Abstract
Introduction
Probiotic Lactiplantibacillus plantarum MC5 produces large amounts of exopolysaccharides (EPS), and its use as a compound fermentor can greatly improve the quality of fermented milk.
Methods
To gain insight into the genomic characteristics of probiotic MC5 and reveal the relationship between its EPS biosynthetic phenotype and genotype, we analyzed the carbohydrate metabolic capacity, nucleotide sugar formation pathways, and EPS biosynthesis-related gene clusters of strain MC5 based on its whole genome sequence. Finally, we performed validation tests on the monosaccharides and disaccharides that strain MC5 may metabolize.
Results
Genomic analysis showed that MC5 has seven nucleotide sugar biosynthesis pathways and 11 sugar-specific phosphate transport systems, suggesting that the strain can metabolize mannose, fructose, sucrose, cellobiose, glucose, lactose, and galactose. Validation results showed that strain MC5 can metabolize these seven sugars and produce significant amounts of EPS (> 250 mg/L). In addition, strain MC5 possesses two typical eps biosynthesis gene clusters, which include the conserved genes epsABCDE, wzx, and wzy, six key genes for polysaccharide biosynthesis, and one MC5-specific epsG gene.
Discussion
These insights into the mechanism of EPS-MC5 biosynthesis can be used to promote the production of EPS through genetic engineering.
Keywords: comparative genomics, bioinformatics analysis, carbohydrate metabolic capacity, sugar nucleotide-forming pathways, eps gene cluster
1. Introduction
Probiotics are a diverse group of microorganisms widely distributed in nature and an important component of the indigenous microbiota of healthy humans and animals, where they regulate host gut flora (Siezen et al., 2010; Kimoto-Nira, 2018). Strains of Lactiplantibacillus plantarum are on the list of probiotic strains available for food and exhibit various probiotic functions such as exopolysaccharide (EPS) production, and antioxidant and antibacterial properties (Peng et al., 2022). EPSs are macromolecular secondary metabolites produced during the growth of lactic acid bacteria (LAB). EPSs produced by LAB are usually divided into homotypic exopolysaccharides (HoPS) and heterotypic exopolysaccharides (HePS) according to their structure (Prete et al., 2021). HoPS are composed of one monosaccharide, whereas HePS are composed of a variety of monosaccharides comprising three to eight repeat units (Feng et al., 2021). EPS-producing LAB are more tolerant to adverse external environments than other LAB and have become the dominant strains during LAB evolution (Fukao et al., 2019a). As EPS produced by LAB can be used as a natural antioxidant, thickener, and stabilizer, EPS-producing LAB (Streptococcus salivarius subsp. thermophilus, Lactobacillus delbrueckii subsp. bulgaricus, Lactococcus lactis subsp. lactis, etc.) are widely used in the dairy industry (Şanlier et al., 2019). In addition, EPS-LAB are reported to play an important role in immune stimulation, anti-inflammation, and cholesterol-lowering (Werning et al., 2022).
Since the first whole-genome sequence of LAB (Lactobacillus lactis subsp. lactis IL1403) was generated in 2001, whole-genome sequencing (WGS) technology has been applied to find correlations between genetic characteristics of LAB and their functions (Huang et al., 2021). The genomic sequences of many probiotic LAB strains are publicly available, and their essential characteristics have been defined using genomic approaches (Panthee et al., 2019). Qureshi et al. (2020) used WGS to identify genes associated with probiotic properties of Lactobacillus paracasei ZFM54 and validated them in vitro. Yi et al. (2020) used WGS to predict the genome sequence of Lp. pentosus DZ35 and identified two previously unknown genes related to bacteriocins (DZ1 and DZ2). However, few studies have systematically reported on the molecular processes involved in the EPS biosynthesis pathway of LAB. To further develop the EPS-producing capacity of LAB, it is essential to gain insight into the genetics, biosynthetic pathways, regulation, structure, and function of LAB based on known biochemical indicators (Zeidan et al., 2017). The cluster of genes associated with EPS biosynthesis in LAB is known as the eps gene cluster (Sun and Zhang, 2021). Typical eps gene clusters are 14–25 kb in size and contain less than 30 open reading frames (ORFs). The eps gene cluster consists of five highly conserved genes, epsABCDE, and a variable region that specifically includes polymerase (wzy), flip-flop (wzx), genes encoding one or more glycosyltransferases (GTF), and other modifier genes (Zeidan et al., 2017). Song et al. (2018) studied the whole genome sequence of Lc. casei LC2W and showed that genes encoding glucose-1-phosphate thymidyltransferase (LC2W_2179), an unidentified EPS biosynthetic protein (LC2W_2188), and an EPS biosynthetic protein (LC2W_2189) were associated with EPS biosynthesis. Xiong et al. (2019) reported that S. salivarius subsp. thermophilus S-3 has a complete cluster of eps genes with 13 ORFs involved in regulation, chain length determination, repeat unit biosynthesis, polymerization, and export during EPS biosynthesis.
High-EPS-producing Lp. plantarum MC5, showing good fermentation characteristics and probiotic properties, was isolated from traditional fermented yak yogurt in the Tibetan area of Gansu. Strain MC5 has great application value for production of fermented milk in the Tibetan area and has been used as a compound fermenter for yogurt production (Zhao and Liang, 2022). High EPS production is an important probiotic property of Lp. plantarum MC5, and the whole-genome sequence of this strain has been predicted and annotated through various databases (COG, KEGG, GO, CAZy, etc.). In this study, we revealed the general characteristics of the genome and the EPS-producing mechanism of this strain at the genetic level, including the sugar phosphotransferase system (PTS), the eps biosynthetic pathways, key enzymes, and gene clusters involved in EPS biosynthesis. The genetic characteristics of strain MC5 are important for the full exploitation and development of its probiotic properties, and also provide a genetic explanation for the high EPS production characteristics of strain MC5.
2. Materials and methods
2.1. Lp. plantarum MC5 culture conditions
Lp. plantarum MC5 was isolated from traditional fermented yak yogurt in the Tibetan region of Gansu and stored in skimmed milk glycerol tubes at −80°C. Strain MC5 was inoculated 4% (w/v) in MRS (Solarbio Technology Co., Beijing, China) broth for two generations (34°C, 30 h), and the suspensions were collected by centrifugation (5,000 r/min, 4°C for 10 min) for subsequent experiments.
2.2. Extraction of genomic DNA from Lp. plantarum MC5
Genomic DNA was extracted from strain MC5 using the CTAB method (cetyltrimethylammonium bromide) (Arseneau et al., 2017). DNA quality control was performed using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) to determine DNA concentration, and using a Qubit 3.0 (Life Technologies, Carlsbad, CA, USA) for DNA quantification, followed by 0.8% (w/v) agarose gel electrophoresis to determine the purity of DNA.
2.3. Identification and whole-genome sequence analysis of Lp. plantarum MC5
2.3.1. Biochemical identification of Lp. plantarum MC5
Gram staining was performed using a Gram staining kit (Solarbio Technology Co., Beijing, China). Chemical reagents used for staining included ammonium oxalate crystalline violet staining solution, iodine solution, 95% (v/v) alcohol (decolorization solution), and red staining solution. Under a fluorescence microscope, Gram-positive bacteria appear purple and Gram-negative bacteria appear red (Asuman and Emin, 2018).
To test for catalase activity, A colony of Lp. plantarum MC5 growing on MRS agar medium was selected and placed onto a slide; 0.5–1.0 ml 3% (v/v) H2O2 solution was added slowly, and the production of bubbles within 30 s was observed. Those colonies with bubbles were positive for catalase, and those colonies without bubbles were negative (Asuman and Emin, 2018).
2.3.2. Whole-genome sequence analysis of Lp. plantarum MC5
The genome of Lp. plantarum MC5 was sequenced using Illumina and PacBio Sequel II platforms (OEbiotech, Qinghai, China). Denovo assembly of triple sequencing data was performed using Wtdbg2 software (Ruan and Li, 2019), which obeys the overlap-layout-consensus model. The algorithm was developed based on the Fuzzy Bruin Graph (FBG) theory.
2.4. Genome annotation of Lp. plantarum MC5
The software, prodigal (full name: Prokaryotic Dynamic Programming Genefinding Algorithm) (Hyatt et al., 2010), was used to predict coding genes. Diamond software (Buchfink et al., 2015) was used to compare predicted coding sequences, taking annotations with e < 1e-5 and filtering for proteins with the highest sequence similarity to obtain functional annotation information. HMMER software (Eddy, 2009) was used to compare these sequences with protein family models to determine the highest-scoring families. The predicted coding sequence regions were then compared against the NR (non-redundant),1 KEGG (Kyoto Encyclopedia of Genes and Genomes),2 COG (Cluster of Orthologous Groups of proteins),3 GO (Gene Ontology)4 and CAZy (Carbohydrate-Active Enzymes Database)5 databases to complete the basic functional prediction and annotation. Circos (v0.69) software (Krzywinski et al., 2009) was used to present a combination of information in a single genomic circle map.
2.5. Comparative genomics analysis of Lp. plantarum MC5
The whole-genome sequence of strain MC5 was compared with those of seven strains of LAB (different number of eps genes) downloaded from the NCBI database (Table 1).6 A whole-genome sequence phylogenetic tree of strain MC5 was reconstructed by Mega 6.4 (Mega Limited, Auckland, New Zealand) using the neighbor-joining method (Davidson and Martín Del Campo, 2020). Protein sequences homologous to the eps biosynthesis-related gene cluster in the Lp. plantarum MC5 genome were analyzed using blast (see text footnote 6).
TABLE 1.
