Medical Molecular Morphology. 2012;45(2):59–65. doi: 10.1007/s00795-012-0571-x. [DOI] [PubMed] [Google Scholar]
- 76.Heiler S, Wang Z, Zöller M. Pancreatic cancer stem cell markers and exosomes - the incentive push. World Journal of Gastroenterology. 2016;22(26):5971–6007. doi: 10.3748/wjg.v22.i26.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bonner-Weir S, Sharma A. Pancreatic stem cells. The Journal of Pathology. 2002;197(4):519–526. doi: 10.1002/path.1158. [DOI] [PubMed] [Google Scholar]
- 78.Zhou Q, Melton DA. Pancreas regeneration. Nature. 2018;557(7705):351–358. doi: 10.1038/s41586-018-0088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vizio B, et al. Comparative evaluation of cancer stem cell markers in normal pancreas and pancreatic ductal adenocarcinoma. Oncology Reports. 2012;27(1):69–76. doi: 10.3892/or.2011.1461. [DOI] [PubMed] [Google Scholar]
- 80.Maruyama T. A revised stem cell theory for the pathogenesis of endometriosis. Journal of Personalized Medicine. 2022;12(2):216. doi: 10.3390/jpm12020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang S, et al. Prostaglandin E2 hydrogel improves cutaneous wound healing via M2 macrophages polarization. Theranostics. 2018;8(19):5348–5361. doi: 10.7150/thno.27385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cheng H, et al. Role of prostaglandin E2 in tissue repair and regeneration. Theranostics. 2021;11(18):8836–8854. doi: 10.7150/thno.63396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nakanishi M, Rosenberg DW. Multifaceted roles of PGE2 in inflammation and cancer. Seminars in Immunopathology. 2013;35(2):123–137. doi: 10.1007/s00281-012-0342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Echizen K, et al. Inflammation in gastric cancer: Interplay of the COX-2/prostaglandin E2 and Toll-like receptor/MyD88 pathways. Cancer Science. 2016;107(4):391–397. doi: 10.1111/cas.12901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang D, et al. Prostaglandin E2 promotes colorectal cancer stem cell expansion and metastasis in mice. Gastroenterology. 2015;149(7):1884–1895.e4. doi: 10.1053/j.gastro.2015.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lichtman MK, Otero-Vinas M, Falanga V. Transforming growth factor beta (TGF-β) isoforms in wound healing and fibrosis. Wound Repair and Regeneration. 2016;24(2):215–222. doi: 10.1111/wrr.12398. [DOI] [PubMed] [Google Scholar]
- 87.Mascharak S, et al. Multiomic analysis reveals divergent molecular events in scarring and regenerative wound healing. Cell Stem Cell. 2022;29(2):315–327.e6. doi: 10.1016/j.stem.2021.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.LeBlanc L, Ramirez N, Kim J. Context-dependent roles of YAP/TAZ in stem cell fates and cancer. Cellular and Molecular Life Sciences. 2021;78(9):4201–4219. doi: 10.1007/s00018-021-03781-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the roots of cancer. Cancer Cell. 2016;29(6):783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dey A, Varelas X, Guan KL. Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nature Reviews Drug Discovery. 2020;19(7):480–494. doi: 10.1038/s41573-020-0070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Park JH, Shin JE, Park HW. The role of hippo pathway in cancer stem cell biology. Molecules and Cells. 2018;41(2):83–92. doi: 10.14348/molcells.2018.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. The New England Journal of Medicine. 1986;315(26):1650–1659. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- 93.Atiya H, et al. Mesenchymal stem cells in the tumor microenvironment. Advances in Experimental Medicine and Biology. 2020;1234:31–42. doi: 10.1007/978-3-030-37184-5_3. [DOI] [PubMed] [Google Scholar]
- 94.Lee YT, Tan YJ, Oon CE. Molecular targeted therapy: Treating cancer with specificity. European Journal of Pharmacology. 2018;834:188–196. doi: 10.1016/j.ejphar.2018.07.034. [DOI] [PubMed] [Google Scholar]
