Abstract
INTRODUCTION
The Centiloid scale aims to harmonize amyloid beta (Aβ) positron emission tomography (PET) measures across different analysis methods. As Centiloids were created using PET/computerized tomography (CT) data and are influenced by scanner differences, we investigated the Centiloid transformation with data from Insight 46 acquired with PET/magnetic resonanceimaging (MRI).
METHODS
We transformed standardized uptake value ratios (SUVRs) from 432 florbetapir PET/MRI scans processed using whole cerebellum (WC) and white matter (WM) references, with and without partial volume correction. Gaussian‐mixture‐modelling–derived cutpoints for Aβ PET positivity were converted.
RESULTS
The Centiloid cutpoint was 14.2 for WC SUVRs. The relationship between WM and WC uptake differed between the calibration and testing datasets, producing implausibly low WM‐based Centiloids. Linear adjustment produced a WM‐based cutpoint of 18.1.
DISCUSSION
Transformation of PET/MRI florbetapir data to Centiloids is valid. However, further understanding of the effects of acquisition or biological factors on the transformation using a WM reference is needed.
HIGHLIGHTS
Centiloid conversion of amyloid beta positron emission tomography (PET) data aims to standardize results.
Centiloid values can be influenced by differences in acquisition.
We converted florbetapir PET/magnetic resonance imaging data from a large birth cohort.
Whole cerebellum referenced values could be reliably transformed to Centiloids.
White matter referenced values may be less generalizable between datasets.
Keywords: Alzheimer's disease, amyloid beta, centiloid, florbetapir, positron emission tomography/magnetic resonance imaging
1. BACKGROUND
In vivo estimation of amyloid beta (Aβ) burden using positron emission tomography (PET) is crucial for accurate clinical diagnosis of Alzheimer's disease (AD), characterizing disease progression, and assessment of eligibility and efficacy in therapeutic trials. 1 , 2
To understand the complex pathologies underlying AD, we must use data from many cohorts. One such study is Insight 46, the neuroimaging substudy of the Medical Research Council National Survey of Health and Development (NSHD), initially comprising 5362 individuals born in mainland Britain during the same week in March 1946. Each participant has rich life course data, which has been coupled with neuroimaging, fluid biomarker, and cognitive assessments from age ≈70 onward. 3 Insight 46 is a single‐site PET/magnetic resonance imaging (MRI) study and standardization of Aβ PET measures would improve comparability between the study and other large datasets such as the Alzheimer's Disease Neuroimaging Initiative (ADNI).
Aβ PET standardization is complicated by the number of different processing methods used. While the standardized uptake value ratio (SUVR) is widely used for analyzing Aβ PET images, 4 this measure is dependent on many factors, including the choice of radiotracer, data acquisition parameters, target and reference regions, and analysis methodology. 5 , 6 , 7 Deviations in these factors can impede comparison of results across studies or between centers. There are multiple approaches for harmonizing datasets that can be applied to Aβ PET; the most widely implemented to date is the Centiloid scale. 8 , 9
The Centiloid Project provides a common scale to standardize Aβ PET measurements using a post hoc linear transformation. 9 Anchor points at 0 and 100 Centiloid units (CL) correspond to the mean SUVR in groups of young healthy controls and patients with typical AD, respectively. However, the Centiloid conversion can be affected by various factors such as image acquisition methodology including differences in scanner type. 10
Here, we explore the implementation of the Centiloid scale for [18F]florbetapir data acquired on a combined PET/MRI system. PET/MRI scanners have only recently become widely available but reduce burden to the participant through simultaneous acquisition. This concerns particularly clinical research studies using advanced imaging protocols with longer acquisition times. However, there are substantial differences between PET/MRI and conventional PET/computed tomography (CT) that could affect the Centiloid transformation, such as how to perform attenuation correction without acquiring CT 11 or the longer axial field of view (FoV). While previous studies have explored PET/MRI differences in SUVRs, 12 , 13 and a study has included a small PET/MRI dataset in Centiloid transformations, 14 we believe this study is the first to assess the effects of Centiloid scale transformation using data from a PET/MRI scanner.
2. METHODS
2.1. Participants
We used three separate datasets. The Centiloid Project “Standard PiB” and “Florbetapir Calibration” datasets were downloaded from the Global Alzheimer's Association Interactive Network website. The Standard PiB dataset is described in detail in Klunk et al. 9 Briefly, the YC‐0 group consists of 34 young cognitively normal (YCN) individuals and the AD‐100 group consists of 45 AD patients. These groups form the anchor points at 0 and 100 Centiloids. The Florbetapir Calibration dataset is described in Navitsky et al. 15 and is made up of 46 participants across the Aβ continuum: a group of YCN (N = 13, aged ≤ 35 years) and elder subjects (ES; N = 33, aged > 50 years) with mixed diagnoses.
PET/MRI data comes from the Insight 46 study. 3 From the larger birth cohort, 502 participants were recruited, 471 of which had PET/MRI data available. After quality control, 432 had both MRI and list‐mode PET data required for this investigation (see Figure S1 in supporting information). Table 1 shows demographic information for all cohorts.
TABLE 1.
Demographic characteristics for each cohort.
| Standard PiB | Florbetapir calibration | ||||
|---|---|---|---|---|---|
| YC‐0 | AD‐100 | YCN | ES | Insight 46 | |
| N | 34 | 45 | 13 | 33 | 432 |
| Female sex % * | — | — | 53 | 36 | 48 |
| APOE ε4: carrier/total (%) a | 8/32 (25) | 28/44 (64) | 1/10 (10) | 14/22 (63) | 129/430 (30) |
| Mean MMSE (SD) b | — | — | 29.5 (0.5) | 24.7 (5.1) | 29.2 (1.0) |
| Mean Age (SD) | 31.5 (6.3) | 67.5 (10.5) | 27.0 (4.3) | 70.2 (9.6) | 70.6 (0.7) |
Abbreviations: AD‐100, Alzheimer's disease group; APOE, apolipoprotein E; ES, elder subject group; MMSE, Mini‐Mental State Examination; PiB, Pittsburgh compound B; SD, standard deviation; YC‐0, young control group; YCN, young cognitively normal group.
a APOE ε4 carrier is defined as carrying at least one APOE ε4 allele and is unknown for the following numbers in each group: YC‐0 = 2, AD‐100 = 1, YCN = 3, ES = 11, Insight 46 = 2.
