Abstract
The coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has posed a global health crisis. Increasing evidence underlines the key role of competent immune responses in resisting SARS-CoV-2 infection and manifests the disastrous consequence of host immune dysregulation. Elucidating the mechanisms responsible for deregulated host immunity in COVID-19 may provide a theoretical basis for further research on new treatment modalities. Gut microbiota comprises trillions of microorganisms colonizing the human gastrointestinal tract and has a vital role in immune homeostasis and the gut-lung crosstalk. Particularly, SARS-CoV-2 infection can lead to the disruption of gut microbiota equilibrium, a condition called gut dysbiosis. Due to its regulatory effect on host immunity, gut microbiota has recently received considerable attention in the field of SARS-CoV-2 immunopathology. Imbalanced gut microbiota can fuel COVID-19 progression through production of bioactive metabolites, intestinal metabolism, enhancement of the cytokine storm, exaggeration of inflammation, regulation of adaptive immunity and other aspects. In this review, we provide an overview of the alterations in gut microbiota in COVID-19 patients, and their effects on individuals’ susceptibility to viral infection and COVID-19 progression. Moreover, we summarize currently available data on the critical role of the bidirectional regulation between intestinal microbes and host immunity in SARS-CoV-2-induced pathology, and highlight the immunomodulatory mechanisms of gut microbiota contributing to COVID-19 pathogenesis. In addition, we discuss the therapeutic benefits and future perspectives of microbiota-targeted interventions including faecal microbiota transplantation (FMT), bacteriotherapy and traditional Chinese medicine (TCM) in COVID-19 treatment.
Keywords: COVID-19, gut microbiota, cytokine storm, inflammation, adaptive immunity, microbiota-targeted interventions
1. Introduction
The coronavirus disease 2019 (COVID-19), an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has been a global pandemic for 3 years (1). Due to reduced viral pathogenicity and widespread immunity from vaccination or natural infection, SARS-CoV-2 becomes endemic in many countries around the world (2). SARS-CoV-2 is likely to constantly circulate among the human population causing an endemic seasonal respiratory infection (3). SARS-CoV-2 is transmitted among human populations through respiratory droplets, aerosols and, to a lesser extent, via the faecal-oral route (4). The clinical presentation of COVID-19 ranges from an asymptomatic form, mild respiratory tract illness to severe pneumonia, multiorgan dysfunction, and even death (5). It is acknowledged that the respiratory system is the primary target of SARS-CoV-2 infection (6). Therefore, the common initial manifestations of COVID-19 are respiratory symptoms such as cough and dyspnea (7). Remarkably, human enterocytes with high expression of angiotensin-converting enzyme 2 (ACE2), an entry receptor for SARS-CoV-2, serve as a site of extrapulmonary viral infection as evidenced by the presence of SARS-CoV-2 RNA in faecal samples from COVID-19 patients even after clearance of viral respiratory infection (8, 9). Moreover, gastrointestinal symptoms such as diarrhoea, nausea and vomiting are common in COVID-19 patients (10). COVID-19 patients with gastrointestinal symptoms are prone to develop severe diseases (e.g., liver injury and acute respiratory distress syndrome (ARDS)) and have poorer clinical outcomes (11). Accordingly, SARS-CoV-2 infection-induced gastrointestinal manifestations have recently gained much attention. It is speculated that dysregulated gut microbiota may contribute to the gastrointestinal symptoms in COVID-19 patients.
Gut microbiota refers to a complex population of microbial communities (e.g., bacteria, fungi, protozoa and viruses) that inhabit the mammalian gastrointestinal tract (12). It has many pivotal functions including nutrient acquisition, metabolism, maintenance of intestinal epithelial barrier, modulation of host immunity and protection against diseases (13). Perturbations of gut microbiota, termed gut dysbiosis, are correlated with the pathogenesis of various diseases, including allergy, cancer, inflammatory bowel disease, metabolic syndrome and obesity (14). The composition of gut microbiota is altered in SARS-CoV-2-infected individuals, alluding to the linkage between intestinal microbes and COVID-19 development (15, 16). With more and more scientific evidence and research reports, the underlying mechanisms are gradually emerging. In this review, we provide an overview of correlations between gut microbiota dysbiosis and SARS-CoV-2 pathogenicity. We also summarize the mechanisms responsible for the implication of gut microbiota in COVID-19 progression. In addition, we highlight manipulation of gut microbiota as a therapeutic approach for COVID-19 treatment.
2. Dynamic alterations of gut microbiota during SARS-CoV-2 infection
The gut microbiota of COVID-19 patients was seriously dysbiotic, characterized by an expansion of conditional pathogens (e.g., Actinomyces, Akkermansia, Collinsella, Enterococcus, Lactobacillus, Lactococcus, Methanobrevibacter, Odoribacter, Parabacteroides, Phascolarctobacterium, Serratia and Staphylococcus) and a reduction in the relative abundance of beneficial microbes (e.g., Bacteroides, Blautia, Coprococcus, Dialister, Faecalibacterium, Lachnospira, Oscillospira, Prevotella, Roseburia, Ruminococcus) (17) ( Table 1 ). Notably, Enterococcus might be clinically associated with illness severity due to its pathogenic capacity. Gut microbiota of COVID-19 patients serves as a reservoir of opportunistic pathogens that may migrate across impaired epithelial barriers into circulation. Secondary Enterococcus infection could be a non-negligible complication associated with poor outcomes in COVID-19 (29). Future research studies are required to delve into whether SARS-CoV-2 infection acts as a predisposing factor to secondary pathogen infections. It is necessary to understand whether coinfection of SARS-CoV-2 and Enterococcus leads to worse clinical outcomes in COVID-19 patients. Another study showed that the opportunistic microorganisms Enterococcus faecium, Escherichia coli, Klebsiella pneumoniae, Salmonella enterica and Staphylococcus auricularis were enriched in COVID-19 patients while the probiotic bacteria Bacteroides vulgatus, Eubacterium eligens and Faecalibacterium prausnitzii were reduced compared with healthy controls (20). It was found that Acinetobacter and Rhodococcus were negatively correlated with the levels of CD3, CD4, CD45, haemoglobins and red blood cells (RBC) ( Table 2 ). Bacteroides and Veillonella had a positive relationship with CD3, haemoglobins and RBC. Enterococcus was positively associated with the plasma concentration of carbon dioxide. These results suggested that gut microbiota alterations might be connected with the immune status and clinical manifestations of inflammation in COVID-19 patients. Nevertheless, the clinical implications of intestinal microbiota deserve further research.
Table 1.
Gut microbiota profiles of SARS-CoV-2-infected subjects.
| Microorganism | Phylum | Family | Relative abundance (SARS-CoV-2-infected subjects vs. healthy controls) | Reference |
|---|---|---|---|---|
| Pathogenic microbes | ||||
| Actinomyces | Actinomycetota | Actinomycetaceae | Enriched | (17–19) |
| Akkermansia | Verrucomicrobiota | Akkermansiaceae | Enriched | |
| Collinsella | Actinomycetota | Coriobacteriaceae | Enriched | |
| Enterococcus | Firmicutes | Enterococcaceae | Enriched | |
| Lactobacillus | Firmicutes | Lactobacillaceae | Enriched | |
| Lactococcus | Firmicutes | Streptococcaceae | Enriched | |
| Methanobrevibacter | Euryarchaeota | Methanobacteriaceae | Enriched | |
| Odoribacter | Bacteroidetes | Odoribacteraceae | Enriched | |
| Parabacteroides | Bacteroidetes | Tannerellaceae | Enriched | |
| Phascolarctobacterium | Firmicutes | Acidaminococcaceae | Enriched | |
| Serratia | Pseudomonadota | Yersiniaceae | Enriched | |
| Staphylococcus | Firmicutes | Staphylococcaceae | Enriched | |
| Enterococcus faecium | Firmicutes | Enterococcaceaes | Enriched | (20) |
| Escherichia coli | Pseudomonadota | Enterobacteriaceae | Enriched | |
| Klebsiella pneumoniae | Pseudomonadota | Enterobacteriaceae | Enriched | |
| Salmonella enterica | Pseudomonadota | Enterobacteriaceae | Enriched | |
| Staphylococcus auricularis | Firmicutes | Staphylococcaceae | Enriched | |
| Acinetobacter | Pseudomonadota | Moraxellaceae | Enriched | |
| Rhodococcus | Actinomycetota | Nocardiaceae | Enriched | |
| Dorea | Firmicutes | Lachnospiraceae | Enriched | (18, 19, 21) |
| Streptococcus | Firmicutes | Streptococcaceae | Enriched | |
| Bacteroides | Bacteroidetes | Bacteroidaceae | Enriched | (19, 21, 22) |
| Corynebacterium | Actinomycetota | Corynebacteriaceae | Enriched | |
| Enterocloster | Firmicutes | Lachnospiraceae | Enriched | |
| Flavonifractor | Firmicutes | Oscillospiraceae | Enriched | |
| Collinsella aerofaciens | Actinomycetota | Coriobacteriaceae | Enriched | (23) |
| Collinsella tanakaei | Actinomycetota | Coriobacteriaceae | Enriched | |
| Morganella morganii | Pseudomonadota | Morganellaceae | Enriched | |
| Streptococcus infantis | Firmicutes | Streptococcaceae | Enriched | |
| Ruminococcus gnavus | Firmicutes | Lachnospiraceae | Enriched | |
| Clostridium hathewayi | Firmicutes | Clostridiaceae | Enriched | |
| Enterococcus avium | Firmicutes | Enterococcaceae | Enriched | |
| Prevotellaceae | Bacteroidetes | – | Enriched | (24) |
| Clostridium aldenense | Firmicutes | Lachnospiraceae | Enriched | (25, 26) |
| Escherichia unclassified | Pseudomonadota | Enterobacteriaceae | Enriched | |
| Intestinibacter bartlettii | Firmicutes | Peptostreptococcaceae | Enriched | |
| Clostridium bolteae | Firmicutes | Lachnospiraceae | Enriched | |
| Flavonifractor plautii | Firmicutes | Oscillospiraceae | Enriched | |
| Clostridium ramosum | Firmicutes | Clostridiaceae | Enriched | |
| Candida albicans | Ascomycota | Debaryomycetaceae | Enriched | (27, 28) |
| Beneficial microbes | ||||
| Blautia | Firmicutes | Lachnospiraceae | Depleted | (17, 20–22, 24) |
| Coprococcus | Firmicutes | Lachnospiraceae | Depleted | |
| Dialister | Firmicutes | Veillonellaceae | Depleted | |
| Faecalibacterium | Firmicutes | Oscillospiraceae | Depleted | |
| Lachnospira | Firmicutes | Lachnospiraceae | Depleted | |
| Oscillospira | Firmicutes | Oscillospiraceae | Depleted | |
| Prevotella | Bacteroidetes | Prevotellaceae | Depleted | |
| Roseburia | Firmicutes | Lachnospiraceae | Depleted | |
| Ruminococcus | Firmicutes | Oscillospiraceae | Depleted | |
| Bacteroides vulgatus | Bacteroidetes | Bacteroidaceae | Depleted | (20, 26) |
| Eubacterium eligens | Firmicutes | Eubacteriaceae | Depleted | |
| Faecalibacterium prausnitzii | Firmicutes | Clostridia | Depleted | |
| Veillonella | Firmicutes | Veillonellaceae | Depleted | |
| Megasphaera | Firmicutes | Veillonellaceae | Depleted | (21) |
| Alistipes onderdonkii | Bacteroidetes | Rikenellaceae | Depleted | (23) |
| Bacteroides stercoris | Bacteroidetes | Bacteroidaceae | Depleted | |
| Lachnospiraceae bacterium 1_1_57FAA | Firmicutes | Lachnospiraceae | Depleted | |
| Parabacteroides merdae | Bacteroidetes | Tannerellaceae | Depleted | |
| Bacteroides uniformis | Bacteroidetes | Bacteroidaceae | Depleted | |
| Parabacteroides distasonis | Bacteroidetes | Tannerellaceae | Depleted | |
| Adlercreutzia | Actinomycetota | Eggerthellaceae | Depleted | (22, 24) |
| Bacteroidaceae | Bacteroidetes | – | Depleted | |
| Eubacterium brachy | Firmicutes | Eubacteriales Family XIII. Incertae Sedis | Depleted | |
| Lachnospiraceae | Firmicutes | – | Depleted | |
| Ruminococcaceae | Firmicutes | – | Depleted | |
| Butyricicoccus pullicaecorum | Firmicutes | Clostridiaceae | Depleted | (25, 26) |
| Eubacterium hallii | Firmicutes | Lachnospiraceae | Depleted | |
| Fusicatenibacter saccharivorans | Firmicutes | Lachnospiraceae | Depleted | |
| Intestinimonas butyriciproducens | Firmicutes | – | Depleted | |
| Roseburia inulinivorans | Firmicutes | Lachnospiraceae | Depleted | |
| Ruminococcus bromii | Firmicutes | Oscillospiraceae | Depleted | |
| Firmicutes | – | – | Depleted | (22) |
| Aspergillus | Ascomycota | Aspergillaceae | Depleted | (27) |
| Candida parapsilosis | Ascomycota | Debaryomycetaceae | Depleted | |
| Malassezia yamatoensis | Basidiomycota | Malasseziaceae | Depleted | |
| Moesziomyces aphidis | Basidiomycota | Ustilaginaceae | Depleted | |
| Mucor racemosus | Mucoromycota | Mucoraceae | Depleted | |
| Penicillium citrinum | Ascomycota | Aspergillaceae | Depleted | |
| Penicillium polonicum | Ascomycota | Aspergillaceae | Depleted | |
| Rhodotorula mucilaginosa | Basidiomycota | Sporidiobolaceae | Depleted | |
| Talaromyces wortmannii | Ascomycota | Trichocomaceae | Depleted | |
| Trechispora sp. | Basidiomycota | Hydnodontaceae | Depleted | |
| Wallemia sebi | Basidiomycota | Wallemiaceae | Depleted | |
Table 2.
