Abstract
Previous studies in our laboratory have shown that the Staphylococcus aureus LytSR two-component regulatory system affects murein hydrolase activity and autolysis. A LytSR-regulated dicistronic operon has also been identified and shown to encode two potential membrane-associated proteins, designated LrgA and LrgB, hypothesized to be involved in the control of murein hydrolase activity. In the present study, a lrgAB mutant strain was generated and analyzed to test this hypothesis. Zymographic and quantitative analysis of murein hydrolase activity revealed that the lrgAB mutant produced increased extracellular murein hydrolase activity compared to that of the wild-type strain. Complementation of the lrgAB defect by providing the lrgAB genes in trans restored the wild-type phenotype, indicating that these genes confer negative control on extracellular murein hydrolase activity. In addition to these effects, the influence of the lrgAB mutation on penicillin-induced lysis and killing was examined. These studies demonstrated that the lrgAB mutation enhanced penicillin-induced killing of cells approaching the stationary phase of growth, the time at which the lrgAB operon was shown to be maximally expressed. This effect of the lrgAB mutation on penicillin-induced killing was shown to be independent of cell lysis. In contrast, the lrgAB mutation did not affect penicillin-induced killing of cells growing in early-exponential phase, a time in which lrgAB expression was shown to be minimal. However, expression of the lrgAB operon in early-exponential-phase cells inhibited penicillin-induced killing, again independent of cell lysis. The data generated by this study suggest that penicillin-induced killing of S. aureus involves a novel regulator of murein hydrolase activity.
Murein hydrolases are a unique family of enzymes that specifically cleave structural components of the bacterial cell wall. They have been shown to participate in a number of important biological processes during cell growth and division, including daughter cell separation, cell wall growth, peptidoglycan recycling, and turnover (1, 17, 28, 34, 35, 40). In addition, these enzymes have been shown to contribute to the pathogenicity of bacteria and are required for susceptibility to antibiotics (17). Biochemical analysis of murein hydrolases reveals that these enzymes have hydrolytic activities that are specific for various structural components of the peptidoglycan. These include N-acetylmuramidase, N-acetylglucosaminidase, N-acetylmuramyl-l-alanine amidase, and endotransglycosidase activities that presumably have specific roles in the biosynthesis and processing of the bacterial cell wall (17, 35, 40). Those murein hydrolases that lead to the destruction of the cell wall and subsequent cell lysis are known as autolysins.
Because of the capacity to destroy the cell wall, the expression and activity of murein hydrolases must be tightly controlled. At the posttranscriptional level, murein hydrolase activity has been shown to be modulated by mechanisms such as substrate modification, selective transport, interactions with lipoteichoic acids and cationic peptides, and cleavage by proteolytic enzymes (17, 35, 40). In addition, a number of investigations have demonstrated that monovalent cations may act as environmental signals that affect murein hydrolase activity (8, 24). Tobin et al. (37) demonstrated that the addition of NaCl to the growth medium has a differential effect on murein hydrolase activity in Staphylococcus aureus. Cultures grown in the presence of 1.5 M NaCl exhibited a higher rate of autolysis. However, this high concentration of NaCl inhibited the activities of cell wall-bound murein hydrolases. In contrast, Gilpin et al. (13) and Wong et al. (41) demonstrated that the addition of 1.0 M NaCl to S. aureus cultures increased cell wall turnover due to the increased activity of the N-acetylmuramyl-l-alanine amidase.
In addition to posttranscriptional control, the expression of some murein hydrolases has been shown to be regulated at the transcriptional level. The expression of the Bacillus subtilis murein hydrolase, cwlB, is dependent upon the alternative sigma factor ςD (10, 22, 26, 38) and the late-growth regulator Sin (22, 32). For S. aureus, Mani et al. (25) reported the isolation of two transposon insertion mutants that produced no detectable murein hydrolase activity and exhibited negligible autolysis rates. Although the genes affected by the transposon insertions have not been characterized, the authors suggested that they lie within a master regulatory gene or a structural gene responsible for the synthesis or processing of staphylococcal murein hydrolases.
Recent studies in our laboratory have demonstrated that the virulence factor regulators Agr and Sar also affect autolysis and murein hydrolase activity in a manner that indicates that they play opposing roles in the regulation of these processes (11). A mutation in the gene encoding Agr resulted in cells that exhibited a reduced rate of autolysis, while mutations in the Sar gene resulted in an increased autolysis rate. Furthermore, pleiotropic effects on different murein hydrolases were observed. These data indicate that, in addition to regulating virulence factor expression, the Agr and Sar regulators could play a significant role in the in vivo susceptibility of S. aureus to penicillin.
Finally, another regulatory system that has been shown to affect murein hydrolase activity is the LytSR regulatory locus of S. aureus (5). The lytS and lytR genes, whose predicted protein products share sequence characteristics with sensor and response regulator proteins, respectively, form a dicistronic operon. A lytS mutant strain exhibited an increased propensity for spontaneous lysis, Triton X-100-induced lysis, and altered murein hydrolase activities (5). The lytSR locus is located immediately upstream of another dicistronic operon containing the lrgA and lrgB genes. Examination of lrgAB expression revealed that transcription was positively regulated by the lytSR regulatory locus (6), leading to the hypothesis that the lrgA and lrgB gene products likely play some role in cell wall metabolism.
In this study, the function of the S. aureus lrgAB operon was examined by constructing an lrgAB null mutant and testing this strain for murein hydrolase activity and penicillin sensitivity. The data generated indicated that the lrgAB gene products inhibit extracellular murein hydrolase activity and promote penicillin tolerance.
MATERIALS AND METHODS
Strains and growth conditions.
The S. aureus strains used in this study were grown in tryptic soy broth (TSB; Difco Laboratories, Detroit, Mich.) or filter-sterilized NZY broth (3% N-Z Amine A [Sigma Chemical Co., St. Louis, Mo.] plus 1% yeast extract [Fisher Scientific, Fair Lawn, N.J.]), and Escherichia coli DH5α was grown in Luria-Bertani medium (Fisher Scientific). All bacterial cultures were grown with shaking (250 rpm) at 37°C. Antibiotics needed for plasmid maintenance were purchased from either Sigma Chemical Co. or Fisher Scientific and were used at the following concentrations: kanamycin, 50 μg/ml; erythromycin, 2 μg/ml; tetracycline, 5 μg/ml; ampicillin, 100 μg/ml; and spectinomycin, 50 μg/ml.