bMMSE was unknown for three participants in the YCN group. Data regarding MMSE for the Standard PiB dataset was not published in Klunk et al. 9
*Data regarding the sex of participants in the Standard PiB dataset was not published in Klunk et al. 9
2.2. Image acquisition
All three datasets contain static Aβ PET images and volumetric T1‐weighted MRI (full acquisition details can be found in Lane et al., 3 Klunk et al., 9 and Navitsky et al. 15 ). The Standard PiB dataset consists of Pittsburgh compound B (PiB) data acquired 50 to 70 minutes post‐injection. The Florbetapir Calibration dataset contains both PiB (50–70 minutes) and florbetapir (50–60 minutes) data, all acquired in 5‐minute frames. Data from the Insight 46 cohort were acquired on a single 3T Siemens Biograph mMR PET/MRI scanner. The MRI sequences included a volumetric T1‐weighted magnetization‐prepared rapid acquisition gradient echo (repetition time [TR] = 2000 ms, inversion time [TI] = 870 ms, FoV = 282 × 282 mm, 1.1 mm isotropic resolution) and a 3D T2‐weighted Turbo Spin Echo (TR = 3200 ms, TE = 409 ms, FoV = 282 × 282 mm, 1.1 mm isotropic resolution). Dynamic PET data were acquired in list mode format after intravenous injection of ≈370 MBq florbetapir. Static PET images from 50 to 60 minutes post‐injection were reconstructed from list mode data on Siemens e7 tools with a 3D ordered‐subset expectation‐maximization algorithm consisting of three iterations and 21 subsets, smoothed with a 4 mm Gaussian kernel. For attenuation correction, pseudo CT (pCT) images were synthesized through a widely validated multi‐atlas approach using a database of paired MRI and CT scans. 11 , 14 For comparison with an alternative approach to PET/MRI attenuation correction that is available from the vendor and thus does not require off‐line processing, PET images were also reconstructed directly on the scanner console at the time of scanning using ultrashort echo‐time (UTE) attenuation correction. The pCT reconstruction was used in the main analysis and is the recommended method, as pCT has previously been shown to produce results most consistent with CT compared to UTE and other methods of attenuation correction for PET/MRI. 13
RESEARCH IN CONTEXT
Systematic Review: We reviewed the literature using PubMed and Google Scholar. Articles relating to the Centiloid standardization of amyloid beta (Aβ) positron emission tomography (PET) were reviewed and relevant publications are cited appropriately. While previous studies have highlighted the effects of acquisition and cohort characteristics on Centiloid transformations, we found no studies focusing on conversion of data acquired on PET/magnetic resonance imaging (MRI) scanners to Centiloids.
Interpretation: Our findings show that the Centiloid method can be applied to florbetapir data acquired on PET/MRI scanners, and we provide Centiloid values for the Insight 46 cohort. White matter–referenced standardized uptake value ratio (SUVR) values may be less generalizable than whole cerebellum referenced SUVRs.
Future Directions: This work allows researchers to draw better comparisons between a rich life‐course dataset and other cohorts, helping to elucidate the role of Aβ in Alzheimer's disease. Future methodological work should further our understanding of the differences in florbetapir SUVRs using a white matter reference region between cohorts.
2.3. Imaging analysis
When a “non‐standard” approach is used to generate a Centiloid transformation, values must first be calibrated to the standard approach (STD) used in Klunk et al. 9 before scaling to CL. We compared our SUVR analysis methods to the STD processing using the Standard PiB dataset. We then used the Florbetapir Calibration dataset to calibrate non‐standard florbetapir SUVRs to PiB SUVRs processed using the standard pipeline. The Standard Centiloid processing method, 9 was reimplemented for processing on the Insight 46 dataset using SPM8 (revision number 4290).
All three datasets were processed with the in‐house Geodesic Information Flows (GIF) pipeline. For each individual, the T1‐weighted image was parcellated with GIF, an automated multi‐atlas propagation algorithm. 16 The T1‐weighted and PET images were then co‐registered using an affine block matching registration algorithm. 17 The GIF parcellations were resampled into PET space using the affine transformations generated by the registration. SUVR images were then created by dividing all voxels by mean uptake in whole cerebellum (WC) or subcortical white matter (WM) with an erosion of one PET voxel. As a common approach in many studies is to combine reference regions, we performed a supplementary analysis using SUVRs calculated with a composite reference consisting of WC and WM regions combined. 18 Another version of the GIF pipeline incorporated partial volume correction (PVC), in which the PET image was resampled to native MR space, and the Iterative Yang PVC algorithm was performed using the T1 parcellation, with parameters optimized for our PET/MRI dataset (Gaussian kernel of 6.8 mm full width half maximum, 10 iterations). 19 , 20 , 21 , 22 For consistency, identical PVC parameters were applied to all Centiloid and ADNI datasets.
In all GIF pipelines, mean SUVR values were extracted from a large cortical composite target region that corresponds to the widely used composite region based on FreeSurfer, 7 including frontal, cingulate, lateral parietal, and lateral temporal cortical regions.
We have adapted the nomenclature set out by the Centiloid project to label results from each methodology, in the following format:
where {RADIOTRACER} is either PiB or florbetapir (FBP); {PIPELINE} is STD or GIF processing; {REFERENCE} is WC, WM, or COMP; {PVC} indicates that PVC is applied; and {UNIT} is SUVR or CL. For example, the standard SUVR approach is PiB_STD_WCSUVR. In total, six variants of the GIF SUVR pipeline were evaluated for calibration to the Centiloid scale: GIF_WCSUVR, GIF_WC_PVCSUVR, GIF_WMSUVR, GIF_WM_PVCSUVR, GIF_COMPSUVR, and GIF_COMP_PVCSUVR. SUVR values that are estimated using linear regression are denoted by calcxSUVR.
2.4. Statistical analysis
First, we investigated whether our in‐house GIF pipeline could be calibrated to the Centiloid scale using the procedure laid out in Klunk et al. 9 for “Level 2” analysis of a non‐standard method. The associations between our non‐standard GIF pipelines (y) and PiB_STD_WCSUVR (x) were assessed using linear regression in the Standard PiB dataset, checking that the reliability threshold specified by the Centiloid project (R 2 > 0.7) was satisfied. 9 We then calculated conversion equations using the paired Florbetapir Calibration dataset, calibrating for differences in both radiotracer (florbetapir to PiB) and processing method (GIF to STD) in a single step. FBP SUVRs from each GIF pipeline (y) were regressed against PiB_STD_WCSUVR (x), and the reliability of the conversion process for each of the pipelines was assessed. The slope and intercept of these relationships were used to transform each set of non‐standard SUVRs to estimated SUVRs for the standard pipeline,
calcPiB_STD_WCSUVR.
calcPiB_STD_WCSUVR values were then scaled to Centiloids using equation 1.3b in Klunk et al., 9 substituting group mean values for YC‐0 = 1.00 and AD‐100 = 2.07 (PiB_STD_WCSUVR anchor points published in Navitsky et al. 15 ). Finally, to derive direct conversion equations, Centiloid values were regressed against original SUVR values in a manner similar to Navitsky et al. 15
The transformation from florbetapir SUVR to Centiloid units for each pipeline was then applied to the Insight 46 florbetapir data. SUVR Aβ positivity cutpoints for the Insight 46 PET/MRI dataset were estimated using Gaussian mixture modeling (GMM) in MATLAB R2018a Statistics and Machine Learning toolbox. Models with one, two, and three Gaussians were compared, and the two‐Gaussian model was selected as the optimal model based on Bayesian information criterion. All other statistical analyses were performed in R version 3.6.3. The cutpoint value was defined as the 99th percentile of the lower (Aβ negative) distribution and the equivalent Centiloid was determined. Fleiss’ Kappa was used to report agreement in Aβ positivity between each non‐standard method and the FBP_STD_WCSUVR.