Effects of dysregulated gut microbiota on the immune status and COVID-19 progression.
| Microbial change | Potential effect | Outcome | Reference |
|---|---|---|---|
| ↑Acinetobacter, Rhodococcus | Inversely correlate with CD3, CD4, CD45, haemoglobins and RBC | Promote COVID-19 severity | (20) |
| ↓Bacteroides, Veillonella | Positively correlate with CD3, haemoglobins and RBC | Lessen COVID-19 severity | |
| ↓Enterococcus | Positively correlate with the plasma concentration of carbon dioxide | Lessen COVID-19 severity | |
| ↑Eubacterium dolichum, Prevotella copri | Positively correlate with SARS-CoV-2 viral load | Promote COVID-19 severity | (30) |
| ↓Alistipes, Bifidobacterium, Clostridium citroniae, Dialister, Haemophilus, Haemophilus parainfluenzae, Ruminococcus, Streptococcus anginosus | Inversely correlate with SARS-CoV-2 viral load | Lessen COVID-19 severity | |
| ↑Actinomyces viscosus, Bacteroides nordii, Clostridium hathewayi | Induce bacteremia | Promote COVID-19 severity | (31) |
| ↑Coprobacillus, Clostridium hathewayi, Clostridium ramosum | Positively correlate with COVID-19 severity | Promote COVID-19 severity | |
| ↓Alistipes onderdonkii, Faecalibacterium prausnitzii | Inversely correlate with COVID-19 severity | Lessen COVID-19 severity | |
| ↑Alistipes finegoldii, Clostridium innocuum, Ruthenibacterium lactatiformans | Positively correlate with inflammatory biomarkers CRP and WBC | Promote COVID-19 severity | (32) |
| ↓Alistipes putredinis, Blautia luti, Dorea longicatena, Faecalibacterium prausnitzii, Gemmiger formicilis | Inversely correlate with COVID-19 severity | Lessen COVID-19 severity | |
| ↑Bacteroides | Positively correlate with COVID-19 severity | Promote COVID-19 severity | (33) |
| ↓Bifidobacterium, Roseburium | Inversely correlate with COVID-19 severity | Lessen COVID-19 severity | |
| ↑Aspergillus | Positively correlate with COVID-19 severity | Promote COVID-19 severity | (34) |
| ↑Blautia, Lactobacillus, Ruminococcus | Positively correlate with proinflammatory cytokines IFN-γ, IL-2, IL-4, IL-6, IL-8, IL-10 and TNF-α | Magnify inflammation | (35) |
| ↓Bacteroides, Clostridiales, Streptococcus | Inversely correlate with proinflammatory cytokines IFN-γ, IL-2, IL-4, IL-6, IL-8, IL-10 and TNF-α | Inhibit inflammation | |
| ↑Burkholderia contaminans | Positively correlate with inflammation biomarkers CRP and IL-6; inversely correlate with the levels of lymphocytes, CD3+ T cells and CD4+ T cells | Magnify inflammation; impair adaptive immune responses | (36) |
| ↑Enterococcus faecalis | Inversely correlate with the count of CD8+ T cells | Impair adaptive immune responses | (37) |
| ↓Lachnospira, Prevetolla, Roseburia | Inversely correlate with IL-21 | Inhibit inflammation | (38) |
| ↑Akkermansia muciniphila, Bacteroides dorei | Positively correlate with IL-6, CXCL8 and IL-1β | Magnify inflammation | (39) |
| ↓Bifidobacterium adolescentis, Collinsella aerofaciens, Coprococcus comes, Dorea longicatena, Eubacterium rectale, Faecalibacterium prausnitzii | Inversely correlate with CXCL10 | Inhibit inflammation | |
| ↓Collinsella aerofaciens, Coprococcus comes, Dorea formicigenerans, Dorea longicatena, Ruminoccocus obeum | Inversely correlate with IL-10 | Inhibit inflammation | |
| ↓Collinsella aerofaciens, Coprococcus comes | Inversely correlate with TNF-α | Inhibit inflammation | |
| ↓Coprococcus comes, Eubacterium rectale | Inversely correlate with CCL2 | Inhibit inflammation | |
| ↓Clostridia | Inversely correlate with IFN-γ | Inhibit inflammation | (40) |
| ↑Actinobacteria | Positively correlate with the gp130/sIL-6Rb level | Magnify inflammation | |
| ↑Citrobacter, Fusobacterium, Peptostreptococcus | Positively correlate with faecal IL-18 level | Facilitate the cytokine storm | (41) |
| ↑Enterococcus faecalis (GroEL) | Positively correlate with IL-6 and IL-10 | Facilitate the cytokine storm | (37) |
| ↑Eubacterium ramulus | Inversely correlate with IL-6 | Prevent the cytokine storm | |
| ↑Blautia obeum, Coprococcus catus, Coprococcus comes | Positively correlate with the number of lymphocytes, CD3+ T cells, CD4+ T cells and CD8+ T cells and lymphocyte proportion | Enhance adaptive immune responses | (42) |
| ↓Roseburia intestinalis | Positively correlate with the number of lymphocytes, CD3+ T cells, CD4+ T cells and CD8+ T cells and lymphocyte proportion | Enhance adaptive immune responses |
↑ represents high abundance; ↓ represents low abundance.
Gut microbiota dysbiosis causes changes in the gut metabolome profiles. Increased abundance of Blautia, Dorea, Parabacteroides and Streptococcus resulted in the upregulation of glycerophospholipid, glycerolipid, linoleic acid and ether lipid metabolism in SARS-CoV-2-infected individuals (18). Lipids could be crucial for viral replication and dissemination (43). These identified bacteria may be potential predictors for COVID-19 severity. Gut microbiota diversity was markedly reduced in COVID-19 patients compared with healthy controls (19). The decreased microbial diversity was associated with poor prognosis in COVID-19 patients. COVID-19 patients exhibited an enrichment of opportunistic pathogenic species including Bacteroides, Corynebacterium, Enterocloster, Enterococcus, Flavonifractor, Parabacteroides and Streptococcus compared with healthy controls. The abundance of anti-inflammatory butyrate-producing bacteria (e.g., Dialister, Faecalibacterium, Lachnospira, Megasphaera, Prevotella, Roseburia and Ruminococcus) was reduced in COVID-19 patients (21). COVID-19 patients showed an imbalanced metabolism characterized by enhanced protein metabolism and suppressed carbohydrate-oriented catabolism, which might impel putrefactive gut microbiota dysbiosis in COVID-19 patients (19). This potentially led to the reinforcement of systemic inflammation. SARS-CoV-2-induced variations in intestinal microorganisms and metabolic capacities warrant further exploration. It is intriguing how SARS-CoV-2 affects microbial metabolism in the gut.
Remarkably, COVID-19 patients were detected positive for SARS-CoV-2 in the faeces during disease course or even after clearance of viral respiratory infection by viral RNA metagenomic sequencing (23). Such prolonged viral existence in patients without gastrointestinal symptoms underscored the possibility of intestinal infection and faecal-oral transmission of SARS-CoV-2. SARS-CoV-2-positive faecal samples were enriched in the opportunistic pathogens Collinsella aerofaciens, Collinsella tanakaei, Morganella morganii and Streptococcus infantis, while SARS-CoV-2-negative samples had an overgrowth of short-chain fatty acid (SCFA)-producing bacteria, Alistipes onderdonkii, Bacteroides stercoris, Lachnospiraceae bacterium 1_1_57FAA and Parabacteroides merdae. A. onderdonkii and L. bacterium were previously reported to play a protective role in ameliorating the severity of COVID-19 (31). These salutary bacteria might combat SARS-CoV-2 infection in the gut. Moreover, SARS-CoV-2-positive faecal samples presented higher functional capacity for amino acid biosynthesis, glycolysis and nucleotide de novo biosynthesis than those with low-to-none virus infectivity. These pathways might be essential for the survival, growth and metabolism of intestinal bacteria under the immunopathological state. Nevertheless, the impact of gut bacterial bioactivities on SARS-CoV-2 pathogenesis needs to be corroborated in future studies. The number of beneficial bacteria Bacteroides uniformis and Parabacteroides distasonis was increased in gut microbiome after viral clearance from faecal samples, whereas that of inflammation-related bacterium Ruminococcus gnavus was decreased. Inflammation-associated bacteria Clostridium hathewayi, Enterococcus avium and R. gnavus were enriched in patients who were constantly positive for SARS-CoV-2 in the faeces. COVID-19 patients exhibited significant alterations in gut microbiota composition with and without the presence of SARS-CoV-2 virus in the faeces, alluding to an ever-changing gut microbial landscape during disease course. Substantial research efforts are necessary to dissect the infectivity and pathogenesis of SARS-CoV-2 in the gut.
Currently, there is controversy concerning how long SARS-CoV-2-induced dysbiosis of gut microbiota will last. Gut microbiota profile of COVID-19 patients was characterized by an expansion of Prevotellaceae and a reduction of Adlercreutzia, Bacteroidaceae, Eubacterium brachy, Faecalibacterium, Lachnospiraceae and Ruminococcaceae compared with healthy and recovered subjects (24). Remarkably, recovered COVID-19 patients exhibited a descending trend in microbial diversity even after three-month recovery (25, 26). Recovered subjects had an enrichment of opportunistic pathogens (e.g., Clostridium aldenense, Escherichia unclassified and Intestinibacter bartlettii), inflammation-relevant pathogens (e.g., Clostridium bolteae and Flavonifractor plautii) and COVID-19-related bacteria Clostridium ramosum as well as a depletion of beneficial commensals including Butyricicoccus pullicaecorum, Eubacterium hallii, F. prausnitzii, Fusicatenibacter saccharivorans, Intestinimonas butyriciproducens, Roseburia inulinivorans and Ruminococcus bromii. Despite recovered patients had an insufficiency of SCFA-producing bacteria E. hallii and Faecalibacterium, the faecal levels of SCFAs in recovered patients was comparable to those in healthy controls, implying the supplementation from other SCFA-secreting bacteria. Escherichia unclassified and I. bartlettii were positively associated with persistent symptoms such as fatigue, myalgia and anorexia. F. prausnitzii and I. butyriciproducens negatively correlated with persistent symptoms (e.g., chest tightness and cough) after discharge. The underlying mechanisms through which gut microbiota influences the recovery of COVID-19 patients deserve in-depth investigation. A prospective study was previously carried out to longitudinally track alterations of gut microbiota composition in COVID-19 patients (44). The richness of gut microbiota was not restored to normal values even after six-month recovery. The significant loss of gut microbiota richness correlated with worse pulmonary functions. COVID-19 patients with attenuated postconvalescence richness presented higher levels of C-reactive protein (CRP) and disease severity during the acute phase, implying a strong association between intestinal dysbiosis and inflammatory responses in COVID-19. As stable ecosystems confer colonization resistance to opportunistic pathogenic microbes (45), constant attenuation of gut microbiota richness may have long-term biological impacts. A large population of COVID-19 patients suffer ongoing symptoms post initial recovery, termed long COVID-19 (46, 47). Persistent gut dysbiosis may contribute to long COVID-19, which needs to be experimentally verified. Disequilibrium of gut microbiota seems to correlate with the convalescence process of COVID-19, but this hypothesis needs further verification in larger cohorts.
Paradoxically, several studies indicated that gut microbiota could be restored with disease resolution. For instance, a longitudinal study showed that SARS-CoV-2 infection induced gut microbiota dysbiosis by reducing the relative abundance of Bacteroidetes and increasing the Firmicutes/Bacteroidetes ratio (48). However, these SARS-CoV-2-assocated variations in gut microbiota were reversed during the recovery phase. Nine recovered patients after at least two-week recovery exhibited a microbiota configuration similar to that of uninfected subjects (49). Gut microbiota richness increased in 31 recovered subjects after six-month recovery compared with patients with non-SARS-CoV-2 pneumonia (22). The relative abundance of Bacteroidetes was increased during SARS-CoV-2 infection and was declined after viral clearance. On the contrary, the number of Firmicutes was decreased in SARS-CoV-2-infected subjects and was elevated after recovery. SARS-CoV-2 positive patients had a surge of Bacteroides and a depletion of Enterococcus. The levels of Blautia, Lachnospiraceae and Ruminococcaceae were enriched after SARS-CoV-2 negativization, leading to the re-equilibrium of gut microbiota composition. Reportedly, Bacteroides had a positive correlation with the cytokine storm (39). Blautia possessed anti-inflammatory activities and was beneficial to the recovery from COVID-19 (50). These intestinal commensals might play an important role in disease resolution in COVID-19 patients. COVID-19 rehabilitation in turn facilitates the restoration of gut microbiota to the pre-infection state. It is speculative how gut microbiota affects the course of COVID-19, calling for subsequent studies in the future. The mechanisms responsible for the dynamic alterations in gut microbiota during SARS-CoV-2 infection warrant greater attention. Further studies are needed to define how long the imbalanced gut microbiota will last. Collectively, modifying gut microbiota may be instrumental in preventing the lasting impacts of SARS-CoV-2 infection and could improve the clinical outcome of SARS-CoV-2-infected subjects.
Gut mycobiota richness was also decreased in COVID-19 patients compared with healthy controls (27, 28). The number of fungi in the faeces of COVID-19 patients was markedly higher than in healthy controls. The association between increased fungal burden and susceptibility to SARS-CoV-2 infection deserves further verification. Aspergillus, Candida parapsilosis, Malassezia yamatoensis, Moesziomyces aphidis, Mucor racemosus, Penicillium citrinum, Penicillium polonicum, Rhodotorula mucilaginosa, Talaromyces wortmannii, Trechispora sp. and Wallemia sebi were depleted in COVID-19 patients. COVID-19 patients showed a predominance of Candida albicans. Aspergillus niger was positively associated with the incidence of diarrhoea. P. citrinum and R. mucilaginosa showed an inverse correlation with the levels of CRP and ACE. The diversity of mycobiota recovered at six months after recovery (28). The effects of gut mycobiota dysbiosis on COVID-19 pathogenesis and severity are worth exploring.
