Abstract
Columbid alphaherpesvirus 1 (CoHV1) is associated with oral or upper respiratory tract lesions, encephalitis, and occasional fatal systemic disease in naive or immunosuppressed pigeons. Clinical disease is often reported with CoHV1 and coinfecting viruses, including pigeon circovirus (PiCV), which may cause host immunosuppression and augment lesion development. A natural outbreak of CoHV1 and PiCV coinfection occurred in a flock of 60 racing rock pigeons (Columba livia), in which 4 pigeons succumbed within 7 d of clinical onset. Lesions included suppurative stomatitis, pharyngitis, cloacitis, meningitis, and tympanitis, with eosinophilic intranuclear inclusion bodies consistent with herpesviral infection. In addition, large numbers of botryoid intracytoplasmic inclusion bodies were present in the skin, oral mucosa, and bursa of Fabricius, suggestive of circoviral infection, which was confirmed by immunohistochemistry. The concurrent viral load of CoHV1 and PiCV was high in liver, oropharynx, and bursa of Fabricius. We found PiCV in oro-cloacal swabs from 44 of 46 additional birds of variable clinical status, PiCV alone in 23 birds, and coinfection with CoHV1 in 21 birds. Viral copy numbers were significantly higher (p < 0.0001) for both viruses in clinically affected pigeons than in subclinical qPCR-positive birds. The CoHV1-induced lesions might have been exacerbated by concomitant PiCV infection.
Keywords: Columba livia, coinfection, columbid herpesvirus 1, pigeon circovirus, pathology, qPCR
Concurrent infections with multiple viruses may accommodate, augment, or suppress replication of coinfecting viruses to modulate clinical outcomes, or they may coexist without exerting any effects. 7 Columbid alphaherpesvirus 1 (CoHV1; Herpesviridae, Mardivirus) is a widely distributed pathogen that usually results in subclinical or latent infection in its natural host, the rock pigeon (Columba livia). 8 The neuropathogenic peracute form of disease, historically known as contagious paralysis, has been recognized for decades. 18 When introduced into a naive or immunosuppressed population, CoHV1 may cause multisystemic disease with high mortality or may remain latent, making pigeons susceptible to secondary infections with other bacterial and viral pathogens. 8 Furthermore, spillover infection of CoHV1 from pigeons into birds of prey (Strigiformes and Falconiformes) can cause lethal disseminated infection, inclusion body hepatitis, and encephalomyelitis. 12
Similarly, pigeon circovirus (PiCV; Circoviridae, Circovirus) causes a ubiquitous viral infection of pigeons worldwide. 6 The characteristic basophilic botryoid intracytoplasmic inclusion bodies (ICIBs) with lymphoid tissue destruction, particularly in the bursa of Fabricius, have led to assumptions that PiCV infection may be immunosuppressive to its hosts. 16 PiCV was widely considered a contributing agent for young pigeon disease syndrome (YPDS), a multifactorial disease condition characterized by lethargy, weight loss, respiratory distress, diarrhea, and poor performance in young racing pigeons 1–4-mo-old. 17 Although the mechanism of immunosuppression induced by PiCV infection has not been thoroughly elucidated, marked apoptosis of bursal lymphocytes might be responsible. 17 However, several studies, including experimental inoculation, failed to demonstrate PiCV infection as a primary cause of immunosuppression. Furthermore, subclinical infection is observed commonly in older birds. 16 Thence, concurrent infections with a diverse range of viral, bacterial, and parasitic agents have been proposed in syndromes such as YPDS. 9 We document here the lesions associated with a naturally occurring outbreak of CoHV1 in a flock of racing pigeons coinfected with PiCV and compare viral loads with the clinical presentation of both viral pathogens.
As part of a routine diagnostic investigation for morbidity and mortality, whole body carcasses of 4 racing pigeons from a loft located in southern NSW, Australia were presented for postmortem examination. The pigeons were vaccinated against pigeon pox, rotavirus, paramyxovirus, and paratyphoid. No antiprotozoal or antimycotic agents were given to the affected flock. Clinical signs included depression, lethargy, anorexia, weight loss, poor racing performance, and dyspnea. Four of 60 birds had died over the 7 d since clinical onset.
