Abstract
The flavodoxins are flavin mononucleotide-containing electron transferases. Flavodoxin I has been presumed to be the only flavodoxin of Escherichia coli, and its gene, fldA, is known to belong to the soxRS (superoxide response) oxidative stress regulon. An insertion mutation of fldA was constructed and was lethal under both aerobic and anaerobic conditions; only cells that also had an intact (fldA+) allele could carry it. A second flavodoxin, flavodoxin II, was postulated, based on the sequence of its gene, fldB. Unlike the fldA mutant, an fldB insertion mutant is a viable prototroph in the presence or absence of oxygen. A high-copy-number fldB+ plasmid did not complement the fldA mutation. Therefore, there must be a vital function for which FldB cannot substitute for flavodoxin I. An fldB-lacZ fusion was not induced by H2O2 and is therefore not a member of the oxyR regulon. However, it displayed a soxS-dependent induction by paraquat (methyl viologen), and the fldB gene is preceded by two overlapping regions that resemble known soxS binding sites. The fldB insertion mutant did not have an increased sensitivity to the effects of paraquat on either cellular viability or the expression of a soxS-lacZ fusion. Therefore, fldB is a new member of the soxRS (superoxide response) regulon, a group of genes that is induced primarily by univalent oxidants and redox cycling compounds. However, the reactions in which flavodoxin II participates and its role during oxidative stress are unknown.
Flavodoxins are small flavin mononucleotide (FMN)-containing electron transferases that are found in bacteria and algae. In Escherichia coli, a flavodoxin is required for the reductive activation of cobalamin-dependent methionine synthase (31), for biotin synthesis (34), and for the anaerobic activation of both ribonucleoside triphosphate reductase (8, 9) and pyruvate formate-lyase (36) through the formation of glycyl free radicals at their active centers. In nitrogen-fixing bacteria, a flavodoxin is the electron donor for the Fe-containing protein of the nitrogenase complex (16). A flavodoxin can often substitute for a ferredoxin, a small electron transfer protein with an iron-sulfur center (reviewed in reference 31). Thus, both a flavodoxin and a ferredoxin are substrates for the NADPH:ferredoxin oxidoreductases of cyanobacteria and of E. coli, for the pyruvate-ferredoxin oxidoreductase of Clostridium pasteurianum, and for the enzymes of dissimilatory sulfate reduction. In iron-poor media, where we should expect a ferredoxin deficiency, the flavodoxins of cyanobacteria and Anacystis nidulans are induced.
Most of the reactions of E. coli flavodoxin were demonstrated in vitro with purified flavodoxin I, the product of the cloned fldA gene, which was presumed to be the only flavodoxin of E. coli. However, a putative second flavodoxin gene, fldB, was discovered during the sequencing of the unrelated neighboring xerD gene (GenBank accession no. AE000373 [F. R. Blattner] and Z48060 [F. Hayes]), which encodes a site-specific recombinase. The deduced structure of flavodoxin II (FldB) has 43% identity with E. coli flavodoxin I (FldA), and it has the characteristic flavodoxin signature, an FMN binding motif near its N terminus.
This study of the flavodoxins was prompted by our interest in the soxRS (superoxide response) regulon because it includes the genes for flavodoxin I (fldA) (49) and for NADPH:ferredoxin (flavodoxin) oxidoreductase (fpr) (27). The soxRS regulon (17, 44) responds to the oxidative stress produced by redox agents that engage mainly in one-electron transfers, agents such as superoxide, nitric oxide, and paraquat (methyl viologen). The sensor for the regulon resides in the [2Fe-2S] centers of the ferredoxin-like SoxR protein. When these centers are oxidized, SoxR becomes a transcriptional activator of soxS, and the newly synthesized SoxS protein (itself a transcriptional activator) then induces other genes of the regulon. In the uninduced cell, SoxR is mainly in its inactive, reduced form, and because it is auto-oxidizable, it must be continually reduced. Through separate NADPH-linked reductases, both SoxR (23) and flavodoxin (11) are in redox equilibrium with NADP+/NADPH. Depletion of NADPH, e.g., during the production of superoxide by the redox cycling of paraquat, activates the regulon. FldA and Fpr may be induced to help restore the redox balance of the oxidatively stressed cell (27, 28).
In this study, we produce insertion mutations of fldA and fldB and we generate an fldB-lacZ gene fusion. These constructs are then used to approach the following questions. What are the phenotypes of the mutants? Is either gene essential? Do flavodoxins I and II have the same functions? Is fldB a member of the soxRS regulon? Does flavodoxin II have a discernible role in protection against oxidative stress?
MATERIALS AND METHODS
Nomenclature.
cat, tet, and bla refer to plasmid- or transposon-derived DNA segments specifying resistance to chloramphenicol (Camr), tetracycline (Tetr), and ampicillin or carbenicillin (Ampr), respectively. λattP and attλ are the preferred DNA sequences in phage λ and E. coli, respectively, at which site-specific integration of phage λ occurs.
Strains and plasmids.