DNA manipulations.
Chromosomal DNA was isolated from S. aureus by the method of Dyer and Iandolo (9). Plasmid DNA was purified using a plasmid isolation kit from Qiagen, Inc. (Chatsworth, Calif.) or Promega, Inc. (Madison, Wis.). Enzymes used in the manipulation of DNA in this study were purchased from either New England Biolabs (Beverly, Mass.) or GIBCO-BRL (Gaithersburg, Md.). Preparation and transformation of E. coli were accomplished using the procedure described by Inoue et al. (18), and electroporation into S. aureus RN4220 was carried out using the method of Kraemer and Iandolo (21). φ11-mediated transduction of plasmids into S. aureus was carried out using the method of Shafer and Iandolo (33).
Northern blot analysis.
RNA was isolated from S. aureus as described previously by Hart et al. (16). Briefly, 10-ml aliquots of S. aureus cultures were removed, added to 10 ml of ice-cold ethanol–acetone (1:1), and stored at −20°C until sampling was complete. This suspension was centrifuged at 6,000 × g (at 4°C) for 15 min, and the pellet was resuspended in 10 ml of TEN buffer (30). The suspension was centrifuged again, and the pellet was then resuspended in 1 ml of TEN buffer containing 2.5 M NaCl. To protoplast the cells, recombinant lysostaphin (AMBI Inc., Tarrytown, N.Y.) was added to a final concentration of 50 μg/ml and incubated at 37°C for 40 min. RNA was purified by utilizing 5 ml of RNAzol-B (Tel-Test, Friendswood, Tex.) in accordance with the manufacturer's directions.
Northern hybridization analysis was performed by denaturing 20 μg of RNA at 65°C and separating it in a 1.0% formaldehyde-agarose gel (30). The RNA was then transferred to a Schleicher & Schuell, Inc. (Keene, N.H.) charged nylon membrane by downward capillary transfer (2). For the dot blot analysis, 20-μg samples of RNA were applied to a charged nylon membrane using a dot blot manifold. Prehybridization was carried out at 65°C in 15 ml of hybridization buffer (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] [30], 0.02% sodium dodecyl sulfate [SDS], 0.1% N-lauroylsarcosine, 1% Boehringer Mannheim [Indianapolis, Ind.] blocking reagent for nucleic acid detection). Digoxigenin (Dig)-labeled lrgAB-specific probes were generated using the primers lrgAB-1 (5′-GCCGGATCCGAAGTGAGCCATCTATA-3′) and lrgAB-2 (5′-GCCGAATTCGATAATAACAATGGCTC-3′) and a Dig-dUTP PCR kit from Boehringer Mannheim. Hybridization to the lrgAB-specific probe was performed at 65°C for 16 h. The membrane was then washed in 2.0× SSC–0.01% SDS at room temperature twice for 15 min each. This was followed by two washes in 0.5× SSC–0.01% SDS at 68°C for 15 min each time. The remainder of the detection procedure followed the protocol supplied with the Digoxigenin kit from Boehringer Mannheim Co.
Allele replacement of the lrgAB operon.
An lrgAB mutation was generated in RN6390 using the following strategy. First, a 760-bp DNA fragment spanning a region 5′ to lrgA was PCR amplified using the primers PRO31 (5′-GCGAATTCGGATGAAAATGGGATCG-3′) and PRO32 (5′-CCGGATCCGCTGGTTTTGATGCGTC-3′) and was ligated into the EcoRI and SmaI sites of plasmid pDG647 (15), upstream of an Em cassette. This recombinant plasmid was designated pSKY14. Next, a 455-bp DNA fragment spanning a region 3′ to lrgB was PCR amplified utilizing the primers Sal-dn (5′-CGGCGTCGACGGTGTCATTATTTATGCCCTAGG-3′) and Pst-dn (5′-CTATAATTGTCTGCAGGTGAACCATGTTTACG-3′) and was ligated into the SalI and PstI sites of pSKY14, downstream of the Em cassette. This plasmid, designated pSKY17, was then digested with EcoRI and PstI to liberate the Em cassette along with the flanking lrgA and lrgB sequences. This 2.8-kb fragment was subsequently ligated into the EcoRI and PstI sites of pCL52.2 (31) to generate pSKY18. This plasmid was then transformed into S. aureus strain RN4220 by electroporation, spread onto tryptic soy agar (TSA) plates containing erythromycin, and incubated at 30°C overnight. The plasmid was then transferred into RN6390 by phage-mediated transduction. This strain, designated KB344, was grown at the nonpermissive temperature (43°C) in the presence of tetracycline to select for cells in which the plasmid had integrated into the chromosome via homologous recombination. To promote a second recombination event, a single colony was inoculated into antibiotic-free TSB medium and grown at 30°C for 5 days with 1:1,000 dilutions into fresh antibiotic-free medium each day. After the 5th day, the culture was diluted and spread on TSA medium to yield isolated colonies. The colonies were then screened for Emr and Tcs. Verification that the lrgA and lrgB genes had been deleted was carried out by PCR amplification and Southern blot analysis. The confirmed mutant strain was designated KB345. Complementation of this strain was achieved by PCR amplifying the lrgAB operon (without its native promoter [see reference 6]) using primers lrgAB-1 and lrgAB-2 (see above) and cloning the resulting fragment into the EcoRI and BamHI sites of the gram-positive expression vector pRB374 (4). This placed the expression of the lrgAB operon under the control of the constitutively expressed, vegetative promoter originating from B. subtilis.
Penicillin sensitivity assays.