At postmortem examination, samples of oropharynx, trachea, liver, lung, brain, kidney, intestine, pre-cloacal skin, vent, and bursa of Fabricius were stored in 10% neutral-buffered formalin, processed routinely, and slides stained with H&E. Some tissues were stored at −80°C for molecular testing. Swabs from the pooled tissue samples were streaked on sheep blood agar, chocolate agar, MacConkey agar, and Columbia nalidixic acid agar with 5% sheep blood (Oxoid) and incubated for 48 h at 37°C under aerobic conditions. Sabouraud dextrose agar plates (Oxoid) were incubated for 21 d at 24°C. Immunohistochemistry of selected deparaffinized histologic sections was conducted using an optimized dilution (1:500) of anti-PiCV polyclonal antibody raised in sheep and a commercial rabbit anti-sheep secondary antibody (Bio-Rad) following a reported protocol. 14 Each tissue section was treated for antigen retrieval by microwaving 3 times for 3 min each time in citrate buffer pH 9. Endogenous peroxidase activity was blocked by incubation with 3% hydrogen peroxide in distilled water for 30 min. To avoid background staining, protein block (Dako) was used for 10 min, and the sections were then washed 3 times with PBS. Primary antibody was then applied to the sections for 4 h at room temperature. The sections were then washed twice with PBS and tapped dry. Horseradish peroxidase conjugate rabbit anti-sheep secondary antibody was then added, and the sections incubated at room temperature for 60 min, after which the sections were washed twice with PBS. Sections were finally incubated in diaminobenzidine (DAB) solution for 10 min, counterstained with Harris hematoxylin, dehydrated, coverslipped, and examined microscopically. Sections from a case of PiCV infection diagnosed previously in our laboratory were used as positive control. Negative controls consisted of archived samples of liver and lymphoid tissues of a pigeon from an experimental farm with no history of PiCV infection.
Total genomic DNA was extracted from the liver, oropharynx, and bursa of Fabricius of the dead birds as well as from the oro-cloacal swabs of 46 live birds using protocols described elsewhere. 3 Initially, conventional PCR was used to detect specific DNA of PiCV, CoHV1, Chlamydia psittaci, or avian paramyxovirus 1 (Avian orthoavulavirus 1) from pooled tissue samples. Later, individual tissue and oro-cloacal swab samples were tested with quantitative PCR (qPCR) using a real-time thermocycler (Rotor-Gene 6000; Qiagen). PCR primers and probes for PiCV and CoHV1 were designed based on all available genome sequences from GenBank utilizing design tools in the Geneious Prime v.2021.1.1 package (Dotmatics). Selected fragments of replication-associated protein coding (Rep) gene for PiCV and DNA-dependent DNA polymerase (DDDP) gene for CoHV1 was amplified for PCR and qPCR assay (Table 1). A synthetic plasmid construct (pMCGS-21) containing inserts of Rep- and DDDP-selected fragments was used as a positive control for the test; nuclease-free sterile water was used as a negative control. Purified amplicons were sequenced at the Australian Genome Research Facility (AGRF-Sydney node; Sanger dideoxy sequencer AB 3730xl, Applied Biosystems). Obtained raw data were trimmed, edited, and deposited in GenBank (OM470912, OM470913, OM470914, OM470915).
Table 1.