Bacterial strains and plasmids used are listed in Table 1. Some of the plasmid constructions are detailed in Fig. 1 and 2. Insertions into attλ were performed as previously described (18), except that a recA+ strain was used because it was a better donor for subsequent transductions. The attλ::(fldA+ bla+) element of BW1527 was prepared from plasmid pWB52 (Table 1), which had been cut with NotI to yield a bla+-fldA+-λattP fragment. This DNA was then circularized by ligation and used to transform strain GC4468(pLDR8) to carbenicillin resistance at 42°C. The cells were grown in Luria-Bertani (LB) broth at 37°C for 1.5 h to allow gene expression before selection. pLDR8 specifies a thermoinducible λ integrase that mediated the insertion of the bla+-fldA+ segment into the attλ site. BW1527 was Kans, indicating that it had been cured of the temperature-sensitive helper plasmid. The attλ::(Φ[soxS′-′lacZ] bla+) element of BW1157 was constructed similarly. λRZ5::fldB was produced by growing λRZ5 on a strain carrying plasmid pWB51; the lysate was used to infect Δlac strain GC4468, and the desired lysogens were selected as red colonies on MacConkey agar (Difco) containing ampicillin.
TABLE 1.
Bacterial strains, phages, and plasmids used
| Strain or plasmid | Descriptiona | Reference or sourceb |
|---|---|---|
| E. coli strains | ||
| BW831 | GC4468 soxS3::Tn10 | 48 |
| BW1157 | GC4468 attλ::(Φ[soxS′-′lacZY] bla+); circularized NotI fragment of pWB53 integrated with the help of pLDR8 | This study |
| BW1485 | JC7623 fldB::tet | pPG5 × JC7623 |
| BW1526 | KL16 attλ::(fldA+ bla+) | P1(BW1527) × KL16 |
| BW1527 | GC4468 attλ::(fldA+ bla+) | See Materials and Methods |
| BW1528 | JC7623 attλ::(fldA+ bla+) | P1(BW1527) × GC4468 |
| BW1529 | JC7623 fldA::cat attλ::(fldA+ bla+) | pPG1::cat × BW1524 |
| BW1530 | JC7623 fldA::cat zbf-3057::Tn10 attλ::(fldA+ bla+) | P1(CAG18433) × BW1529 |
| BW1531 | W3110 fldB::tet | P1(BW1485) × W3110 |
| BW1534 | BW1157 fldB::tet | P1(BW1485) × BW1157 |
| BW1535 | GC4468(λRZ5::fldB)soxS3::Tn10 | P1(BW831) × GC4468(λRZ5::fldB) |
| CAG18433 | zbf-3057::Tn10 | 40 |
| GC4468 | Δ(argF-lac)169 rpsL179 IN(rrnD-rrnE) | 12 |
| JC7623c | recB21 recC22 sbcB15 sbcC201 | 25 |
| KL16 | Hfr PO-45 spoT1 rel-1 thi-1 | 4 |
| W3110 | Prototroph; IN(rrnD-rrnE) | 4 |
| Plasmids | ||
| pBR322 | oriColE1bla tet; cloning vector | GenBank accession no. J01749 |
| pLDR8 | pSC101(Ts) derivative containing λpRint+ cI857 segment; Kanr | 18 |
| pLDR10 | Cloning vector containing attλ; Ampr Camr | 18 |
| pMOB02 | pBR322 bla::Tn9; Camr Tetr Amps | 10 |
| pPG1 | fldA+; Ampr; partial deletion of pRMEcoRI | Fig. 1 |
| pPG1::cat | fldA::cat; Ampr | Fig. 1 |
| pPG2 | fldB+; Ampr; high copy number | Fig. 2 |
| pPG5 | pPG2 fldB::tet | Fig. 2 |
| pRMEcoRI | fldA gene on 5.8-kb insert in phagemid pBluescript II SK+; Ampr | 31; Fig. 1 |
| pRS414 | Protein fusion vector containing, in sequence, bla+, T14, EcoRI site, SmaI site, and ′lacZ | 39 |
| pWB51 | pRS414 containing an fldB′-′lacZ protein fusion; EcoRI-MscI piece of pPG2 (Fig. 2) replacing EcoRI-SmaI segment of pRS414. | This study |
| pWB52 | pLDR10::fldA; 1-kb DraI fragment of pRMEcoRI (Fig. 1) cloned into SmaI site of pLDR10 | This study |
| pWB53 | pLDR10::Φ(soxS′-′lacZ); λJW2 piece from ScaI (in J) to HindIII (in kan) replacing SmaI-HindIII segment of pLDR10 | This study |
| Phages | ||
| λ469 | Kohara phage containing the fldB region | 24 |
| λJW2 | Φ(soxS′-′lacZ) bla+kan+ | 48 |
| λRZ5 | c+ ′bla ′lacZ lacY+; prophage vector for acquiring lacZ fusions from plasmids by recombination | 32; from R. Zagursky |
| λRZ5::fldB | λRZ5 Φ(fldB′-′lacZ) bla+ | λRZ5 × pWB51 |
Bacterial strains are derivatives of E. coli K-12 λ−. Unless otherwise noted, the strains are F−, and genetic descriptions are complete. T14 is a set of four tandem transcriptional terminators (39).
P1 transductional crosses are described as follows: P1(donor) × recipient.
For a complete listing of markers, see reference 25.
FIG. 1.
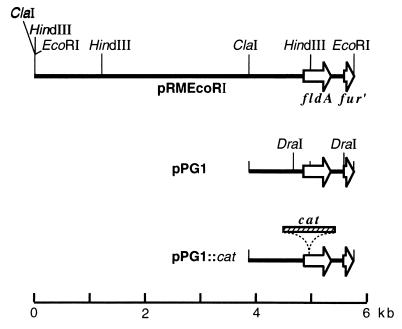
fldA plasmids. Only the inserted E. coli DNA is shown, together with part of the multiple cloning site. pRMEcoRI was digested with ClaI and religated, thereby removing all but one HindIII site and producing plasmid pPG1. A chloramphenicol resistance (cat) element from Tn9 was excised from plasmid pMOB02 as a 1.9-kb FspI fragment and ligated into the HindIII site of pPG1 to produce pPG1::cat. For clarity, the DraI sites that were used for subcloning fldA in pWB52 are shown on only one of the plasmids.