2.5. Complementary analysis in ADNI dataset
The Florbetapir Calibration dataset differs from the Insight 46 dataset in both image acquisition and sample characteristics. To examine the Centiloid conversions in an independent age‐matched PET/CT dataset, T1‐weighted MRI and florbetapir images from 93 controls aged 68 to 72 years were downloaded from ADNI (adni.loni.usc.edu) and processed with GIF pipelines. The conversion equations were then applied to SUVRs in ADNI and Centiloid results compared.
3. RESULTS
UTE console reconstruction produced results that were highly correlated with pCT (see Figure S2 in supporting information). The Insight 46 dataset was also processed with a local implementation of the STD_WCSUVR pipeline, which was validated through replication of the “Level 1” analysis using the Standard PiB dataset (R 2 = 0.9994; see Figure S3 in supporting information).
3.1. Reliability of non‐standard approaches
3.1.1. Standard PiB dataset
Strong correlations (R 2 between 0.91 and 0.99, Figure 1A‐D) were observed between the non‐standard GIF pipelines and the STD_WCSUVR pipeline in the Standard PiB dataset, well above the established Centiloid criteria of R 2 > 0.7. Information on the coefficient of variation (CoV) and effect size of each method is provided in Table S3.
FIGURE 1.
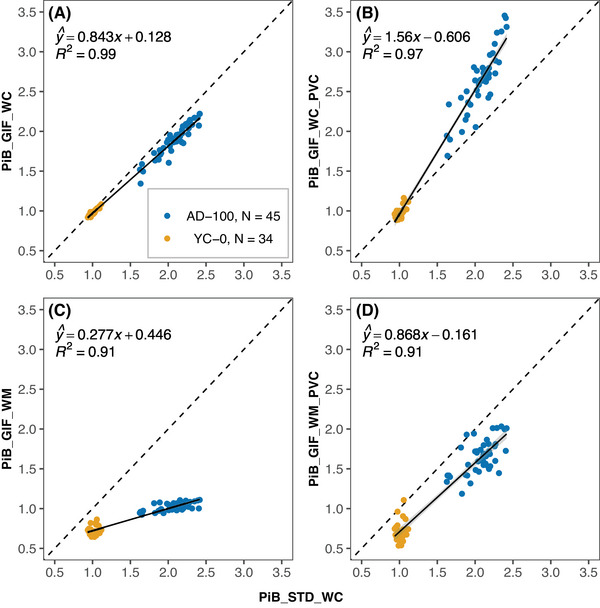
The relationship between non‐standard SUVRs (y‐axis) with GIF_WC (A), GIF_WC_PVC (B), GIF_WM (C), and GIF_WM_PVC (D) processing and standard processing PiB SUVRs (all x‐axis) in the Standard PiB dataset. The dashed line represents x = y and the black line is the linear regression fit with gray area representing 95% confidence interval. AD‐100, Alzheimer's disease group; GIF, Geodesic Information Flows‐based pipeline; PiB, Pittsburgh compound B; PVC, partial volume corrected; STD, standard Centiloid pipeline; SUVR, standardized uptake value ratio; WC, whole cerebellum reference; WM, eroded white matter reference; YC‐0, young control group.
3.1.2. Florbetapir calibration dataset
All GIF pipelines using florbetapir data reached the pre‐specified Centiloid criteria for reliability (all R 2 > 0.7, Figure 2). The second equation in parts A‐D of Figure 2 was used to convert SUVRs from each approach to calcPiB_STD_WCSUVR values, which were then scaled to Centiloids. The relative variance of Centiloid values in young controls are presented in Table S4 in supporting information.
FIGURE 2.
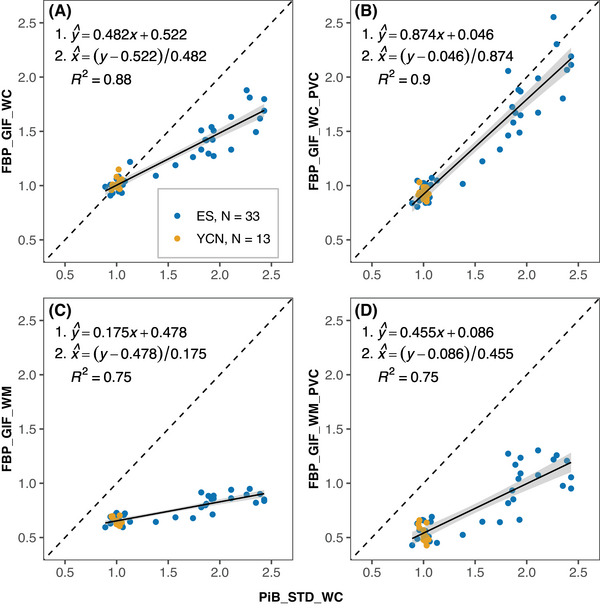
Paired FBP and PiB SUVR data from the Florbetapir Calibration dataset. Plots show the relationship between FBP SUVRs (y‐axis) processed using GIF_WC (A), GIF_WC_PVC (B), GIF_WM (C), and GIF_WM_PVC (D) pipelines and PiB SUVRs with STD_WC processing (all x‐axes). The dashed line represents x = y and the black line is the linear regression fit with gray area representing 95% confidence interval. Conversion equations and R 2 are displayed on the plots. All non‐standard methods exceed the reliability threshold (R 2 > 0.7) set by Klunk et al. 9 and are therefore suitable for Centiloid conversion. ES, elder subjects group; FBP, florbetapir; GIF, Geodesic Information Flows‐based pipeline; PiB, Pittsburgh compound B; PVC, partial volume corrected; STD, standard Centiloid pipeline; SUVR, standardized uptake value ratio; WC, whole cerebellum reference; WM, eroded white matter reference; YCN, young cognitively normal group.
3.2. Centiloid conversion of PET/MR data
3.2.1. Whole cerebellum reference region
In Insight 46 (N = 432), the SUVR cutpoint, Centiloid cutpoint, and Aβ positivity rates for each method were, respectively: 1.150, 19.2, 15.7% (FBP_STD_WCSUVR, Figure 3A); 1.077, 14.2, 16.2% (FBP_GIF_WCSUVR, Figure 3B); and 1.031, 11.8, 23.8% (FBP_GIF_WC_PVCSUVR, Figure 3C). All participants that were positive with FBP_GIF_WC were also positive with FBP_GIF_WC_PVC. Agreement (Kappa scores) in Aβ status between each method compared to the FBP_STD_WC were 0.95 for FBP_GIF_WCSUVR and 0.75 for FBP_GIF_WC_PVCSUVR. Figure 3A‐C shows the direct transformation equations and the resulting distribution of Insight 46 SUVRs and Centiloids are presented in Figure 3D‐E.