Genomic information of other lactic acid bacteria.
| Strains | GenBank ID | eps genes |
| Lactiplantibacillus plantarum WCFS1 | NC_004567.2 | epsG |
| Lactiplantibacillus plantarum subsp. plantarum ST-III | CP002222.1 | eps2ABCE, wzx, eps3ABDEFHIJ, eps4ABCEFGHIJ |
| Lactiplantibacillus plantarum JDM1 | NC_012984.1 | epsD/epsB, epsF |
| Lactiplantibacillus plantarum subsp. plantarum NC8 | AGRI00000000.1 | – |
| Lactiplantibacillus plantarum subsp. plantarum ATCC 14917 | NZ_ACGZ00000000.2 | – |
| Lactiplantibacillus plantarum 19L3 | NZ_AWTS00000000.1 | – |
| Lactiplantibacillus plantarum strain SMB758 | CP118222.1 | epsD/epsB, epsF, epsG |
2.6. Determination of the EPS content of Lp. plantarum MC5
Lp. plantarum MC5 was inoculated at 4% (v/v) into MRS broth with glucose, lactose, galactose, sucrose, fructose, cellobiose, mannose, trehalose, or arabinose as the sole carbon source, respectively, for 30 h of fermentation. Cultures were then centrifuged (8,000 r/min, 4°C for 20 min), and the supernatant was collected, respectively. The supernatant (5 ml) was mixed with 20% (w/v) trichloroacetic acid (TCA), allowed to stand at 4°C for 24 h, and then centrifuged again (8,000 r/min, 4°C for 20 min). The supernatant was collected, mixed with 95% alcohol (3:1, v/v), and allowed to stand again at 4°C for 24 h. The mixture was centrifuged (8,000 r/min, 4°C for 20 min), and the pellet was suspended in deionized water and dialyzed at 4°C for 2 days using dialysis bags (molecular weights of 8–14 kDa). Afterward, the dialysate was concentrated and vacuum freeze-dried. The EPS concentration in the suspension after dialysis was quantified using the phenol-sulfuric method of Charchoghlyan et al. (2017) and expressed as glucose equivalent with glucose as the standard (Panthavee et al., 2017).
2.7. Statistical analysis
All experiments were repeated three times. SPSS 22.0 (Statistical Package for the Social Sciences, Chicago, IL, USA) was used for statistical analysis. Experimental differences were determined using ANOVA and Duncan’s test at 0.05 levels. Figures were drawn using Origin 8.0 software (Statistical Package for the Social Sciences, Northampton, MA, USA).
3. Results
3.1. Genomic characteristics of Lp. plantarum MC5
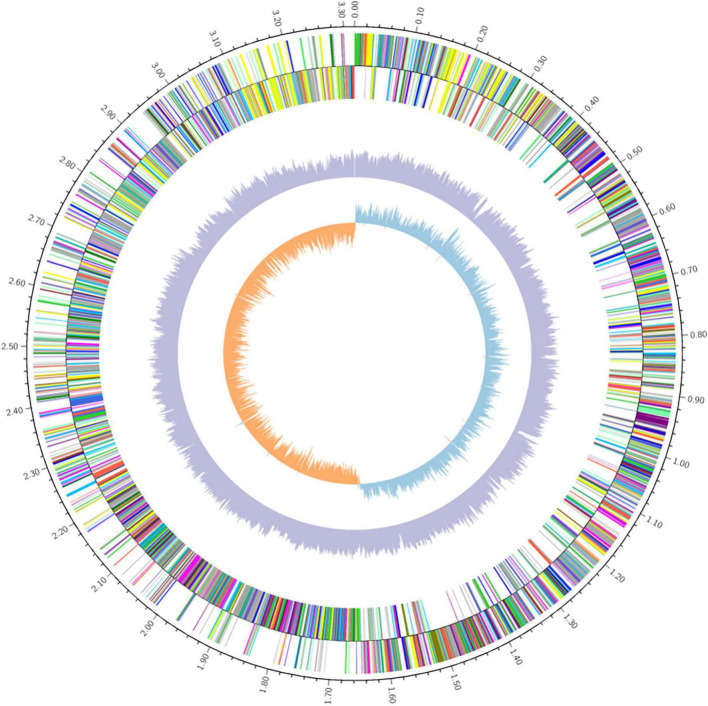
The whole-genome sequence of Lp. plantarum MC5 was obtained using PacBio Sequel II sequencing technology. The genome sequence of MC5 was submitted to NCBI (accession ID: PRJNA897707). The genome of MC5 was 3,317,904 bp in total length, consisting of one circular chromosome of 2,791,014 bp, with a GC content of 45.55%, and two plasmids, plasmid 1 (60,060 bp with 38.79% GC content), and plasmid 2 (15,567 bp with 36.82% GC content). We predicted 3,138 genes, 15 rRNAs (5S, 16S, 23S), 74 tRNAs, and 38 sRNAs (Table 2). The whole-genome circle map of strain MC5 is shown in Figure 1, which comprehensively shows the characteristics of the genome, such as the distribution of genes on the forward and antisense strands, the COG functional classification of genes, GC content, genomic islands, homologous genes, etc.
TABLE 2.
Assembly statistics of the Lp. plantarum MC5 genome.
| Sample | Genome size (bp) | Gene number | GC (%) | Gene length/ genome size (%) |
| Chromosome | 2,791,014 | 3138 | 45.55 | 84.12 |
| Plasmid 1 | 60,060 | 82 | 38.79 | 78.09 |
| Plasmid 2 | 15,567 | 46 | 36.82 | 56.54 |
FIGURE 1.
Whole genome circle map of Lp. plantarum MC5. From inside to outside: GC skew, GC content, non-coding RNA (rRNA red, tRNA blue, sRNA green), lagging strand COG annotations, leading strand COG annotations.
3.2. Functional annotation of the Lp. plantarum MC5 genome
The predicted protein sequences encoded by the MC5 genome were annotated using NR, KEGG, COG, eggNOG, GO, CAZy, and other databases. If there was more than one annotation result, the one with the best ratio was selected as the gene annotation.
3.2.1. Identification of Lp. plantarum MC5
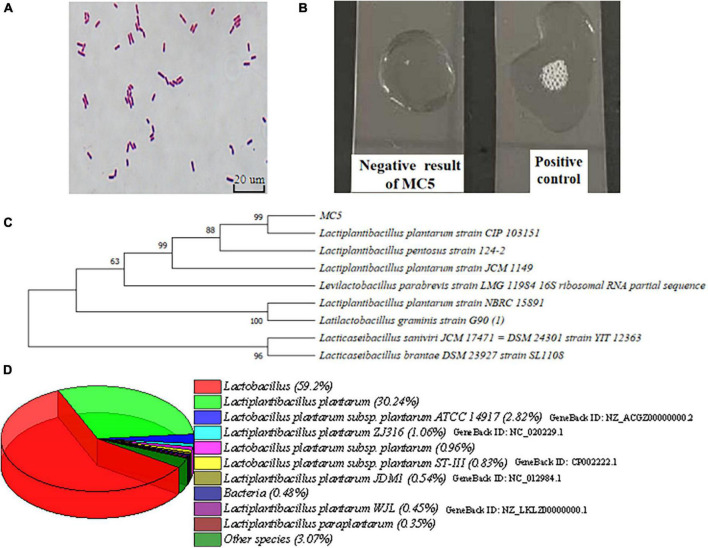
The Gram stain of strain MC5 was purple, and the catalase test did not produce bubbles, indicating that strain MC5 is Gram-positive and catalase-negative. MC5 produced small white colonies on MRS agar and appeared as short rods under fluorescence microscopy (Figures 2A, B). Strain MC5 shared the highest sequence similarity of the 16S rRNA gene with Lp. plantarum CIP 103151 (99%) and Lp. plantarum JCM 1149 (92%), so strain MC5 was identified as Lp. plantarum (Figure 2C, Zhao and Liang, 2023). The NR (non-redundant) database excludes the most significant sources of redundant sequences (EST, STS, GSS, HTGS) and contains protein sequences translated from DNA sequences preserved throughout NCBI GenBank; making the information in this database is very comprehensive. Annotation results from this database contained information on the species annotated according to genes, and statistics on the number of species and genes annotated. Species identification results of MC5 protein sequences in the NR database included Lactobacillus (59.20%), Lp. plantarum (30.24%), and Lp. plantarum ZJ316 (1.06%). NR database annotation of strain MC5 indicated that the strain was most closely related to Lp. plantarum, which verified the above identification results (Figure 2D).
FIGURE 2.
(A) Morphology of strain MC5 by fluorescence microscopy; (B) catalase test of MC5; (C) phylogenetic tree of 16S rRNA gene sequences, and numbers at nodes represent represent homology; and (D) species identification of MC5 using annotations from the NR database.
3.2.2. KEGG and COG database annotation of Lp. plantarum MC5
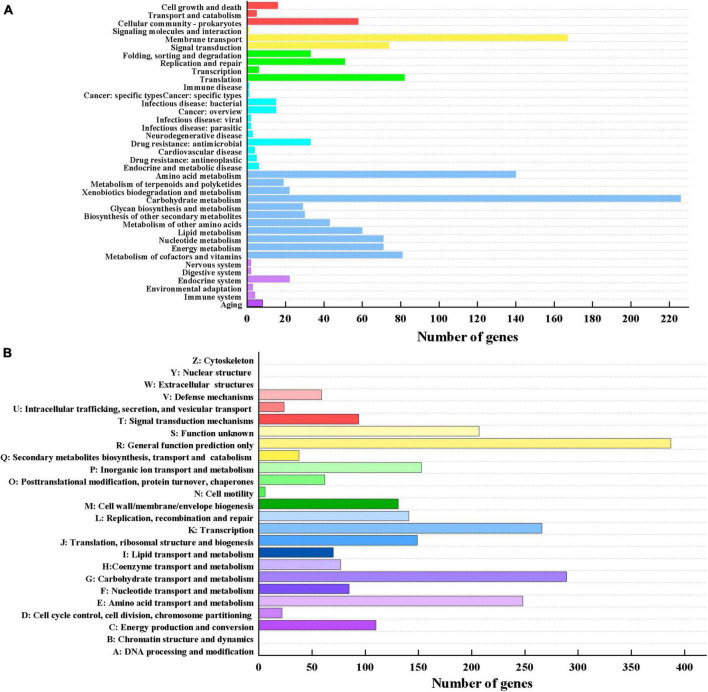
A total of 1,413 genes were annotated in the KEGG database (Figure 3A), classified into six major categories (38 subcategories). There were 79, 242, 172, 87, 792, and 41 annotated genes associated with cellular processes, environmental information processing, genetic information processing, human diseases, metabolism, and organismal system, respectively. The greatest proportion of annotated genes (792) was in the metabolic category, where 226 and 140 genes were related to carbohydrate metabolism and amino acid metabolism, respectively. Genes associated with membrane transport had the highest abundance (167) among genes annotated in the environmental information processing category. Most of the functional genes in the eps gene cluster were annotated with membrane transport functions. Genes associated with translation comprised the highest proportion (82) of genes annotated in the genetic information processing category. The above results indicated that MC5 has strong capacity for carbohydrate metabolism, amino acid metabolism, and membrane transportation.