The sensitivity of S. aureus strains to penicillin was assessed by inoculating single colonies into 3 ml of NZY medium and incubating them at 37°C with antibiotic selection for the pRB374 plasmid constructs. Following overnight growth, the cells were pelleted at 6,000 × g for 5 min, washed twice in 3 ml of NZY medium, and then resuspended in 3 ml of NZY medium. This antibiotic-free culture was diluted 1:100 in NZY medium and grown at 37°C with shaking at 250 rpm to early-exponential phase (∼60 Klett units) or late-exponential phase (∼300 Klett units). Penicillin G (Sigma Chemical Co.) was added to a concentration of 0.4 μg/ml, equivalent to 20 times the MIC for RN6390. Incubation of the cultures was continued, and the culture turbidity was measured using a Klett-Summerson colorimeter (filter no. 60) every 30 min for a total of 8 h. In addition, 100-μl aliquots were taken every hour and diluted, and viable cells were quantified using the track dilution method (19).
Zymographic analysis.
Zymographic analysis of extracellular, cell wall-associated, and intracellular murein hydrolases from strains grown in filter-sterilized NZY medium was carried out essentially as described by Qoronfleh and Wilkinson (29). Extracellular murein hydrolases were isolated by pelleting 15 ml of a 17-h culture at 6,000 × g for 15 min at 4°C. The supernatant was filter sterilized and concentrated 100-fold using a Centricon-3 concentrator (Amicon, Beverly, Mass.). To obtain cell wall-associated and intracellular murein hydrolases, the pellet obtained as described above was resuspended in 25 ml of 0.01 M KPO4 (pH 7.0) and split into two portions. The cell wall-associated murein hydrolases were isolated by first washing one of the above cell suspensions twice in 25 ml of 0.01 M KPO4 (pH 7.0) and then centrifuging at 6,000 × g for 15 min at 4°C. This washed pellet was then resuspended in 25 ml of 0.01 M KPO4 (pH 7.0) and stored overnight at −20°C. Upon thawing at room temperature, the suspension was centrifuged at 16,000 × g for 15 min at 4°C. The pellet obtained was then resuspended in a 3 M LiCl2 solution and shaken at 300 rpm at 4°C for 10 min. The cells were pelleted again by centrifugation at 27,000 × g for 10 min. The supernatant was then dialyzed overnight at 4°C against 0.01 M KPO4 (pH 7.0) and concentrated 10-fold in a Centricon-3 concentrator. Intracellular murein hydrolases were obtained by pelleting the other cell portion as described above and then resuspending in 2.5 ml of SMMP (7) in a 15-ml Falcon tube. The cells were protoplasted by adding lysostaphin to a final concentration of 100 μg/ml and incubating at 37°C for 30 min. To eliminate any residual lysostaphin, the protoplasts were further diluted with an additional 2.5 ml of SMMP and centrifuged at room temperature for 10 min at 2,000 × g. The cells were gently resuspended in 5 ml of SMMP and pelleted again as above. The cells were then resuspended in 2.5 ml of 0.01 M KPO4 (pH 7.0) and lysed by passing several times through an 18-gauge hypodermic needle. The cellular debris was removed by centrifugation at 16,000 × g for 15 min at 4°C.
The concentration of total proteins present in each preparation was determined using the Bradford assay (Bio-Rad, Hercules, Calif.) according to the manufacturer's directions. A 15-μg aliquot of each preparation was diluted in an equal volume of SDS reducing sample buffer (Bio-Rad) and loaded onto an SDS–15% polyacrylamide gel containing either Micrococcus luteus (Sigma Chemical Co.) or autoclaved and lyophilized S. aureus cells at a concentration of 1.0 mg/ml. Following electrophoretic separation of the murein hydrolases, the proteins were allowed to hydrolyze the embedded bacterial cells by incubation of the gel in a 1% Triton X-100–25 mM Tris-HCl (pH 8.0) buffer overnight at 37°C. After this incubation, the gel was stained using a 1% methylene blue solution and destained in water. Following destaining, the gel was photographed over a light box. White bands in the gel (zones of hydrolysis) indicated regions of murein hydrolase activity.
Cell wall hydrolysis assays.
To quantify the amount of hydrolysis observed in the zymographic analysis, cell wall hydrolysis assays were performed essentially as described by Mani et al. (25). Briefly, 100 μg of enzyme extract was added to a suspension of autoclaved and lyophilized S. aureus cells (1.0 mg/ml) in 100 mM Tris-HCl (pH 8.0) and incubated at 37°C with shaking (250 rpm). Turbidity measurements were taken at 30-min intervals with a Klett-Summerson colorimeter utilizing a no. 60 red filter.
RESULTS
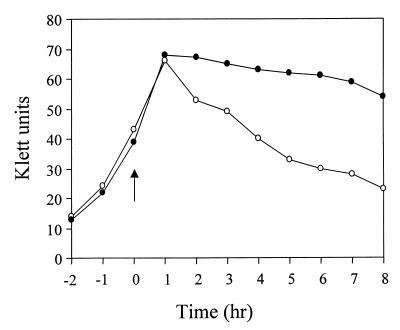
Studies in our laboratory have revealed the presence of an S. aureus regulatory system, termed LytSR, which is involved in the regulation of murein hydrolase activity (5). The consequences of disrupting the LytSR genes include increased spontaneous and Triton X-100-induced lysis, altered murein hydrolase activity, and loss of expression of a dicistronic operon containing the lrgA and lrgB genes (5, 6). As shown in Fig. 1, the LytSR system also affects lysis induced by penicillin. S. aureus strains 8325-4 and KB300 were grown to early-exponential phase and then treated with 0.4 μg of penicillin/ml at a concentration equivalent to 20 times the MIC for 8325-4. The turbidity of both cultures declined shortly after the addition of penicillin, but that of the KB300 culture declined to a much greater extent. Measurements of culture viability corresponded with the increased level of penicillin-induced lysis exhibited by KB300 (unpublished results). These data demonstrate that, in addition to exhibiting increased Triton X-100-induced lysis (5), the KB300 strain also exhibits increased penicillin-induced lysis compared to the parental strain.
FIG. 1.
Penicillin-induced lysis of KB300. S. aureus strain 8325-4 (filled circles) and KB300 (open circles) were grown to early-exponential phase and treated with penicillin (arrow). Turbidity measurements of the cultures were performed every hour for 8 h after the addition of penicillin. Data are representative of experiments performed three times.
Generation of an lrgAB mutant.