FIGURE 3.
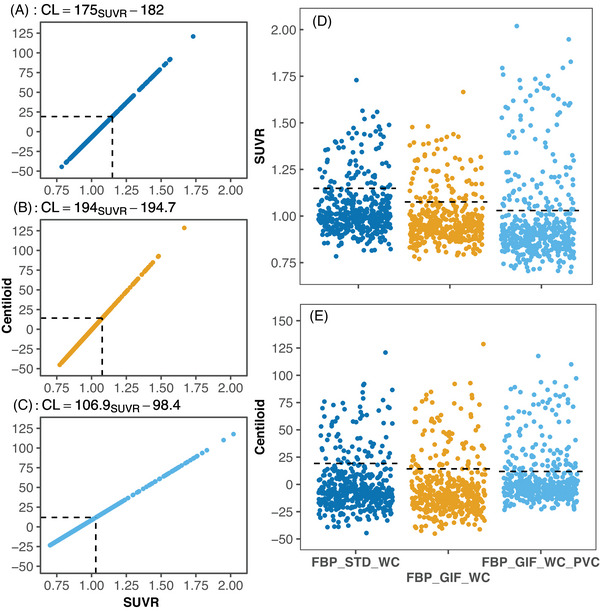
The Centiloid conversion of florbetapir PET/MRI data from Insight 46 (N = 432) processed using the whole cerebellum reference region. The direct linear transformation from SUVR method to Centiloid values and conversion equations are shown for FBP_STD_WC (A), FBP_GIF_WC (B), and FBP_GIF_WC_PVC (C). For comparison, the distribution of SUVR (D), and Centiloid (E) are shown for each processing method. Dashed lines represent cutpoints derived using Gaussian mixture modelling in Insight 46. CL, Centiloid units; FBP, florbetapir; GIF, Geodesic Information Flows‐based pipeline; MRI, magnetic resonance imaging; PET, positron emission tomography; PVC, partial volume corrected; SD, standard deviation; STD, standard Centiloid pipeline; SUVR, standardized uptake value ratio; WC, whole cerebellum reference.
3.2.2. Eroded WM reference region
The Aβ positivity rates were 18.3% (FBP_GIF_WMSUVR, Kappa = 0.75) and 18.1% (FBP_GIF_WM_PVCSUVR, Kappa = 0.75). Converting the FBP_GIF_WMSUVR Insight 46 data, the SUVR cutpoint of 0.610 corresponded to –23.0 CL and the mean (SD) Centiloid value was –48.3 (39.5) (see Figure 4A). For FBP_GIF_WM_PVCSUVR, the SUVR cutpoint of 0.671 equated to +26.7 CL, with a mean (SD) Centiloid value of +10.5 (30.0; see Figure 4C). Post hoc analyses were performed to investigate these unexpected results.
FIGURE 4.
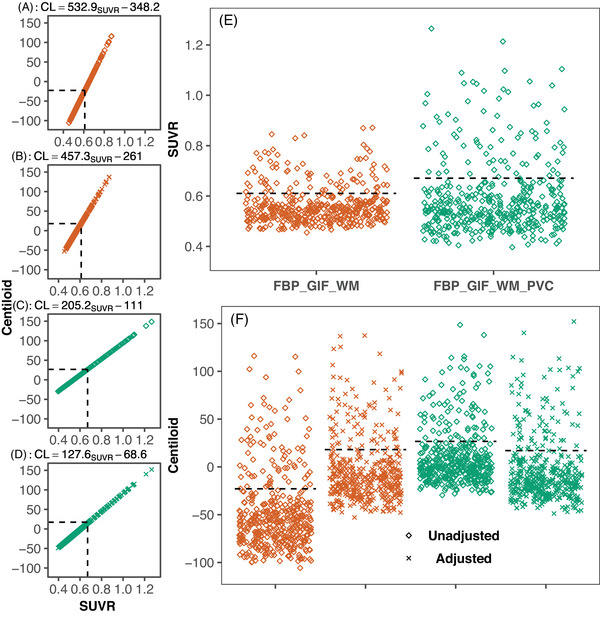
The Centiloid conversion of florbetapir PET/MRI SUVRs from Insight 46 (N = 432) processed with an eroded white matter reference region, without (A‐B) and with PVC (C‐D), with both unadjusted (A, C) and adjusted Centiloid values (B, D), conversion equations and cutpoints are presented. SUVR values (E) can be compared to both adjusted and unadjusted Centiloid values (F). Dashed black lines represent cutpoint values derived using Gaussian mixture modelling in Insight 46. Dashed lines represent cutpoints derived using Gaussian mixture modelling in Insight 46. CL, Centiloid units; FBP, florbetapir; GIF, Geodesic Information Flows‐based pipeline; MRI, magnetic resonance imaging; PET, positron emission tomography; PVC, partial volume corrected; SD, standard deviation; STD, standard Centiloid pipeline; SUVR, standardized uptake value ratio; WC, whole cerebellum reference.
Further analyses indicated a differential relationship between WM and WC uptake in Insight 46 compared to the Florbetapir Calibration dataset. The regression line between FBP_GIF_WMSUVR (y) and FBP_GIF_WCSUVR (x) had a smaller slope and higher intercept in the Florbetapir Calibration compared to the Insight 46 dataset (see Figure S5 in supporting information). Therefore, the linear equation from the Florbetapir Calibration dataset gives higher estimates of calcFBP_GIF_WMSUVR than is appropriate for the Insight 46 dataset. As a result, the reverse transformation from FBP_GIF_WMSUVR to calcFBP_GIF_WCSUVR leads to underestimated Centiloid values in Insight 46. To adjust for this, we implemented a dataset‐specific adjustment to convert WM normalized SUVRs from Insight 46 to Centiloids. To bring the WM values in line with the GIF_WCSUVR, we added an initial step to convert WM‐referenced SUVRs to estimated WC values. Figure 5 outlines the process for conversion without (Figure 5A) and with the initial adjustment (Figure 5B). The adjustment equations for non‐PVC and PVC SUVRs were as follows: calcFBP_GIF_WCSUVR = (FBP_GIF_WMSUVR ‐ 0.145)/0.424 and calcFBP_GIF_WCSUVR = (FBP_GIF_WM_PVCSUVR + 0.234)/0.838. After this adjustment, the FBP_GIF_WCSUVR to Centiloid equation was applied to adjusted values (CL = calcFBP_GIF_WCSUVR × 194.0 – 194.7). The adjusted cutpoints were 18.1 CL and 17.0 CL for FBP_GIF_WMSUVR and FBP_GIF_WM_PVCSUVR, respectively (see Figure 4B and 4D). The distibution of SUVRs and Centiloids (with and without adjustment) are presented in Figure 4E‐F.