- 95.Xu Y, et al. Technological advances in cancer immunity: From immunogenomics to single-cell analysis and artificial intelligence. Signal Transduction and Targeted Therapy. 2021;6(1):312. doi: 10.1038/s41392-021-00729-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jiang T, et al. Tumor neoantigens: From basic research to clinical applications. Journal of Hematology & Oncology. 2019;12(1):93. doi: 10.1186/s13045-019-0787-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sterner RC, Sterner RM. CAR-T cell therapy: Current limitations and potential strategies. Blood Cancer Journal. 2021;11(4):69. doi: 10.1038/s41408-021-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sancho P, Barneda D, Heeschen C. Hallmarks of cancer stem cell metabolism. British Journal of Cancer. 2016;114(12):1305–1312. doi: 10.1038/bjc.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wyld L, Audisio RA, Poston GJ. The evolution of cancer surgery and future perspectives. Nature Reviews Clinical Oncology. 2015;12(2):115–124. doi: 10.1038/nrclinonc.2014.191. [DOI] [PubMed] [Google Scholar]
- 100.Nie Y, et al. Surgical prognosis of synchronous multiple primary lung cancer: Systematic review and meta-analysis. Clinal Lung Cancer. 2021;22(4):341–350.e3. doi: 10.1016/j.cllc.2020.10.022. [DOI] [PubMed] [Google Scholar]
- 101.Zhang F, et al. Conversion surgery for stage IV gastric cancer. Frontiers in Oncology. 2019;9:1158. doi: 10.3389/fonc.2019.01158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Han D, et al. Analysis of radiotherapy impact on survival in resected stage I/II pancreatic cancer patients: A population-based study. BMC Cancer. 2021;21(1):560. doi: 10.1186/s12885-021-08288-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gemenetzis G, et al. Survival in locally advanced pancreatic cancer after neoadjuvant therapy and surgical resection. Annals of Surgery. 2019;270(2):340–347. doi: 10.1097/SLA.0000000000002753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen Z, et al. Surgical stress and cancer progression: The twisted tango. Molecular Cancer. 2019;18(1):132. doi: 10.1186/s12943-019-1058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hao Y, Baker D, Ten Dijke P. TGF-β-Mediated epithelial-mesenchymal transition and cancer metastasis. International Journal of Molecular Science. 2019;20(11):2767. doi: 10.3390/ijms20112767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang YM, Kim SY, Seki E. Inflammation and liver cancer: Molecular mechanisms and therapeutic targets. Seminars in Liver Disease. 2019;39(1):26–42. doi: 10.1055/s-0038-1676806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Siersbæk R, et al. IL6/STAT3 signaling hijacks estrogen receptor α enhancers to drive breast cancer metastasis. Cancer Cell. 2020;38(3):412–423.e9. doi: 10.1016/j.ccell.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fang T, et al. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nature Communication. 2018;9(1):191. doi: 10.1038/s41467-017-02583-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu W, et al. IL-6 promotes metastasis of non-small-cell lung cancer by up-regulating TIM-4 via NF-κB. Cell Proliferation. 2020;53(3):e12776. doi: 10.1111/cpr.12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Taniguchi K, Karin M. IL-6 and related cytokines as the critical lynchpins between inflammation and cancer. Seminars in Immunology. 2014;26(1):54–74. doi: 10.1016/j.smim.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 111.Goumas FA, et al. Inhibition of IL-6 signaling significantly reduces primary tumor growth and recurrencies in orthotopic xenograft models of pancreatic cancer. International Journal of Cancer. 2015;137(5):1035–1046. doi: 10.1002/ijc.29445. [DOI] [PubMed] [Google Scholar]
- 112.Wakabayashi H, et al. Interleukin-6 receptor inhibitor suppresses bone metastases in a breast cancer cell line. Breast Cancer. 2018;25(5):566–574. doi: 10.1007/s12282-018-0853-9. [DOI] [PubMed] [Google Scholar]