Primers and probes used in quantitative PCR assays.
| Organism | Target gene | Primer/probe | Sequence (5′–3′) | Amplicon size, bp | Reference |
|---|---|---|---|---|---|
| PiCV | Rep | PiCV SD-F | GTGAAAGCCGGAAGAGCAATG | 139 | Our study |
| PiCV SD-R | GTGATGACGATGACTTCCGTTTTGAAG | ||||
| PiCV-probe | 6 FAM-CGAGACTTCAGTGAGATATACGTCAA-BHQ 1 | Our study | |||
| CoHV1 | DDDP | CoHV1 Phalen-F | 5′-TCTGTACCCCAGCATCATCC-3′ | 480 | 12 |
| CoHV1 Phalen-R | 5′-AGTAGTCCTCCCCCGCCTCC-3′ | ||||
| CoHV1 SD-F | GCTTTTCGCATAGCCAGC | 100 | Our study | ||
| CoHV1 SD-R | CAGATCAACGTGAACGG | ||||
| CoHV1-probe | 6 HEX-CTGTACTTTGTGAAAAAGCACGTTC-BHQ 1 | Our study | |||
| BFDV | Rep | Primer 2 | AACCCTACAGACGGCGAG | 717 | 19 |
| Primer 4 | GTCACAGTCCTCCTTGTACC | ||||
| C. psittaci | pmp | CpsiA | ATGAAACATCCAGTCTACTGG | ~300 | 13 |
| CpsiB | TTGTGTAGTAATATTATCAAA |
BFDV = beak and feather disease virus; C. psittaci = Chlamydia psittaci; CoHV1 = columbid herpesvirus 1; DDDP = DNA-dependent DNA polymerase gene; PiCV = pigeon circovirus; Rep = replication-associated gene.
Maximum likelihood (ML)-based phylogenetic reconstruction was used to determine the genealogic relatedness of the PiCV and CoHV1 amplicons with reported genotypes or strains in GenBank. ML trees were reconstructed with 100 bootstrap replicates in PhyML 3.3 implemented in T-REX (http://www.trex.uqam.ca) using the above parameters. The final tree was visualized and edited in FigTree 1.4.
Two separate TaqMan-based qPCR protocols were developed and validated for PiCV and CoHV1 (Table 1). Sensitivity, specificity, and limits of detection of the tests were calculated using synthetic plasmid constructs with target inserts as positive control, and sterile water and specific-pathogen–free template DNA as negative control. A 10-fold dilution between 2.5 × 109 and 1 copies/μL was used for assay optimization and performance evaluation. The cycle threshold (Ct) values of the known-copy-number standards were graphed against time to construct a standard curve using the software supplied with the Rotor-Gene real-time thermocycler. The Ct of each sample was then compared against the graph to give an estimate of viral load/μL of extracted DNA. Association between clinical disease and viral genome copy number/DNA μL was assessed using the unpaired t-test with the Prism 9.2.0 software package (GraphPad). Statistical significance was set at p < 0.05.
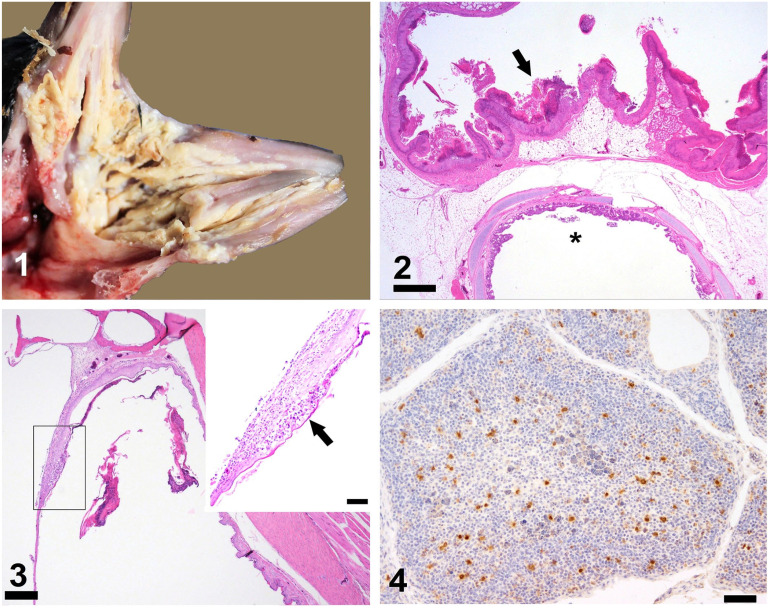
Gross lesions in all clinically affected birds included chemosis and hyperemia of the conjunctiva, diffuse facial subcutaneous edema, peri-cloacal skin thickening with surface crust, and diffusely thickened diphtheritic membranes in the oral cavity, tongue, and pharynx (Fig. 1; Suppl. Figs. 1–4). Wet preparations of oral and crop fluid from the dead birds were negative for Trichomonas spp. There was moderate-to-marked hepatomegaly; the air sacs, intestine, and kidneys appeared relatively normal. Similarly, no evidence of endoparasites was seen grossly or by wet preparation of duodenal or colonic contents. Cultures of the pooled tissues yielded no significant growth of bacteria or fungi. Histologic examination of a wide range of tissue sections also indicated that there were no other bacterial or mycotic pathogens contributing to lesions. Pooled tissue samples from dead birds tested negative by PCR for C. psittaci and avian paramyxovirus 1.