Altogether, SARS-CoV-2 infection exerts a prolonged adverse effect on gut microbiota composition. ACE2, an important receptor of SARS-CoV-2, can regulate amino acid malnutrition to affect gut microbiota and intestinal inflammation (51). Specifically, ACE2 affects the absorption of neutral amino acids, especially tryptophan, in the small intestine as a chaperone of amino acid transporters. ACE2 downregulation can inhibit the uptake of tryptophan, which reduces the antimicrobial peptide formation by Paneth cells, culminating in gut microbiota dysbiosis and gastrointestinal disorders related to intestinal microbiota imbalance (52). Previous reports showed that SARS-CoV-2 could decrease ACE2 expression in intestinal epithelial cells (53–55). It is thus inferred that SARS-CoV-2 infection contributes to gut microbiota dysfunction by downregulating ACE2. Nevertheless, this hypothesis remains to be validated in further studies. Diverse pathological variations including hypoxia and inflammation could be significant factors contributing to imbalanced gut microbiota (56). Moreover, the intake of antibiotics also causes gut microbiota dysbiosis. The use of unnecessary antibiotics should be avoided in COVID-19 treatment. More studies are required to achieve a deeper understanding of the exact mechanisms underlying dysregulated gut microbiota during COVID-19 pathogenesis. Dysbiotic gut microbiota can affect the response and susceptibility to viral infection in COVID-19 patients. Characterizing gut microbiota composition in COVID-19 patients can enrich our knowledge of the role of intestinal microorganisms in COVID-19 pathogenesis and lead to the identification of new therapeutic targets for COVID-19 treatment. The dynamic changes in gut microbiome during SARS-CoV-2 infection remain to be in-depth investigated. The exact effects of intestinal commensals on COVID-19 progression necessitate further study. Gut microbiota plays an important role in intestinal function and immunity. It is unknown whether the reduction of anti-inflammatory bacteria could give rise to chronic intestinal inflammation. Due to the persistence of abnormal gut microbiota, it is of great importance to continually monitor the intestinal health in recovered COVID-19 patients. Further clinical studies should be directed to clarify the long-term impact of SARS-CoV-2-induced gut microbiota dysbiosis on human health.
3. The antagonistic effects of commensal microbiota against SARS-CoV-2
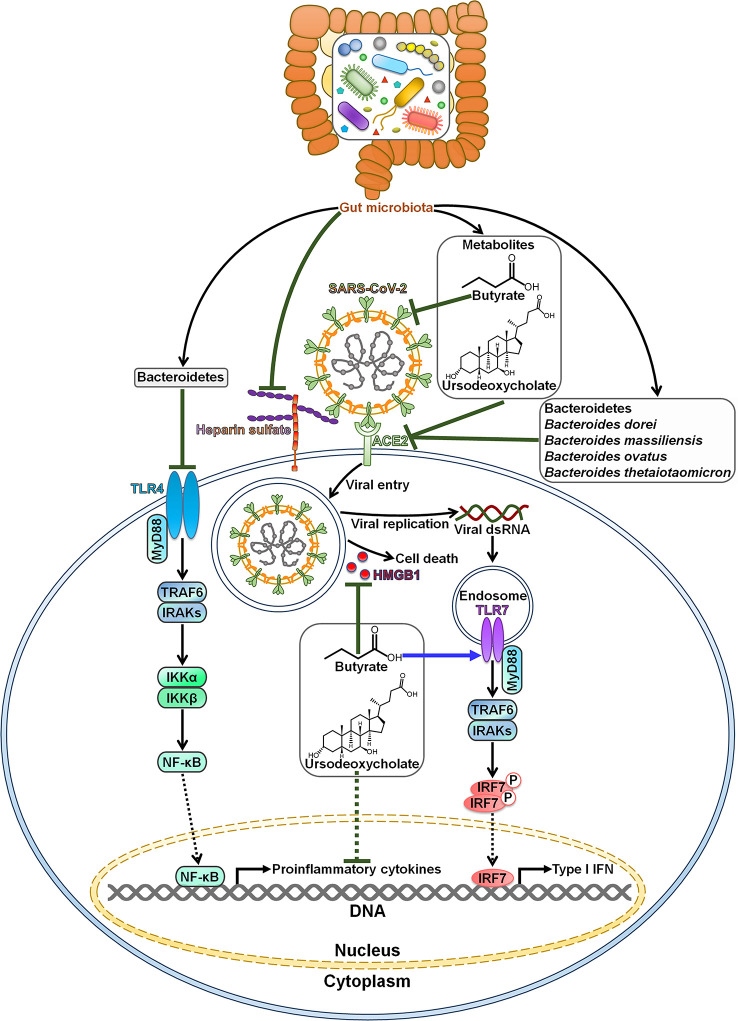
Gut microbiota is capable of impeding SARS-CoV-2 infection and pathogenesis ( Figure 1 ). Commensal host bacteria interfered with SARS-CoV-2 adhesion to target cells by regulating heparin sulfate (57). Depletion of these microbes rendered individuals susceptible to SARS-CoV-2 infection. A sophisticated understanding of the effect of microbiota composition alterations on SARS-CoV-2 infection may be helpful in predicting susceptibility to viral infections and open up new opportunities for the treatment of COVID-19. COVID-19 patients had an enrichment of Bacteroidetes that was diminished after recovery (58). Bacteroidetes was reported to inhibit toll-like receptor 4 (TLR4) and ACE2-related signaling pathways (59, 60). The expansion of Bacteroidetes may constitute a component of host immune defense system, which remains to be further investigated. Furthermore, Bacteroides dorei, Bacteroides massiliensis, Bacteroides ovatus and Bacteroides thetaiotaomicron, known to reduce colonic ACE2 expression in vivo, showed negative correlation with faecal SARS-CoV-2 load in COVID-19 patients (31). These Bacteroides species might play a protective role in fighting SARS-CoV-2 infection by blocking ACE2-mediated viral entry.
Figure 1.
Roles of gut microbiota in resisting SARS-CoV-2 infection and pathogenesis. Bacteroidetes can block SARS-CoV-2-induced cytokine storm by targeting the TLR4 signaling pathway. Gut microbiota inhibits SARS-CoV-2 adhesion to target cells by regulating heparin sulfate. Gut microbiota-derived metabolite butyrate reduces the expression of membrane ACE2 and suppresses the activation of viral spike protein. Moreover, butyrate represses SARS-CoV-2-induced cell death by downregulating HMGB1. In addition, butyrate initiates antiviral immune responses by motivating the TLR7 signaling cascade. The metabolite ursodeoxycholate also exerts anti-SARS-CoV-2 effects. It prevents SARS-CoV-2 infection by blocking viral attachment to ACE2. Ursodeoxycholate has the ability to restrict the expression of pro-inflammatory cytokines, thereby ameliorating SARS-CoV-2-induced pathology. Species belonging to the Bacteroidetes phylum, such as Bacteroides dorei, Bacteroides massiliensis, Bacteroides ovatus and Bacteroides thetaiotaomicron, are found to restrain ACE2-mediated viral entry. TLR4, toll-like receptor 4; MyD88, myeloid differentiation factor 88; TRAF6, tumor necrosis factor receptor-associated factor 6; IRAKs, interleukin-1 receptor-associated kinases; IKKα, inhibitor of κB kinase α; IKKβ, inhibitor of κB kinase β; NF-κB, nuclear factor-κB; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; ACE2, angiotensin-converting enzyme 2; HMGB1, high mobility group protein 1; dsRNA, double-stranded RNA; TLR7, toll-like receptor 7; IRF7, interferon regulatory factor 7; IFN, interferon.
Intestinal microbiota-derived metabolite butyrate could inhibit SARS-CoV-2 infectivity through downregulation of membrane ACE2 and inactivation of viral spike protein in gut epithelium (61). It also restricted SARS-CoV-2 replication by reducing the expression of high mobility group protein 1 (HMGB1). Butyrate could activate TLR signaling pathway. Butyrate might represent a preventive agent against SARS-CoV-2 infection. Nevertheless, the role of commensal-produced butyrate in opposing SARS-CoV-2 infection remains to be confirmed in preclinical and clinical studies. There was a negative relationship between intestinal Collinsella and COVID-19 mortality (62). Collinsella-derived ursodeoxycholate could prevent SARS-CoV-2 infection by blocking viral attachment to ACE2 and downregulating pro-inflammatory cytokines (interleukin-1β (IL-1β), IL-2, IL-4, IL-6 and tumor necrosis factor-α (TNF-α)) (63, 64). Ursodeoxycholate might relieve ARDS in COVID-19 by repressing cytokine storm syndrome (65). However, it will be necessary to ascertain whether this microbial product has beneficial effects against SARS-CoV-2 infection and COVID-19 development.
4. Effects of gut microbiota on SARS-CoV-2-related pathology
4.1. The role of gut microbiota in regulating susceptibility to SARS-CoV-2 infection
It is proposed that gut microbiota contributes to the inter-individual variability in susceptibility to SARS-CoV-2 infection. The COVID-19 mortality and morbidity in the elderly are higher than in younger adults. The elderly gut microbiota was characterized by reduced microbial diversity, an enrichment of Alistipes and Parabacteroides and an underrepresentation of Firmicutes (66). Significant inter-individual differences in the amount of Faecalibacterium and Ruminococcus were observed among the elderly. These gut microbiota signatures may be an explanation for the high level of susceptibility to SARS-CoV-2 infection in this population. Gut microbiota could be a significant factor affecting individuals’ susceptibility to SARS-CoV-2 infection ( Figure 2 ). However, there is a scarcity of direct evidence regarding the effect of gut microbiota composition on susceptibility to SARS-CoV-2 infection in healthy individuals. Further study is needed to compare the gut microbiota profiles among healthy individuals with different susceptibility toward SARS-CoV-2 infection. Gut microbiota dysbiosis may enhance the risk of SARS-CoV-2 infection by coordinating the expression of the viral entry receptor ACE2 in the gut (67). Supplementation of Bifidobacterium longum could elevate the expression of ACE2 receptor in mice, suggesting that altered B. longum might be a pivotal factor linked to COVID-19 susceptibility (68). The mechanisms employed by gut microbiota to regulate colonic ACE2 expression remain elusive and deserve further research. The use of broad-spectrum antibiotics could induce variations in gut microbiota, which may influence colonic ACE2 expression (69, 70). Therefore, substantial research efforts should be directed toward the interrelation between antibiotic-induced dysbiosis and COVID-19 infectivity. The colonic ACE2 expression was downregulated in germ-free (GF) rats co-housed with conventional rats compared with GF rats (70). This conventionalization caused enhanced systemic inflammation in conventionalized GF (GFC) rats, as evidenced by markedly increased levels of lipocalin 2 and neutrophilia. Metabolomic analysis demonstrated that the levels of hydroxy kynurenine, kynurenic acid and tryptophan metabolites were elevated in GFC rats, suggesting that altered expression of ACE2 might have an impact on intestinal microbiota metabolism. A better understanding of the reciprocal impact between gut microbiota and ACE2 expression in COVID-19-susceptible and -resistant patients will provide valuable insights into new treatment modalities for COVID-19. The intestinal microbe Bacteroides that was reduced in COVID-19 patients might impede viral invasion by regulating heparin sulfate, implying a potential interrelation between Bacteroides and susceptibility to SARS-CoV-2 infection (57). In addition, gut microbiota-derived signals have the ability to dominate immune cells for pro- and anti-inflammatory reactions, hence influencing the host’s susceptibility to SARS-CoV-2 infection (71). The association of indigenous microbes with susceptibility to SARS-CoV-2 infection is poorly understood. The mechanism through which gut microbiota affects COVID-19 susceptibility should be revealed in future studies.
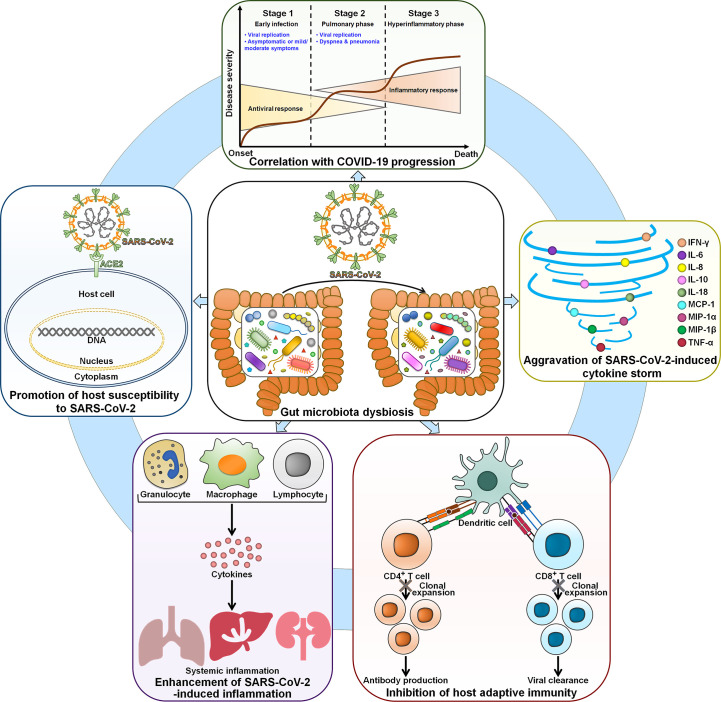
Figure 2.
The involvement of dysbiotic gut microbiota in COVID-19 physiopathology. Gut microbiota affects the susceptibility of host cells to SARS-CoV-2 infection. SARS-CoV-2-induced gut microbiota perturbation contributes to COVID-19 deterioration. Thus, the gut microbiota signature is closely associated with clinical outcomes in COVID-19 patients. In terms of mechanism, gut microbiota exacerbates SARS-CoV-2-induced cytokine storm and leads to hyperinflammation. Moreover, gut microbiota is capable of abrogating host adaptive immune responses by inhibiting CD4+ T and CD8+ T cell expansion. SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; ACE2, angiotensin-converting enzyme 2; IFN-γ, interferon-γ; IL-6, interleukin-6; IL-8, interleukin-8; IL-10, interleukin-10; IL-18, interleukin-18; MCP-1, monocyte chemoattractant protein-1; MIP-1α, macrophage inflammatory protein-1α; MIP-1β, macrophage inflammatory protein-1β; TNF-α, tumor necrosis factor-α.