FIGURE 3.
(A) KEGG functional annotation of Lp. plantarum MC5. The six different colors from top to bottom represent cellular processes, environmental information processing, genetic information processing, human diseases, metabolism, and organismal systems, respectively. (B) COG functional annotation of Lp. plantarum MC5.
A total of 2,618 functional genes were annotated in the COG database (Figure 3B). Figure 3B shows that a large proportion of proteins were identified in the categories of general function prediction only (R, 387), carbohydrate transport and metabolism (G, 289), transcription (K, 266), and amino acid transport and metabolism (E, 248), while a small proportion were identified in the category of cell motility (N, 6). Notably, among the annotated carbohydrate transport and metabolism genes were a large number of genes related to the biosynthesis of sugar nucleotides, such as those encoding lactose/galactose permease, 1-phosphofructokinase, fructokinase, UDP-glucose-hexose-1-phosphate uridylyltransferase, fructose-bisphosphate aldolase, glucosamine-6-phosphate deaminase, and glucose-6-phosphate-1-dehydrogenase. These results suggested that this strain has a strong capacity for sugar biosynthesis and transport.
3.3. CAZy database annotation of Lp. plantarum MC5
The above findings revealed that strain MC5 has strong carbohydrate metabolism and EPS biosynthesis potential. To investigate the EPS biosynthesis mechanism, we analyzed the carbohydrate-active enzymes, sugar transport systems, and nucleotide sugar biosynthesis pathways of strain MC5 at the genomic level.
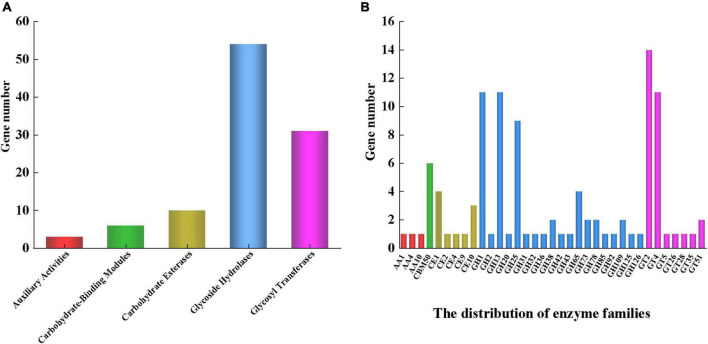
We identified five major classes of genes for MC5 in the CAZy database, of which glycoside hydrolase (GH, 54) and glycosyltransferase (GTF, 31) related genes were the most abundant (Figure 4A). The most abundant GH genes annotated in strain MC5 were GH1, GH13, and GH25, with 11, 11, and 9, respectively. This could be because the isolation source of strain MC5 (yak yogurt) is rich in carbohydrates. CAZy annotation results showed that GH1 family genes mainly encode beta-glucosidase, beta-galactosidase, and lactase; GH13 family genes mainly encode pullulanase, maltotriose-forming alpha-amylase, and amylosucrase. Thus, the abundance of genomic GH means that strain MC5 can efficiently utilize metabolically different carbon sources, allowing the formation of nucleotide sugar bases. In addition, we annotated in seven classes of GTs family genes in strain MC5, with 14 and 11 genes belonging to GT2 and GT4, respectively (Figure 4B). The above results indicated that strain MC5 has a high capacity for sugar biosynthesis. Notably, the only carbohydrate-binding modules (CBM) family gene annotated in the CAZy results was CBM50 (6), which primarily encodes modules found attached to various enzymes from families GH18, GH19, GH23, GH24, GH25, and GH73, i.e., enzymes cleaving either chitin or peptidoglycan. CBM50 modules are also found in a multitude of other enzymes targeting the peptidoglycan such as peptidases and amidases (Sofie et al., 2021). Ten genes of the carbohydrate esterase (CE) family were annotated (including CE1, CE2, CE9, and CE10), which primarily encode acetyl xylan esterase, diacylglycerol O-acyltransferase, and N-acetylglucosamine 6-phosphate deacetylase.
FIGURE 4.
(A) CAZy functional annotation of Lp. plantarum MC5. (B) Distribution of related genes. Red, green, yellow, blue, and purple represent auxiliary activities, carbohydrate-binding modules, carbohydrate esterases, glycoside hydrolases, and glycosyl transferases, respectively.
3.4. The sugar transport system of Lp. plantarum MC5
The annotation results of this experiment showed that strain MC5 has a total of 61 PTS-type sugar transporter proteins associated with a variety of sugars, including 11 sugar-specific transporters such as sucrose PTS EIIBCA or EIIBC components, mannitol PTS EIICBA or EIICB components, cellobiose PTS EIIC component, and fructose PTS EIIBC or EIIC components (Supplementary Table 1). According to the transporter database, these transporters can be classified into thirteen PTS systems with the following distributions: cellobiose PTS (16), beta-glucoside PTS (8), mannose PTS (6), glucitol/sorbitol PTS (4), fructose PTS (3), ascorbate PTS (3), galactitol PTS (3), galactosamine PTS (2), sucrose PTS (2), mannitol PTS (2), sugar PTS (2), N-acetylglucosamine PTS (1), and fructoselysine/glucoselysine PTS (1).
The above results indicated that strain MC5 can utilize cellobiose, glucoside, mannose/mannitol, fructose, sucrose, ascorbate, galactitol/galactosamine, glucitol/sorbitol, and N-acetylglucosamine. In addition, we found that some of the oligosaccharide phosphate transfer system genes (for cellobiose, mannose, fructose, etc.) in the genome of strain MC5 occurred in multiple copies, and this coding redundancy could enhance the ability of MC5 to utilize these carbon sources. These functional proteins would provide a competitive advantage for the survival of strain MC5 in complex environments.
3.5. Nucleotide sugar biosynthesis pathway of Lp. plantarum MC5
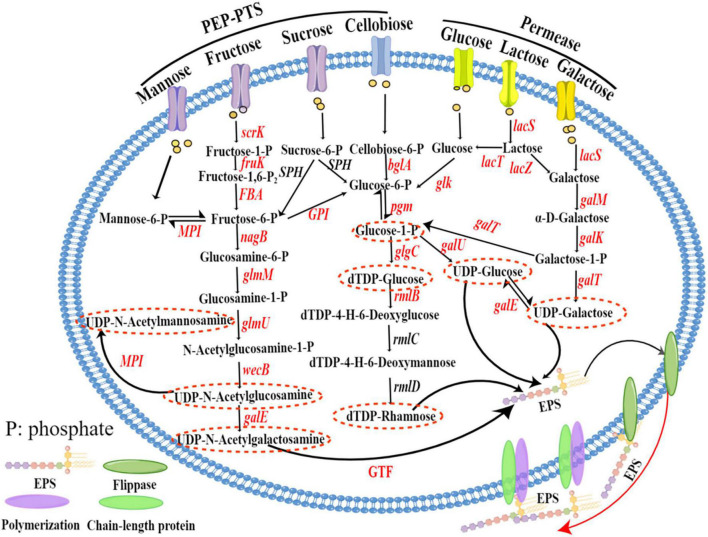
Nucleotide sugars are precursors in the biosynthesis of EPS. Table 3 shows the key enzymes in EPS biosynthesis annotated in the KEGG database that convert different carbon sources into nucleotide sugars. The sugar sources available to the strain can be inferred from these enzymes. We identified a total of 21 key enzymes involved in the EPS biosynthesis process of strain MC5, involved in the metabolism of mannose, fructose, sucrose, cellobiose, glucose, lactose, and galactose, respectively. The nucleotide sugar biosynthesis pathways of strain MC5 were predicted according to the KEGG metabolic pathways (Figure 5). In LAB, the PTS and permease pathways are essential for the transport of monosaccharides and disaccharides. As shown in Figure 5, mannose, fructose, sucrose, and cellobiose are transported through the PEP-PTS from outside the cell into the cell, where they are phosphorylated to produce mannose 6-phosphate, fructose 1-phosphate, sucrose 6-phosphate, and cellobiose 6-phosphate, respectively. These are then converted to fructose 6-phosphate and glucose 6-phosphate by related enzymes (fruK, FBA, GPI, bglA, SPH) and are eventually involved in the biosynthesis of UDP-N-acetylglucosamine, dTDP-rhamnose, UDP-glucose, and dTDP-glucose. Importantly, glucose 6-phosphate and glucose 1-phosphate are converted by the catalytic reaction of pgm, which is involved in the biosynthesis of dTDP-glucose, UDP-glucose, and dTDP-rhamnose. Glucose 1-phosphate is catalyzed by enzymes encoded by glgC, galU, galE, rmlB, rmlC, and rmlD to biosynthesize dTDP-glucose, UDP-glucose, UDP-galactose, and dTDP-rhamnose, respectively. Lactose is converted to glucose 6-phosphate by the catalytic reaction of glucokinase (glk) and 6-phosphate-beta-glycosidase, and is eventually converted to UDP-glucose, dTDP-glucose, and dTDP-rhamnose by the catalytic reactions of glucose-6-phosphate isomerase, UTP-glucose-1-phosphate uridylyltransferase, glucose-1-phosphate adenylyltransferase (glgC), dTDP-glucose-4,6-dehydratase (rmlC), and dTDP-4-dehydrorhamnorea reductase (rmlD).