Figures 1–4.
Clinical signs and histopathologic findings in affected young pigeons. Figure 1. Severe diphtheritic stomatitis. Figure 2. Oropharynx, esophagus, and trachea (*), with severe diffuse thickening of the oral mucosa (arrow). Bar = 1 mm. Figure 3. External ear canal and tympanic membrane with layers of serocellular exudate in the lumen as well as focal-to-diffuse thickening. Bar = 200 μm. Inset: infiltration of the tympanic membrane with heterophils and macrophages (arrow). Bar = 200 μm. Figure 4. Widespread pigeon circovirus (PiCV) antigen in the bursa of Fabricius. Polyclonal anti-PiCV capsid protein antibody immunohistochemistry. Bar = 50 μm.
Histologically, pharynx, oral cavity, and laryngeal mucosa had severe diffuse infiltrates of heterophils and formation of thick serocellular crusts including aggregates of bacteria and yeasts (likely secondary infections; Fig. 2, Suppl. Fig. 5). In all carcasses, the splenic parenchyma had marked lymphoid depletion with reticuloendothelial proliferation. There was marked diffuse ballooning degeneration of the keratinocytes with eosinophilic intranuclear inclusion bodies (INIBs) in the peri-cloacal skin and feather follicular infundibulum. Heterophilic inflammation was noted in the cloacal lips, with subcorneal vesicles filled with cellular debris and heterophils (Suppl. Figs. 6, 7).
Sections of the bursa of Fabricius from the 4 dead birds had massive numbers of amphophilic, botryoid ICIBs within B lymphocytes and macrophages (Suppl. Fig. 8). There were foci of intense heterophilic inflammation in the peri-cloacal skin (Suppl. Fig. 9). In all dead birds, there were bilateral diffuse mixed-inflammatory cellular infiltrates, dominated by macrophages and heterophils, into the epithelium of the external ear canal, tympanic membrane, and inner ear (Fig. 3). There was severe thickening of the meninges with edema, hemorrhage, and a diffuse infiltrate of macrophages and heterophils in the brain (Suppl. Fig. 10). Immunohistochemically, PiCV antigen was widespread in the bursa of Fabricius (Fig. 4).
Conventional PCR on tissue homogenates from pool 1 (oropharynx) and pool 2 (liver, spleen, bursa of Fabricius) combining all 4 dead birds were positive for PiCV and CoHV1 DNA, and negative for Chlamydia spp., pigeonpox virus, pigeon paramyxovirus 1 (PPMV1; Avian orthoavulavirus 1), and beak and feather disease virus (BFDV) infection. Sanger-based sequencing and a BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi) revealed that the CoHV1 amplicons were 935-bp and 597-bp DDDP gene fragments obtained from pool 1 and 2 respectively, and both had 100% sequence identity with reported CoHV1 genomes sequenced in a feral pigeon (KX589235, KJ995972) and a falcon (KJ668231). A ML tree based on DDDP sequences of publicly available avian herpesviruses positioned these 2 sequences in a strongly supported monophyletic clade comprised of CoHV1 in different host species (Suppl. Fig. 11). On the other hand, PiCV amplicons were 137-bp Rep gene sequences with 135 of 137 (99%) nucleotide identity with a full-genome PiCV (MF136686–MF136691) sequenced from clinically normal rock pigeons in Australia. The ML phylogenetic tree also positioned these sequences into a monophyletic clade populated by PiCV sequences from Australia, suggesting an endemic strain (Suppl. Fig. 12).