FIGURE 5.
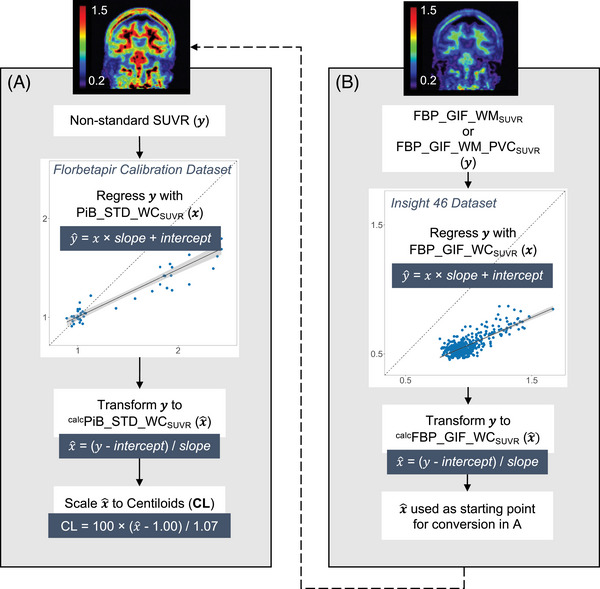
Diagram outlining the steps for calibration of non‐standard whole cerebellum referenced SUVRs to Centiloids (A). An additional adjustment is required for conversion of SUVRs using a white matter reference region (B). Dashed lines represent cutpoints derived using Gaussian mixture modelling in Insight 46. CL, Centiloid units; FBP, florbetapir; GIF, Geodesic Information Flows‐based pipeline; PVC, partial volume corrected; STD, standard Centiloid pipeline; SUVR, standardized uptake value ratio; WC, whole cerebellum reference.
Using the composite reference region (GIF_COMP), Centiloid values fell between values from pipelines using either region separately, both with and without adjustment (see Figure and Text S6 in supporting information).
3.3. Complementary analysis in ADNI dataset
In the ADNI dataset, the GMM‐derived cutpoint for FBP_GIF_WCSUVR was 1.123, which scaled to 23 CL. For FBP_GIF_WMSUVR, the SUVR cutpoint was 0.691, which equated to 20.3 CL using the Centiloid transformation. The relationship between WM and WC uptake in ADNI was similar to the Florbetapir Calibration dataset (see Figure S7 in supporting information).
4. DISCUSSION
In this study, we found that florbetapir data acquired on a combined PET/MRI scanner—in particular SUVR using a WC reference—can successfully be transformed to the Centiloid scale. After calibration of four non‐standard SUVR methods to Centiloids, including the use of an eroded WM reference region and PVC, we converted florbetapir PET/MRI data from a large cohort of ≈70‐year‐old individuals. For SUVRs using a WC reference, Aβ positivity cutpoints fell within a range of 11.8 to 19.2 Centiloids. These data‐driven cutpoints are consistent with other studies using different methodologies. 23 , 24 , 25 , 26 However, the same conversion process when applied to WM‐referenced SUVRs resulted in unexpected cutpoint values of –23.0 Centiloids without and +26.7 Centiloids with PVC. These results are likely due to a differential relationship between WM and WC uptake between the Florbetapir Calibration and Insight 46 datasets. We therefore introduced an adjustment step based on this relationship before scaling Insight 46 data to Centiloids. This resulted in more plausible cutpoints of 18.1 Centiloids without and 17.0 Centiloids with PVC.
The Centiloid method compresses or stretches results into a similar range, and it is important to report information regarding the reliability of the conversion and precision of each non‐standard approach. 9 In the current study, we applied the Centiloid conversion to SUVRs with an eroded WM reference region and with PVC applied. These extra degrees of separation from the Standard Centiloid processing approach could reduce the reliability of transformations. In the Standard PiB dataset, SUVRs from all GIF pipelines were strongly associated with the Standard Centiloid processing (all R 2 > 0.90), indicating a reliable conversion between methods. The linear association between values from GIF WC and Standard Centiloid pipelines was particularly strong (R 2 = 0.99), with a slight underestimation for the GIF pipeline compared to the Standard Centiloid processing approach (Figure 1A). This underestimation could be due to greater inclusion of WM within the Montreal Neurological Institute space Standard Centiloid target region in the AD group (due to gray matter atrophy), compared to the subject‐specific cortical GIF target region. In the Florbetapir Calibration dataset, the conversion across both radiotracer and processing method were highly reliable for WC reference (R 2 > 0.87, Figures 2A and 2B) and lower, but still above the Centiloid threshold for WM reference (R 2 = 0.75, Figures 2C and 2D). In young controls presumed to have no Aβ accumulation, the relative variance between non‐standard Centiloids compared to Standard PiB Centiloids reflects both the relative dynamic range and precision of the non‐standard method. 9 Similar to Navitsky et al. 15 we found that florbetapir SUVRs had a dynamic range of about half that of PiB when using the Standard Centiloid processing (slope = 0.482 in Figure 3A), resulting in a doubling of the variance ratio between florbetapir and PiB SUVRs when scaling to Centiloids. We found that a WM reference reduced the dynamic range of florbetapir further (slope = 0.175 in Figure 2C) and PVC increased it for both WC (slope = 0.874 in Figure 2B) and WM referenced values (slope = 0.455 in Figure 2D), although PVC also increased variability in the YCN group.