FIG. 2.
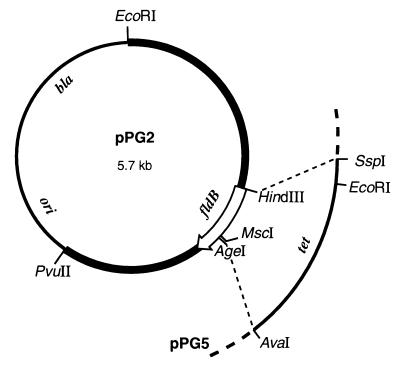
fldB plasmids. Plasmid pPG2, an fldB+ plasmid with a high copy number (due to a partial rop deletion), was produced by replacing the EcoRI-PvuII region of pBR322 with a 3.4-kb EcoRI-PvuII fragment of Kohara phage λ469. Plasmid pPG5 is a derivative of pPG2 in which most of the fldB gene was replaced by a 1.6-kb segment of pBR322 containing the tet gene. The HindIII site is in the ribosome-binding site for fldB. The thin lines represent pBR322 DNA.
Media and growth conditions.
LB media (29) were used for the routine growth of E. coli. The minimal medium used for aerobic growth (VB medium) was medium E described by Vogel and Bonner (43) that was supplemented with 0.4% glucose and 1 μg of thiamine/ml. The minimal medium used under anaerobic conditions was a glycerol-fumarate medium without Casamino Acids (41). For anaerobic growth in rich solid media, the cells were suspended in 15 ml of nutrient agar (Difco) containing an E. coli membrane preparation (EC Oxyrase; Oxyrase, Inc.), overlaid with a barrier of 2% agar (in H2O), and incubated in air (1). Alternatively, the cells were grown on the surfaces of agar plates in an AnaeroPak chamber (Mitsubishi Gas Chemical America, Inc.) under 80% N2–20% CO2. Growth in broth under stringent anaerobic conditions was performed in Na2S-supplemented media (20). Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 25 μg/ml; tetracycline, 5 or 15 μg/ml. The lower concentration of tetracycline was required for the selection of single copies of fldB::tet. Carbenicillin (50 μg/ml) was used for the selection of single copies of bla.
Gene transfers.
Generalized transductions were performed with bacteriophage P1 dam rev6 (42). Bacterial transformations were performed as previously described (14, 22). Transfers of fldA and fldB insertion mutations from plasmid to chromosome (by double crossovers) were accomplished as previously described (30) with a recBC sbcBC strain (JC7623), which does not support the growth of ColE1-derived replicons (6).
Computer analysis.
Sequence similarity searches were performed with the BLAST, version 2.0, program at the National Center for Biotechnology Information website. FMN binding sites were detected with the MOTIFS program of the Wisconsin Package, version 9.1 (Genetics Computer Group, Madison, Wis.).
Other methods.
PCR and general cloning methods were as described previously (3). DNA fragments with incompatible ends were blunted with bacteriophage T4 DNA polymerase before being joined. Curing of the attλ::fldA element was accomplished by infection with λc+ phage at a multiplicity of 10 (38). PCR amplifications of the fldA and fldB regions were performed with Taq DNA polymerase and the following primers: 5′-GAAGAAGTCATCCCAGTCACA-3′ and 5′-ACCCCCATTTCAATAAGTTTC-3′ for fldA and 5′-TTAGTTTCATCCAGCGCC-3′ and 5′-CCATTATGCCTTATTGTGCC-3′ for fldB. Treatments of growing cells with paraquat or H2O2 were as previously described (13). After 1 h, the cultures were chilled, and 0.1 volume of ethanol (37) was added to each. Assays for β-galactosidase were performed by the method of Miller (29) on cells that were permeabilized with polymyxin B (37); specific activity is reported in Miller units. Catalase assays were performed as previously described (13).
RESULTS
fldA insertion mutation.
The fldA::cat mutation was constructed on a plasmid (Fig. 1). However, we were unable to transfer the mutation to the chromosome by common methods of transformation with linear DNA (33, 45). This result suggested that the mutation might be lethal, in which case the host chromosome should accept fldA::cat by substitutive recombination only if the cell has a second copy of fldA+. We introduced a second copy of fldA+ as part of a nonreplicating circular element that integrated into attλ (see Materials and Methods). The cloned DNA was a DraI segment of pRMEcoR1 (Fig. 1) in which fldA was the only open reading frame (GenBank accession no. AE000372 [F. R. Blattner]). The attλ::(fldA+ bla) element was then transduced into JC7623, a recBC sbcBC strain that can undergo chromosomal transformation. The resulting fldA+ diploid, BW1528, could now be transformed to Camr by a double crossover with plasmid pPG1::cat (containing fldA::cat) to yield strain BW1529. The Camr trait of BW1529 could be crossed out in transductions with strain CAG18433, which contains a Tn10 near fldA; the linkage was 50% (24 of 48). Therefore, in strain BW1529, the insertion mutation is in the normal chromosomal fldA locus; it is not on a plasmid or in the inserted attλ::fldA+ element.
fldA is a vital gene.