TABLE 3.
Key enzymes of the nucleotide sugar biosynthesis process of Lp. plantarum MC5 annotated in the KEGG database.
| Locus tag | Gene name | Encoded protein |
| OOD38_0176 | scrK | Fructokinase |
| OOD38_2099 | GPI | Glucose-6-phosphate isomerase |
| OOD38_2320 | bglA | 6-Phosphate-β-glucosidase |
| OOD38_0673 | pgm | Phosphoglucomutase |
| OOD38_0661 | galU | UTP-glucose-1-phosphate uridylyltransferase |
| OOD38_1828 | galE | UDP-glucose 4-epimerase |
| OOD38_0020 | glgC | Glucose-1-phosphate adenylyltransferase |
| OOD38_1337 | glk | Glucokinase |
| OOD38_2959 | lacS | Lactose/raffinose/galactose permease |
| OOD38_2973 | lacZ | Beta-galactosidase |
| OOD38_2977 | galM | Aldose 1-epimerase |
| OOD38_2972 | galK | Galactokinase |
| OOD38_2970 | galT | UDP-glucose–hexose-1-phosphate uridylyltransferase |
| OOD38_1819 | fruK | 1-phosphofructokinase |
| OOD38_0304 | FBA | Fructose-bisphosphate aldolase |
| OOD38_0210 | nagB | Glucosamine-6-phosphate deaminase |
| OOD38_0721 | glmM | Phosphoglucosamine mutase |
| OOD38_0422 | glmU | Glucosamine-1-phosphate N-acetyltransferase |
| OOD38_1001 | wecB | UDP-N-acetylglucosamine 2-epimerase |
| OOD38_2069 | MPI | Mannose-6-phosphate isomerase |
| OOD38_1828 | rmlB | dTDP-glucose-4,6-dehydratase |
FIGURE 5.
Nucleotide sugar biosynthesis pathways of Lp. plantarum MC5. Nucleotide sugar biosynthesis pathways mapped by Figdraw (https://www.figdraw.com/static/index.html#/). Annotated genes are marked in red, and the nucleotide sugars are in red dashed ovals. scrK, fructokinase; SPH, sucrose-6-phosphate hydrolase; GPI, glucose-6-phosphate isomerase; bglA, 6-phosphate-β-glucosidase; pgm, phosphoglucomutase, galU, UTP-glucose-1-phosphate uridylyltransferase; galE, UDP-glucose 4-epimerase; glgC, glucose-1-phosphate adenylyltransferase; rmlB, dTDP-glucose-4,6-dehydratase; rmlC, dTDP-4-dehydrorhamnosac-3,5-epimerase; rmlD, dTDP-4-dehydrorhamnorea reductase; glk, glucokinase; lacS, lactose/galactose permease; lacZ, β-galactosidase; galM, aldose 1-epimerase; galK, galactokinase; galT, UDP-glucose-hexose-1-phosphate uridylyltransferase; fruK, 1-phosphofructokinase; FBA, fructose-bisphosphate aldolase; nagB, glucosamine-6-phosphate deaminase; glmM, phosphoglucosamine mutase; glmU, glucosamine-1-phosphate N-acetyltransferase; wecB, UDP-N-acetylglucosamine 2-epimerase (non-hydrolyzing); MPI, mannose-6-phosphate isomerase; GTF, glycosyltransferase.
Therefore, analysis of the predicted genes indicated that strain MC5 can biosynthesize seven sugar nucleotides: UDP-glucose, UDP-galactose, dTDP-glucose, dTDP-rhamnose, UDP-N-acetylglucosamine, UDP-N-acetylgalactosamine, and UDP-N-acetylmannosamine. The genes encoding all of these enzymes were identified in strain MC5, indicating strong tolerance and adhesion of MC5.
3.6. Comparative genomic analysis of different eps gene strains
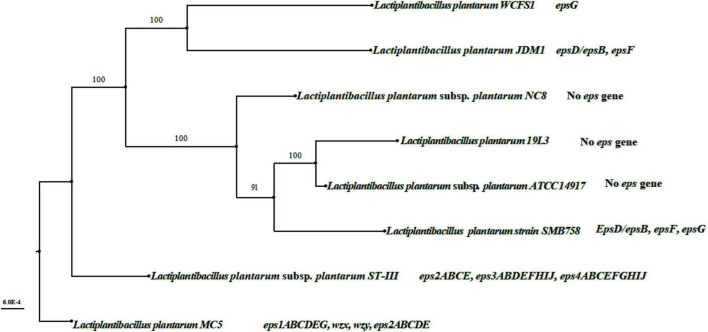
Sequences of seven Lactiplantibacillus strains containing different numbers of eps genes were downloaded from the NCBI database; the names, GenBank IDs, and eps genes of these seven strains are shown in Table 1 and Supplementary Table 2. Strains Lp. plantarum subsp. plantarum NC8, Lp. plantarum 19L3, and Lp. plantarum subsp. plantarum ATCC 14917 had no eps genes, strains Lp. plantarum WCFS1 and Lp. plantarum JDM1 contained very few eps genes (epsB/epsD, epsG), and strain Lp. plantarum SMB758 contained epsB/epsD, epsF, and epsG. Notably, strain Lp. plantarum subsp. plantarum ST-III contained three eps gene clusters, eps2ABCEFGHI, eps3ABDEFHIJ, and eps4ABCEFGHIJ, respectively (Table 1). Figure 6 compares the clustering relationships of LAB containing different numbers of eps genes. The three strains without eps genes (Lp. plantarum subsp. plantarum NC8, Lp. plantarum 19L3, and Lp. plantarum subsp. plantarum ATCC 14917) were clustered together, the two strains with few eps genes (Lp. plantarum WCFS1 and Lp. plantarum JDM1) were clustered together, and the strain with a complete eps gene cluster (Lp. plantarum subsp. plantarum ST-III) formed a separate branch. The strains with different numbers of eps genes were sister branches of each other. Lp. plantarum MC5 and Lp. plantarum subsp. plantarum ST-III were the closest relatives, as these two strains contained the most eps gene clusters and had the highest EPS production capacity.
FIGURE 6.
Whole-genome phylogenetic tree of Lp. plantarum strains with different eps genes. The whole genome sequence of the 7 strains of LAB in Figure 6 was downloaded from the NCBI database, and the GenBank ID of these 7 strains were shown in Table 1. Mega 6.4 (Mega Limited, Auckland, New Zealand) software done their whole genome phylogenetic tree.
3.7. Comparative analysis of eps gene clusters of different lactic acid bacteria
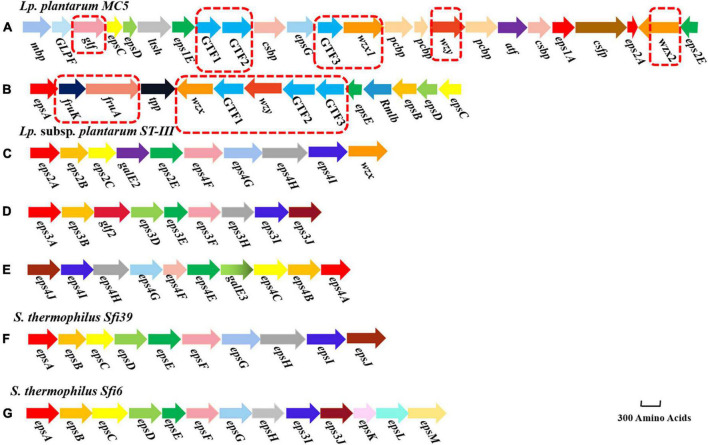
It has been shown that there is a cluster of related genes located on plasmids or chromosomes that control extracellular polysaccharide biosynthesis; these are organized into four parts: a gene regulatory region (epsAB), genes encoding a related protein involved in the detection of polymeric chain length (epsCD), genes encoding a glycosyltransferase (GTF) that biosynthesizes extracellular polysaccharide repeat units, and genes for polysaccharide polymerization, transport, and export functions (wzx and wzy) (Soumya and Nampoothiri, 2021). Lp. plantarum subsp. plantarum ST-III (GenBank ID: CP002222.1), S. salivarius subsp. thermophilus Sfi39 (GenBank ID: AF373595.1), and S. salivarius subsp. thermophilus Sfi6 (GenBank ID: U40830.1) were used as controls for comparing known eps gene clusters with those of strain MC5 (Figure 7). Figure 6 shows that genes encoding eps in LAB include epsA, epsB, epsC, epsD, epsE, epsF, epsG, epsH, epsI, glf, GTF, galE, wzx, and wzy. The genes epsA, epsB, epsC, epsD, and epsE were common among the seven eps gene clusters (a-g), indicating that they are highly conserved and stable in the eps gene clusters of LAB. However, epsF, epsG, epsH, epsI, epsJ, and epsK were unique to S. salivarius subsp. thermophilus and encode mainly glycosyltransferases and membrane transfer-associated proteins in this species. The genes fruK, fruA, glf, GTF, wzx, and wzy were unique to Lp. plantarum MC5. Glycosyltransferase genes differed in type, number, location, and order among the seven eps gene clusters (a-g), making them specific and variable.
FIGURE 7.
Structure of the eps gene clusters of Lp. plantarum MC5 (A: eps cluster 1, B: eps cluster 2), Lp. plantarum subsp. plantarum ST-III (C: eps cluster 1, D: eps cluster 2, and E: eps cluster 3), S. salivarius subsp. thermophilus Sfi39 (F: eps cluster 1), and S. salivarius subsp. thermophilus Sfi6 (G: eps cluster 1). Different colors indicate the different functions of genes. Genes in red dashed boxes are MC5-specific genes.