While Centiloid values can appear more consistent than SUVR, this harmonization process is still affected by the analysis technique and cohort. 10 In the current study, we could not generalize between datasets when converting SUVRs with an eroded WM reference region without implementing a dataset‐specific linear adjustment. This adjustment does not change Aβ positivity rates but brings WM‐referenced SUVRs into the same range as WC‐referenced SUVRs before scaling to Centiloids. We hypothesize two potential sources of variation that could be contributing to the differences between datasets: (1) the method of image acquisition or reconstruction and (2) the biological characteristics of the cohorts. Neither of these are accounted for with the Centiloid approach, in which equations are calculated on a calibration dataset and applied to an independent dataset. Studies have previously observed lower longitudinal intra‐individual variability when using a WM reference region compared to the cerebellum. This, in part, could be due to increased noise and signal dropout that occurs in peripheral brain structures, such as the cerebellum, which are positioned near the edge of the FoV. 5 , 27 , 28 , 29 , 30 , 31 The PET/MRI scanner used in Insight 46 has a large axial FoV (25.8 cm) compared to the scanners used for the Florbetapir Calibration and ADNI datasets, which were typically ≈16 cm. 32 It is possible that these differences in axial FoV contribute to the differing relationships between reference region uptake observed in PET/MRI and PET/CT datasets (see WM to WC ratios in Text S7). However, given that the WC‐referenced SUVRs appear more similar across cohorts compared to WM‐referenced SUVRs, we believe the differences between datasets are driven mostly by WM signal, rather than the WC. Differences in attenuation correction between PET/MRI and PET/CT datasets could be a contributing factor, although differences in attenuation correction are mainly found in the cerebellum, so we do not expect this to be contributing to the observed bias within the WM values. Biological characteristics of the cohorts, including age and disease status, can affect radiotracer dynamics through changes in cerebral blood flow, which differentially affects cerebellum and WM regions. 33 , 34 There are differences between the Florbetapir Calibration dataset (a wide range of age and disease status from control to AD) and Insight 46 (community aging cohort in tight age range 68–72), and these difference in biological characteristics could contribute to the differential relationship between WM and cerebellum uptake. 35 We aimed to address some of these differences with a subset of ADNI (PET/CT) data consisting of controls matched in age to Insight 46. The relationship between WM and WC uptake in the ADNI dataset was more similar to the Florbetapir Calibration dataset than the Insight 46 dataset, suggesting that PET/MR– differences may be playing a role. However, the WM‐referenced mean Centiloid (–3.1) is lower than that of WC‐referenced values (12.9) in the ADNI dataset, suggesting there may also be some underlying difference between Florbetapir Calibration and ADNI. ADNI has stricter recruitment criteria against WM disease compared to the Insight 46 community sample, which could result in differences in florbetapir WM uptake. 36 , 37 Several studies have attributed high Aβ tracer binding in WM to an affinity for myelin basic protein and have explored their use in multiple sclerosis. 37 , 38 , 39 , 40 , 41 , 42 This myelin involvement is a potential confounder of WM as a reference region and could lead to differences in SUVR dependent on sex, age, and WM disease. 43 Furthermore, as WM uptake is higher in 18F‐labelled tracers compared to PiB, these differences could affect the Centiloid conversion of WM‐referenced results. 44
A limitation of this paper is that we are unable to identify the exact source of variation between datasets, which leads to the lack of generalizable WM‐referenced SUVRs. Ideally sources of variation from acquisition and sample could be characterized with further paired (PiB and florbetapir) calibration datasets controlling for all other factors; however, it is unfeasible, due to PiB radiotracer availability, to collect these data. We found a balance between logistics and generalizability by adjusting GIF WM‐referenced SUVR values to GIF WC‐referenced values within the Insight 46 dataset, then linking these values to the Centiloid scale using the WC‐referenced equations from the independent calibration dataset. Another limitation is that the PVC parameters in the pipeline were kept consistent between datasets, using the parameters optimized for the PET/MRI florbetapir dataset. 20 While the Siemens Biograph mMR PET/MRI scanner used in the current study has similar spatial resolution to other modern PET/CT equivalents, 45 the older scanners (e.g., Siemens HR+) used to acquire the Florbetapir Calibration dataset data have lower spatial resolution and are therefore undercorrected for partial volume effects. The reliability of the Centiloid conversion of PVC values may also be reduced due to use of PET images from three different scanners in the Florbetapir Calibration dataset. The PiB scans will also have slightly poorer resolution compared to florbetapir due to the higher energy of Carbon‐11 positrons. 46 We also note that the correlation between FBP_GIF_WC and FBP_GIF_WM SUVRs in the Insight 46 dataset (R 2 = 0.6 for PVC and non‐PVC, Figure S5) is slightly lower than in the calibration set (R 2 = 0.71 for non‐PVC and 0.68 for PVC). This relationship is used for the adjustment of WM Centiloids and should be considered when using adjusted WM Centiloid values.
Future work will explore the Centiloid implementation with longitudinal follow‐up data in this PET/MRI dataset. The relationship between WM and cerebellum florbetapir uptake will be examined further, which will be important for the standardization of SUVR results using a WM reference region.
In summary, we implemented the Centiloid scale in a large florbetapir PET/MRI dataset from a community birth cohort data with full life course data. Our results suggest that the Centiloid conversion of WC‐referenced SUVRs can be generalized to PET/MRI datasets. However, careful consideration to underlying differences between datasets must be given, as they can produce implausible conversions to Centiloids, particularly when using a WM reference region. We show that a linear dataset‐specific adjustment can facilitate conversion of values should differences between datasets arise.
AUTHOR CONTRIBUTIONS
William Coath: conceptualization, methodology, visualization, formal analysis, writing—original draft; Marc Modat: methodology, software, writing—review & editing; M. Jorge Cardoso: methodology, software, writing—review & editing; Pawel J. Markiewicz: methodology, software, writing—review & editing; Christopher A. Lane: investigation (data acquisition), writing—review & editing; Thomas D. Parker: investigation (data acquisition), writing—review & editing; Ashvini Keshavan: investigation (data acquisition), writing—review & editing; Sarah M. Buchanan: investigation (data acquisition), writing—review & editing; Sarah E. Keuss: investigation (data acquisition), writing—review & editing; Matthew J. Harris: investigation (data acquisition), writing—review & editing; Ninon Burgos: methodology, software, writing—review & editing; John Dickson: resources, methodology, writing—review & editing; Anna Barnes: methodology, writing—review & editing; David L. Thomas: methodology, writing—review & editing; Daniel Beasley: software, writing—review & editing; Ian B. Malone: software, methodology, writing—review & editing; Andrew Wongh: project administration, writing—review & editing; Kjell Erlandsson: methodology, software, writing—review & editing; Benjamin A. Thomas: software, writing—review & editing; Michael Schöll: supervision, writing—review & editing; Sebastien Ourselin: conceptualization, writing—review & editing; Marcus Richards: conceptualization, funding acquisition, writing—review & editing; Nick C. Fox: resources, conceptualization, funding acquisition, writing—review & editing; Jonathan M. Schott: resources, conceptualization, supervision, funding acquisition, writing—review & editing; David M. Cash: conceptualization, methodology, supervision, writing—review & editing.
CONFLICT OF INTEREST STATEMENT
NCF's research group has received payment for consultancy or for conducting studies from Biogen, Eli Lilly Research Laboratories, GE Healthcare, and Roche. NCF receives no personal compensation for the aforementioned activities. JMS has received research funding from Avid Radiopharmaceuticals (a wholly owned subsidiary of Eli Lilly); has consulted for Roche Pharmaceuticals, Biogen, Merck, and Eli Lilly; given educational lectures sponsored by GE Healthcare, Eli Lilly, and Biogen; and serves on a Data Safety Monitoring Committee for Axon Neuroscience SE. All other authors report no competing interests.Author disclosures are available in the supporting information.
CONSENT STATEMENT
Written informed consent was obtained from all individual participants included in the study.