An fldA::cat insertion mutation was readily transduced into a strain carrying two copies of fldA+, but it could not be efficiently transferred to a wild-type strain under aerobic or anaerobic conditions (Table 2). This result suggested that the insertion mutation is lethal. A nearby Tn10 served as a control for the efficiency of transduction. It was easily transferred to strains carrying either one or two intact copies of fldA. If the fldA::cat mutation is lethal, the number of Tetr recombinants of the wild-type strain should be reduced by the number that would have coinherited fldA::cat. Compared to the fldA+ diploid, the wild-type strain had 71% fewer Tetr recombinants aerobically and 48% fewer anaerobically (Table 2). These results, too, are consistent with the inviability of the Camr cotransductants. fldA is the only open reading frame from its region that is contained within the DraI fragment that was cloned in the attλ element (Fig. 1) (GenBank accession no. AE000372). The fact that this element complements the lethality of fldA::cat indicates that the defect is due solely to the loss of fldA function and not to a polar effect of the insertion mutation.
TABLE 2.
P1-mediated transduction of fldA::cat and a nearby Tn10 into strains carrying either one or two copies of fldA+
| Recipient | Genotype | No. of transductantsa
|
|||
|---|---|---|---|---|---|
| Aerobic
|
Anaerobicb
|
||||
| Camr | Tetr | Camr | Tetr | ||
| KL16 | fldA+ | 4 | 252 | 0 | 33 |
| BW1526 | fldA+ attλ::(fldA+) | 1,331 | 866 | 176 | 63 |
Transductions were each performed with equal amounts a lysate of the donor strain, BW1530 [fldA::cat zbf-3507::Tn10 attλ::(fldA+ bla+)]. After 1 h of incubation to permit gene expression, each cell suspension was divided into four equal portions and plated.
Anaerobic growth was for 3 days at 37°C in nutrient agar (Difco) containing Oxyrase.
Four fldA::cat recombinants in an apparently haploid strain were observed (Table 2). Although this result might seem to argue against the lethality of the mutation, it was expected because E. coli acquires tandem duplications of any chromosomal gene at a frequency of about 1% (2); substitution of one tandem fldA+ allele by fldA::cat would leave the second copy of fldA functional. The presence of intact fldA genes was confirmed by PCR. Aerobic transduction of the haploid strain was repeated on a larger scale, and eight Camr recombinants were analyzed. The PCR primers were complementary to sequences that flank the cat insertion site. All eight transductants yielded the 248-bp products that were expected from templates containing uninterrupted fldA genes.
The following experiment produced independent evidence for the lethality of fldA::cat. If fldA is vital, we should not be able to eliminate the attλ::fldA+ element from a cell bearing the fldA::cat mutation. To cure strains of this element, they were infected with λc+ bacteriophage at a high multiplicity. During lysogenization, the transient production of the phage-encoded Int and Xis proteins should lead to the excision and subsequent loss of the nonreplicating attλ::(fldA+ bla+) element. The method was the same as that for the curing of λ prophages by superinfection (38). After infection of BW1528 [fldA+ attλ::(fldA+ bla+), 24% (23 of 96) of the tested colonies lost the attλ element (i.e., became Amps). However, none (0 of 89) of the tested colonies of BW1529 [fldA::cat attλ::(fldA+ bla+)] were cured, indicating that a functional fldA gene is essential for viability.
An fldB insertion mutant is viable.
In plasmid pPG5 (Fig. 2), a tetracycline resistance element replaced most of the fldB gene. The mutation was transferred to the chromosome of strain JC7623 (see Materials and Methods). All twenty of the Tetr recombinants examined were Amps, i.e., plasmid free. Four of the transformants were used as PCR templates in reactions with primers that bracketed the insertion site (see Materials and Methods). The mutant DNAs generated the 1.6-kb products expected for the interrupted gene, and they failed to yield the 0.6-kb product that was obtained from fldB+ chromosomal DNA (results not shown). Although the recombinants no longer had an intact copy of fldB, their colony size on LB agar was observed to be equal to that of their fldB+ parents under both aerobic and anaerobic conditions. Their growth rate and viability in LB broth under stringent anaerobic conditions (with Na2S) were also the same as those of their fldB+ parent. Therefore, unlike the fldA insertion mutant, the fldB mutant is viable.
fldB mutants are still prototrophs.
The fldB::tet mutation was transduced from strain BW1485 into strain W3110, an ancestral K-12 (F−λ−) prototroph. Three transductants were plated on a glycerol-fumarate minimal medium under both aerobic and anaerobic (80% N2–20% CO2) conditions and on VB (glucose) minimal medium under aerobic conditions. Their growth was indistinguishable from that of the fldB+ parental strain grown on the same media.
Multiple copies of fldB cannot substitute for fldA.
Why is an fldB mutation not lethal whereas an fldA mutation is lethal? Either flavodoxins I and II are needed for different reactions or, if they participate in the same reactions, the activity of FldB may be too low to permit an fldA mutant to survive. To see if overproduced FldB could substitute for FldA, we attempted to transduce an fldA::cat mutation into a strain carrying pPG2, a high-copy-number fldB plasmid. The plasmid contains a functional promoter for fldB, which was used to construct an fldB′-′lacZ gene fusion, as described in the next section. The results (Table 3) were similar to those obtained with a wild-type plasmid-free strain (Table 2): whereas a Tn10 marker was readily transferred, the closely linked fldA::cat mutation was not. In the control experiment, both markers were transferred efficiently to a strain carrying two copies of fldA+. Therefore, even a high gene dose of fldB cannot prevent the lethality of an fldA mutation. These results suggest that in E. coli there is at least one vital reaction that specifically requires FldA.