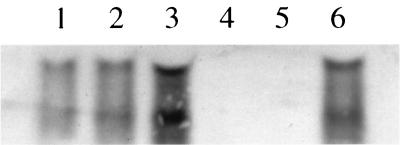
Based on previous data (6) and on the findings described above, it was hypothesized that the lrgA and lrgB gene products affect, in some way, the activity of murein hydrolases produced by S. aureus and/or the sensitivity of the bacteria to penicillin. To test this hypothesis, an lrgAB deletion mutant derivative of RN6390 (designated KB345) was generated by replacing these genes with an erythromycin resistance cassette. As shown in Fig. 2 (lanes 4 and 5), the deletion of the lrgAB operon in strain KB345 resulted in the loss of production of the previously identified 1.2- and 0.8-kb lrgAB- and lrgB-specific transcripts, respectively (6). The production of these transcripts could be restored by introducing an lrgAB expression plasmid, pRB-lrgAB, into KB345 (Fig. 2, lane 6). Furthermore, the presence of pRB-lrgAB in RN6390 increased the lrgAB transcripts (Fig. 2, lane 3) to levels greater than that observed in either of the control strains (Fig. 2, lanes 1 and 2), indicating that the lrgAB transcripts are overexpressed in this strain.
FIG. 2.
Northern blot analysis of the lrgAB mutant and complemented strains. RNA was isolated from S. aureus strains, separated in a 1.0% formaldehyde-agarose gel, and subjected to Northern blot analysis using an lrgAB-specific probe. Lanes: 1, RN6390; 2, RN6390(pRB374); 3, RN6390(pRB-lrgAB); 4, KB345; 5, KB345(pRB374); 6, KB345(pRB-lrgAB). The RNAs detected correspond to the 1.2-kb lrgAB-encoding transcripts and the 0.8-kb lrgB-encoding transcripts as previously described (6).
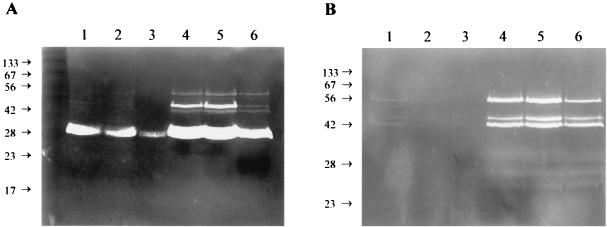
Initial testing of the KB345 strain revealed that the lrgAB mutation did not alter the sensitivity of the strain to Triton X-100-induced lysis or the morphological features of individual cells (unpublished results) as observed with the lytSR mutant, KB300 (5). Furthermore, the growth rate of KB345 was identical to that of the parental strain (unpublished results). To determine if the lrgAB mutation affects murein hydrolase activity, proteins were isolated from stationary-phase cultures and analyzed by zymography using either M. luteus (Fig. 3A) or S. aureus (Fig. 3B) cells as a substrate. Interestingly, the KB345 strain was shown to produce increased levels of several extracellular murein hydrolases (Fig. 3, lanes 4) compared to the parental strain, RN6390 (Fig. 3, lanes 1), using either substrate. As shown in Fig. 3, complementation of this mutation by supplying the lrgAB operon in trans resulted in a reduction in the overall murein hydrolase activity produced (lanes 6) compared to that in the KB345 control strain (lanes 5); this reduced level was similar to that produced by RN6390 (lanes 1) and RN6390(pRB374) (lanes 2). The overexpression of lrgAB in RN6390 (Fig. 3, lanes 3) resulted in decreased extracellular murein hydrolase activity compared to that in the RN6390 control strains (Fig. 3, lanes 1 and 2). No differences between the activities of murein hydrolases isolated from the cell wall-associated fraction and those isolated from the intracellular fraction were observed (unpublished results).
FIG. 3.
Zymographic analysis of the lrgAB mutant. Extracellular proteins were isolated, and 15 μg of each was separated in SDS-polyacrylamide gel electrophoresis gels containing 1.0 mg of either M. luteus (A) or S. aureus (B) cells/ml. Murein hydrolase activity was detected by incubation overnight at 37°C in a buffer containing Triton X-100, followed by staining with methylene blue. Lanes: 1, RN6390; 2, RN6390(pRB374); 3, RN6390(pRB-lrgAB); 4, KB345; 5, KB345(pRB374); 6, KB345(pRB-lrgAB). Molecular size markers (in kilodaltons) are indicated to the left of each gel.
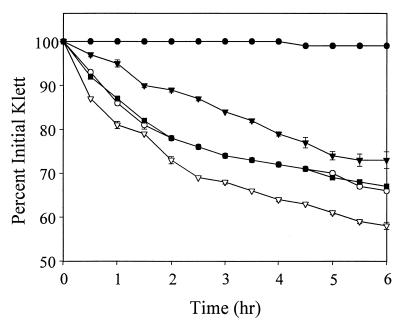
To quantify the observations of the zymographic analysis, cell wall hydrolysis assays were performed by preparing a suspension of killed S. aureus cells in a Tris-HCl buffer, adding 100-μg portions of extracellular murein hydrolase extracts, and monitoring the decrease in the suspension's turbidity over time. As shown in Fig. 4, KB345 extracellular murein hydrolases caused a 42% decrease in turbidity after 6 h of incubation, compared to a 34% decrease with RN6390 extracellular murein hydrolases. Furthermore, the expression of pRB-lrgAB in KB345 caused a 33% decrease in turbidity after 6 h, similar to that in the parental strain and consistent with the zymographic analysis. These hydrolysis assays also confirmed the observation that the presence of pRB-lrgAB in RN6390 reduced the level of extracellular murein hydrolase secreted by this strain. Extracellular murein hydrolases from RN6390(pRB-lrgAB) were capable of causing a 27% decrease in turbidity after 6 h, compared to a reduction of 34% with extracellular murein hydrolases from RN6390 containing the control plasmid. These results, along with the zymographic analysis, demonstrate that expression of the lrgAB operon results in a reduction in extracellular murein hydrolase activity produced by S. aureus.
FIG. 4.