Supporting information
Supp Information 1
Supp Information 2
ACKNOWLEDGMENTS
The authors are very grateful to those study members who helped in the design of the study through focus groups, and to the participants for their contributions to Insight 46 and their commitment to research over the past seven decades. The authors are grateful to the radiographers and nuclear medicine physicians at the University College London Institute of Nuclear Medicine; the staff at the Leonard Wolfson Experimental Neurology Centre at University College London; the neuroradiologists Dr. Chandrashekar Hoskote and Dr. Sachit Shah for providing clinical reads for the MRI scans; the DRC trials team for assistance with imaging QC; Dan Marcus and Rick Herrick for assistance with XNAT; and Dr. Philip Curran for assistance with data sharing with the MRC Unit for Lifelong Health and Ageing. The authors are also particularly indebted to the support of the late Chris Clark of Avid Radiopharmaceuticals who championed this study from its outset.
This study is principally funded by grants from Alzheimer's Research UK (ARUK‐PG2014‐1946, ARUK‐PG2017‐1946), the Medical Research Council Dementias Platform UK (CSUB19166), and the Wolfson Foundation (PR/ylr/18575). Florbetapir amyloid tracer is provided by AVID Radiopharmaceuticals (a wholly owned subsidiary of Eli Lilly), which had no part in the design, conduct, or analysis of the study. The National Survey of Health and Development is funded by the Medical Research Council (MC_UU_12019/1, MC_UU_12019/3). AK was supported by a Wolfson Clinical Research Fellowship and a Weston Brain Institute and Selfridges Group Foundation award (UB17005). TDP was supported by a Wellcome Trust Clinical Research Fellowship (200109/Z/15/Z). The NSHD, MR, and AW are funded by the Medical Research Council (MC_UU_00019/1, MC_UU_00019/3, and additionally MC_UU_12019/3 for MR). NCF is supported by UK Dementia Research Institute at University College London, Medical Research Council, National Institute for Health Research (Senior Investigator award), and Engineering and Physical Sciences Research Council. DMC is supported by the UK Dementia Research Institute, which receives its funding from DRI Ltd, funded by the UK Medical Research Council, Alzheimer's Society and Alzheimer's Research UK, as well as Alzheimer's Research UK (ARUK‐PG2017‐1946) and the UCL/UCLH NIHR Biomedical Research Centre. JMS is supported by University College London Hospitals Biomedical Research Centre, Engineering and Physical Sciences Research Council (EP/J020990/1), British Heart Foundation (PG/17/90/33415), and EU's Horizon 2020 research and innovation programme (666992). NCF and JMS are supported by the National Institute for Health Research Queen Square Dementia Biomedical Research Unit and the Leonard Wolfson Experimental Neurology Centre. Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI; National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH‐12‐2‐0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie; Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol‐Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann‐La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC; Johnson & Johnson Pharmaceutical Research & Development LLC; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
COLLABORATORS
ACKNOWLEDGEMENT LIST FOR ADNI PUBLICATIONS
The Data and Publications Committee, in keeping with the publication policies adopted by the ADNI Steering Committee, here provides lists for standardized acknowledgement. The list consists of three parts: I. ADNI Infrastructure Investigators and Site Investigators, II. DOD ADNI Infrastructure Investigators and Site Investigators, and III. ADNI Depression Infrastructure Investigators and Site Investigators. Infrastructure Investigators represent the names responsible for leadership and infrastructure. Site Investigators represent the names of individuals at each recruiting site. All papers, including methodological papers, should have an acknowledgement list that consists of Infrastructure Investigators plus the FULL list.
I. ADNI I, GO, II and III
Part A: Leadership and Infrastructure
Principal Investigator
| Michael W. Weiner, MD | University of California, San Francisco |
ATRI PI and Director of Coordinating Center Clinical Core
| Paul Aisen, MD | University of Southern California |
Co PI of Clinical Core
| Ronald Petersen, MD, PhD | Mayo Clinic, Rochester |
Executive Committee
| Michael W. Weiner, MD | University of California, San Francisco |
| Paul Aisen, MD | University of Southern California |
| Ronald Petersen, MD, PhD | Mayo Clinic, Rochester |
| Clifford R. Jack, Jr., MD | Mayo Clinic, Rochester |
| William Jagust, MD | University of California, Berkeley |
| John Q. Trojanowki, MD, PhD | University of Pennsylvania |
| Arthur W. Toga, PhD | University of Southern California |
| Laurel Beckett, PhD | University of California, Davis |
| Robert C. Green, MD, MPH | Brigham and Women's Hospital/Harvard Medical School |
| Andrew J. Saykin, PsyD | Indiana University |
| John C. Morris, MD | Washington University St. Louis |
| Richard J. Perrin, MD, PhD | Washington University St. Louis |
| Leslie M. Shaw, PhD | University of Pennsylvania |
ADNI External Advisory Board (ESAB)
| Zaven Khachaturian, PhD | Prevent Alzheimer's Disease 2020 (Chair) |
| Maria Carrillo, PhD | Alzheimer's Association |
| William Potter, MD | National Institute of Mental Health |
| Lisa Barnes, PhD | Rush University |
| Marie Bernard, MD | NIA |
| Hector González | University of California, San Diego |
| Carole Ho | Denali Therapeutics |
| John K. Hsiao, MD | NIH |
| Jonathan Jackson, PhD | Massachusetts General Hospital |
| Eliezer Masliah, MD | NIA |
| Donna Masterman, MD | Biogen |
| Ozioma Okonkwo, PhD | University of Wisconsin, Madison |
| Richard Perrin, MD | Washington University |
| Laurie Ryan, PhD | NIA |
| Nina Silverberg, PhD | NIA |
ADNI 3 Private Partner Scientific Board (PPSB)
| Adam Fleisher, MD | Eli Lilly (Chair) |
Administrative Core ‐ Northern California Institute for Research & Education (NCIRE / The Vererans Health Research Institute)
| Michael W. Weiner, MD | University of California, San Francisco |
| Diana Truran Sacrey | NCIRE / The Vererans Health Research Institute |
| Juliet Fockler | University of California, San Francisco |
| Cat Conti, BA | NCIRE / The Vererans Health Research Institute |
| Dallas Veitch, PhD | NCIRE / The Vererans Health Research Institute |
| John Neuhaus, PhD | University of California, San Francisco |
| Chengshi Jin, PhD | University of California, San Francisco |
| Rachel Nosheny, PhD | University of California, San Francisco |
| Miriam Ashford, PhD | NCIRE / The Vererans Health Research Institute |