4.2. The association between gut microbiota dysbiosis and COVID-19 severity
There is a significant correlation between gut microbiota complexity and COVID-19 severity ( Table 2 ). SARS-CoV-2-driven gut dysbiosis may cause intestinal barrier disruption that contributes to the migration or passage of extra-intestinal bacteria (e.g., Granulicatella and Rothia Mucilaginosa) into the gut (30). Moreover, Eubacterium dolichum and Prevotella copri had a positive correlation with the faecal levels of SARS-CoV-2 while Alistipes, Bifidobacterium, Clostridium citroniae, Dialister, Haemophilus, Haemophilus parainfluenzae, Ruminococcus and Streptococcus anginosus showed an inverse association with the SARS-CoV-2 viral load. Antibiotic-naïve COVID-19 patients presented a higher abundance of opportunistic pathogens that could induce bacteremia such as Actinomyces viscosus, Bacteroides nordii and C. hathewayi than healthy controls and pneumonia controls (31). It was thus inferred that severe COVID-19 might be the outcome of secondary bacterial infection. Several members belonging to the Firmicutes phylum, Coprobacillus, C. hathewayi and C. ramosum, showed a positive correlation with COVID-19 severity (31). Reportedly, Coprobacillus was able to increase ACE2 expression in murine gut (60). C. hathewayi and C. ramosum were revealed to be connected with human infection and bacteremia (72, 73). In contrast, baseline faecal abundance of beneficial species A. onderdonkii and F. prausnitzii was inversely associated with COVID-19 severity (31). Alistipes species played a critical role in preserving intestinal immune homeostasis, while F. prausnitzii possessed anti-inflammatory abilities (74, 75). Nevertheless, substantial work remains to address the direct causality between gut microbiota dysbiosis and COVID-19 deterioration and prognosis.
The relative abundance of Alistipes finegoldii, Clostridium innocuum and Ruthenibacterium lactatiformans positively correlated with inflammatory biomarkers (e.g., CRP and WBC) and tended to increase with COVID-19 progression (32). Conversely, Alistipes putredinis, Blautia luti, Dorea longicatena, F. prausnitzii and Gemmiger formicilis were remarkably reduced in severe/critical patients and post COVID-19 patients. Parabacteroides was enriched while F. prausnitzii was depleted in patients with complications (e.g., hemodialysis). These intestinal microorganisms may be involved in the occurrence of complications in COVID-19. COVID-19 severity was correlated with increased abundance of Bacteroides and reduced abundance of Bifidobacterium and Roseburium (33). The immunoregulatory Bifidobacterium appeared to play a role in inhibiting SARS-CoV-2-induced cytokine storm. The loss of Bifidobacterium may be predisposing factor for severe disease. Additional studies are required to confirm and expand this assumption. Aspergillus flavus, C. albicans and Candida auris were strikingly enriched in COVID-19 patients but could not be detected in healthy controls (34). The expansion of pathogenic fungi might lead to gut mycobiota imbalance in COVID-19 patients. The persistent existence of these fungi probably caused a long-term detrimental effect on human health. Reportedly, C. albicans colonization exacerbated inflammation in the gut and extra-gut tissues (76). Aspergillus infection was capable of inducing pulmonary and respiratory manifestations (77). Thus, cough was prevalent in COVID-19 patients with an enrichment of Aspergillus species, implying Aspergillus-induced systemic effect and the perplexing connection between the gastrointestinal and respiratory systems. Consistently, the abundance of Aspergillus species seemed to be positively associated with disease severity (34). The variation in gut microbiota composition might be attributable to SARS-CoV-2 infection. C. albicans and Aspergillus species potentially resulted in disease progression in COVID-19 patients. These mutual effects are largely unknown and need more detailed investigation.
4.3. Gut microbiota-mediated immune dysregulation in COVID-19
Gut microbiota performs a critical role in the development and functionality of host immune system (35). It is thus supposed that gut microbiota could affect COVID-19 severity through actions on the host immune system ( Figure 2 ).
4.3.1. The reciprocal impacts between gut microbiota and the immune system
Gut microbiota is implicated in host inflammation and disease progression in COVID-19. Commensal and pathogenic microbes act as a vital source of microbe-associated molecular patterns (MAMPs) and pathogen-associated molecular patterns (PAMPs) (78). SARS-CoV-2-induced direct damage of enterocytes and gut microbiota dysbiosis could enhance intestinal permeability, facilitating the translocation of microbe-derived signals into the circulating system (79). Pattern recognition receptors (PRRs), such as TLRs, can sense these microbial factors and initiate distinct immunologic reactions depending on the type of cell, ligand or receptor. Due to the breach of intestinal barrier, opportunistic pathogens could enter the circulation and result in systemic inflammation (80). Thus, gut barrier dysfunction interrupts the critical equilibrium between intestinal microbes and host immunity in COVID-19.
SARS-CoV-2 can translocate from the lungs to the gastrointestinal tract (81). Once SARS-CoV-2 reaches intestinal epithelial cells, the virus invades the enterocytes via its receptor ACE2. After numerous viruses are secreted from epithelial cells, innate immune cells (e.g., dendritic cells and macrophages) then detect and bind viral PAMPs, and this eventually results in the excessive release of proinflammatory cytokines. The inflammatory cascade further drives gut microbiota dysbiosis (82). Host immune response should be effective at antagonizing SARS-CoV-2 infection. In some circumstances, an over-reactive immune reaction may occur, resulting in extensive lung or multiorgan damage (83). Reportedly, the levels of various chemokines and cytokines, such as basic fibroblast growth factor (FGF), granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), monocyte chemoattractant protein-1 (MCP-1), vascular endothelial growth factor (VEGF), IL-2, IL-7, IL-10 and TNF-α were higher in COVID-19 patients than healthy controls (84). Particularly, the levels of some factors (e.g., G-CSF, MCP-1, IL-2, IL-7, IL-10 and TNF-α) were increased in severe patients relative to nonsevere patients. An integrative analysis of multi-omics data from patients with gastrointestinal manifestations revealed that Blautia, Lactobacillus and Ruminococcus positively correlated with proinflammatory cytokines, including interferon-γ (IFN-γ), IL-2, IL-4, IL-6, IL-8, IL-10 and TNF-α (35). In contrast, Bacteroides, Clostridiales and Streptococcus were inversely associated with these proinflammatory cytokines. Gut microbiota composition of COVID-19 patients was found to be connected with plasma concentrations of aspartate aminotransferase (AST), C-X-C motif ligand 10 (CXCL10), CRP and lactate dehydrogenase (LDH) (39). COVID-19-depleted Bifidobacterium adolescentis, Eubacterium rectale and F. prausnitzii could regulate the gastrointestinal immune response (85–87). It is thus conceivable that the reduction of these microorganisms could disrupt the intestinal immune homeostasis and lead to exuberant inflammation. Furthermore, correlative blood samples indicated an interrelation between gut microbiota dysbiosis, multiplication of inflammatory mediators and severity of systemic inflammation in COVID-19 (39).
Gut microbiota plays an important role in the evocation and training of the host immune system (88). Immunoregulatory signals and metabolites, such as SCFAs and bile acids, produced by intestinal commensals (e.g., Bacteroides, Bifidobacteria and Lactobacillus) can bind to the corresponding receptors on innate immune cells, thus regulating their metabolism and functions (89). The balance between proinflammatory cells (e.g., T helper 17 (Th17)) and inflammatory regulatory cells (regulatory T cells (Tregs)) is essential for intestinal health and homeostasis, which is ultimately controlled by intestinal microorganisms (78). Gut microbiota dysbiosis is a contributing factor to immune dysfunction. For instance, SARS-CoV-2-induced overgrowth of opportunistic pathogens including Burkholderia contaminans and Enterococcus faecalis could contribute to the suppression of T cell immunity (36, 37). Impaired immune responses could lead to an increased susceptibility to pathogen infection, which fosters the perturbation in intestinal flora. Collectively, there is a complex interplay between gut microbiota and systemic immunity in COVID-19. However, further research in this emerging field is warranted.
4.3.2. Gut microbiota-induced inflammation and cytokine storm
Gut dysbiosis can disturb the homeostatic relationship between resident microbiota and intestinal immune system, leading to chronic inflammatory diseases (90). COVID-19 microbiota was characterized by a reduction of anti-inflammatory bacteria including Bacteroides plebeius, F. prausnitzii, Lachnospira, Prevetolla and Roseburia (38). Decreased levels of Lachnospira, Prevetolla and Roseburia were associated with enhancement of inflammatory responses (91). Consistently, the level of the inflammatory cytokine IL-21 was elevated in COVID-19 patients. Gut microbiota dysbiosis might be linked to the exacerbation of inflammatory responses in COVID-19. The loss of butyrate-producing anti-inflammatory bacteria (e.g., Faecalibacterium and Roseburia) as well as the upregulation of proinflammatory mediators (e.g., CRP, IL-6 and soluble IL-2 receptor (sIL2R)) was more pronounced in severe/critical COVID-19 patients than mild patients (92). The depletion of anti-inflammatory bacteria is likely to lead to excessive inflammation in severe/critical patients. The correlation between gut microbiota and inflammatory factors has been preliminarily established. For instance, two COVID-19-enriched bacteria Akkermansia muciniphila and B. dorei exhibited a positive relationship with IL-6, CXCL8 and IL-1β (39). Conversely, six depleted gut commensals in COVID-19 including B. adolescentis, C. aerofaciens, Coprococcus comes, D. longicatena, E. rectale and F. prausnitzii were negatively connected with CXCL10. C. aerofaciens, C. comes, D. formicigenerans, D. longicatena and Ruminoccocus obeum negatively correlated with IL-10. C. aerofaciens and C. comes were negatively associated with TNF-α, and C. comes and E. rectale inversely correlated with C-C motif chemokine ligand 2 (CCL2) (39). The relative abundance of COVID-19-depleted Clostridia showed an inverse association with the IFN-γ level, while the count of COVID-19-enriched Actinobacteria was positively connected with the glycoprotein 130 (gp130)/soluble IL-6 receptor subunit β (sIL-6Rb) level (40). Therefore, intestinal flora disturbance might lead to microbial-mediated immune dysregulation ( Figure 2 ). Future research efforts should be made to verify the causal relationship between gut microbiota and excessive inflammation in COVID-19 patients.
The impairment of intestinal barrier integrity is deemed as critical inducer of systemic inflammation in COVID-19. The existence of intestinal microbiota in the plasma as well as increased plasma levels of gut permeability markers fatty acid binding protein 2 (FABP2), lipopolysaccharides (LPS) and peptidoglycan (PGN) indicated the intestinal barrier dysfunction in COVID-19 (93). Severe COVID-19 was associated with higher levels of intestinal barrier integrity marker (zonulin) and microbial translocation markers (β-glucan and lipopolysaccharide binding protein (LBP)) (94). Zonulin, β-glucan and LBP positively correlated with the factors of systemic inflammation and immune activation including CRP, IL-6 and IL-10. Enhancement of tight junction permeability and microbial translocation could contribute to microbiota-mediated myeloid inflammation. As expected, the levels of monocyte and neutrophil inflammation markers (soluble CD14 (sCD14) and myeloperoxidase (MPO)) were increased in the severe group compared with mild and control groups, alluding to the association between microbial translocation and COVID-19 severity. Citrulline, a marker of intestinal function, was remarkably decreased in severe COVID-19, while the marker of intestinal dysbiosis succinic acid showed an opposite trend (94). The kynurenine/tryptophan ratio was increased in severe patients. The citrulline level was inversely associated with IL-6, whereas the level of succinic acid and the kynurenine/tryptophan ratio were positively correlated with the concentration of IL-6. It was plausible to assume that disrupted gut functions concomitant with dysregulated metabolic activities could be potential forces contributing to COVID-19-associated inflammation. The loss of gut barrier integrity in severe COVID-19 may be attributed to dysregulated gut microbiota. Reportedly, COVID-19-enriched Eggerthella could enhance intestinal permeability, which might allow the transfer of intestinal microbes and toxins to the circulation and exacerbate inflammation-induced injuries (40). It will be important to adequately reveal the mechanisms underlying intestinal barrier impairment. The potential effect of microbial translocation on mucosal and systemic immune responses must be a research priority. In addition, intestinal microbe-produced metabolites may act as regulators of the inflammatory response in COVID-19. COVID-19 patients exhibited reduced capacity for SCFA production and L-isoleucine biosynthesis due to the loss of F. prausnitzii (95). Impaired SCFA and L-isoleucine biosynthesis showed an intimate association with elevated plasma levels of CRP and CXCL10 (96, 97). The inverse association between these microbial products and proinflammatory cytokines underscored their significance in COVID-19 pathology and disease outcome. Collectively, the linkage between altered microbial functions and COVID-19 severity opens new horizons for dissecting the role of gut microbiota in COVID-19 development. The long-term implications of disrupted metabolic pathways during COVID-19 should be an important subject of future studies.
Increasing evidence has indicated the bidirectional crosstalk between gut microbiota and lung, called the gut-lung axis (78). On the one side, the gut can affect the progression of lung disorders. Under pathologic conditions, gut microbiota dysbiosis damages intestinal barrier integrity, which enables microbial components and metabolites to translocate into the lung tissues via the bloodstream. This effect can lead to sepsis and even ARDS (98). On the other side, pulmonary inflammation (e.g., respiratory viral infections) induces the perturbation of gut microbiota. It is reasonable to propose that the bidirectional gut-lung crosstalk has a pivotal role in SARS-CoV-2 pathogenesis and disease severity in COVID-19 patients. The gut microbiota in COVID-19 is characterized by a remarkable depletion of beneficial species and an enrichment of opportunistic pathogens. These variations eventually contribute to intestinal disruption, migration of pathogenic microorganisms across the intestinal mucosa, secondary microbial infections, increased inflammatory responses, multiorgan failure and unsatisfactory clinical outcome (99). SARS-CoV-2-induced gut dysbiosis may be a fundamental pathophysiological process contributing to COVID-19-associated hyperinflammation. The role of gut microbiota in mediating the interaction between SARS-CoV-2 and host innate immunity should be explicitly defined in future studies.