Tissues of the dead pigeons had substantial variation in viral copy numbers. PiCV load was much higher than CoHV1 in all organs; highest copies were detected in liver (2.38 × 1011 copies/μL; Suppl. Table 1). In contrast, the highest number of viral copies for CoHV1 were in the oropharynx (2.68 × 106 copies/μL; Suppl. Table 1). Of 46 live pigeons tested, 44 of 46 (96%) were PiCV positive; 23 of 46 (50%) were positive for PiCV alone; coinfection with CoHV1 was detected in 21 of 46 (46%) birds. Interestingly, CoHV1 infection was not detected alone but was always coupled with PiCV infection. Coinfection was detected both in 39 clinically healthy pigeons as well as in the 4 dead pigeons (Suppl. Table 1); however, in both cases, PiCV viral copy numbers were significantly higher (p < 0.0001) compared to CoHV1 (Table 2). In diseased birds with clinical signs, the viral loads of CoHV1 and PiCV were much higher (p < 0.0001) compared to the healthy pigeons, subclinical carriers, or clinically recovered birds (Suppl. Figs. 13–15).
Table 2.
Viral load of pigeon circovirus (PiCV) and columbid herpesvirus 1 (CoHV1) in oro-cloacal swabs from subclinical pigeons.
| Description | PiCV | CoHV1 | PiCV alone | CoHV1 alone | Coinfection |
|---|---|---|---|---|---|
| Cloacal swab of 46 live birds, n | 44 | 21 | 23 | 0 | 21 |
| % Frequency | 96 | 46 | 50 | 0 | 46 |
| Copy no./µL DNA | 10–8.98 × 108 | 11–1.86 × 105 | 10–2.8 × 108 | 0 | PiCV = 18–8.98 × 108 CoHV1 = 11–1.86 × 105 |
Histologic lesions in the dead pigeons indicated a tropism for mucocutaneous junction epithelium with herpesviral INIBs in the peri-cloacal skin, and oral–respiratory and middle ear–meningeal transition zones. Given that CoHV1 most likely shares morphologic and physicochemical properties with other alphaherpesviruses, tropism for epithelial cells and nerve tissue is not surprising. Previous studies also noted the oral cavity wall, esophagus, salivary gland, nasal passages, and trachea as primary predilection sites for CoHV1 infection, with resultant respiratory diseases, rhinitis, and conjunctivitis in young pigeons. 2 Apart from the aforesaid epithelial lesions, some authors noted multifocal-to-diffuse necrosis of the pancreas, spleen, liver, and kidneys with eosinophilic INIBs in infected birds. 20 However, hepatic lesions are not always consistent and may be present only in young naive pigeons lacking maternal immunity or in aberrant hosts that have preyed upon infected pigeons. 11
Concurrent infection was widespread in organs of the dead birds, with the highest concentrations detected in oropharynx for CoHV1 and in liver samples for PiCV. Our qPCR assay could detect variable quantities of both viral DNAs in oro-cloacal swabs of the live birds, which is consistent with observations of high sensitivity of molecular tools for screening CoHV1 infection in oral swabs. 12 Interestingly, in every sample including visceral organs of the dead birds, the viral load of PiCV was significantly higher than CoHV1. This is probably the result of fundamental biological differences, such as the genome size, replication strategy, and the length of replication cycle of these 2 distantly related viruses. Mechanistically, the large (~200 kb) double-stranded DNA genome of herpesviruses utilizes high-fidelity DNA polymerase and energy-expensive, error-correcting mechanisms culminating in much slower replication cycle and low mutation rate in the range of 1 × 10−7 to 1 × 10−8 subs/site/year. 5 On the contrary, circovirus possess a far simpler compact single-stranded DNA (~2 kb) genome that mutates much faster (3.41 × 10−3 subs/site/year) and likely adapted a fast but error-prone replication mechanism, as seen with RNA viruses. 15 With short genomes and large burst sizes, a faster replicator such as PiCV could produce more copies of progeny viruses per replication cycle, and thereby might have a substantial copy number advantage in concurrent infection with CoHV1, as seen in our case.