Quantitative murein hydrolase assays of the lrgAB mutant. Aliquots (100 μg) of the extracellular proteins used in Fig. 3 were added to a 1-mg/ml suspension of S. aureus cells, and the turbidity was monitored for 6 h. Strains used in this analysis were RN6390(pRB374) (○), RN6390(pRB-lrgAB) (▾), KB345(pRB374) (▿), and KB345(pRB-lrgAB) (■). A negative control (●) in which no murein hydrolase was added to the cell suspension was also performed.
Penicillin sensitivity assays.
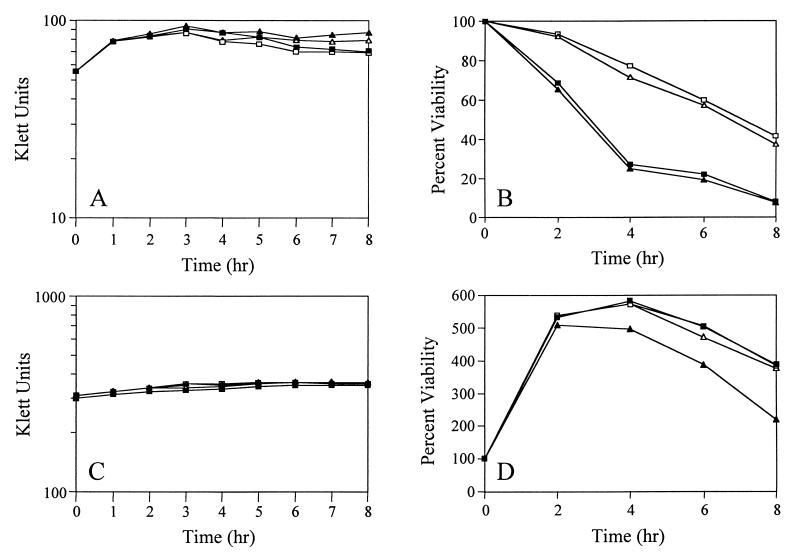
As demonstrated in Fig. 1, the LytSR regulatory system affected the sensitivity of S. aureus to penicillin. Thus, to examine the role that the LytSR-regulated lrgAB operon might have in response to exposure to penicillin, lysis and viability measurements of the KB345 strain after exposure to this antibiotic were performed. In these assays, penicillin G (20 times the MIC) was added to early-exponential-phase S. aureus cultures (∼60 Klett units) and the culture lysis and viability were monitored over 8 h. As shown in Fig. 5, neither the lytic (Fig. 5A) nor the killing (Fig. 5B) effects of penicillin were affected by the absence of the lrgAB operon. The mutant and wild-type strains exhibited similarly reduced viability over the course of this assay, resulting in a reduction of approximately 75% in viability after 4 h. However, the presence of pRB-lrgAB in RN6390 or KB345 resulted in a marked increase in these strains' tolerance to penicillin (Fig. 5B); they exhibited only an approximately 25% decrease in viability after 4 h. In contrast, the presence of this plasmid had no effect on the lytic response to penicillin (Fig. 5A).
FIG. 5.
Effects of penicillin on S. aureus strains. (A and B) Penicillin was added to early-exponential-phase S. aureus cultures (∼60 Klett units), and turbidity measurements (A) were performed every hour for 8 h. Viable cell counts (B) were determined by diluting aliquots of the cultures and plating them on TSA medium. (C and D) Penicillin was added to late-exponential-phase S. aureus cultures (∼300 Klett units), followed by turbidity (C) and viable cell count (D) measurements as described above. The S. aureus strains used in this analysis are RN6390(pRB374) (■), RN6390(pRB-lrgAB) (□), KB345(pRB374) (▴), and KB345(pRB-lrgAB) (▵). Data are representative of experiments performed at least three times.
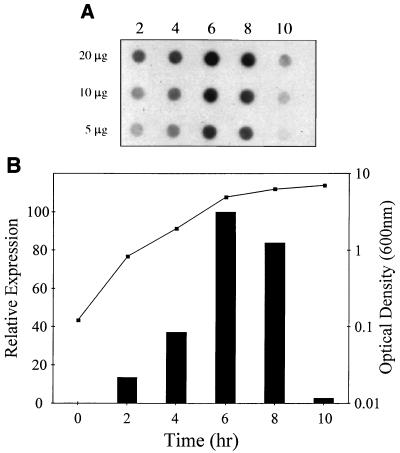
Based on the observations that overexpression of the lrgAB operon affected penicillin tolerance while the deletion of this operon had no apparent effect, we hypothesized that these genes may not be expressed efficiently during the early-exponential phase of growth, when penicillin was added. If this hypothesis is correct, the KB345 mutant would not be expected to exhibit differences in penicillin sensitivity during this growth phase, as observed. In fact, the results of a primer extension analysis of the lrgAB transcription start site suggested that more lrgAB transcripts are present in stationary-phase RNA than in exponential-phase RNA (6). To confirm these results, RNA was collected from growing cultures of S. aureus RN6390 at various time points and subjected to dot blot analysis using an lrgAB-specific probe (Fig. 6A). A significant increase in the amount of lrgAB transcripts was detected between 4 and 6 h (Fig. 6), indicating that these genes are maximally expressed as the cells approach stationary phase (Fig. 6B). Quantification of the signals generated revealed that lrgAB expression in early-exponential-phase cells was only 15% that of cells in the late-exponential phase (Fig. 6B).
FIG. 6.
Dot blot analysis of lrgAB expression. (A) RNA was isolated from S. aureus RN6390 grown to various times throughout the growth cycle and applied to nitrocellulose using a dot blot apparatus. The RNA was hybridized to an lrgAB-specific probe. (B) Data from the dot blot analysis shown in panel A were quantified and plotted next to the S. aureus growth curve. Maximal lrgAB expression was observed at the transition between the exponential and stationary phases of growth (6 h).