Given the linkage between gut microbiota and proinflammatory cytokines, the cytokine storm may result from SARS-CoV-2-dependent shifts in gut microbiota composition ( Figure 2 ). The faecal level of IL-18, a proinflammatory cytokine produced by intestinal cells, was higher in COVID-19 patients than in healthy controls (41). Faecal IL-18 levels showed a positive correlation with the abundance of Citrobacter, Fusobacterium and Peptostreptococcus. Intestinal epitopes derived from E. faecalis GroEL were enriched in COVID-19 patients with fever and showed a positive relationship with IL-6 and IL-10 (37). In contrast, the anti-inflammatory bacterium Eubacterium ramulus that was overabundant in patients with non-fever was negatively linked to IL-6. SARS-CoV-2-induced intestinal dysbiosis resulted in the alterations in gut metabolites including amino acids, carbohydrates and neurotransmitters (100). Microbe-mediated amino acids were positively associated with increased levels of inflammatory cytokines (CXCL9, CXCL10, IFN-γ and IL-6) and negatively correlated with decreased levels of cytokines (IL-9 and IL-17) in COVID-19. COVID-19-depleted carbohydrate metabolites and neurotransmitters had a positive correlation with reduced levels of cytokines (e.g., CCL22, IL-12 and IL-13) while negatively correlated with increased levels of inflammatory cytokines (e.g., IL-6 and IL-10). Thus, gut metabolites might be implicated in cytokine dynamics in COVID-19. The contribution of gut microbiota and its derived metabolites to COVID-19 development should be corroborated in in vivo experimental models. Disrupted gut barrier favored the translocation of intact microbes or microbial components (e.g., LPS) into the systemic circulation, which impelled the secretion of proinflammatory cytokines, resulting in the exacerbation of cytokine storm in COVID-19 patients (93, 101). As expected, the plasma concentrations of proinflammatory cytokines IFN-γ, IL-6, IL-8, MCP-1, macrophage inflammatory protein (MIP)-1α, MIP-1β and TNF-α were elevated in COVID-19 subjects (93). The mechanisms by which gut microbiota modulates the cytokine response in COVID-19 are still equivocal and should be a subject of future studies.
4.3.3. The interaction between gut microbiota and adaptive immunity
Mild/moderate COVID-19-enriched Blautia obeum, Coprococcus catus and C. comes and severe/critical COVID-19-depleted Roseburia intestinalis showed a positive relationship with the number of lymphocytes, CD3+ T cells, CD4+ T cells and CD8+ T cells and lymphocyte proportion (42). Altered gut microbiota led to inhibition of superpathways of polyamine biosynthesis II and sulfur oxidation in severe/critical COVID-19. The suppression of polyamine biosynthesis may contribute to reduced T cell proliferation and uncontrolled production of cytokines, which necessitates further verification. Further metabolomics studies are required to comprehensively delineate the crosstalk among gut microbiota, microbial metabolites and host adaptive immune system. The opportunistic pathogen E. faecalis that was overabundant in COVID-19 patients had a negative correlation with CD8+ T cells (37). COVID-19-enriched B. contaminans was negatively associated with the levels of lymphocytes, CD3+ T cells and CD4+ T cells (36). The microbial metabolic activity (e.g., glucose metabolism) might mediate the effect of gut microbiota dysbiosis on immune reaction in COVID-19. The circulating level of the microbial translocation marker LBP, which was markedly elevated in severe patients, had a correlation with lymphocytes. It is thus inferred that the impairment of gut barrier function may mediate the crosstalk between gut microbiota and T cell immunity in COVID-19. The effects of gut microbiota on adaptive immunity during COVID-19 development are worthy of greater attention.
4.4. Gut microbiota as potential therapeutic targets for COVID-19
A growing body of evidence has suggested the dynamic variation of gut microbiota composition during SARS-CoV-2 infection. It was reported that COVID-19 patients had an expansion of Streptococcus and a contraction of Ruminococcaceae, Agathobacter, Anaerostipes, E. hallii, Fusicatenibacter, unclassified Lachnospiraceae and Roseburia when compared to healthy controls (102). Compared with non-COVID-19 subjects, COVID-19 patients had a depletion of Bariatricus comes, Blautia_A obeum, Blautia_A wexlerae, D. formicigenerans, F. prausnitzii_D, F. saccharivorans and Faecalibacterium sp900539945 (103). Importantly, COVID-19 patients at different severity stages (mild, moderate and severe) show varied gut microbiota signatures. A previous study showed that moderate and severe COVID-19 patients presented a lower Firmicutes/Bacteroidetes ratio, a higher abundance of Proteobacteria and a lower proportion of beneficial bacteria including Lachnospira and Roseburia than mild patients (104). Severe patients has an overrepresentation of Actinomyces naeslundii and S. salivarius, while recovered patients had a high abundance of B. luti and C. comes (105). Agathobacter rectalis, Alistipes senegalensis, Bacteroides finegoldii, Bacteroides xylanisolvens, B. adolescentis, E. coli, Gemmiger formicillis, Pantoea agglomerans, Rodentibacter trehalosifermentans and Streptococcus parasanguinis were only present in mild patients. Moreover, the microbiota signatures were correlated with common symptoms in COVID-19, such as dry cough, dyspnea, fever and headache. Manipulation of gut microbiota may be an auxiliary treatment option to mitigate the clinical presentations in COVID-19 patients. However, gut microbiota signatures in COVID-19 patients await further verification in larger cohorts. Longitudinal studies are needed to explore the changes in gut microbiota composition at distinct disease stages in COVID-19 patients.
The relative abundance of probiotic bacteria (Bifidobacterium and Lactobacillus) and butyrate-producing bacteria (Clostridium butyricum, Clostridium leptum, E. rectale and F. prausnitzii) markedly diminished as disease severity increased (106). The number of conditional pathogenic bacteria Enterococcus and Enterobacteriaceae was elevated with COVID-19 severity. Therefore, gut microbiota composition may reflect disease severity in COVID-19 patients. The perturbation of gut microbiota may increase the risk of mortality in COVID-19 patients. Increased abundance of Proteobacteria and decreased levels of secondary bile acids and desaminotyrosine were found to correlate with mortality in critical COVID-19 patients (107). These microbial metabolites were also connected with the occurrence of respiratory failure resulting in mechanical ventilation. The genera Bacteroides and Parabacteroides were decreased in non-survivor patients with COVID-19 when compared to survivors (108). Hence, further study of intestinal microbiota relevant to better clinical outcomes will facilitate the development of effective therapeutic options for COVID-19. Although several registered clinical trials involving regulation of gut microbiota for COVID-19 management are in progress (109), more clinical evidence is still required to substantiate the safety and efficacy of microbiota-directed therapies.
5. Potential therapeutic role of microbiota-targeted treatments in COVID-19
5.1. Faecal microbiota transplantation
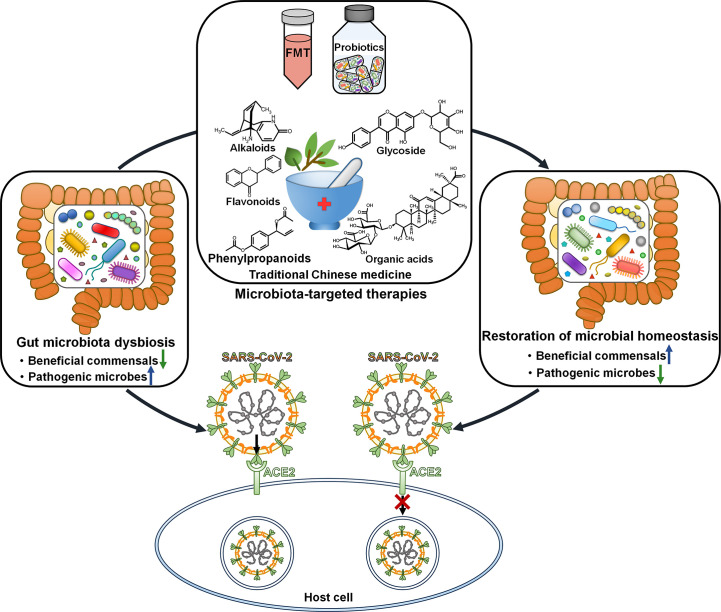
Faecal microbiota transplantation (FMT) from healthy donors could mitigate gastrointestinal symptoms in discharged COVID-19 patients (110). The richness of gut microbiota was significantly increased in COVID-19 patients after FMT. Moreover, FMT treatment partially ameliorated gut dysbiosis by ascending the level of Actinobacteria, Bifidobacterium and Faecalibacterium and descending the level of Proteobacteria. Manipulating gut microbiota via FMT may represent a promising therapeutic intervention for COVID-19 ( Figure 3 ). Gut microbiota dysbiosis in discharged COVID-19 patients awaits further validation through large-scale studies. Sustained efforts should be dedicated to understanding the impact of altered gut microbiota on post-infection recovery.
Figure 3.
Therapeutic potential of microbiota-directed treatments in COVID-19. Microbiota-targeted therapies, including FMT, probiotics and traditional Chinese medicine, may reverse gut microbiota dysbiosis and play a protective role in antagonizing SARS-CoV-2 infection and pathogenesis. FMT, faecal microbiota transplantation; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; ACE2, angiotensin-converting enzyme 2.
5.2. Bacteriotherapy
A retrospective, observational cohort study that included 200 patients with severe COVID-19 were conducted to assess the efficacy of oral bacteriotherapy composed of Bifidobacterium lactis DSM 32246, B. lactis DSM 32247, Lactobacillus acidophilus DSM 32241, Lactobacillus brevis DSM 27961, Lactobacillus helveticus DSM 32242, Lactobacillus paracasei DSM 32243, Lactobacillus plantarum DSM 32244 and Streptococcus thermophilus DSM 32245 (111). It turned out that the best available therapy (BAT) plus oral bacteriotherapy could effectively prevent disease progression in COVID-19 patients. Moreover, the mortality was lower in patients treated with BAT plus oral bacteriotherapy than those who only received BAT. Further analysis showed that oral bacteriotherapy served as an independent parameter related to a decreased risk for death. Larger prospective clinical trials are still necessary to verity the aforementioned results. Twelve COVID-19 patients were treated with probiotics that included B. lactis subsp. HNO19, Lactobacillus casei subsp. Lc-11, L. plantarum subsp. Lp-15, B. lactis subsp. B420, B. longum subsp. BL05, Lactobacillus format subsp. Lg-36, Lactobacillus rhamnosus subsp. Lr-32, L. paracasei subsp. Lpc-37, and Lactobacillus salivariu (20). The use of probiotics could partially reverse COVID-19-related gut microbiota dysbiosis. The dominant bacteria shifted from Escherichia-Shigella to Bacteroides, Enterococcus and Veillonella after treatment. The probiotics group had a reduction of Rhodococcus and conditional pathogenic bacteria E. coli and K. pneumonia and an enrichment of Clostridium XlVa, F. prausnitzii and R. hominis. The gut microbiota profiles may be varied among different COVID-19 patients or during the courses of probiotic-based treatment, calling for continual research efforts to comprehensively reveal the characteristics of gut microbiota in COVID-19. A total of 375 COVID-19 patients were enrolled in a retrospective clinical trial to investigate the therapeutic efficacy of a probiotic combination that included Bifidobacterium, Enterococcus and Lactobacillus (112). Of these, 179 patients were treated with standard care plus probiotics. The administration of probiotics was correlated with improved clinical outcomes in COVID-19 patients, as evidenced by dramatic reductions in duration of fever, viral shedding and hospital stay. These findings may open new avenues toward the development of innovative COVID-19 therapies. It is worth mentioning that more randomized controlled trials are necessary to verify the clinical benefit of probiotics in treating COVID-19.
A randomized, quadruple-blinded, placebo-controlled trial was conducted to assess the therapeutic potential of a probiotic formula that included Lactiplantibacillus plantarum strains KABP022, KABP023 and KABP033 and Pediococcus acidilactici strain KABP021 in 300 COVID-19 patients (113). This probiotic formula was well-tolerated and reduced nasopharyngeal viral load, lung infiltrates as well as duration of clinical symptoms (e.g., diarrhoea, dyspnea, headache and myalgia) compared with placebo. Moreover, the probiotic group had a higher serum level of virus-specific IgG and IgM than the placebo group. Probiotic treatment had no significant effect on gut microbiota composition. It was proposed that this probiotic formula might act on the gut-lung axis via interaction with host immune system. More clinical studies are warranted to testify these findings and uncover the mechanism of action of probiotic. A formula of Bifidobacteria strains, galactooligosaccharides, resistant dextrin and xylooligosaccharide (SIM01) efficiently diminished nasopharyngeal SARS-CoV-2 viral load and fostered the generation of SARS-CoV-2 antibody in COVID-19 patients (114). SIM01 formula also reduced the plasma levels of proinflammatory markers IL-1 receptor antagonist (IL-1RA), IL-6, MCP-1, macrophage colony-stimulating factor (M-CSF) and TNF-α. It also restored gut microbiota dysbiosis in COVID-19 patients by increasing commensal bacteria (e.g., Bifidobacterium spp., Eubacterium spp. and F. prausnitzii) and decreasing opportunistic pathogens (e.g., Bacteroides spp. and E. coli). Thus, manipulation of gut microbiota can be beneficial to strengthen host immune responses against COVID-19.
5.3. Traditional Chinese medicine
Compelling evidence has demonstrated that traditional Chinese medicine (TCM) can help to alleviate clinical symptoms and improve prognosis in COVID-19 patients (115). For instance, Huashi Baidu granules, Lianhua Qingwen granules and Qingfei Paidu decoction were reported to relieve the main clinical presentations of cough, chest tightness, fatigue and fever (116–118). Particularly, Lianhua Qingwen granules, Qingfei Paidu decoction and Shuanghuanglian oral liquid facilitated the absorption of pulmonary inflammation and reduced nucleic acid negative time and the average length of hospitalization (119–121). Furthermore, TCM can inhibit the inflammatory response and enhance antiviral immune function. Reportedly, Xuanfei Baidu decoction reduced the levels of CRP and erythrocyte sedimentation rate (ESR) and increased lymphocyte amount (122). Maxing Shigan decoction diminished the levels of alanine aminotransferase (ALT), AST, CRP and IL-6 and elevated the counts of CD4+ and CD8+ T cells (123). Tanreqing capsule elevated the number of CD3+ T cells (124).