TABLE 3.
A high gene dose of fldB does not prevent the lethality of an fldA mutation
| Recipient | Genotype | No. of transductantsa
|
|||
|---|---|---|---|---|---|
| Aerobic
|
Anaerobicb
|
||||
| Camr | Tetr | Camr | Tetr | ||
| BW1527 | fldA+ attλ::(fldA+) | 288 | 179 | 159 | 145 |
| BW1527(pPG2) | pPG2 (fldB+)/fldA+ | 0 | 155 | 1 | 70 |
Transductions were performed with equal amounts of a lysate of the donor strain, BW1530 [attλ::(fldA+ bla+) fldA::cat zbf-3507::Tn10]. The selective media contained carbenicillin in addition to chloramphenicol or tetracycline.
Anaerobic growth was for 3 days at 37°C under 80% N2–20% CO2 on LB agar containing 40 mM glycerol and 40 mM sodium lactate.
fldB belongs to the soxRS regulon.
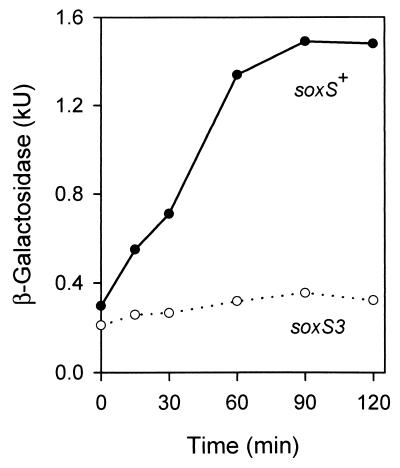
An fldB′-′lacZ protein fusion was constructed on a plasmid (pWB51; Table 1) and transferred by recombination to λRZ5. The λRZ5::fldB′ prophage contained most of fldB and 1.7 kb of the upstream chromosomal region including the divergently transcribed xerD gene. The cloned sequence was preceded by transcriptional terminators from vector pRS414. The construction fused the promoter, ribosome binding site, and first 356 nucleotides (nt) of fldB to a 5′-truncated lacZ gene. It is unlikely that the cloned portion of fldB contained a promoter for a downstream gene because in the chromosome the next two open reading frames are transcribed in the opposite direction (GenBank accession no. AE000373). The expression of the fldB′-′lacZ fusion was determined by measuring the β-galactosidase activity in a lac deletion mutant (GC4468) containing the prophage. It was not significantly affected (≤20%) by anaerobic growth in liquid media. Treatment with 1 mM H2O2 resulted in an induction of catalase activity (7.5-fold) but not of fldB expression (≤20%). Therefore, fldB does not belong to the OxyR regulon. However, fldB expression was induced fivefold by paraquat (methyl viologen), and the induction was blocked by a mutation in the soxS gene (Fig. 3). The unresponsiveness of the soxS mutant was not caused by a general inhibition of protein synthesis: the cell mass increased 7.5-fold during the 2-h treatment. These results indicate that fldB is a member of the soxRS regulon.
FIG. 3.
SoxS-dependent induction of an fldB′-lacZ fusion by paraquat. At zero time, paraquat was added to aerated log-phase cultures to a final concentration of 0.2 mg/ml. Samples were removed periodically, and the specific activity of β-galactosidase was measured in Miller units (29). The strains used were GC4468(λRZ5::fldB) and its soxS3::Tn10 derivative, BW1535.
soxS expression is not affected by fldB.
If FldB is required to maintain the normal redox balance of the cell, then an fldB mutant might constitutively overexpress soxS, the transcription of which is activated by the oxidized form of SoxR (19, 21). It might at least sensitize soxS to induction by a redox cycling agent such as paraquat, which depletes the cell of reducing equivalents while forming superoxide. To test this hypothesis, the fldB::tet insertion/deletion mutation was transduced into a soxS′-′lacZ fusion strain. The specific activities of β-galactosidase in the fldB+ and fldB mutant strains (BW1157 and BW1534) were the same (86 U). Inducibility by paraquat was tested at concentrations of from 0.01 to 1.0 mg/ml. Similar levels of induction were observed in both strains. For example, at 0.01 mg/ml, which produced about 40% of maximum induction, values for the wild type and mutant were 2,626 and 2,418 U, respectively.
The gene dose of fldB does not affect paraquat sensitivity.
The soxRS regulon protects the cell against univalent redox cycling compounds. Thus, soxS mutants display an increased sensitivity to killing by paraquat (47). This is also a property of an fpr mutant (7) which lacks a soxS-regulatable NADPH:ferredoxin (flavodoxin) reductase. It was therefore reasonable to suspect that the copy number of fldB might affect paraquat sensitivity. The gradient plate technique (15) was used to test strain W3110 (fldB+) together with three of its derivatives: BW1531 (fldB::tet), W3110(pPG2[fldB+]), and W3110(pPG5[fldB::tet]). The LB agar contained a gradient of from 0 to 120 μg of paraquat/ml and, for the plasmid-bearing strains, a uniform concentration of ampicillin. All four strains showed the same degree of sensitivity: 60 to 70 mm of growth along the gradient.