In light of the finding that the lrgAB operon is maximally expressed as the cells are approaching stationary phase, it was of interest to ask whether phenotypic differences in penicillin sensitivity could be detected in late-exponential-phase cells. Although bacteria are intrinsically more resistant to penicillin as they enter the stationary phase of growth (20), reproducible differences in sensitivity were observed. As shown in Fig. 5D, the RN6390 culture exhibited a viability 8 h after the addition of penicillin that was 383% that of the viability when penicillin was added. In contrast, the mutant, KB345, exhibited a viability that was only 220% higher 8 h after the addition of penicillin. As shown in Fig. 5D, the decreased tolerance of KB345 to the killing effects of penicillin could be increased to wild-type levels by the presence of pRB-lrgAB in this strain, indicating that the mutation could be functionally complemented. Again, the presence of lrgA and lrgB did not affect the lytic response of the cells to penicillin (Fig. 5C), indicating that the effects of these genes on penicillin-induced killing were independent of murein hydrolase activity.
DISCUSSION
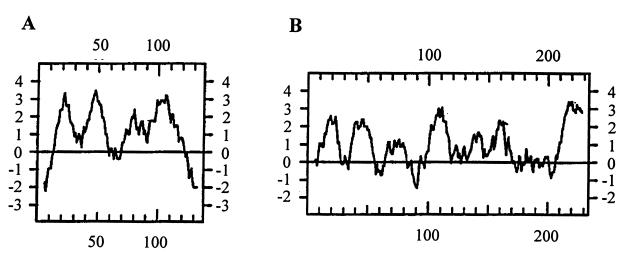
In this study, the function of the S. aureus lrgAB operon was investigated. An earlier investigation by this laboratory suggested the involvement of lrgA and lrgB in autolysis and murein hydrolase activity (6). Based on this information, it was hypothesized that these genes encode murein hydrolases and/or proteins that affect murein hydrolase activity (6). Given the predicted extremely hydrophobic nature of the lrgA and lrgB gene products (Fig. 7), it is unlikely that either of these proteins contains murein hydrolase activity itself. Consistent with this is the inability to demonstrate increased murein hydrolase activity in E. coli or B. subtilis strains expressing either the lrgA or the lrgB gene (unpublished results). Thus, it is more likely that the lrgA and lrgB gene products are involved in controlling murein hydrolase activity at some level. In the study reported here, an lrgAB null mutant, designated KB345, was generated to test this hypothesis. Quantification of the extracellular murein hydrolase activity produced by these strains demonstrated that KB345 produced increased overall activity compared to that of the parental strain (Fig. 4). Reintroduction of the lrgA and lrgB genes into KB345 restored the murein hydrolase activities to wild-type levels. It was also found that the overexpression of lrgA and lrgB in wild-type cells resulted in a significant decrease in extracellular murein hydrolase activities. Taken together, these analyses suggest that the lrgA and lrgB gene products may be acting in some capacity to reduce extracellular murein hydrolase activity.
FIG. 7.
Hydropathy analysis of LrgA (A) and LrgB (B) according to Kyte and Doolittle (23) with an 11-amino-acid window.
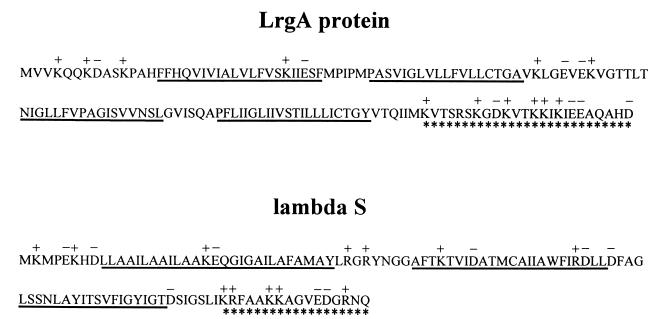
An insight into the possible role of LrgA comes from the observation that it shares many sequence characteristics with the bacteriophage holin family of proteins (Fig. 8). Although there is great divergence in the amino acid sequences of holins, several features tie the members of this group together. For example, holins are typically small proteins comprised of 60 to 145 amino acids; LrgA is slightly larger at 148 amino acids. Members of this family also have a charge-rich, polar C terminus, a hydrophilic N terminus, and two or three membrane-spanning domains linked by beta turns (42). LrgA shares all of these characteristics except that the predicted amino acid sequence indicates four membrane-spanning domains (Fig. 7A). Holins are known to play a crucial role in the life cycles of most lytic bacteriophages by providing a timing mechanism for release of newly formed bacteriophage particles. The prototypical holin, λS, allows the bacteriophage-encoded murein hydrolases to gain access to their substrate, the cell wall peptidoglycan (42). This function is believed to be carried out by small, nonspecific channels formed as a result of the oligomerization of individual holin subunits in the cytoplasmic membrane of the host. Transport is Sec independent (many bacteriophage-encoded murein hydrolases lack signal peptides), and without an active holin, the murein hydrolases would simply accumulate in the cytosol of the host (12).
FIG. 8.
Structural similarities between LrgA and the prototypical holin, bacteriophage lambda S protein. The symbols used are as previously defined by Young and Blasi (42), as follows. The predicted charge pattern is shown above the amino acid sequence, with K and R residues shown as positively charged and E and D residues shown as negatively charged. Potential membrane-spanning domains are underlined, and the highly charged carboxyl termini are underscored with asterisks.
Regulation of holin activity is crucial, since unchecked production of these proteins would lead to early death of the host cell by way of cytoplasmic leakage and loss of transmembrane potential. This control is accomplished by the utilization of a “dual-start motif” (42). Translation of the holin mRNA can occur from either one of two methionine codons that are separated by only one or two lysine or arginine codons. Although the two mature forms of λS differ by only two amino-terminal amino acids (105 versus 107 residues), the proteins carry out completely different functions. The 105-residue polypeptide functions as the “effector” holin, while the 107-residue protein has an antagonistic activity and is termed an “inhibitor” holin, or “antiholin.” A proposed model of holin activity has the effector holin forming a ring in the host cell membrane by oligomerization of individual subunits (36). Oligomerization of the effector holin subunits into a completed hole is effectively “poisoned” by the presence of antiholins. As the time for host cell lysis approaches, it is thought that the inhibitory effects of antiholins are relaxed (and even converted to effector holin activity), resulting in an increase in murein hydrolase export and host cell lysis (3, 14).