Whole-genome analysis of Lp. plantarum MC5 showed that its chromosome encodes two complete eps genes clusters, eps C1 and eps C2 (Figures 7A, B). The two eps gene clusters were 26 and 14 kb in length and consisted of 24 and 14 ORFs, respectively (Table 4). These two eps genes clusters contained epsABCDE. The epsCD in eps cluster 1 was located in the conserved region at the beginning of the cluster and was mainly involved in chain-length regulation of EPS biosynthesis. The first gene in the cluster, epsA, has homology to the family of transcriptional regulators, in Streptococcus pneumoniae deletion of a similar gene cpsIaA caused a reduction in capsule production (Dertli et al., 2016). The amino acid sequence identity of eps1A in eps C1 with GenBank ID: MCG0826270.1 was 99.60%, and eps2A in eps C1 with GenBank ID: WP_151502160.1 was 99.32%, respectively. The amino acid sequence homology of the epsA gene of S. salivarius subsp. thermophilus Sfi39 with ID: MCE2268317.1 was 99.38%. The amino acid sequence homology of the epsA gene of S. salivarius subsp. thermophilus Sfi6 with ID: MCE2192772.1 was 98.35%. This suggested that eps1A and eps2A have been relatively conserved during evolution. Because the epsA gene is mainly responsible for the transcription and regulation of EPS genes during EPS biosynthesis, it determines the direction and degree of expression of eps genes. Interestingly, eps cluster 1 contained two epsE genes, indicating that strain MC5 has a strong ability to start glycosyl transfer. The amino acid sequence identity of eps1E and eps2E encoded by eps C1 was 60.16% (GenBank ID: KRN47847.1) and 100.00% (GenBank ID: ACT61928.1), respectively (Table 4). Although these two epsA genes were located at the end of cluster 1, they play a major role in the transcriptional regulation of EPS biosynthesis. In addition, eps cluster 1 contained four conserved polysaccharide biosynthesis proteins (amino acid sequence identity of both genes is > 90%) and two capsular biosynthesis proteins that may play a key role in the biosynthesis of EPS. Both proteins were also annotated in strain Lactobacillus plantarum subsp. plantarum ST-III (GenBank ID: CP002222.1). Furthermore, strain MC5 eps C1 contained a unique epsG gene that encodes a membrane transporter protein. By contrast, the conserved gene in eps C2, located at the beginning of the cluster, was epsA, while epsBCDE was located at the end of the cluster and had a negative transcriptional orientation. In eps C2, the amino acid sequence identity of epsA with GenBank ID: OEZ35589.1 was 99.60%, the amino acid sequence identity of epsB with GenBank ID: MCG0716518.1 was 99.63%, the amino acid sequence identity of epsC with GenBank ID: MCG0585072.1 was 97.70%, the amino acid sequence identity of epsD with GenBank ID: WP_213474994.1 was 99.57%, and the amino acid sequence identity of epsE with GenBank ID: ADN98952.1 was 100.00%, respectively (Table 4).
TABLE 4.
Information about the eps gene clusters and homologous sequences of Lp. plantarum MC5.
| Locus tag | Sequence length of AA | Gene name | Gene direction | Gene function | GenBank ID | Homology (%) |
| eps genes cluster 1 | ||||||
| OOD38_1002 | 309 | mbp | + | Integral membrane protein | MCG0749228.1 | 99.68 |
| OOD38_1003 | 238 | GLPF | + | Glycerol uptake facilitator protein | KRO27638.1 | 96.49 |
| OOD38_1004 | 372 | glf | + | UDP-galactopyranose mutase | WP_063490764.1 | 99.73 |
| OOD38_1005 | 217 | epsC | + | Polysaccharide biosynthesis protein | MCG0678676.1 | 37.91 |
| OOD38_1006 | 194 | epsD | + | CpsD family tyrosine-protein kinase | WP_181589845.1 | 31.34 |
| OOD38_1007 | 371 | ltsh | + | Class A beta-lactamase-related serine hydrolase | MCT3250941.1 | 99.73 |
| OOD38_1008 | 244 | eps1E | + | Lipopolysaccharide biosynthesis glycosyltransferase | KRN47847.1 | 60.16 |
| OOD38_1009 | 298 | GTF1 | + | Glycosyltransferase | WP_195631017.1 | 99.66 |
| OOD38_1010 | 354 | GTF2 | + | Glycosyltransferase | WP_063490758.1 | 99.72 |
| OOD38_1011 | 332 | csbp | + | Capsular biosynthesis protein | MPQ37623.1 | 55.45 |
| OOD38_1012 | 365 | epsG | + | Transmembrane protein epsG | MBP5842394.1 | 99.45 |
| OOD38_1013 | 297 | GTF3 | + | Glycosyltransferase | AUV72009.1 | 44.37 |
| OOD38_1014 | 473 | wzx1 | + | Flippase | WP_260203456.1 | 78.30 |
| OOD38_1015 | 295 | pcbp | + | Polysaccharide biosynthesis protein | WP_237421427.1 | 99.66 |
| OOD38_1016 | 207 | pcbp | + | Polysaccharide biosynthesis protein | WP_262339904.1 | 99.52 |
| OOD38_1017 | 398 | wzy | + | Polysaccharide polymerase | ERO40262.1 | 99.50 |
| OOD38_1018 | 369 | pcbp | + | Polysaccharide biosynthesis protein | WP_088467704.1 | 99.19 |
| OOD38_1019 | 359 | atf | + | O-acetyltransferase | MCG0909952.1 | 99.72 |
| OOD38_1020 | 258 | csbp | + | Capsular biosynthesis protein | KRK22728.1 | 86.67 |
| OOD38_1021 | 248 | eps1A | − | Transcription regulator, AraC family | MCG0826270.1 | 99.60 |
| OOD38_1022 | 1133 | csfp | + | Cell surface protein precursor | KRL33646.1 | 93.88 |
| OOD38_1023 | 146 | eps2A | + | MarR family transcriptional regulator | WP_151502160.1 | 99.32 |
| OOD38_1024 | 473 | wzx2 | − | Flippase | WP_087613142.1 | 99.79 |
| OOD38_1025 | 222 | eps2E | − | Priming glycosyltransferase | ACT61928.1 | 100.00 |
| eps genes cluster 2 | ||||||
| OOD38_1818 | 251 | epsA | + | DeoR/GlpR transcriptional regulator | OEZ35589.1 | 99.60 |
| OOD38_1819 | 305 | fruK | + | 1-phosphofructokinase | WP_003640776.1 | 100.00 |
| OOD38_1820 | 655 | fruA | + | Fructose PTS, EIIABC | WP_128536963.1 | 99.85 |
| OOD38_1821 | 440 | Tpp | + | Serine/threonine protein phosphatase family protein | ETF10433.1 | 99.55 |
| OOD38_1822 | 483 | wzx | − | Oligosaccharide flippase family protein | WP_061871654.1 | 100.00 |
| OOD38_1823 | 322 | GTF1 | − | Glycosyltransferase | WP_133279738.1 | 99.69 |
| OOD38_1824 | 424 | wzy | − | Polysaccharide polymerase | WP_128468359.1 | 99.76 |
| OOD38_1825 | 342 | GTF2 | − | Glycosyltransferase | WP_112297297.1 | 99.71 |
| OOD38_1826 | 363 | GTF3 | − | Glycosyltransferase | WP_003640783.1 | 99.72 |
| OOD38_1827 | 221 | epsE | − | Priming glycosyltransferase | ADN98952.1 | 100.00 |
| OOD38_1828 | 313 | rmlB | − | dTDP-glucose 4 6-dehydratase | KZU50552.1 | 99.25 |
| OOD38_1829 | 273 | epsB | − | Tyrosine protein phosphatase | MCG0716518.1 | 99.63 |
| OOD38_1830 | 235 | epsD | − | CpsD family tyrosine-protein kinase | WP_213474994.1 | 99.57 |
| OOD38_1831 | 252 | epsC | − | Tyrosine-protein kinase transmembrane modulator epsC | MCG0585072.1 | 97.70 |
Glycosyltransferases (GTFs) can transfer different nucleotide sugars, and GTF and EPS molecular diversity are closely related. Strain MC5 eps C1 and C2 had abundant genes encoding GTFs (6), indicating a high polymorphism of GTFs. We also found that the GTF genes of MC5 mainly encoded galactosyltransferase, glucosyltransferase, and D-mannosaminyltransferase, leading to the assumption that this strain can biosynthesize EPS of different structures. In eps C1 the amino acid sequence identity of GTF1 with GenBank ID: WP_195631017.1 was 99.66%, the amino acid sequence identity of GTF2 with GenBank ID: WP_063490758.1 was 99.72%, and the amino acid sequence identity of GTF3 with GenBank ID: AUV72009.1 was 44.37%, respectively (Table 4). In eps C2, the amino acid sequence homology of GTF1 with GenBank ID: WP_133279738.1 was 99.69%, the amino acid sequence homology of GTF2 with GenBank ID: WP_112297297.1 was 99.71%, and the amino acid sequence homology of GTF3 with GenBank ID: WP_003640783.1 was 99.72%, respectively (Table 4). In addition, the GTF genes included genes encoding furanyltransferase, mannosyltransferase, and α-glucosyltransferase. Both eps C1 and C2 of strain MC5 contained genes encoding wzx and wzy, located in the middle region of the cluster (both of which have > 90% amino acid sequence identity, Table 4). Notably, eps C1 encoded two wzx genes, indicating that cluster 1 has a strong repeat unit transport capacity. The above findings suggested that strain MC5 has a unique EPS biosynthesis mechanism and a strong EPS production capacity.
3.8. Validation of the EPS-producing capacity of Lp. plantarum MC5
Sugar source utilization capacity of MC5 and the number of key enzymes that metabolize the sugar were predicted from KEGG database annotation results (Table 5). MC5 was predicted to contain key enzymes for metabolism of nine sugar sources (glucose, lactose, sucrose, arabinose, trehalose, fructose, cellobiose, galactose, and mannose), indicating that strain MC5 can metabolize these nine sugar sources. However, key metabolic enzymes of five sugar sources (mannitol, xylose, raffinose, rhamnose, and sorbitol) were not predicted, indicating that strain MC5 cannot metabolize these five sugars. We predicted two permeases for raffinose metabolism and three PTS transport systems for sorbitol metabolism, indicating that these two sugars might enter the cells of strain MC5 but not be metabolized.