DISCUSSION
fldA is 348 nt upstream of fur, which encodes an iron-responsive regulatory protein. The two genes form an operon belonging to the soxRS regulon, and fur is also transcribed independently from an OxyR-regulated promoter (49). In our experiments, the fldA::cat mutation was complemented by an attλ element that contained fldA. Therefore, the lethality of fldA::cat must be directly due to a loss of fldA function. However, we have not excluded the formal possibility that the lethal effect is due to the loss of fldA combined with the noninducibility of fur.
In in vitro reactions, the anaerobic ribonucleoside triphosphate reductase (encoded by nrdD and nrdG) requires FldA; ferredoxin cannot be substituted for it (9). It should be an essential enzyme for anaerobic growth. However, nrdD and nrdG mutants fail to grow only under the most stringent anaerobic conditions, in a low-redox-potential broth medium containing Na2S (20). These were conditions that we could not apply to our plating experiments (Tables 2 and 3). The nrd mutants grew well on solid media in oxygen-depleted chambers. Therefore, the lethality of an fldA::cat mutation under our plating conditions cannot be attributed to its effect on anaerobic nucleotide reduction alone; there must be at least one other essential pathway requiring FldA. Similarly, inviability could not be explained by the requirement of pyruvate formate-lyase for flavodoxin (36). A mutant lacking the lyase gene grows anaerobically, displaying only a mild requirement for acetate (35). The results of anaerobic transductions, similar to those in Table 2, were not significantly altered by the addition of 5 mM acetate to the medium (results not shown).
Although there is as yet no known phenotype associated with an fldB mutation, there is strong evidence that the wild-type allele is a functional gene in E. coli. Our fldB′-′lacZ fusion depended for its expression on both the fldB promoter and fldB ribosome binding site, and the hybrid protein was expressed constitutively at a high level. In addition, its expression was regulated by SoxS (Fig. 3). Even if the gene product itself is not functional in E. coli, it is likely to be closely derived from one that is functional in another organism. Salmonella enterica serovar Typhimurium, a close relative of E. coli, also has an fldB gene next to xerD (BLAST program, version 2.0).
Apart from S. enterica serovar Typhimurium FldB, there are no known or predicted proteins that have a degree of identity with E. coli FldB that exceeds that of FldA (43%). There are, however, known or predicted proteins from other organisms that are about 39 to 49% homologous to both FldA and FldB of E. coli. They include putative flavodoxins from Anabaena sp., Synechoccus sp., Klebsiella aerogenes, Azotobactor sp., and Haemophilus influenzae. In the paralogs, the regions of sequence similarities appear to be distributed throughout the length of the polypeptides. In summary, the available evidence based on current sequencing data does not indicate that FldB is a member of a distinct subclass of flavodoxins found among distantly related bacteria.
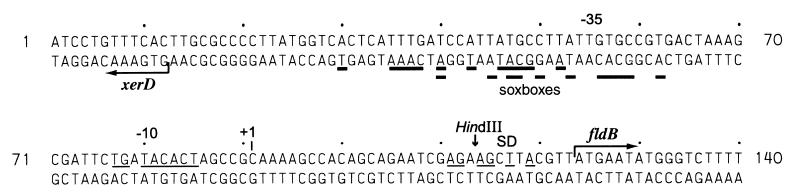
The xerD-fldB intergenic region (Fig. 4) contains sequences that are similar to those of known regulatory regions. Two sources using different algorithms predicted an fldB promoter with the same transcriptional start site (Fig. 4) (M. G. Reese, Neural network promoter prediction tool, http://www.fruitfly.org/seq_tools/promoter.html [revision date, 18 December 1999; last date accessed, 29 December 1999]; GenBank accession no. AE000373) predict an fldB promoter with the same transcriptional start site (Fig. 4). The putative −10 hexamer 5′-TACACT-3′ is preceded by a 5′-TGN-3′ sequence characteristic of “extended −10” regions that are found in promoters lacking recognizable −35 hexamers (5). Near the −35 region, on the antisense strand, are two overlapping sequences that resemble a SoxS-binding site, or “soxbox” (26, 46). Although they might regulate the xerD gene, they appear to be too close to it, and there is no apparent reason why xerD should belong to an oxidative stress regulon. Physical and genetic studies are needed to confirm that these are indeed regulatory sequences.
FIG. 4.
Putative regulatory sequences upstream of fldB. The sequence shown begins at nt 3037752 of the E. coli genome (GenBank accession no. AE000373). The −10 hexamer of the putative ς70 promoter of fldB is lightly underlined, as are the bases complementary to the end of 16S rRNA in the putative Shine-Dalgarno sequence (SD). The solid bars underline nucleotides that match the consensus soxbox (SoxS binding site) sequence, 5′-AX2GCA(C/T)X2(T/A)2(G/A)XCAAAX3(A/T)(A/T)-3′ (46). +1, predicted transcriptional start site for fldB.
The inclusion of fldB together with fldA and fpr in the soxRS regulon underscores the importance of flavodoxins in this global response to oxidative stress. The soxRS regulon is induced by the depletion of reducing equivalents through univalent redox reactions. The induced flavodoxins together with their reductase may facilitate the restoration of the redox equilibrium that must accompany recovery from oxidative stress.
ACKNOWLEDGMENTS
We acknowledge the valuable technical assistance of Fred Kung and Jason Kron.
This work was supported by National Science Foundation Research Grant MCB-9996231.