Based on the data presented in this report, we propose that LrgA, and possibly LrgB, functions in a manner analogous to that of an antiholin to inhibit murein hydrolase export. Although the possibility that LrgA and LrgB are acting in another capacity (e.g., in the capacity of a protease or a transcription factor) to reduce murein hydrolase activity cannot be ruled out, the predicted extremely hydrophobic nature of these proteins (see Fig. 7) suggests that these possibilities are unlikely. If the hypothesis that LrgA and LrgB are functioning in the capacity of an antiholin is correct, one must also predict the presence of an effector holin in S. aureus. Since no dual-start motif is present in lrgA, the putative effector holin would most likely be encoded by an entirely separate gene.
An understanding of the functions of the lrgAB gene products is particularly significant, since it would provide important new information regarding the regulation of murein hydrolase activity. Another aspect of lrgAB function to be considered is their potential involvement in the bactericidal effects of penicillin. Genetic analysis of the factors necessary for penicillin-induced killing has revealed that a complex system of physiological effects is involved in this process. Genetic studies have revealed the participation of two independent factors in the penicillin-induced killing of Streptococcus pneumoniae (27). The first, an amidase, encoded by the lytA gene, was found to be responsible for a 1-log-unit loss of culture viability every 6 h upon exposure of exponentially growing cultures of S. pneumoniae to penicillin. An additional 3 to 4 log units of killing were dependent on a second, yet to be identified factor that was defective in so-called “cid” mutants (27). The cid mutation was able to dramatically reduce the amount of penicillin-induced killing in both wild-type and lytA mutant strains of S. pneumoniae (27), underscoring the murein hydrolase-independent nature of this phenotype. Although the gene(s) responsible for the cid phenotype has never been reported, it was hypothesized that it encodes a protein analogous to bacteriophage-encoded holins. Interestingly, similar S. aureus mutants, which are tolerant to the killing effects of penicillin, have been previously reported (39) and also generated by our laboratory (unpublished results).
In the present study, the expression of the lrgAB operon was shown to inhibit penicillin-induced killing of S. aureus, independent of cell lysis. This effect was shown to be growth phase dependent and could be complemented by expressing lrgAB using a constitutive promoter. It is envisioned that the effect that lrgAB expression has on penicillin-induced killing could occur via a mechanism directly involving the lrgAB gene products similar to that previously proposed by Moreillon et al. (27). Alternatively, an indirect effect of these genes on lysis-independent killing cannot be ruled out. For example, the murein hydrolase activity affected by lrgAB expression could be inducing cell wall defects that lead to nonlytic death.
Finally, our laboratory is interested in identifying additional LytSR-regulated genes, or genes that encode proteins with effector holin-like activity. Recent studies in our laboratory have revealed the presence of additional S. aureus genes that enhance extracellular murein hydrolase activity and penicillin-induced killing. Whether these genes encode proteins that contain effector holin activity and interact with the lrgAB gene products is currently under investigation.
ACKNOWLEDGMENTS
This work was funded by NIH grant R29-AI38901 and NSF-Idaho EPSCoR grant EPS-9720634.
We thank Chia-Yen Lee for providing us with plasmid pCL52.2.
REFERENCES
- 1.Archibald A R, Hancock I C, Harwood C R. Cell wall structure, synthesis, and turnover. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C.: American Society for Microbiology; 1993. pp. 381–410. [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1991. [Google Scholar]
- 3.Bonovich M T, Young R. Dual start motif in two lambdoid S genes unrelated to lambda S. J Bacteriol. 1991;173:2897–2905. doi: 10.1128/jb.173.9.2897-2905.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruckner R. A series of shuttle vectors for Bacillus subtilis and Escherichia coli. Gene. 1992;122:187–192. doi: 10.1016/0378-1119(92)90048-t. [DOI] [PubMed] [Google Scholar]
- 5.Brunskill E W, Bayles K W. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J Bacteriol. 1996;178:611–618. doi: 10.1128/jb.178.3.611-618.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunskill E W, Bayles K W. Identification of LytSR-regulated genes from Staphylococcus aureus. J Bacteriol. 1996;178:5810–5812. doi: 10.1128/jb.178.19.5810-5812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang S, Cohen S N. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol Gen Genet. 1979;168:111–115. doi: 10.1007/BF00267940. [DOI] [PubMed] [Google Scholar]
- 8.Cheung H, Freese E. Monovalent cations enable cell wall turnover of the turnover-deficient lyt-15 mutant of Bacillus subtilis. J Bacteriol. 1985;161:1222–1225. doi: 10.1128/jb.161.3.1222-1225.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyer D W, Iandolo J J. Rapid isolation of DNA from Staphylococcus aureus. Appl Environ Microbiol. 1983;46:283–285. doi: 10.1128/aem.46.1.283-285.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster S J. Analysis of the autolysins of Bacillus subtilis 168 during vegetative growth and differentiation by using renaturing polyacrylamide gel electrophoresis. J Bacteriol. 1992;174:464–470. doi: 10.1128/jb.174.2.464-470.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujimoto D F, Bayles K W. Opposing roles of the Staphylococcus aureus virulence regulators, Agr and Sar, in Triton X-100- and penicillin-induced autolysis. J Bacteriol. 1998;180:3724–3726. doi: 10.1128/jb.180.14.3724-3726.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garrett J M, Young R. Lethal action of bacteriophage lambda S gene. J Virol. 1982;44:886–892. doi: 10.1128/jvi.44.3.886-892.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilpin R W, Chatterjee A N, Young F E. Autolysis of microbial cells: salt activation of autolytic enzymes in a mutant of Staphylococcus aureus. J Bacteriol. 1972;111:272–283. doi: 10.1128/jb.111.1.272-283.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graschopf A, Blasi U. Molecular function of the dual-start motif in the lambda S holin. Mol Microbiol. 1999;33:569–582. doi: 10.1046/j.1365-2958.1999.01501.x. [DOI] [PubMed] [Google Scholar]