TABLE 5.
Prediction of sugar source utilization capacity of MC5 and number of key enzymes that metabolize the sugar based on KEGG database annotation results (number of enzymes).
| Items | Glucose | Trehalose | Lactose | Mannitol | Sucrose | Arabinose | Xylose |
| Number of enzymes | 6 | 3 | 9 | 0 | 4 | 2 | 0 |
| Result | + | + | + | − | + | + | − |
| Items | Raffinose | Fructose | Cellobiose | Rhamnose | Sorbitol | Galactose | Mannose |
| Number of enzymes | 2 permeases | 8 | 4 | 0 | 3 PTS | 8 | 6 |
| Result | − | + | + | − | − | + | + |
“+” means that the sugar can be utilized; “−” means that the sugar cannot be utilized.
Based on the predicted nucleotide sugar biosynthesis pathways of strain MC5 at the gene level, we verified the sugar metabolism capacity of MC5 and the corresponding EPS yield (Figure 8). Using Lp. plantarum CD11 and Lp. plantarum CD36 as controls, we predicted that these three strains of Lp. plantarum can metabolize glucose, lactose, galactose, sucrose, fructose, cellobiose, mannose, trehalose, and arabinose. However, the EPS yield of MC5 was significantly higher than that of CD11 and CD36, which was consistent with the predicted results of the PTS transport systems and sugar metabolism pathways above (sections “3.4. The sugar transport system of Lp. plantarum MC5 and 3.5. Nucleotide sugar biosynthesis pathway of Lp. plantarum MC5”). Furthermore, when these nine sugars were used as the sole carbon source for the biosynthesis of EPS, the EPS yields of the seven sugars predicted by the genomic sequence were all greater than 250 mg/L, indicating that all these sugars are involved in the biosynthesis of EPS. The highest EPS yields were achieved when glucose and lactose were the only carbon sources, at 345.72 and 327.30 mg/L. Although only some of the enzymes of the trehalose and arabinose metabolic pathways were identified in the genome prediction results, the validation results showed that strain MC5 could metabolize both sugars normally, mainly due to the complexity of the EPS biosynthesis pathways and the possible existence of other gene loci and enzymes involved in the metabolism of trehalose and arabinose. The high EPS yield of strain MC5 results from it having two eps gene clusters and the highest number of predicted genes related to carbohydrate metabolism (226) in the KEGG database. The above results showed that strain MC5 had a strong capacity for sugar metabolism and can produce large amounts of EPS.
FIGURE 8.
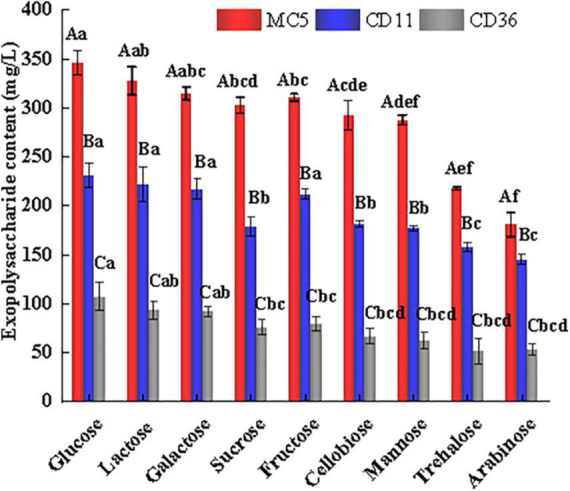
Sugar metabolism types and EPS production of Lp. plantarum MC5 (red column). Experiments were repeated three times. Lp. plantarum CD11 and Lp. plantarum CD36 (blue and gray columns) were the control groups for moderate EPS production and low EPS production, respectively. Capital letters (A, B, and C), respectively, represent the differences in EPS produced by different strains using the same carbon source (p < 0.05). Lowercase letters (a, b, c, d, e, and f) represent the differences between EPS produced by the same strain using different carbon sources (p < 0.05). ANOVA tests were used to determine significant differences between treatments with a significance level of p < 0.05 by the SPSS 22.0 package program. Error bars represent the standard errors (SE) of the mean value (n = 3), and the experiment was repeated three times.
In addition, all three strains produced the highest amount of EPS when glucose and lactose were used as carbon sources and the lowest amount when trehalose and arabinose were used as carbon sources, which may be because all three strains were isolated from a dairy source (traditional fermented yak milk in Gansu) and have undergone evolutionary adaptation to dairy polysaccharides. Among them, Lp. plantarum CD36 produced the lowest EPS content, probably due to the low expression of the eps gene cluster in this strain.
4. Discussion
We studied the genomic features of MC5 and the biosynthetic pathways of EPS from both genomic and phenotypic aspects. Whole-genome sequence results indicated that probiotic MC5 has a strong metabolism of carbohydrates and amino acids. Genes identified in MC5 are chiefly involved in the metabolism of sucrose, galactose, fructose, mannose, and starch, and some intermediates of their metabolic pathways may be involved in the formation of nucleotide sugars (Xiong et al., 2019). MC5 can grow and reproduce on MRS agar containing glucose, sucrose, fructose, or lactose, respectively, validating the annotation results from the KEGG database. Lactic acid bacteria are reported to be rich in carbohydrate metabolism genes and can use a variety of carbohydrates, which might also provide energy for growth (Ghattargi et al., 2018). Our results indicate that strain MC5 has a strong capacity for sugar biosynthesis and transport. Huang et al. (2021) reported that strains with many genes related to amino acid and carbohydrate transporter metabolism (Lp. plantarum DMDL 9010) are highly resistant to the outside environment and have good proliferation capacity, supporting the results of this study. Pessione (2012) also reported that some genes related to carbohydrate metabolism and amino acid decarboxylation play an important regulatory role for LAB in the host gut.
CAZy annotation results showed that GH1 family genes mainly encode beta-glucosidase, beta-galactosidase, and lactase; GH13 family genes mainly encode pullulanase, maltotriose-forming alpha-amylase, and amylosucrase. Lammens et al. (2009) reported that the GH family of genes plays an important role in a variety of biological processes including sugar biosynthesis, cellular metabolism, and signaling. Graebin et al. (2016) and Liang et al. (2022) reported that the glucosidases encoded by GH1 and GH13 family genes produce monosaccharides by hydrolyzing glucosidic bonds, providing precursors for the biosynthesis of EPS. The main component of EPS (nucleotide sugars) might be an important control factor for EPS production levels; this has been evaluated extensively in Lactobacillus cells that produced EPS by regulating nucleotide sugar biosynthetic pathways (Kleerebezem and Hugenholtz, 2003). The abundance of genomic GH in the genome of strain MC5 suggests that it can efficiently utilize different carbon sources, thus allowing the formation of nucleotide sugar bases. In addition, the GT family is one of the key gene families for EPS biosynthesis, but there is a high variability of GT family genes in EPS-producing LAB (Zeidan et al., 2017). GT family genes can catalyze the formation of various sugar conjugates such as oligosaccharides and polysaccharides, as well as the biosynthesis of biologically important carbohydrates (Ibrahim et al., 2016). GT2 and GT4 enzymes are associated with the biosynthesis of sugars such as sucrose, cellulose, lipopolysaccharides, and chitosan, which (especially polysaccharide compounds) may be related to the final viscosity, water-holding capacity, and other qualities of fermented milk (Oehme et al., 2019). The above results indicate that strain MC5 has a high capacity for sugar biosynthesis. Our results also show that strain MC5 used as a compound starter can largely improve the EPS content and rheological properties of yogurt (Zhao and Liang, 2022), verifying the functions of genes predicted in strain MC5.
Genomic analysis showed that MC5 has 11 sugar-specific PTS and seven nucleotide sugar biosynthesis pathways. The PTS system is one of the major functional components of the intracellular transport of glycosides (Morabbi Heravi and Altenbuchner, 2018). It generally consists of the non-specific energy-coupled protease (EI) and phosphocarrier protein (HPr) and the sugar-specific PTS transporter protease complex (EII). The histidine residue at position 15 of the HPr protein is a phosphorylation site dependent on phosphoenolpyruvate, which is catalyzed by EI to form His-P-HPr and functions as a transmitting phosphate group during sugar uptake (Liu et al., 2019). Fagbemigun et al. (2021) reported that four strains have mannose PTS transport systems, with two strains having additional glucitol/sorbitol PTS transport systems, after analyzing their whole-genome sequences, and found that these two strains had greater sugar utilization capacity. Sugar-specific PTS analysis indicated that strain MC5 can utilize cellobiose, glucoside, mannose/mannitol, fructose, sucrose, ascorbate, galactitol/galactosamine, glucitol/sorbitol, and N-acetylglucosamine.
Endogenous levels of the major component of EPS, nucleotide sugars, can be an important control on the level of EPS production (Hugenholtz et al., 2000; Kleerebezem and Hugenholtz, 2003), which has been evaluated extensively in Lactobacillus. Hamon et al. (2014) identified the gene encoding rmlB (Locus tag: Gene-1828) as being associated with low pH tolerance in Lp. plantarum. Adhesion to the epithelium is crucial for a probiotic strain, and genes encoding lactate dehydrogenase (7, Locus tags: Gene-0322, 0469, 0765, 0944, 1038, 1733, 2041) and 6-phosphogluconate dehydrogenase (2, Locus tags: Gene-1043, 1310) have been identified, which are known to promote bacterial adhesion to mucin and epithelial cells (Izquierdo et al., 2009). Previous studies have identified glutathione peroxidase (Locus tag: Gene-0206) as playing an important role in the acid stress response of LAB (Lee et al., 2010). In addition, Fukao et al. (2019b) reported that EPS consisting of glucose and N-acetylglucosamine in Levilactobacillus brevis KB290 is associated with its wrinkled colony morphology, suggesting that EPS is responsible for the formation of wrinkled colonies. Although the presence of some precursor biosynthetic genes in the eps cluster indicates the presence of this sugar in EPS, the deletion of this gene cannot be taken as an indicator of the absence of this sugar in EPS (Dimopoulou et al., 2014). Therefore, it is necessary to use WGS techniques to reveal the relationship between the genotypic and phenotypic profiles of probiotics.