REFERENCES
- 1.Adler H I. The use of microbial membranes to achieve anaerobiosis. Crit Rev Biotechnol. 1990;10:119–127. doi: 10.3109/07388559009068263. [DOI] [PubMed] [Google Scholar]
- 2.Anderson R P, Roth J R. Tandem genetic duplications in phage and bacteria. Annu Rev Microbiol. 1977;31:473–505. doi: 10.1146/annurev.mi.31.100177.002353. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1989. [Google Scholar]
- 4.Bachmann B J. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2460–2488. [Google Scholar]
- 5.Barne K A, Bown J A, Busby S J, Minchin S D. Region 2.5 of the Escherichia coli RNA polymerase sigma70 subunit is responsible for the recognition of the 'extended-10' motif at promoters. EMBO J. 1997;16:4034–4040. doi: 10.1093/emboj/16.13.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bassett C L, Kushner S R. Exonucleases I, III, and V are required for stability of ColE1-related plasmids in Escherichia coli. J Bacteriol. 1984;157:661–664. doi: 10.1128/jb.157.2.661-664.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bianchi V, Haggård-Ljungquist E, Pontis E, Reichard P. Interruption of the ferredoxin (flavodoxin) NADP+ oxidoreductase gene of Escherichia coli does not affect anaerobic growth but increases sensitivity to paraquat. J Bacteriol. 1995;177:4528–4531. doi: 10.1128/jb.177.15.4528-4531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bianchi V, Reichard P, Eliasson R, Pontis E, Krook M, Jörnvall H, Haggård-Ljungquist E. Escherichia coli ferredoxin NADP+ reductase: activation of E. coli anaerobic ribonucleotide reduction, cloning of the gene (fpr), and overexpression of the protein. J Bacteriol. 1993;175:1590–1595. doi: 10.1128/jb.175.6.1590-1595.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bianchi V, Eliasson R, Fontecave M, Mulliez E, Hoover D M, Matthews R G, Reichard P. Flavodoxin is required for the activation of the anaerobic ribonucleotide reductase. Biochem Biophys Res Commun. 1993;197:792–797. doi: 10.1006/bbrc.1993.2548. [DOI] [PubMed] [Google Scholar]
- 10.Bittner M, Vapnek D. Versatile cloning vectors derived from the runaway-replication plasmid pKN402. Gene. 1981;15:319–329. doi: 10.1016/0378-1119(81)90175-x. [DOI] [PubMed] [Google Scholar]
- 11.Blaschkowski P, Neuer G, Ludwig-Festl M, Knappe J. Routes of flavodoxin and ferredoxin reduction in Escherichia coli. CoA-acylating pyruvate:flavodoxin and NADPH:flavodoxin oxidoreductases participating in the activation of pyruvate formate-lyase. Eur J Biochem. 1982;123:563–569. [PubMed] [Google Scholar]
- 12.Carlioz A, Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan E, Weiss B. Endonuclease IV of Escherichia coli is induced by paraquat. Proc Natl Acad Sci USA. 1987;84:3189–3193. doi: 10.1073/pnas.84.10.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung C T, Niemela S L, Miller R H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cunningham R P, Saporito S M, Spitzer S G, Weiss B. Endonuclease IV (nfo) mutant of Escherichia coli. J Bacteriol. 1986;168:1120–1127. doi: 10.1128/jb.168.3.1120-1127.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deistung J, Thorneley R N. Electron transfer to nitrogenase. Characterization of flavodoxin from Azotobacter chroococcum and comparison of its redox potentials with those of flavodoxins from Azotobacter vinelandii and Klebsiella pneumoniae (nifF-gene product) Biochem J. 1986;239:69–75. doi: 10.1042/bj2390069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Demple B. Redox signaling and gene control in the Escherichia coli soxRS oxidative stress regulon—a review. Gene. 1996;179:53–57. doi: 10.1016/s0378-1119(96)00329-0. [DOI] [PubMed] [Google Scholar]
- 18.Diederich L, Rasmussen L J, Messer W. New cloning vectors for integration in the λ attachment site attB of the Escherichia coli chromosome. Plasmid. 1992;28:14–24. doi: 10.1016/0147-619x(92)90032-6. [DOI] [PubMed] [Google Scholar]
- 19.Ding H, Hidalgo E, Demple B. The redox state of the [2Fe-2S] clusters in SoxR protein regulates its activity as a transcription factor. J Biol Chem. 1996;271:33173–33175. doi: 10.1074/jbc.271.52.33173. [DOI] [PubMed] [Google Scholar]
- 20.Garriga X, Eliasson R, Torrents E, Jordan A, Barbe J, Gibert I, Reichard P. nrdD and nrdG genes are essential for strict anaerobic growth of Escherichia coli. Biochem Biophys Res Commun. 1996;229:189–192. doi: 10.1006/bbrc.1996.1778. [DOI] [PubMed] [Google Scholar]
- 21.Gaudu P, Weiss B. SoxR, a [2Fe-2S] transcription factor, is active only in its oxidized form. Proc Natl Acad Sci USA. 1996;93:10094–10098. doi: 10.1073/pnas.93.19.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 23.Kobazyashi K, Tagawa S. Isolation of reductase for SoxR that governs an oxidative response regulon from Escherichia coli. FEBS Lett. 1999;451:227–230. doi: 10.1016/s0014-5793(99)00565-7. [DOI] [PubMed] [Google Scholar]
- 24.Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 25.Kushner S R, Nagaishi H, Clark A J. Indirect suppression of recB and recC mutations by exonuclease I deficiency. Proc Natl Acad Sci USA. 1972;69:1366–1370. doi: 10.1073/pnas.69.6.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z Y, Demple B. Sequence specificity for DNA binding by Escherichia coli SoxS and Rob proteins. Mol Microbiol. 1996;20:937–945. doi: 10.1111/j.1365-2958.1996.tb02535.x. [DOI] [PubMed] [Google Scholar]