- 15.Guerout-Fleury A M, Shazand K, Frandsen N, Stragier P. Antibiotic-resistance cassettes for Bacillus subtilis. Gene. 1995;167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- 16.Hart M E, Smeltzer M S, Iandolo J J. The extracellular protein regulator (xpr) affects exoprotein and agr mRNA levels in Staphylococcus aureus. J Bacteriol. 1993;175:7875–7879. doi: 10.1128/jb.175.24.7875-7879.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holtje J-V, Tuomanen E I. The murein hydrolases of Escherichia coli: properties, functions and impact on the course of infections in vivo. J Gen Microbiol. 1991;137:441–454. doi: 10.1099/00221287-137-3-441. [DOI] [PubMed] [Google Scholar]
- 18.Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 19.Jett B D, Hatter K L, Huycke M M, Gilmore M S. Simplified agar plate method for quantifying viable bacteria. BioTechniques. 1997;23:648–650. doi: 10.2144/97234bm22. [DOI] [PubMed] [Google Scholar]
- 20.Kim K S, Anthony B F. Importance of bacterial growth phase in determining minimal bactericidal concentrations of penicillin and methicillin. Antimicrob Agents Chemother. 1981;19:1075–1077. doi: 10.1128/aac.19.6.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kraemer G R, Iandolo J J. High-frequency transformation of Staphylococcus aureus by electroporation. Curr Microbiol. 1990;21:373–376. [Google Scholar]
- 22.Kuroda A, Asami Y, Sekiguchi J. Molecular cloning of a sporulation-specific cell wall hydrolase gene of Bacillus subtilis. J Bacteriol. 1993;175:6260–6268. doi: 10.1128/jb.175.19.6260-6268.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 24.Madiraju M V, Brunner D P, Wilkinson B J. Effects of temperature, NaCl, and methicillin on penicillin-binding proteins, growth, peptidoglycan synthesis, and autolysis in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1987;31:1727–1733. doi: 10.1128/aac.31.11.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mani N, Tobin P, Jayaswal R K. Isolation and characterization of autolysis-defective mutants of Staphylococcus aureus created by Tn917-lacZ mutagenesis. J Bacteriol. 1993;175:1493–1499. doi: 10.1128/jb.175.5.1493-1499.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marquez L M, Helmann J D, Ferrari E, Parker H M, Ordal G W, Chamberlin M J. Studies of ςD-dependent functions in Bacillus subtilis. J Bacteriol. 1990;172:3435–3443. doi: 10.1128/jb.172.6.3435-3443.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreillon P, Markiewicz Z, Nachman S, Tomasz A. Two bactericidal targets for penicillin in pneumococci: autolysis-dependent and autolysis-independent killing mechanisms. Antimicrob Agents Chemother. 1990;34:33–39. doi: 10.1128/aac.34.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perkins H R. The bacterial autolysins. In: Rogers H J, Perkins H R, Ward J B, editors. Microbial cell walls. London, United Kingdom: Chapman and Hall; 1980. pp. 437–456. [Google Scholar]
- 29.Qoronfleh M W, Wilkinson B J. Effects of growth of methicillin-resistant and -susceptible Staphylococcus aureus in the presence of β-lactams on peptidoglycan structure and susceptibility to lytic enzymes. Antimicrob Agents Chemother. 1986;29:250–257. doi: 10.1128/aac.29.2.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Sau S, Sun J, Lee C Y. Molecular characterization and transcriptional analysis of type 8 capsule genes in Staphylococcus aureus. J Bacteriol. 1997;179:1614–1621. doi: 10.1128/jb.179.5.1614-1621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sekiguchi J, Ezaki B, Kodama K, Akamatsu T. Molecular cloning of a gene affecting the autolysin level and flagellation in Bacillus subtilis. J Gen Microbiol. 1988;134:1611–1621. doi: 10.1099/00221287-134-6-1611. [DOI] [PubMed] [Google Scholar]
- 33.Shafer M W, Iandolo J J. Genetics of staphylococcus enterotoxin B in methicillin-resistant isolates of Staphylococcus aureus. Infect Immun. 1979;25:902–911. doi: 10.1128/iai.25.3.902-911.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shockman G D, Barrett J F. Structure, function, and assembly of cell walls of gram-positive bacteria. Annu Rev Microbiol. 1983;37:501–527. doi: 10.1146/annurev.mi.37.100183.002441. [DOI] [PubMed] [Google Scholar]
- 35.Shockman G D, Holtje J-V. Microbial peptidoglycan (murein) hydrolases. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Vol. 27. Amsterdam, The Netherlands: Elsevier Science B.V.; 1994. pp. 131–166. [Google Scholar]
- 36.Smith D L, Chang C Y, Young R Y. The lambda holin accumulates beyond the lethal triggering concentration under hyperexpression conditions. Gene Expr. 1998;7:39–52. [PMC free article] [PubMed] [Google Scholar]
- 37.Tobin P J, Mani N, Jayaswal R K. Effect of physiological conditions on the autolysis of Staphylococcus aureus strains. Antonie Leeuwenhoek. 1994;65:71–78. doi: 10.1007/BF00878281. [DOI] [PubMed] [Google Scholar]
- 38.Tokunaga T, Rashid M H, Kuroda A, Sekiguchi J. Effect of degS-degU mutations on the expression of sigD, encoding an alternative sigma factor, and autolysin operon of Bacillus subtilis. J Bacteriol. 1994;176:5177–5180. doi: 10.1128/jb.176.16.5177-5180.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomasz A, de Vegvar M L. Construction of a penicillin-tolerant laboratory mutant of Staphylococcus aureus. Eur J Clin Microbiol. 1986;5:710–713. doi: 10.1007/BF02013310. [DOI] [PubMed] [Google Scholar]
- 40.Ward J B, Williamson R. Bacterial autolysins: specificity and function. In: Nombela C, editor. Microbial cell wall synthesis and autolysis. Amsterdam, The Netherlands: Elsevier Science Publishers; 1985. pp. 159–166. [Google Scholar]
- 41.Wong W, Chatterjee A N, Young F E. Regulation of bacterial cell walls: correlation between autolytic activity and cell wall turnover in Staphylococcus aureus. J Bacteriol. 1978;134:555–561. doi: 10.1128/jb.134.2.555-561.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young R, Blasi U. Holins: form and function in bacteriophage lysis. FEMS Microbiol Rev. 1995;17:191–205. doi: 10.1111/j.1574-6976.1995.tb00202.x. [DOI] [PubMed] [Google Scholar]