The viral loads of both coinfecting viruses were significantly higher in dead birds with clinical signs and gross lesions compared to subclinical but PCR-positive pigeons. Interestingly, in this flock, single infection with CoHV1 was not detected and was always accompanied with high viral load of PiCV. This suggests that the clinical presentation of CoHV1 might have been exacerbated by concomitant PiCV infection. Although the effect of coinfection in increased or decreased disease severity for any given natural infection is difficult to assess, clinical records of increased severity by coinfecting viral pathogens are common. 4 Aggravation of disease in concurrent infections by ≥2 viral pathogens in mammalian and avian species highlights the role of decreasing host immunity. 10 A study noted that, if the flock is immunosuppressed, experimental inoculation of CoHV1 can spread either by tissue contiguity or by viremia. 18 This might be highly relevant in our case, given that all clinically affected pigeons had high viral loads of PiCV, a pathogen widely documented for its immunosuppressive role. 1 It was not possible to determine whether the flock has been latently infected with CoHV1 and PiCV was introduced later or vice versa. A concurrently high PiCV load alongside herpesvirus-associated lesions suggests a scenario of enhanced disease severity of CoHV1 in its reservoir host. Our findings suggest that the CoHV1-induced lesions may have been augmented by concomitant PiCV infection.
Supplemental Material
Supplemental material, sj-pdf-1-vdi-10.1177_10406387231156839 for Lesions and viral loads in racing pigeons naturally coinfected with pigeon circovirus and columbid alphaherpesvirus 1 in Australia by Babu K. Nath, Shubhagata Das, Naomie Tidd, Tridip Das, Jade K. Forwood and Shane R. Raidal in Journal of Veterinary Diagnostic Investigation
Acknowledgments
We thank the referring veterinary clinicians who not only recognized the history and clinical condition as a unique opportunity to thoroughly investigate but also collected and sent samples for analysis
Footnotes
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: Babu K. Nath received an Australian Government Research Training Program (AGRTP) International Scholarship for Doctoral Research.
ORCID iDs: Babu K. Nath  https://orcid.org/0000-0003-3620-1181
https://orcid.org/0000-0003-3620-1181
Tridip Das  https://orcid.org/0000-0001-5642-4150
https://orcid.org/0000-0001-5642-4150
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Babu K. Nath, School of Agricultural, Environmental and Veterinary Sciences, Faculty of Science and Health, Charles Sturt University, Wagga Wagga, New South Wales, Australia Department of Dairy and Poultry Science, Chattogram Veterinary and Animal Sciences University, Chattogram, Bangladesh.
Shubhagata Das, School of Agricultural, Environmental and Veterinary Sciences, Faculty of Science and Health, Charles Sturt University, Wagga Wagga, New South Wales, Australia.
Naomie Tidd, Veterinary Diagnostic Laboratory, Charles Sturt University, Wagga Wagga, New South Wales, Australia.
Tridip Das, School of Agricultural, Environmental and Veterinary Sciences, Faculty of Science and Health, Charles Sturt University, Wagga Wagga, New South Wales, Australia.