Gut microbiota in COVID-19 patients is disordered, leading to pathogen infection and disease deterioration (115). Gut microbiota can adjust the host immune system and has emerged as a turning point of the prognosis of COVID-19 patients. TCM may be effective in treating COVID-19 by correcting gut microbiota imbalances and improving the host immune reactions. Most TCM is taken orally, enabling TCM to interact with numerous microbes in the intestine. Gut microbiota plays a vital role in the pharmacological effects of TCM by affecting the metabolic transformation and bioavailability of TCM. TCM in turn alters gut microbiota composition by favoring the growth of beneficial microbes and suppressing the reproduction of harmful species, contributing to the preservation of a healthy intestinal environment and the improvement of body immunity (125). The interplay between the effective components of TCM and gut microbiota has been investigated. Alkaloids, flavonoids, glycoside, organic acids and phenylpropanoids are the primary categories of TCM ingredients that have the ability to alter gut microbiota (126). Alkaloids are a class of nitrogen-containing organic compounds and possess important physiological functions (127). The structural features of alkaloids are commonly small molecules, ether bonds and coordination bonds, which may undergo hydrolysis and dehydration reactions under the action of intestinal microorganisms (128). The majority of flavonoids (except flavanols) naturally bind to sugars to form β-glycosides, thus they are not readily absorbed by the small intestine (129). Most glycosylated flavonoids reach the intestine, where intestinal microorganisms can metabolize the flavonoids to generate phenolic acids or other metabolites. Flavonoids exert a modulatory effect on gut microbiota, while intestinal commensals can regulate flavonoid activity and bioavailability (130). Intestinal microbes produce esterases, glycoside hydrolases and lyases to break down sugar chains to produce energy (131). The important end products generated by polyglycolysis are SCFAs that include acetic acid, butyric acid and propionic acid (132). TCM may enhance the population of SCFA-producing bacteria (e.g., anaerobic bacteria and Bifidobacterium) to strengthen the barrier function of the intestine and restrict the inflammatory response of the intestinal mucosa (133). Intestinal microbes secrete esterase to hydrolyze organic acids in the intestine. Particularly, lactic acid, an organic acid produced by anaerobic glucose metabolism, is capable of suppressing the growth of pathogenic bacteria. Moreover, specific microbes (e.g., Clostridium, E. coli and Lactobacillus) can convert carbohydrates or polyphenols into organic acids (134). Due to a lactone structure, phenylpropanoids are prone to demethylation and lactone hydrolysis by intestinal microbiota (135). Future work is necessary to clarify the interaction between TCM and gut microbiota, which will provide new ideas and new targets for the management of COVID-19. It is conceivable that TCM could alleviate COVID-19-associated gut microbiota dysbiosis, hence ameliorating the pathological conditions in patients. However, the mechanisms underlying TCM-mediated regulation of gut microbiota need to be further revealed.
6. Conclusions and future perspectives
Gut microbiota exhibits dynamic alterations during the course of COVID-19. The exact effect of SARS-CoV-2 infection on gut microbiota composition should be determined. Since therapeutic drugs could cause the variations in gut microbiota composition, it is essential to explore the gut microbiota configuration in treatment-naïve patients. The underlying mechanism of gut microbiota dysbiosis remains largely unknown. Continued research efforts are required to ascertain whether the enrichment of specific gut microorganisms is delicately coordinated or occasionally occurs owing to the decrement of other bacterial commensals. It is conjectured that SARS-CoV-2 affects gut microbiota composition via direct and indirect manners. SARS-CoV-2 may interrupt gut microbiota homeostasis through downregulation of ACE2. SARS-CoV-2-induced inflammation also leads to gut microbiota dysbiosis. Follow-up studies are required to elucidate the mechanisms exploited by SARS-CoV-2 to orchestrate the host gut microbiota. The duration of altered gut microbiota remains to be determined in further studies. An in-depth investigation on gut microbiome in convalescent patients would be conducive to unearthing suitable nutritional interventions for COVID-19 patients during the recovery phase. Furthermore, it is critical to address whether gut microbiota dysbiosis predisposes recovered subjects to future health problems.
The gut microbiome signatures serve as prospective biomarkers for risk assessment and prognostic prediction in COVID-19 patients. However, different treatments can affect gut microbiota configuration. Properly controlled studies including different severity groups of COVID-19 patients with minimal variations across clinical managements are needed to verify the clinical value of gut commensals. Gut microbiota disturbances may be a contributor to COVID-19 progression. Nevertheless, the causative linkage between gut dysbiosis and disease severity in COVID-19 necessitates additional studies. The perplexing role of gut microbiota in COVID-19 progression has not been fully understood yet. Given the association of the cytokine profile with hyperinflammation in COVID-19, intensive effort should be directed to characterize the interplay between gut microbiota and host cytokine responses. It is still intriguing which bacterial species are involved in COVID-19-associated pathologies. Some microorganisms induce protective immune response against SARS-CoV-2 infection, while pathogenic bacteria act to antagonize host defensive machineries. The overall impact of gut microbiota on host antiviral immunity is worthy of in-depth investigation. Owing to intestinal barrier impairment, commensal microbes and their metabolites can enter the circulation and augment inflammation-induced damages. Great attention must be concentrated on the complex intercourse between gut microbiota and host immune system during COVID-19 pathogenesis. Given the close relationship between gut microbiota and COVID-19 physiopathology, manipulation of gut microbiota may represent an alternative strategy for COVID-19 treatment. Preliminary clinical studies have proven the therapeutic efficacy of FMT, live biotherapeutic products and probiotics in COVID-19 treatment. Since the majority of these studies are retrospective, it should be cautious to draw the final conclusion. Further preclinical and clinical studies are urgently needed to reveal the mechanistic links underlying the therapeutic potential of these microbiota-supportive interventions. Our expanding knowledge of gut microbiome, metabolomics and viral immunology will provide a tremendous opportunity to drive microbiota-related discoveries towards clinical applications.
Author contributions
MW and CL conceived and supervised the project. MW wrote the manuscript and drew the figures. YZ and WC searched and analyzed the literatures. CL and LZ revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding Statement
This work was supported by the Natural Science Foundation of Shandong Province, China (Grant No. ZR2021MH018).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Li Q, Wang Y, Sun Q, Knopf J, Herrmann M, Lin L, et al. Immune response in covid-19: what is next? Cell Death Differ (2022) 29(6):1107–22. doi: 10.1038/s41418-022-01015-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Covid is here to stay: countries must decide how to adapt. Nature (2022) 601(7892):165. doi: 10.1038/d41586-022-00057-y [DOI] [PubMed] [Google Scholar]
- 3. Torjesen I. Covid-19 will become endemic but with decreased potency over time, scientists believe. Bmj (2021) 372:n494. doi: 10.1136/bmj.n494 [DOI] [PubMed] [Google Scholar]
- 4. Harrison AG, Lin T, Wang P. Mechanisms of sars-Cov-2 transmission and pathogenesis. Trends Immunol (2020) 41(12):1100–15. doi: 10.1016/j.it.2020.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Synowiec A, Szczepanski A, Barreto-Duran E, Lie LK, Pyrc K. Severe acute respiratory syndrome coronavirus 2 (Sars-Cov-2): a systemic infection. Clin Microbiol Rev (2021) 34(2):e00133–20. doi: 10.1128/CMR.00133-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chu H, Chan JF, Yuen TT, Shuai H, Yuan S, Wang Y, et al. Comparative tropism, replication kinetics, and cell damage profiling of sars-Cov-2 and sars-cov with implications for clinical manifestations, transmissibility, and laboratory studies of covid-19: an observational study. Lancet Microbe (2020) 1(1):e14–23. doi: 10.1016/s2666-5247(20)30004-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhou A, Song Q, Peng Y, Liao X, Huang P, Liu W, et al. Symptoms at disease onset predict prognosis in covid-19 disease. Libyan J Med (2022) 17(1):2010338. doi: 10.1080/19932820.2021.2010338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for gastrointestinal infection of sars-Cov-2. Gastroenterology (2020) 158(6):1831–3.e3. doi: 10.1053/j.gastro.2020.02.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gupta A, Madhavan MV. Extrapulmonary manifestations of covid-19. Nat Med (2020) 26(7):1017–32. doi: 10.1038/s41591-020-0968-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cheung KS, Hung IFN, Chan PPY, Lung KC, Tso E, Liu R, et al. Gastrointestinal manifestations of sars-Cov-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology (2020) 159(1):81–95. doi: 10.1053/j.gastro.2020.03.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ye Q, Wang B, Zhang T, Xu J, Shang S. The mechanism and treatment of gastrointestinal symptoms in patients with covid-19. Am J Physiol Gastrointestinal liver Physiol (2020) 319(2):G245–G52. doi: 10.1152/ajpgi.00148.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barko PC, McMichael MA, Swanson KS, Williams DA. The gastrointestinal microbiome: a review. J Vet Intern Med (2018) 32(1):9–25. doi: 10.1111/jvim.14875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thursby E, Juge N. Introduction to the human gut microbiota. Biochem J (2017) 474(11):1823–36. doi: 10.1042/BCJ20160510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carding S, Verbeke K, Vipond DT, Corfe BM, Owen LJ. Dysbiosis of the gut microbiota in disease. Microbial Ecol Health Dis (2015) 26:26191. doi: 10.3402/mehd.v26.26191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hussain I, Cher GLY, Abid MA, Abid MB. Role of gut microbiome in covid-19: an insight into pathogenesis and therapeutic potential. Front Immunol (2021) 12:765965. doi: 10.3389/fimmu.2021.765965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu Q, Su Q, Zhang F, Tun HM, Mak JWY, Lui GC, et al. Multi-kingdom gut microbiota analyses define covid-19 severity and post-acute covid-19 syndrome. Nat Commun (2022) 13(1):6806. doi: 10.1038/s41467-022-34535-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gaibani P, D'Amico F, Bartoletti M, Lombardo D, Rampelli S, Fornaro G, et al. The gut microbiota of critically ill patients with covid-19. Front Cell Infect Microbiol (2021) 11:670424. doi: 10.3389/fcimb.2021.670424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Al Bataineh MT, Henschel A, Mousa M, Daou M, Waasia F, Kannout H, et al. Gut microbiota interplay with covid-19 reveals links to host lipid metabolism among middle Eastern populations. Front Microbiol (2021) 12:761067. doi: 10.3389/fmicb.2021.761067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mancabelli L, Milani C, Fontana F, Lugli GA, Tarracchini C, Viappiani A, et al. Untangling the link between the human gut microbiota composition and the severity of the symptoms of the covid-19 infection. Environ Microbiol (2022) 24(12):6453–62. doi: 10.1111/1462-2920.16201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu C, Xu Q, Cao Z, Pan D, Zhu Y, Wang S, et al. The volatile and heterogeneous gut microbiota shifts of covid-19 patients over the course of a probiotics-assisted therapy. Clin Trans Med (2021) 11(12):e643. doi: 10.1002/ctm2.643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cheng X, Zhang Y, Li Y, Wu Q, Wu J, Park SK, et al. Meta-analysis of 16s rrna microbial data identified alterations of the gut microbiota in covid-19 patients during the acute and recovery phases. BMC Microbiol (2022) 22(1):274. doi: 10.1186/s12866-022-02686-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. De Maio F, Ianiro G, Coppola G, Santopaolo F, Abbate V, Bianco DM, et al. Improved gut microbiota features after the resolution of Sars-Cov-2 infection. Gut Pathog (2021) 13(1):62. doi: 10.1186/s13099-021-00459-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zuo T, Liu Q, Zhang F, Lui GC, Tso EY, Yeoh YK, et al. Depicting sars-Cov-2 faecal viral activity in association with gut microbiota composition in patients with covid-19. Gut (2021) 70(2):276–84. doi: 10.1136/gutjnl-2020-322294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yin YS, Minacapelli CD, Parmar V, Catalano CC, Bhurwal A, Gupta K, et al. Alterations of the fecal microbiota in relation to acute covid-19 infection and recovery. Mol Biomed (2022) 3(1):36. doi: 10.1186/s43556-022-00103-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tian Y, Sun KY, Meng TQ, Ye Z, Guo SM, Li ZM, et al. Gut microbiota may not be fully restored in recovered covid-19 patients after 3-month recovery. Front Nutr (2021) 8:638825. doi: 10.3389/fnut.2021.638825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhou Y, Zhang J, Zhang D, Ma WL, Wang X. Linking the gut microbiota to persistent symptoms in survivors of covid-19 after discharge. J Microbiol (2021) 59(10):941–8. doi: 10.1007/s12275-021-1206-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lv L, Gu S, Jiang H, Yan R, Chen Y, Chen Y, et al. Gut mycobiota alterations in patients with covid-19 and H1n1 infections and their associations with clinical features. Commun Biol (2021) 4(1):480. doi: 10.1038/s42003-021-02036-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maeda Y, Motooka D, Kawasaki T, Oki H, Noda Y, Adachi Y, et al. Longitudinal alterations of the gut mycobiota and microbiota on covid-19 severity. BMC Infect Dis (2022) 22(1):572. doi: 10.1186/s12879-022-07358-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Giacobbe DR, Labate L, Tutino S, Baldi F, Russo C, Robba C, et al. Enterococcal bloodstream infections in critically ill patients with covid-19: a case series. Ann Med (2021) 53(1):1779–86. doi: 10.1080/07853890.2021.1988695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu Y, Cheng X, Jiang G, Tang H, Ming S, Tang L, et al. Altered oral and gut microbiota and its association with sars-Cov-2 viral load in covid-19 patients during hospitalization. NPJ Biofilms Microbiomes (2021) 7(1):61. doi: 10.1038/s41522-021-00232-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zuo T, Zhang F, Lui GCY, Yeoh YK, Li AYL, Zhan H, et al. Alterations in gut microbiota of patients with covid-19 during time of hospitalization. Gastroenterology (2020) 159(3):944–55 e8. doi: 10.1053/j.gastro.2020.