- 27.Liochev S I, Hausladen A, Beyer W F, Jr, Fridovich I. NADPH:ferredoxin oxidoreductase acts as a paraquat diaphorase and is a member of the soxRS regulon. Proc Natl Acad Sci USA. 1994;91:1328–1331. doi: 10.1073/pnas.91.4.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liochev S I, Fridovich I. Fumarase C, the stable fumarase of Escherichia coli, is controlled by the soxRS regulon. Proc Natl Acad Sci USA. 1992;89:5892–5896. doi: 10.1073/pnas.89.13.5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller J H. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 30.Oden K L, DeVeaux L C, Vibat C R T, Cronan J E, Jr, Gennis R B. Genomic replacement in Escherichia coli K-12 using covalently closed circular plasmid DNA. Gene. 1990;96:29–36. doi: 10.1016/0378-1119(90)90337-q. [DOI] [PubMed] [Google Scholar]
- 31.Osborne C, Chen L M, Matthews R G. Isolation, cloning, mapping, and nucleotide sequencing of the gene encoding flavodoxin in Escherichia coli. J Bacteriol. 1991;173:1729–1737. doi: 10.1128/jb.173.5.1729-1737.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ostrow K S, Silhavy T J, Garrett S. cis-acting sites required for osmoregulation of ompF expression in Escherichia coli K-12. J Bacteriol. 1986;168:1165–1171. doi: 10.1128/jb.168.3.1165-1171.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russell C B, Thaler D S, Dahlquist F W. Chromosomal transformation of Escherichia coli recD strains with linearized plasmids. J Bacteriol. 1989;171:2609–2613. doi: 10.1128/jb.171.5.2609-2613.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanyal I, Gibson K J, Flint D H. Escherichia coli biotin synthase: an investigation into the factors required for its activity and its sulfur donor. Arch Biochem Biophys. 1996;326:48–56. doi: 10.1006/abbi.1996.0045. [DOI] [PubMed] [Google Scholar]
- 35.Sawers G, Böck A. Anaerobic regulation of pyruvate formate-lyase from Escherichia coli K-12. J Bacteriol. 1988;170:5330–5336. doi: 10.1128/jb.170.11.5330-5336.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawers G, Watson G. A glycyl radical solution: oxygen-dependent interconversion of pyruvate formate-lyase. Mol Microbiol. 1998;29:945–954. doi: 10.1046/j.1365-2958.1998.00941.x. [DOI] [PubMed] [Google Scholar]
- 37.Schupp J M, Travis S E, Price L B, Shand R F, Keim P. Rapid bacterial permeabilization reagent useful for enzyme assays. BioTechniques. 1995;19:18–20. [PubMed] [Google Scholar]
- 38.Shimada K, Weisberg R A, Gottesman M E. Prophage lambda at unusual chromosomal locations. I. Location of the secondary attachment sites and the properties of the lysogens. J Mol Biol. 1972;63:483–503. doi: 10.1016/0022-2836(72)90443-3. [DOI] [PubMed] [Google Scholar]
- 39.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 40.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spencer M E, Guest J R. Isolation and properties of fumarate reductase mutants of Escherichia coli. J Bacteriol. 1973;114:563–570. doi: 10.1128/jb.114.2.563-570.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sternberg N L, Maurer R. Bacteriophage-mediated generalized transduction in Escherichia coli and Salmonella typhimurium. Methods Enzymol. 1991;204:18–43. doi: 10.1016/0076-6879(91)04004-8. [DOI] [PubMed] [Google Scholar]
- 43.Vogel H J, Bonner D M. Acetylornithase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 44.Weiss B. Regulation of endonuclease IV as part of an oxidative stress response in Escherichia coli. In: Nickoloff J A, Hoekstra M F, editors. DNA damage and repair. Vol. 1. Totowa, N.J: Humana Press; 1998. pp. 85–96. [Google Scholar]
- 45.Winans S C, Elledge S J, Krueger J H, Walker G C. Site-directed insertion and deletion mutagenesis with cloned fragments in Escherichia coli. J Bacteriol. 1985;161:1219–1221. doi: 10.1128/jb.161.3.1219-1221.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wood T I, Griffith K L, Fawcett W P, Jair K-W, Schneider T D, Wolf R E., Jr Interdependence of the position and orientation of SoxS binding sites in the transcriptional activation of the class I subset of Escherichia coli superoxide-inducible promoters. Mol Microbiol. 1999;34:414–430. doi: 10.1046/j.1365-2958.1999.01598.x. [DOI] [PubMed] [Google Scholar]
- 47.Wu J, Weiss B. Two divergently transcribed genes, soxR and soxS, control a superoxide response regulon of Escherichia coli. J Bacteriol. 1991;173:2864–2871. doi: 10.1128/jb.173.9.2864-2871.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu J, Weiss B. Two-stage induction of the soxRS (superoxide response) regulon of Escherichia coli. J Bacteriol. 1992;174:3915–3920. doi: 10.1128/jb.174.12.3915-3920.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng M, Doan B, Schneider T D, Storz G. OxyR and SoxRS regulation of fur. J Bacteriol. 1999;181:4639–4643. doi: 10.1128/jb.181.15.4639-4643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]