- 113.Oguro T, et al. Humanised antihuman IL-6R antibody with interferon inhibits renal cell carcinoma cell growth in vitro and in vivo through suppressed SOCS3 expression. European Journal of Cancer. 2013;49(7):1715–1724. doi: 10.1016/j.ejca.2012.11.038. [DOI] [PubMed] [Google Scholar]
- 114.Yanaihara N, et al. Antitumor effects of interleukin-6 (IL-6)/interleukin-6 receptor (IL-6R) signaling pathway inhibition in clear cell carcinoma of the ovary. Molecular Carcinogenesis. 2016;55(5):832–841. doi: 10.1002/mc.22325. [DOI] [PubMed] [Google Scholar]
- 115.Rossi JF, et al. Interleukin-6 as a therapeutic target. Clinical Cancer Research. 2015;21(6):1248–1257. doi: 10.1158/1078-0432.CCR-14-2291. [DOI] [PubMed] [Google Scholar]
- 116.Liu T, et al. Circulating glioma cells exhibit stem cell-like properties. Cancer Research. 2018;78(23):6632–6642. doi: 10.1158/0008-5472.CAN-18-0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Aceto N, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158(5):1110–1122. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Andrade F, et al. Polymeric micelles targeted against CD44v6 receptor increase niclosamide efficacy against colorectal cancer stem cells and reduce circulating tumor cells in vivo. Journal of Controlled Release. 2021;331:198–212. doi: 10.1016/j.jconrel.2021.01.022. [DOI] [PubMed] [Google Scholar]
- 119.Sell S. Stem cell origin of cancer and differentiation therapy. Critical Reviews in Oncology/hematology. 2004;51(1):1–28. doi: 10.1016/j.critrevonc.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 120.Dekoninck S, Blanpain C. Stem cell dynamics, migration and plasticity during wound healing. Nature Cell Biology. 2019;21(1):18–24. doi: 10.1038/s41556-018-0237-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tacke F. Targeting hepatic macrophages to treat liver diseases. Journal of Hepatology. 2017;66(6):1300–1312. doi: 10.1016/j.jhep.2017.02.026. [DOI] [PubMed] [Google Scholar]
- 122.Enane FO, Saunthararajah Y, Korc M. Differentiation therapy and the mechanisms that terminate cancer cell proliferation without harming normal cells. Cell Death & Disease. 2018;9(9):912. doi: 10.1038/s41419-018-0919-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jögi A, et al. Cancer cell differentiation heterogeneity and aggressive behavior in solid tumors. Upsala Journal of Medical Sciences. 2012;117(2):217–224. doi: 10.3109/03009734.2012.659294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lu C, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ohm JE, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nature Genetics. 2007;39(2):237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.de Thé H. Differentiation therapy revisited. Nature Reviews Cancer. 2018;18(2):117–127. doi: 10.1038/nrc.2017.103. [DOI] [PubMed] [Google Scholar]
- 127.Holleczek B, Schöttker B, Brenner H. Helicobacter pylori infection, chronic atrophic gastritis and risk of stomach and esophagus cancer: Results from the prospective population-based ESTHER cohort study. International Journal of Cancer. 2020;146(10):2773–2783. doi: 10.1002/ijc.32610. [DOI] [PubMed] [Google Scholar]
- 128.Llovet JM, et al. Hepatocellular carcinoma. Nature Reviews Disease Primers. 2021;7(1):6. doi: 10.1038/s41572-020-00240-3. [DOI] [PubMed] [Google Scholar]
- 129.Olusola P, et al. Human papilloma virus-associated cervical cancer and health disparities. Cells. 2019;8(6):622. doi: 10.3390/cells8060622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kirkegård J, Mortensen FV, Cronin-Fenton D. Chronic pancreatitis and pancreatic cancer risk: A systematic review and meta-analysis. The American Journal of Gastroenterology. 2017;112(9):1366–1372. doi: 10.1038/ajg.2017.218. [DOI] [PubMed] [Google Scholar]
- 131.Singh VK, Yadav D, Garg PK. Diagnosis and management of chronic pancreatitis: A review. JAMA. 2019;322(24):2422–2434. doi: 10.1001/jama.2019.19411. [DOI] [PubMed] [Google Scholar]