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schult D, Reitmeier S, Koyumdzhieva P, Lahmer T, Middelhoff M, Erber J, et al. Gut bacterial dysbiosis and instability is associated with the onset of complications and mortality in covid-19. Gut Microbes (2022) 14(1):2031840. doi: 10.1080/19490976.2022.2031840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hazan S, Stollman N, Bozkurt HS, Dave S, Papoutsis AJ, Daniels J, et al. Lost microbes of covid-19: bifidobacterium, faecalibacterium depletion and decreased microbiome diversity associated with sars-Cov-2 infection severity. BMJ Open Gastroenterol (2022) 9(1):e000871. doi: 10.1136/bmjgast-2022-000871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zuo T, Zhan H, Zhang F, Liu Q, Tso EYK, Lui GCY, et al. Alterations in fecal fungal microbiome of patients with covid-19 during time of hospitalization until discharge. Gastroenterology (2020) 159(4):1302–10 e5. doi: 10.1053/j.gastro.2020.06.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nejadghaderi SA, Nazemalhosseini-Mojarad E, Asadzadeh Aghdaei H. Fecal microbiota transplantation for covid-19; a potential emerging treatment strategy. Med Hypotheses (2021) 147:110476. doi: 10.1016/j.mehy.2020.110476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sun Z, Song ZG, Liu C, Tan S, Lin S, Zhu J, et al. Gut microbiome alterations and gut barrier dysfunction are associated with host immune homeostasis in covid-19 patients. BMC Med (2022) 20(1):24. doi: 10.1186/s12916-021-02212-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou Y, Shi X, Fu W, Xiang F, He X, Yang B, et al. Gut microbiota dysbiosis correlates with abnormal immune response in moderate covid-19 patients with fever. J Inflamm Res (2021) 14:2619–31. doi: 10.2147/JIR.S311518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Khan M, Mathew BJ, Gupta P, Garg G, Khadanga S, Vyas AK, et al. Gut dysbiosis and il-21 response in patients with severe covid-19. Microorganisms (2021) 9(6):1292. doi: 10.3390/microorganisms9061292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yeoh YK, Zuo T, Lui GC, Zhang F, Liu Q, Li AY, et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with covid-19. Gut (2021) 70(4):698–706. doi: 10.1136/gutjnl-2020-323020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mizutani T, Ishizaka A, Koga M, Ikeuchi K, Saito M, Adachi E, et al. Correlation analysis between gut microbiota alterations and the cytokine response in patients with coronavirus disease during hospitalization. Microbiol Spectr (2022) 10(2):e0168921. doi: 10.1128/spectrum.01689-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tao W, Zhang G, Wang X, Guo M, Zeng W, Xu Z, et al. Analysis of the intestinal microbiota in covid-19 patients and its correlation with the inflammatory factor il-18. Med Microecology (2020) 5:100023. doi: 10.1016/j.medmic.2020.100023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu X, Zhang W, Guo M, Xiao C, Fu Z, Yu S, et al. Integrated analysis of gut microbiome and host immune responses in covid-19. Front Med (2022) 16(2):263–75. doi: 10.1007/s11684-022-0921-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yan B, Chu H, Yang D, Sze KH, Lai PM, Yuan S, et al. Characterization of the lipidomic profile of human coronavirus-infected cells: implications for lipid metabolism remodeling upon coronavirus replication. Viruses (2019) 11(1):73. doi: 10.3390/v11010073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen Y, Gu S, Chen Y, Lu H, Shi D, Guo J, et al. Six-month follow-up of gut microbiota richness in patients with covid-19. Gut (2022) 71(1):222–5. doi: 10.1136/gutjnl-2021-324090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol (2013) 13(11):790–801. doi: 10.1038/nri3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang C, Huang L, Wang Y, Li X, Ren L, Gu X, et al. 6-month consequences of covid-19 in patients discharged from hospital: a cohort study. Lancet (2021) 397(10270):220–32. doi: 10.1016/S0140-6736(20)32656-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nabavi N. Long covid: how to define it and how to manage it. Bmj (2020) 370:m3489. doi: 10.1136/bmj.m3489 [DOI] [PubMed] [Google Scholar]
- 48. Kim HN, Joo EJ, Lee CW, Ahn KS, Kim HL, Park DI, et al. Reversion of gut microbiota during the recovery phase in patients with asymptomatic or mild covid-19: longitudinal study. Microorganisms (2021) 9(6):1237. doi: 10.3390/microorganisms9061237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Newsome RC, Gauthier J, Hernandez MC, Abraham GE, Robinson TO, Williams HB, et al. The gut microbiome of covid-19 recovered patients returns to uninfected status in a minority-dominated united states cohort. Gut Microbes (2021) 13(1):1–15. doi: 10.1080/19490976.2021.1926840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu X, Mao B, Gu J, Wu J, Cui S, Wang G, et al. Blautia-a new functional genus with potential probiotic properties? Gut Microbes (2021) 13(1):1–21. doi: 10.1080/19490976.2021.1875796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, et al. Ace2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature (2012) 487(7408):477–81. doi: 10.1038/nature11228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. de Oliveira AP, Lopes ALF, Pacheco G, de Sa Guimaraes Noleto IR, Nicolau LAD, Medeiros JVR. Premises among sars-Cov-2, dysbiosis and diarrhea: walking through the Ace2/Mtor/Autophagy route. Med Hypotheses (2020) 144:110243. doi: 10.1016/j.mehy.2020.110243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lu Y, Zhu Q, Fox DM, Gao C, Stanley SA, Luo K. Sars-Cov-2 down-regulates Ace2 through lysosomal degradation. Mol Biol Cell (2022) 33(14):ar147. doi: 10.1091/mbc.E22-02-0045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yamaguchi T, Hoshizaki M, Minato T, Nirasawa S, Asaka MN, Niiyama M, et al. Ace2-like carboxypeptidase B38-cap protects from sars-Cov-2-Induced lung injury. Nat Commun (2021) 12(1):6791. doi: 10.1038/s41467-021-27097-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lei Y, Zhang J, Schiavon CR, He M, Chen L, Shen H, et al. Sars-Cov-2 spike protein impairs endothelial function Via downregulation of ace 2. Circ Res (2021) 128(9):1323–6. doi: 10.1161/CIRCRESAHA.121.318902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Trottein F, Sokol H. Potential causes and consequences of gastrointestinal disorders during a sars-Cov-2 infection. Cell Rep (2020) 32(3):107915. doi: 10.1016/j.celrep.2020.107915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Martino C, Kellman BP, Sandoval DR, Clausen TM, Marotz CA, Song SJ, et al. Bacterial modification of the host glycosaminoglycan heparan sulfate modulates sars-Cov-2 infectivity. bioRxiv (2020) 2020:238444. doi: 10.1101/2020.08.17.238444 [DOI] [Google Scholar]
- 58. Babszky G, Torma F, Aczel D, Bakonyi P, Gombos Z, Feher J, et al. Covid-19 infection alters the microbiome: elite athletes and sedentary patients have similar bacterial flora. Genes (2021) 12(10):1577. doi: 10.3390/genes12101577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vatanen T, Kostic AD, d'Hennezel E, Siljander H, Franzosa EA, Yassour M, et al. Variation in microbiome lps immunogenicity contributes to autoimmunity in humans. Cell (2016) 165(4):842–53. doi: 10.1016/j.cell.2016.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Geva-Zatorsky N, Sefik E, Kua L, Pasman L, Tan TG, Ortiz-Lopez A, et al. Mining the human gut microbiota for immunomodulatory organisms. Cell (2017) 168(5):928–43 e11. doi: 10.1016/j.cell.2017.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li J, Richards EM, Handberg EM, Pepine CJ, Raizada MK. Butyrate regulates covid-19-Relevant genes in gut epithelial organoids from normotensive rats. Hypertension (2021) 77(2):e13–e6. doi: 10.1161/HYPERTENSIONAHA.120.16647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hirayama M, Nishiwaki H, Hamaguchi T, Ito M, Ueyama J, Maeda T, et al. Intestinal collinsella may mitigate infection and exacerbation of covid-19 by producing ursodeoxycholate. PloS One (2021) 16(11):e0260451. doi: 10.1371/journal.pone.0260451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Poochi SP, Easwaran M, Balasubramanian B, Anbuselvam M, Meyyazhagan A, Park S, et al. Employing bioactive compounds derived from ipomoea obscura (L.) to evaluate potential inhibitor for sars-Cov-2 main protease and Ace2 protein. Food Front (2020) 1(2):168–79. doi: 10.1002/fft2.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ko WK, Kim SJ, Jo MJ, Choi H, Lee D, Kwon IK, et al. Ursodeoxycholic acid inhibits inflammatory responses and promotes functional recovery after spinal cord injury in rats. Mol Neurobiol (2019) 56(1):267–77. doi: 10.1007/s12035-018-0994-z [DOI] [PubMed] [Google Scholar]
- 65. Abdulrab S, Al-Maweri S, Halboub E. Ursodeoxycholic acid as a candidate therapeutic to alleviate and/or prevent covid-19-Associated cytokine storm. Med Hypotheses (2020) 143:109897. doi: 10.1016/j.mehy.2020.109897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kalantar-Zadeh K, Ward SA, Kalantar-Zadeh K, El-Omar EM. Considering the effects of microbiome and diet on sars-Cov-2 infection: nanotechnology roles. ACS Nano (2020) 14(5):5179–82. doi: 10.1021/acsnano.0c03402 [DOI] [PubMed] [Google Scholar]
- 67. Sarkar A, Harty S, Moeller AH, Klein SL, Erdman SE, Friston KJ, et al. The gut microbiome as a biomarker of differential susceptibility to sars-Cov-2. Trends Mol Med (2021) 27(12):1115–34. doi: 10.1016/j.molmed.2021.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Machado AS, Oliveira JR, Lelis DF, de Paula AMB, Guimaraes ALS, Andrade JMO, et al. Oral probiotic bifidobacterium longum supplementation improves metabolic parameters and alters the expression of the renin-angiotensin system in obese mice liver. Biol Res Nurs (2021) 23(1):100–8. doi: 10.1177/1099800420942942 [DOI] [PubMed] [Google Scholar]
- 69. Modi SR, Collins JJ, Relman DA. Antibiotics and the gut microbiota. J Clin Invest (2014) 124(10):4212–8. doi: 10.1172/JCI72333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yang T, Chakraborty S, Saha P, Mell B, Cheng X, Yeo JY, et al. Gnotobiotic rats reveal that gut microbiota regulates colonic mrna of Ace2, the receptor for sars-Cov-2 infectivity. Hypertension (2020) 76(1):e1–3. doi: 10.1161/HYPERTENSIONAHA.120.15360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Souza DG, Vieira AT, Soares AC, Pinho V, Nicoli JR, Vieira LQ, et al. The essential role of the intestinal microbiota in facilitating acute inflammatory responses. J Immunol (2004) 173(6):4137–46. doi: 10.4049/jimmunol.173.6.4137 [DOI] [PubMed] [Google Scholar]
- 72. Elsayed S, Zhang K. Human infection caused by clostridium hathewayi. Emerging Infect Dis (2004) 10(11):1950–2. doi: 10.3201/eid1011.040006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Forrester JD, Spain DA. Clostridium ramosum bacteremia: case report and literature review. Surg Infect (2014) 15(3):343–6. doi: 10.1089/sur.2012.240 [DOI] [PubMed] [Google Scholar]
- 74. Gao J, Xu K, Liu H, Liu G, Bai M, Peng C, et al. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front Cell infection Microbiol (2018) 8:13. doi: 10.3389/fcimb.2018.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Miquel S, Martin R, Rossi O, Bermudez-Humaran LG, Chatel JM, Sokol H, et al. Faecalibacterium prausnitzii and human intestinal health. Curr Opin Microbiol (2013) 16(3):255–61. doi: 10.1016/j.mib.2013.06.003 [DOI] [PubMed] [Google Scholar]
- 76. Sonoyama K, Miki A, Sugita R, Goto H, Nakata M, Yamaguchi N. Gut colonization by candida albicans aggravates inflammation in the gut and extra-gut tissues in mice. Med Mycology (2011) 49(3):237–47. doi: 10.3109/13693786.2010.511284 [DOI] [PubMed] [Google Scholar]
- 77. Kosmidis C, Denning DW. The clinical spectrum of pulmonary aspergillosis. Thorax (2015) 70(3):270–7. doi: 10.1136/thoraxjnl-2014-206291 [DOI] [PubMed] [Google Scholar]