In addition, the EPS biosynthesis capacity of Lactobacillus is controlled by a cluster of eps genes located on chromosomes or plasmids (Deo et al., 2019). Strain MC5 has two complete eps biosynthesis gene clusters (epsABCDE, wzx, wzy, epsG). These two eps genes clusters contain epsABCDE (key genes for EPS biosynthesis) (Kanmani et al., 2018). The expression of epsCD regulatory modules is reported to be coordinated in LAB (Deo et al., 2019). The most critical gene in EPS biosynthesis is epsE (priming glycosyltransferase), which primarily encodes a membrane-associated glycosyltransferase that cannot catalyze the attachment of glycosidic bonds but can transfer 1-phospho-glucose to an undecaprenylphosphate lipid carrier on the cytoplasmic surface of the membrane (Horn et al., 2013). Previous studies have reported that epsA is required for the attachment of EPS to the cell wall (Chan et al., 2014). In addition, eps cluster 1 contains six conserved EPS biosynthetic proteins (> 90% homology) that may play a key role in the biosynthesis of EPS (Song et al., 2018); however, their specific role needs to be further investigated. Kanmani et al. (2018) and Deo et al. (2019) also reported eps genes clusters, wzx, wzy, and GTF in LAB, and the structure of the conserved genes (epsABCDE) in strain MC5 was similar to that of other EPS-producing LAB, suggesting that the prediction was reliable. The wzx and wzy genes are mainly involved in glycosyltransport and polysaccharide polymerization in EPS biosynthesis (Xu et al., 2019). In L. lactis subsp. cremoris SMQ-461, genes with the same function as the wzy polymerase and wzx flippase genes are named epsH and epsM, respectively (Dabour and LaPointe, 2005). Zeidan et al. (2017) reported LAB containing genes associated with EPS production that are capable of producing biofilms and attaching to mucosal surfaces. Martino et al. (2016) compared the eps gene clusters of several Lp. plantarum strains and showed that they were highly variable, while both eps gene clusters of strain MC5 have a highly conserved phenotype. This difference may be caused by the different isolation sources of these Lp. plantarum strains and the sequencing technology employed.
Previous studies also reported monosaccharides and oligosaccharides that can be widely utilized by LAB (Hugenholtz et al., 2000). Our sugar metabolism verification showed that MC5 is able to metabolize the seven sugars (mannose, fructose, sucrose, cellobiose, glucose, lactose, and galactose) predicted by the genomic sequence and is able to produce significant amounts of EPS. Notably, MC5 could additionally metabolize trehalose and arabinose, but EPS production was lower than those of the other seven sugars, probably due to lower transcript levels of the relevant genes when MC5 metabolized these two sugars. Xiong et al. (2019) also confirmed the ability of S. salivarius subsp. thermophilus S-3 containing the eps gene cluster to metabolize lactose, galactose, and glucose. In addition, genes for the uptake and utilization of glucan, lactose, and galactose and the biosynthesis of glucan also have been identified in the genome of L. delbrueckii TUA4408L (Kanmani et al., 2018), providing support for the results of this study. The above results provide better insight into the mechanism of EPS biosynthesis at the levels of biosynthesis pathways, gene clusters, and EPS production.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI, PRJNA897707.
Author contributions
XZ: conceptualization, methodology, validation, formal analysis, data curation, writing—original draft preparation, and writing—editing. QL: conceptualization, methodology, investigation, resources, writing—review and editing, supervision, and funding acquisition. XS: writing—review and editing, supervision, and funding acquisition. YZ: conceptualization, supervision, and funding acquisition. All authors have read and agreed to the published version of the manuscript.
Acknowledgments
We would like to thank Xiangzhu Wang and Ying Liu from Gansu Liaoyuan Dairy Group for their help with funding acquisition and resources.
Funding Statement
This research was funded by the National Natural Science Foundation of China (grant number: 31660468).
Footnotes
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1146566/full#supplementary-material
References
- Arseneau J., Steeves R., Laflamme M. (2017). Modified low-salt CTAB extraction of high-quality DNA from contaminant-rich tissues. Mol. Ecol. Resour. 17 686–693. 10.1111/1755-0998.12616 [DOI] [PubMed] [Google Scholar]
- Asuman K. S., Emin K. (2018). Isolation, identification and technological properties of lactic acid bacteria from raw cow milk. Biosci. J. 34 985–999. 10.14393/BJ-v34n2a2018-34517 33406856 [DOI] [Google Scholar]
- Buchfink B., Xie C., Huson D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12 59–60. 10.1038/nmeth.3176 [DOI] [PubMed] [Google Scholar]
- Chan Y. G., Kim H. K., Schneewind O., Missiakas D. (2014). The capsular polysaccharide of Staphylococcus aureus is attached to peptidoglycan by the LytR-CpsA-Psr (LCP) family of enzymes. J. Biol. Chem. 289 15680–15690. 10.1074/jbc.M114.567669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charchoghlyan H., Bae J., Kwon H., Kim M. (2017). Rheological properties and volatile composition of fermented milk prepared by exopolysaccharide-producing Lactobacillus acidophilus n.v. Er2 317/402 strain Narine. Biotechnol. Bioprocess Eng. 22 327–338. 10.1007/s12257-017-0065-8 [DOI] [Google Scholar]
- Dabour N., LaPointe G. (2005). Identification and molecular characterization of the chromosomal exopolysaccharide biosynthesis gene cluster from Lactococcus lactis subsp. cremoris SMQ-461. Appl. Envrion. Microbiol. 71 7414–7425. 10.1128/AEM.71.11.7414-7425.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson R., Martín Del Campo A. (2020). Combinatorial and computational investigations of neighbor-Joining bias. Front. Genet. 11:584785. 10.3389/fgene.2020.584785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo D., Davray D., Kulkarni R. (2019). A diverse repertoire of exopolysaccharide biosynthesis gene glusters in Lactobacillus revealed by comparative analysis in 106 sequenced genomes. Microorganisms 7:444. 10.3390/microorganisms7100444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dertli E., Mayer M. J., Colquhoun I. J., Narbad A. (2016). EpsA is an essential gene in exopolysaccharide production in Lactobacillus johnsonii FI9785. Microb. Biotechnol. 9 496–501. 10.1111/1751-7915.12314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimopoulou M., Vuillemin M., Campbell-Sills H., Lucas P. M., Ballestra P., Miot-Sertier C., et al. (2014). Exopolysaccharide (EPS) synthesis by Oenococcus oeni: from genes to phenotypes. PLoS One 9:e98898. 10.1371/journal.pone.0098898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S. R. (2009). A new generation of homology search tools based on probabilistic inference. Genome Inform. 23 205–211. 10.1142/9781848165632_0019 [DOI] [PubMed] [Google Scholar]
- Fagbemigun O., Cho G., Rösch N., Brinks E., Schrader K., Bockelmann W., et al. (2021). Isolation and characterization of potential starter cultures from the nigerian fermented milk product nono. Microorganisms 9:640. 10.3390/microorganisms9030640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X. W., Zhang H., Lai P. F. H., Xiong Z. Q., Ai L. Z. (2021). Structure characterization of a pyruvated exopolysaccharide from Lactobacillus plantarum AR307. Int. J. Biol. Macromol. 178 113–120. 10.1016/j.ijbiomac.2021.02.119 [DOI] [PubMed] [Google Scholar]
- Fukao M., Zendo T., Inoue T., Fuke N., Moriuchi T., Yamane Y., et al. (2019a). Relation between cell-bound exopolysaccharide production via plasmid-encoded genes and rugose colony morphology in the probiotic Lactobacillus brevis KB290. Anim. Sci. J. 90 1575–1580. 10.1111/asj.13297 [DOI] [PubMed] [Google Scholar]
- Fukao M., Zendo T., Inoue T., Nakayama J., Suzuki S., Fukaya T., et al. (2019b). Plasmid-encoded glycosyltransferase operon is responsible for exopolysaccharide production, cell aggregation, and bile resistance in a probiotic strain, Lactobacillus brevis KB290. J. Biosci. Bioeng. 128 391–397. 10.1016/j.jbiosc.2019.04.008 [DOI] [PubMed] [Google Scholar]
- Ghattargi V. C., Nimonkar Y. S., Burse S. A., Davray D., Kumbhare S. V., Shetty S. A., et al. (2018). Genomic and physiological analyses of an indigenous strain, Enterococcus faecium 17OM39. Funct. Integr. Genomics 18 385–399. 10.1007/s10142-018-0596-x [DOI] [PubMed] [Google Scholar]
- Graebin N., Schöffer J., Andrades D., Hertz P., Ayub M., Rodrigues R. (2016). Immobilization of glycoside hydrolase families GH1, GH13, and GH70: state of the art and perspectives. Molecules 21:1074. 10.3390/molecules21081074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon E., Horvatovich P., Marchioni E., Aoude-Werner D., Ennahar S. (2014). Investigation of potential markers of acid resistance in Lactobacillus plantarum by comparative proteomics. J. Appl. Microbiol. 116 134–144. 10.1111/jam.12339 [DOI] [PubMed] [Google Scholar]