- 132.Guerra C, et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11(3):291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 133.Tannapfel A, Witzigmann H, Wittekind C. Pankreatische intraduktale Neoplasien bei chronischer Pankreatitis [Pancreatic intraepithelial neoplasia in chronic pancreatitis] Zentralblatt fur Chirurgie. 2001;126(11):879–883. doi: 10.1055/s-2001-19155. [DOI] [PubMed] [Google Scholar]
- 134.Kopp JL, et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;22(6):737–750. doi: 10.1016/j.ccr.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.von Figura G, et al. Nr5a2 maintains acinar cell differentiation and constrains oncogenic Kras-mediated pancreatic neoplastic initiation. Gut. 2014;63(4):656–664. doi: 10.1136/gutjnl-2012-304287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Giroux V, Rustgi AK. Metaplasia: Tissue injury adaptation and a precursor to the dysplasia-cancer sequence. Nature Reviews Cancer. 2017;17(10):594–604. doi: 10.1038/nrc.2017.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kidd S, et al. Origins of the tumor microenvironment: Quantitative assessment of adipose-derived and bone marrow-derived stroma. PLoS ONE. 2012;7(2):e30563. doi: 10.1371/journal.pone.0030563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Piersma B, Hayward MK, Weaver VM. Fibrosis and cancer: A strained relationship. Biochimica Biophysica Acta Reviews on Cancer. 2020;1873(2):188356. doi: 10.1016/j.bbcan.2020.188356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 140.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 141.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell. 2010;141(1):52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lei X, et al. Immune cells within the tumor microenvironment: Biological functions and roles in cancer immunotherapy. Cancer Letters. 2020;470:126–133. doi: 10.1016/j.canlet.2019.11.009. [DOI] [PubMed] [Google Scholar]
- 143.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ferrari SM, et al. Immune and inflammatory cells in thyroid cancer microenvironment. International Journal of Molecular Sciences. 2019;20(18):4413. doi: 10.3390/ijms20184413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lonardo E, et al. Nodal/Activin signaling drives self-renewal and tumorigenicity of pancreatic cancer stem cells and provides a target for combined drug therapy. Cell Stem Cell. 2011;9(5):433–446. doi: 10.1016/j.stem.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 146.Bajaj S, et al. Targeting telomerase for its advent in cancer therapeutics. Medicinal Research Reviews. 2020;40(5):1871–1919. doi: 10.1002/med.21674. [DOI] [PubMed] [Google Scholar]
- 147.Das PK, Islam F, Lam AK. The roles of cancer stem cells and therapy resistance in colorectal carcinoma. Cells. 2020;9(6):1392. doi: 10.3390/cells9061392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lindeman GJ, Visvader JE. Insights into the cell of origin in breast cancer and breast cancer stem cells. Asia-Pacific Journal of Clinical Oncology. 2010;6(2):89–97. doi: 10.1111/j.1743-7563.2010.01279.x. [DOI] [PubMed] [Google Scholar]
- 149.Beck B, Blanpain C. Unravelling cancer stem cell potential. Nature Reviews Cancer. 2013;13(10):727–738. doi: 10.1038/nrc3597. [DOI] [PubMed] [Google Scholar]
- 150.Hu L, et al. Single-cell RNA sequencing reveals the cellular origin and evolution of breast cancer in BRCA1 mutation carriers. Cancer Research. 2021;81(10):2600–2611. doi: 10.1158/0008-5472.CAN-20-2123. [DOI] [PubMed] [Google Scholar]
- 151.Bruce WR, Van Der Gaag H. A quantitative assay for the number of murine lymphoma cells capable of proliferation in vivo. Nature. 1963;199:79–80. doi: 10.1038/199079a0. [DOI] [PubMed] [Google Scholar]
- 152.Park CH, Bergsagel DE, McCulloch EA. Mouse myeloma tumor stem cells: A primary cell culture assay. Journal of the National Cancer Institute. 1971;46(2):411–422. [PubMed] [Google Scholar]