- 78. Dhar D, Mohanty A. Gut microbiota and covid-19- possible link and implications. Virus Res (2020) 285:198018. doi: 10.1016/j.virusres.2020.198018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Uzzan M, Corcos O, Martin JC, Treton X, Bouhnik Y. Why is sars-Cov-2 infection more severe in obese men? the gut lymphatics - lung axis hypothesis. Med Hypotheses (2020) 144:110023. doi: 10.1016/j.mehy.2020.110023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wang B, Zhang L, Wang Y, Dai T, Qin Z, Zhou F, et al. Alterations in microbiota of patients with covid-19: potential mechanisms and therapeutic interventions. Signal Transduct Target Ther (2022) 7(1):143. doi: 10.1038/s41392-022-00986-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Qian Q, Fan L, Liu W, Li J, Yue J, Wang M, et al. Direct evidence of active sars-Cov-2 replication in the intestine. Clin Infect Dis (2021) 73(3):361–6. doi: 10.1093/cid/ciaa925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Katz-Agranov N, Zandman-Goddard G. Autoimmunity and covid-19 - the microbiotal connection. Autoimmun Rev (2021) 20(8):102865. doi: 10.1016/j.autrev.2021.102865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tay MZ, Poh CM, Renia L, MacAry PA, Ng LFP. The trinity of covid-19: immunity, inflammation and intervention. Nat Rev Immunol (2020) 20(6):363–74. doi: 10.1038/s41577-020-0311-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in wuhan, China. Lancet (2020) 395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of crohn disease patients. Proc Natl Acad Sci USA (2008) 105(43):16731–6. doi: 10.1073/pnas.0804812105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. van den Munckhof ICL, Kurilshikov A, Ter Horst R, Riksen NP, Joosten LAB, Zhernakova A, et al. Role of gut microbiota in chronic low-grade inflammation as potential driver for atherosclerotic cardiovascular disease: a systematic review of human studies. Obes Rev (2018) 19(12):1719–34. doi: 10.1111/obr.12750 [DOI] [PubMed] [Google Scholar]
- 87. Parada Venegas D, de la Fuente MK, Landskron G, Gonzalez MJ, Quera R, Dijkstra G, et al. Short chain fatty acids (Scfas)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol (2019) 10:277. doi: 10.3389/fimmu.2019.00277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell (2012) 149(7):1578–93. doi: 10.1016/j.cell.2012.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Jia W, Xie G, Jia W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol (2018) 15(2):111–28. doi: 10.1038/nrgastro.2017.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Belkaid Y, Harrison OJ. Homeostatic immunity and the microbiota. Immunity (2017) 46(4):562–76. doi: 10.1016/j.immuni.2017.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Litvak Y, Byndloss MX, Baumler AJ. Colonocyte metabolism shapes the gut microbiota. Science (2018) 362(6418):eaat9076. doi: 10.1126/science.aat9076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Reinold J, Farahpour F, Fehring C, Dolff S, Konik M, Korth J, et al. A pro-inflammatory gut microbiome characterizes sars-Cov-2 infected patients and a reduction in the connectivity of an anti-inflammatory bacterial network associates with severe covid-19. Front Cell Infect Microbiol (2021) 11:747816. doi: 10.3389/fcimb.2021.747816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Prasad R, Patton MJ, Floyd JL, Fortmann S, DuPont M, Harbour A, et al. Plasma microbiome in covid-19 subjects: an indicator of gut barrier defects and dysbiosis. Int J Mol Sci (2022) 23(16):9141. doi: 10.3390/ijms23169141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Giron LB, Dweep H, Yin X, Wang H, Damra M, Goldman AR, et al. Plasma markers of disrupted gut permeability in severe covid-19 patients. Front Immunol (2021) 12:686240. doi: 10.3389/fimmu.2021.686240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Zhang F, Wan Y, Zuo T, Yeoh YK, Liu Q, Zhang L, et al. Prolonged impairment of short-chain fatty acid and l-isoleucine biosynthesis in gut microbiome in patients with covid-19. Gastroenterology (2022) 162(2):548–61 e4. doi: 10.1053/j.gastro.2021.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yao Y, Cai X, Fei W, Ye Y, Zhao M, Zheng C. The role of short-chain fatty acids in immunity, inflammation and metabolism. Crit Rev Food Sci Nutr (2022) 62(1):1–12. doi: 10.1080/10408398.2020.1854675 [DOI] [PubMed] [Google Scholar]
- 97. Mao X, Gu C, Ren M, Chen D, Yu B, He J, et al. L-isoleucine administration alleviates rotavirus infection and immune response in the weaned piglet model. Front Immunol (2018) 9:1654. doi: 10.3389/fimmu.2018.01654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Dickson RP. The microbiome and critical illness. Lancet Respir Med (2016) 4(1):59–72. doi: 10.1016/S2213-2600(15)00427-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Angurana SK, Bansal A. Probiotics and coronavirus disease 2019: think about the link. Br J Nutr (2021) 126(10):1564–70. doi: 10.1017/S000711452000361X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Nagata N, Takeuchi T, Masuoka H, Aoki R, Ishikane M, Iwamoto N, et al. Human gut microbiota and its metabolites impact immune responses in covid-19 and its complications. Gastroenterology (2023) 164(2):272–88. doi: 10.1053/j.gastro.2022.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Verdam FJ, Fuentes S, de Jonge C, Zoetendal EG, Erbil R, Greve JW, et al. Human intestinal microbiota composition is associated with local and systemic inflammation in obesity. Obesity (2013) 21(12):E607–15. doi: 10.1002/oby.20466 [DOI] [PubMed] [Google Scholar]
- 102. Gu S, Chen Y, Wu Z, Chen Y, Gao H, Lv L, et al. Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1n1 influenza. Clin Infect Dis (2020) 71(10):2669–78. doi: 10.1093/cid/ciaa709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ke S, Weiss ST, Liu YY. Dissecting the role of the human microbiome in covid-19 Via metagenome-assembled genomes. Nat Commun (2022) 13(1):5235. doi: 10.1038/s41467-022-32991-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Moreira-Rosario A, Marques C, Pinheiro H, Araujo JR, Ribeiro P, Rocha R, et al. Gut microbiota diversity and c-reactive protein are predictors of disease severity in covid-19 patients. Front Microbiol (2021) 12:705020. doi: 10.3389/fmicb.2021.705020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Nobre JG, Delgadinho M, Silva C, Mendes J, Mateus V, Ribeiro E, et al. Gut microbiota profile of covid-19 patients: prognosis and risk stratification (Microcovid-19 study). Front Microbiol (2022) 13:1035422. doi: 10.3389/fmicb.2022.1035422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Tang L, Gu S, Gong Y, Li B, Lu H, Li Q, et al. Clinical significance of the correlation between changes in the major intestinal bacteria species and covid-19 severity. Engineering (2020) 6(10):1178–84. doi: 10.1016/j.eng.2020.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Stutz MR, Dylla NP, Pearson SD, Lecompte-Osorio P, Nayak R, Khalid M, et al. Immunomodulatory fecal metabolites are associated with mortality in covid-19 patients with respiratory failure. Nat Commun (2022) 13(1):6615. doi: 10.1038/s41467-022-34260-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Wu D, Zhou K, Xiao M, Liu Z, Zhang S. Differences in intestinal microbiome are associated with the mortality of covid-19 patients in intensive care units. Sci China Life Sci (2022) 65(5):1040–2. doi: 10.1007/s11427-021-2072-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Zhang F, Lau RI, Liu Q, Su Q, Chan FKL, Ng SC. Gut microbiota in covid-19: key microbial changes, potential mechanisms and clinical applications. Nat Rev Gastroenterol Hepatol (2023) 20:323–37. doi: 10.1038/s41575-022-00698-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Liu F, Ye S, Zhu X, He X, Wang S, Li Y, et al. Gastrointestinal disturbance and effect of fecal microbiota transplantation in discharged covid-19 patients. J Med Case Rep (2021) 15(1):60. doi: 10.1186/s13256-020-02583-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Ceccarelli G, Borrazzo C, Pinacchio C, Santinelli L, Innocenti GP, Cavallari EN, et al. Oral bacteriotherapy in patients with covid-19: a retrospective cohort study. Front Nutr (2020) 7:613928. doi: 10.3389/fnut.2020.613928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Zhang L, Han H, Li X, Chen C, Xie X, Su G, et al. Probiotics use is associated with improved clinical outcomes among hospitalized patients with covid-19. Ther Adv Gastroenterol (2021) 14:17562848211035670. doi: 10.1177/17562848211035670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Gutierrez-Castrellon P, Gandara-Marti T, Abreu YAAT, Nieto-Rufino CD, Lopez-Orduna E, Jimenez-Escobar I, et al. Probiotic improves symptomatic and viral clearance in Covid19 outpatients: a randomized, quadruple-blinded, placebo-controlled trial. Gut Microbes (2022) 14(1):2018899. doi: 10.1080/19490976.2021.2018899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Zhang L, Xu Z, Mak JWY, Chow KM, Lui G, Li TCM, et al. Gut microbiota-derived synbiotic formula (Sim01) as a novel adjuvant therapy for covid-19: an open-label pilot study. J Gastroenterol Hepatol (2022) 37(5):823–31. doi: 10.1111/jgh.15796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Yang Z, Liu Y, Wang L, Lin S, Dai X, Yan H, et al. Traditional Chinese medicine against covid-19: role of the gut microbiota. Biomed Pharmacother (2022) 149:112787. doi: 10.1016/j.biopha.2022.112787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Liu J, Yang W, Liu Y, Lu C, Ruan L, Zhao C, et al. Combination of hua shi bai Du granule (Q-14) and standard care in the treatment of patients with coronavirus disease 2019 (Covid-19): a single-center, open-label, randomized controlled trial. Phytomedicine (2021) 91:153671. doi: 10.1016/j.phymed.2021.153671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Fan SJ, Liao JK, Wei L, Wang BY, Kai L, Tan DX. Treatment efficacy of lianhua qingwen capsules for eraly-stage covid-19. Am J Trans Res (2022) 14(2):1332–8. [PMC free article] [PubMed] [Google Scholar]
- 118. Zhang L, Ma Y, Shi N, Tong L, Liu S, Ji X, et al. Effect of qingfei paidu decoction combined with Western medicine treatments for covid-19: a systematic review and meta-analysis. Phytomedicine (2022) 102:154166. doi: 10.1016/j.phymed.2022.154166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Zhang L, Wu L, Xu X, Yuan Y, Jiang R, Yan X, et al. Effectiveness of lianhua qingwen capsule in treatment of asymptomatic covid-19 patients: a randomized, controlled multicenter trial. J Integr Complement Med (2022) 28(11):887–94. doi: 10.1089/jicm.2021.0352 [DOI] [PubMed] [Google Scholar]
- 120. Xin S, Cheng X, Zhu B, Liao X, Yang F, Song L, et al. Clinical retrospective study on the efficacy of qingfei paidu decoction combined with Western medicine for covid-19 treatment. Biomed Pharmacother (2020) 129:110500. doi: 10.1016/j.biopha.2020.110500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Ni L, Wen Z, Hu X, Tang W, Wang H, Zhou L, et al. Effects of shuanghuanglian oral liquids on patients with covid-19: a randomized, open-label, parallel-controlled, multicenter clinical trial. Front Med (2021) 15(5):704–17. doi: 10.1007/s11684-021-0853-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Xiong WZ, Wang G, Du J, Ai W. Efficacy of herbal medicine (Xuanfei baidu decoction) combined with conventional drug in treating covid-19:A pilot randomized clinical trial. Integr Med Res (2020) 9(3):100489. doi: 10.1016/j.imr.2020.100489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Wu J, Tang F, Zhang XQ, Fu ZL, Fu S. Application of jiawei maxing shigan tang in the treatment of covid-19: an observational study. Front Med (2022) 9:1028171. doi: 10.3389/fmed.2022.1028171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Zhang X, Xue Y, Chen X, Wu JM, Su ZJ, Sun M, et al. Effects of tanreqing capsule on the negative conversion time of nucleic acid in patients with covid-19: a retrospective cohort study. J Integr Med (2021) 19(1):36–41. doi: 10.1016/j.joim.2020.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Jia Q, Wang L, Zhang X, Ding Y, Li H, Yang Y, et al. Prevention and treatment of chronic heart failure through traditional Chinese medicine: role of the gut microbiota. Pharmacol Res (2020) 151:104552. doi: 10.1016/j.phrs.2019.104552 [DOI] [PubMed] [Google Scholar]
- 126. Chen Z, Lv Y, Xu H, Deng L. Herbal medicine, gut microbiota, and covid-19. Front Pharmacol (2021) 12:646560. doi: 10.3389/fphar.2021.646560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Ain QU, Khan H, Mubarak MS, Pervaiz A. Plant alkaloids as antiplatelet agent: drugs of the future in the light of recent developments. Front Pharmacol (2016) 7:292. doi: 10.3389/fphar.2016.00292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Sun Y, Zhang HG, Shi XG, Duan MY, Zhong DF. [Study on metabolites on aconitine in rabbit urine]. Acta Pharm Sin (2002) 37(10):781–3. [PubMed] [Google Scholar]
- 129. Kopustinskiene DM, Jakstas V, Savickas A, Bernatoniene J. Flavonoids as anticancer agents. Nutrients (2020) 12(2):457. doi: 10.3390/nu12020457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Zhou J, Chen Y, Wang Y, Gao X, Qu D, Liu C. A comparative study on the metabolism of epimedium koreanum nakai-prenylated flavonoids in rats by an intestinal enzyme (Lactase phlorizin hydrolase) and intestinal flora. Molecules (2013) 19(1):177–203. doi: 10.3390/molecules19010177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Wang J, Liang Q, Zhao Q, Tang Q, Ahmed AF, Zhang Y, et al. The effect of microbial composition and proteomic on improvement of functional constipation by chrysanthemum morifolium polysaccharide. Food Chem Toxicol (2021) 153:112305. doi: 10.1016/j.fct.2021.112305 [DOI] [PubMed] [Google Scholar]
- 132. Blaak EE, Canfora EE, Theis S, Frost G, Groen AK, Mithieux G, et al. Short chain fatty acids in human gut and metabolic health. Beneficial Microbes (2020) 11(5):411–55. doi: 10.3920/BM2020.0057 [DOI] [PubMed] [Google Scholar]
- 133. Shao S, Wang D, Zheng W, Li X, Zhang H, Zhao D, et al. A unique polysaccharide from hericium erinaceus mycelium ameliorates acetic acid-induced ulcerative colitis rats by modulating the composition of the gut microbiota, short chain fatty acids levels and Gpr41/43 respectors. Int Immunopharmacol (2019) 71:411–22. doi: 10.1016/j.intimp.2019.02.038 [DOI] [PubMed] [Google Scholar]
- 134. Kim BG, Jung WD, Mok H, Ahn JH. Production of hydroxycinnamoyl-shikimates and chlorogenic acid in escherichia coli: production of hydroxycinnamic acid conjugates. Microb Cell Factories (2013) 12:15. doi: 10.1186/1475-2859-12-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Jan KC, Hwang LS, Ho CT. Biotransformation of sesaminol triglucoside to mammalian lignans by intestinal microbiota. J Agric Food Chem (2009) 57(14):6101–6. doi: 10.1021/jf901215j [DOI] [PubMed] [Google Scholar]