- Horn N., Wegmann U., Dertli E., Mulholland F., Collins S. R., Waldron K. W., et al. (2013). Spontaneous mutation reveals influence of exopolysaccharide on Lactobacillus johnsonii surface characteristics. PLoS One 8:e59957. 10.1371/journal.pone.0059957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Liu D., Jia X., Liang M., Lu Y., Liu J. (2021). Whole genome sequencing of Lactobacillus plantarum DMDL 9010 and its effect on growth phenotype under nitrite stress. LWT Food Sci. Technol. 149:111778. 10.1016/j.lwt.2021.111778 [DOI] [Google Scholar]
- Hugenholtz J., Looijesteijn E., Starrenburg M., Dijkema C. (2000). Analysis of sugar metabolism in an EPS producing Lactococcus lactis by super 31P NMR. J. Biotechnol. 77 17–23. 10.1016/S0168-1656(99)00204-7 [DOI] [PubMed] [Google Scholar]
- Hyatt D., Chen G. L., Locascio P. F., Land M. L., Larimer F. W., Hauser L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformat. 11:119. 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim M. K., Salama H., Abd El Rahman M., Dawood R. M., Bader, El Din N. G., et al. (2016). Three gene signature for predicting the development of hepatocellular carcinoma in chronically infected hepatitis C virus patients. J. Interferon Cytokine Res. 36 698–705. 10.1089/jir.2016.0042 [DOI] [PubMed] [Google Scholar]
- Izquierdo E., Horvatovich P., Marchioni E., Aoude-Werner D., Sanz Y., Ennahar S. D. (2009). 2-DE and MS analysis of key proteins in the adhesion of Lactobacillus plantarum, a first step toward early selection of probiotics based on bacterial biomarkers. Electrophoresis 30 949–956. 10.1002/elps.200800399 [DOI] [PubMed] [Google Scholar]
- Kanmani P., Albarracin L., Kobayashi H., Hebert E. M., Saavedra L. (2018). Genomic characterization of Lactobacillus delbrueckii TUA4408L and evaluation of the antiviral activities of its extracellular polysaccharides in porcine intestinal epithelial cells. Front. Immunol. 9:2178. 10.3389/fimmu.2018.02178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimoto-Nira H. (2018). New lactic acid bacteria for skin health via oral intake of heat-killed or live cells. Anim. Sci. J. 89 835–842. 10.1111/asj.13017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleerebezem M., Hugenholtz J. (2003). Metabolic pathway engineering in lactic acid bacteria michiel kleerebezem and jeroen hugenholtzy. Curr. Opin. Biotechnol. 14 232–237. 10.1016/S0958-1669(03)00033-8 [DOI] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19 1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammens W., Le Roy K., Schroeven L., Van Laere A., Rabijns A., Van den Ende W. (2009). Structural insights into glycoside hydrolase family 32 and 68 enzymes: functional implications. J. Exp. Bot. 60 727–740. 10.1093/jxb/ern333 [DOI] [PubMed] [Google Scholar]
- Lee K., Pi K., Kim E. B., Rho B., Kang S., Lee H. G., et al. (2010). Glutathione-mediated response to acid stress in the probiotic bacterium, Lactobacillus salivarius. Biotechnol. Lett. 32 969–972. 10.1007/s10529-010-0244-6 [DOI] [PubMed] [Google Scholar]
- Liang S., Wang F., Granato D., Zhong X., Xiao A., Ye Q., et al. (2022). Effect of β-glucosidase on the aroma of liquid-fermented black tea juice as an ingredient for tea-based beverages. Food Chem. 402:134201. 10.1016/j.foodchem.2022.134201 [DOI] [PubMed] [Google Scholar]
- Liu X., Zeng J., Huang K., Wang J. (2019). Structure of the mannose transporter of the bacterial phosphotransferase system. Cell Res. 29 680–682. 10.1038/s41422-019-0194-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino M. E., Bayjanov J. R., Caffrey B. E., Wels M., Joncour P., Hughes S., et al. (2016). Nomadic lifestyle of Lactobacillus plantarum revealed by comparative genomics of 54 strains isolated from different habitats. Environ. Microbiol. 18 4974–4989. 10.1111/1462-2920.13455 [DOI] [PubMed] [Google Scholar]
- Morabbi Heravi K., Altenbuchner J. (2018). Cross talk among transporters of the phosphoenolpyruvate-dependent phosphotransferase system in bacillus subtilis. J. Bacteriol. 200 e213–e218. 10.1128/JB.00213-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehme D. P., Shafee T., Downton M. T., Bacic A., Doblin M. S. (2019). Differences in protein structural regions that impact functional specificity in GT2 family β-glucan synthases. PLoS One 14:e224442. 10.1371/journal.pone.0224442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panthavee W., Noda M., Danshiitsoodol N., Kumagai T., Sugiyama M. (2017). Characterization of exopolysaccharides produced by thermophilic lactic acid bacteria isolated from tropical fruits of Thailand. Biol. Pharm. Bull. 40 621–629. 10.1248/bpb.b16-00856 [DOI] [PubMed] [Google Scholar]
- Panthee S., Paudel A., Blom J., Hamamoto H., Sekimizu K. (2019). Complete genome sequence of Weissella hellenica 0916-4-2 and its comparative genomic analysis. Front. Microbiol. 10:1619. 10.3389/fmicb.2019.01619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X. Q., -Dra A., Yue M. (2022). Whole genome sequencing for the risk assessment of probiotic lactic acid bacteria. Crit. Rev. Food Sci. Nutr. 34 1–19. 10.1080/10408398.2022.2087174 [DOI] [PubMed] [Google Scholar]
- Pessione E. (2012). Lactic acid bacteria contribution to gut microbiota complexity: lights and shadows. Front. Cell. Infect. Microbiol. 2:86. 10.3389/fcimb.2012.00086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prete R., Alam M. K., Perpetuini G., Perla C., Pittia P., Corsetti A. (2021). Lactic acid bacteria exopolysaccharides producers: a sustainable tool for functional foods. Foods 10:1653. 10.3390/foods10071653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi N., Gu Q., Li P. (2020). Whole genome sequence analysis and in vitro probiotic characteristics of a Lactobacillus strain Lactobacillus paracasei ZFM54. J. Appl. Microbiol. 129 422–433. 10.1111/jam.14627 [DOI] [PubMed] [Google Scholar]
- Ruan J., Li H. (2019). Fast and accurate long-read assembly with wtdbg2. Nat. Methods 17 155–158. 10.1038/s41592-019-0669-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Şanlier N., GÖkcen B. B., Sezgİn A. C. (2019). Health benefits of fermented foods. Crit. Rev. Food Sci. Nutr. 59 506–527. 10.1080/10408398.2017.1383355 [DOI] [PubMed] [Google Scholar]
- Siezen R. J., Tzeneva V. A., Castioni A., Wels M., Phan H. T. K., Rademaker J. L. W., et al. (2010). Phenotypic and genomic diversity of Lactobacillus plantarum strains isolated from various environmental niches. Environ. Microbiol. 12 758–773. 10.1111/j.1462-2920.2009.02119.x [DOI] [PubMed] [Google Scholar]
- Sofie M., Bouaballati S. E., Henrissat B., Svensson B. (2021). Functional diversity of three tandem C-terminal carbohydrate-binding modules of a β-mannanase. J. Biol. Chem. 296 100638–100651. 10.1016/j.jbc.2021.100638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Xiong Z., Kong L., Wang G., Ai L. (2018). Relationship between putative eps genes and production of exopolysaccharide in Lactobacillus casei LC2W. Front. Microbiol. 9:1882. 10.3389/fmicb.2018.01882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumya M. P., Nampoothiri K. M. (2021). An overview of functional genomics and relevance of glycosyltransferases in exopolysaccharide production by lactic acid bacteria. Int. J. Biol. Macromol. 184 1014–1025. 10.1016/j.ijbiomac.2021.06.131 [DOI] [PubMed] [Google Scholar]
- Sun X., Zhang J. (2021). Bacterial exopolysaccharides: chemical structures, gene clusters and genetic engineering. Int. J. Biol. Macromol. 173 481–490. 10.1016/j.ijbiomac.2021.01.139 [DOI] [PubMed] [Google Scholar]
- Werning M. L., Hernández-Alcántara A. M., Ruiz M. J., Soto L. P., Dueñas M. T., López P. (2022). Biological functions of exopolysaccharides from lactic acid bacteria and their potential benefits for humans and farmed animals. Foods 11:1284. 10.3390/foods11091284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z., Kong L., Lai P. F. H., Xia Y., Liu J., Li Q., et al. (2019). Genomic and phenotypic analyses of exopolysaccharide biosynthesis in Streptococcus thermophilus S-3. J. Dairy Sci. 102 4925–4934. 10.3168/jds.2018-15572 [DOI] [PubMed] [Google Scholar]
- Xu Y. M., Cui Y. L., Yue F. F., Liu L. H., Shan Y. Y. (2019). Exopolysaccharides produced by lactic acid bacteria and Bifidobacteria: structures, physiochemical functions and applications in the food industry. Food Hydrocolloids 94 475–499. 10.1016/j.foodhyd.2019.03.032 [DOI] [Google Scholar]
- Yi L., Qi T., Hong Y., Deng L., Zeng K. (2020). Screening of bacteriocin-producing lactic acid bacteria in Chinese homemade pickle and dry-cured meat, and bacteriocin identification by genome sequencing. LWT-Food Sci. Technol. 125 109177–109210. 10.1016/j.lwt.2020.109177 [DOI] [Google Scholar]
- Zeidan A. A., Poulsen V. K., Janzen T., Buldo P., Derkx P. M. F., Øregaard G., et al. (2017). Polysaccharide production by lactic acid bacteria: from genes to industrial applications. Fems Microbiol. Rev. 41 S168–S200. 10.1093/femsre/fux017 [DOI] [PubMed] [Google Scholar]
- Zhao X. F., Liang Q. (2022). EPS-producing Lactobacillus plantarum MC5 as a compound starter improves rheology, texture, and antioxidant activity of yogurt during storage. Foods 11:1660. 10.3390/foods11111660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X. F., Liang Q. (2023). Optimization, probiotic characteristics, and rheological properties of exopolysaccharides from Lactiplantibacillus plantarum MC5. Molecules 28:2463. 10.3390/molecules28062463 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI, PRJNA897707.