- 153.Velten L, et al. Identification of leukemic and pre-leukemic stem cells by clonal tracking from single-cell transcriptomics. Nature Communications. 2021;12(1):1366. doi: 10.1038/s41467-021-21650-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yamashita Y, et al. Array-based genomic resequencing of human leukemia. Oncogene. 2010;29(25):3723–3731. doi: 10.1038/onc.2010.117. [DOI] [PubMed] [Google Scholar]
- 155.Tadokoro Y, et al. De novo DNA methyltransferase is essential for self-renewal, but not for differentiation, in hematopoietic stem cells. Journal of Experimental Medicine. 2007;204(4):715–722. doi: 10.1084/jem.20060750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Islam F, et al. Cancer stem cell: Fundamental experimental pathological concepts and updates. Experimental and Molecular Pathology. 2015;98(2):184–191. doi: 10.1016/j.yexmp.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 157.Singh AK, et al. Tumor heterogeneity and cancer stem cell paradigm: Updates in concept, controversies and clinical relevance. International Journal of Cancer. 2015;136(9):1991–2000. doi: 10.1002/ijc.28804. [DOI] [PubMed] [Google Scholar]
- 158.McDonald D, et al. Defining the teratoma as a model for multi-lineage human development. Cell. 2020;183(5):1402–1419.e18. doi: 10.1016/j.cell.2020.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Atwi D, et al. Malignant transformation of mature cystic teratoma of the ovary. The Journal of Obstetrics and Gynaecology Research. 2022;48(12):3068–3076. doi: 10.1111/jog.15409. [DOI] [PubMed] [Google Scholar]
- 160.Matsumura H, et al. Hair follicle aging is driven by transepidermal elimination of stem cells via COL17A1 proteolysis. Science. 2016;351(6273):aad4395. doi: 10.1126/science.aad4395. [DOI] [PubMed] [Google Scholar]
- 161.Fuchs E, Blau HM. Tissue stem cells: Architects of their niches. Cell Stem Cell. 2020;27(4):532–556. doi: 10.1016/j.stem.2020.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tsang JY, et al. Cancer stem cell markers are associated with adverse biomarker profiles and molecular subtypes of breast cancer. Breast Cancer Research and Treatment. 2012;136(2):407–417. doi: 10.1007/s10549-012-2271-6. [DOI] [PubMed] [Google Scholar]
- 163.Gnerlich JL, et al. Elevated breast cancer mortality in women younger than age 40 years compared with older women is attributed to poorer survival in early-stage disease. Journal of the American College of Surgeons. 2009;208(3):341–347. doi: 10.1016/j.jamcollsurg.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhang W, et al. Metastasis patterns and prognosis in young breast cancer patients: A SEER database analysis. Frontiers in Oncology. 2022;12:872862. doi: 10.3389/fonc.2022.872862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Acharki A, et al. Cancer of the uterine cervix in young women. A retrospective study of 337 cases. Bull Cancer. 1997;84(4):373–8. [PubMed] [Google Scholar]
- 166.Cangkrama M, Wietecha M, Werner S. Wound repair, scar formation, and cancer: Converging on Activin. Trends in Molecular Medicine. 2020;26(12):1107–1117. doi: 10.1016/j.molmed.2020.07.009. [DOI] [PubMed] [Google Scholar]
- 167.Ge Y, et al. Stem cell lineage infidelity drives wound repair and cancer. Cell. 2017;169(4):636–650.e14. doi: 10.1016/j.cell.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Scott RE. Differentiation, differentiation/gene therapy and cancer. Pharmacology & Therapeutics. 1997;73(1):51–65. doi: 10.1016/S0163-7258(96)00120-9. [DOI] [PubMed] [Google Scholar]
- 169.Solé R, Aguadé-Gorgorió G. The ecology of cancer differentiation therapy. Journal of Theoretical Biology. 2021;511:110552. doi: 10.1016/j.jtbi.2020.110552. [DOI] [PubMed] [Google Scholar]
- 170.Springfeld C, et al. Chemotherapy for pancreatic cancer. Presse Medicale. 2019;48(3 Pt 2):e159–e174. doi: 10.1016/j.lpm.2019.02.025. [DOI] [PubMed] [Google Scholar]
- 171.Neoptolemos JP, et al. Therapeutic developments in pancreatic cancer: Current and future perspectives. Nature Reviews Gastroenterology & Hepatology. 2018;15(6):333–348. doi: 10.1038/s41575-018-0005-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not available.