Jade K. Forwood, School of Dentistry and Medical Sciences, Faculty of Science and Health, Charles Sturt University, Wagga Wagga, New South Wales, Australia
Shane R. Raidal, School of Agricultural, Environmental and Veterinary Sciences, Faculty of Science and Health, Charles Sturt University, Wagga Wagga, New South Wales, Australia
References
- 1.Abadie J, et al. Pigeon circovirus infection: pathological observations and suggested pathogenesis. Avian Pathol 2001;30:149–158. [DOI] [PubMed] [Google Scholar]
- 2.Callinan RB, et al. An outbreak of disease in pigeons associated with a herpesvirus. Aust Vet J 1979;55:339–341. [DOI] [PubMed] [Google Scholar]
- 3.Das S, et al. A novel pathogenic aviadenovirus from red-bellied parrots (Poicephalus rufiventris) unveils deep recombination events among avian host lineages. Virology 2017;502:188–197. [DOI] [PubMed] [Google Scholar]
- 4.Díaz-Muñoz SL, et al. Sociovirology: conflict, cooperation, and communication among viruses. Cell Host Microbe 2017;22:437–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drake JW, Hwang CBC. On the mutation rate of herpes simplex virus type 1. Genetics 2005;170:969–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freick M, et al. Rapid detection of pigeon herpesvirus, fowl adenovirus and pigeon circovirus in young racing pigeons by multiplex PCR. J Virol Methods 2008;148:226–231. [DOI] [PubMed] [Google Scholar]
- 7.Kumar N, et al. Virological and immunological outcomes of coinfections. Clin Microbiol Rev 2018;31:e00111–e00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marlier D, Vindevogel H. Viral infections in pigeons. Vet J 2006;172:40–51. [DOI] [PubMed] [Google Scholar]
- 9.McCowan C, et al. A novel group A rotavirus associated with acute illness and hepatic necrosis in pigeons (Columba livia), in Australia. PLoS One 2018;13:e0203853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niczyporuk JS, et al. Fowl adenovirus strains 1/A and 11/D isolated from birds with reovirus infection. PLoS One 2021;16:e0256137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phalen DN, et al. Fatal columbid herpesvirus-1 infections in three species of Australian birds of prey. Aust Vet J 2011;89:193–196. [DOI] [PubMed] [Google Scholar]
- 12.Phalen DN, et al. Prevalence of columbid herpesvirus infection in feral pigeons from New South Wales and Victoria, Australia, with spillover into a wild powerful owl (Ninox struena). J Wildl Dis 2017;53:543–551. [DOI] [PubMed] [Google Scholar]
- 13.Piasecki T, et al. Detection and identification of Chlamydophila psittaci in asymptomatic parrots in Poland. BMC Vet Res 2012;8:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Portas T, et al. Beak and feather disease virus carriage by Knemidocoptes pilae in a sulphur-crested cockatoo (Cacatua galerita). Aust Vet J 2017;95:486–489. [DOI] [PubMed] [Google Scholar]
- 15.Sarker S, et al. Mutability dynamics of an emergent single stranded DNA virus in a naïve host. PLoS One 2014;9:e85370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt V, et al. Experimental infection of domestic pigeons with pigeon circovirus. Avian Dis 2008;52:380–386. [DOI] [PubMed] [Google Scholar]
- 17.Stenzel T, et al. The clinical infection with pigeon circovirus (PiCV) leads to lymphocyte B apoptosis but has no effect on lymphocyte T subpopulation. Pathogens 2020;9:632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vindevogel H, Pastoret PP. Pathogenesis of pigeon herpesvirus infection. J Comp Pathol 1981;91:415–426. [DOI] [PubMed] [Google Scholar]
- 19.Ypelaar I, et al. A universal polymerase chain reaction for the detection of psittacine beak and feather disease virus. Vet Microbiol 1999;68:141–148. [DOI] [PubMed] [Google Scholar]
- 20.Zhao P, et al. Isolation and characterization of a herpesvirus from feral pigeons in China. Vet J 2015;206:417–419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-vdi-10.1177_10406387231156839 for Lesions and viral loads in racing pigeons naturally coinfected with pigeon circovirus and columbid alphaherpesvirus 1 in Australia by Babu K. Nath, Shubhagata Das, Naomie Tidd, Tridip Das, Jade K. Forwood and Shane R. Raidal in Journal of Veterinary Diagnostic Investigation