| Derek Flenniken | NCIRE / The Vererans Health Research Institute |
| Adrienne Kormos | NCIRE / The Vererans Health Research Institute |
Data and Publications Committee
| Robert C. Green, MD, MPH | BWH/HMS (Chair) |
Resource Allocation Review Committee
| Tom Montine, MD, PhD | University of Washington (Chair) |
| Cat Conti, BA | NCIRE / The Vererans Health Research Institute |
Clinical Core Leaders and Key Personnel
| Ronald Petersen, MD, PhD | Mayo Clinic, Rochester (Core PI) |
| Paul Aisen, MD | University of Southern California (Core PI) |
| Michael Rafii, MD, PhD | University of Southern California |
| Rema Raman, PhD | University of Southern California |
| Gustavo Jimenez, MBS | University of Southern California |
| Michael Donohue, PhD | University of Southern California |
| Devon Gessert, BS | University of Southern California |
| Jennifer Salazar, MBS | University of Southern California |
| Caileigh Zimmerman, MS | University of Southern California |
| Yuliana Cabrera, BS | University of Southern California |
| Sarah Walter, MSc | University of Southern California |
| Garrett Miller, MS | University of Southern California |
| Godfrey Coker, MBA, MPH | University of Southern California |
| Taylor Clanton, MPH | University of Southern California |
| Lindsey Hergesheimer, BS | University of Southern California |
| Stephanie Smith, BS | University of Southern California |
| Olusegun Adegoke, MSc | University of Southern California |
| Payam Mahboubi, MPH | University of Southern California |
| Shelley Moore, BA | University of Southern California |
| Jeremy Pizzola, BA | University of Southern California |
| Elizabeth Shaffer, BS | University of Southern California |
| Brittany Sloan, BA | University of Southern California |
Biostatistics Core Leaders and Key Personnel
| Laurel Beckett, PhD | University of California, Davis (Core PI) |
| Danielle Harvey, PhD | University of California, Davis |
| Michael Donohue, PhD | University of Southern California |
MRI Core Leaders and Key Personnel
| Clifford R. Jack, Jr., MD | Mayo Clinic, Rochester (Core PI) |
| Arvin Forghanian‐Arani, PhD | Mayo Clinic |
| Bret Borowski, RTR | Mayo Clinic |
| Chad Ward, | Mayo Clinic |
| Christopher Schwarz, PhD | Mayo Clinic |
| David Jones, MD | Mayo Clinic |
| Jeff Gunter, PhD | Mayo Clinic |
| Kejal Kantarci, MD | Mayo Clinic |
| Matthew Senjem, MS | Mayo Clinic |
| Prashanthi Vemuri, PhD | Mayo Clinic |
| Robert Reid, PhD | Mayo Clinic |
| Nick C. Fox, MD | University College London |
| Ian Malone, PhD | University College London |
| Paul Thompson, PhD | University of Southern California School of Medicine |
| Sophia I. Thomopoulos, BS | University of Southern California School of Medicine |
| Talia M. Nir, PhD | University of Southern California School of Medicine |
| Neda Jahanshad, PhD | University of Southern California School of Medicine |
| Charles DeCarli, MD | University of California, Davis |
| Alexander Knaack, MS | University of California, Davis |
| Evan Fletcher, PhD | University of California, Davis |
| Danielle Harvey, PhD | University of California, Davis |
| Duygu Tosun‐Turgut, PhD | University of California, San Francisco |
| Stephanie Rossi Chen, BA | NCIRE / The Vererans Health Research Institute |
| Mark Choe, BS | NCIRE / The Vererans Health Research Institute |
| Karen Crawford | University of Southern California School of Medicine |
| Paul A. Yushkevich, PhD | University of Pennsylvania |
| Sandhitsu Das, PhD | University of Pennsylvania |
PET Core Leaders and Key Personnel
| William Jagust, MD | University of California, Berkeley (Core PI) |
| Robert A. Koeppe, PhD | University of Michigan |
| Eric M. Reiman, MD | Banner Alzheimer's Institute |
| Kewei Chen, PhD | Banner Alzheimer's Institute |
| Chet Mathis, MD | University of Pittsburgh |
| Susan Landau, PhD | University of California, Berkeley |
Neuropathology Core Leaders and Key Personnel
| John C. Morris, MD | Washington University St. Louis |
| Richard Perrin MD | Washington University St. Louis |
| Nigel J. Cairns, PhD, FRCPath | Washington University St. Louis—Past Investigator |
| Erin Householder, MS | Washington University St. Louis |
| Erin Franklin, MS | Washington University St. Louis |
| Haley Bernhardt, BA, R. EEG T | Washington University St. Louis |
| Lisa Taylor‐Reinwald, BA, HTL | Washington University St. Louis (ASCP) – Past Investigator |
Biomarkers Core Leaders and Key Personnel
| Leslie M. Shaw, PhD | Perelman School of Medicine, University of Pennsylvania (co‐PI) |
| John Q. Trojanowki, MD, PhD | Perelman School of Medicine, University of Pennsylvania (co‐PI) |
| Magdalena Korecka, PhD | Perelman School of Medicine, University of Pennsylvania |
| Michal Figurski, PhD | Perelman School of Medicine, University of Pennsylvania |
Informatics Core Leaders and Key Personnel
| Arthur W. Toga, PhD | University of Southern California (Core PI) |
| Karen Crawford | University of Southern California |
| Scott Neu, PhD | University of Southern California |
Genetics Core Leaders and Key Personnel
| Andrew J. Saykin, PsyD | Indiana University School of Medicine (Core PI) |
| Kwangsik Nho, PhD | Indiana University School of Medicine |
| Shannon L. Risacher, PhD | Indiana University School of Medicine |
| Liana G. Apostolova, MD | Indiana University School of Medicine |
| Li Shen, PhD | UPenn School of Medicine |
| Tatiana M. Foroud, PhD | NCRAD/Indiana University School of Medicine |
| Kelly Nudelman, PhD | NCRAD/Indiana University School of Medicine |
| Kelley Faber, MS, CCRC | NCRAD/Indiana University School of Medicine |
| Kristi Wilmes, MS, CCRP | NCRAD/Indiana University School of Medicine |
Initial Concept Planning & Development
| Michael W. Weiner, MD | University of California, San Francisco |
| Leon Thal, MD – Past Investigator | University of California, San Diego |
| Zaven Khachaturian, PhD | Prevent Alzheimer's Disease 2020 |
NIA
| John K. Hsiao, MD | National Institute on Aging |
Part B: Investigators By Site
| Oregon Health & Science University: | Lon S. Schneider, MD |
| Lisa C. Silbert, MD | Sonia Pawluczyk, MD |
| Betty Lind, BS | Mauricio Becerra, MD |
| Rachel Crissey | Liberty Teodoro, RN |
| Jeffrey A. Kaye, MD, A – Past Investigator | Karen Dagerman, MS |
| Raina Carter, BA – Past Investigator | Bryan M. Spann, DO, PhD – Past Investigator |
| Sara Dolen, BS – Past Investigator | |
| Joseph Quinn, MD – Past Investigator | University of California–San Diego: |
| James Brewer, MD, PhD | |
| University of Southern California: | Helen Vanderswag, RN |
Adam Fleisher, MD – Past Investigator
University of Michigan:
Jaimie Ziolkowski, MA, BS, TLLP
Judith L. Heidebrink, MD, MS
Lisa Zbizek‐Nulph, MS
Joanne L. Lord, LPN, BA, CCRC – Past
Investigator
Lisa Zbizek‐Nulph, MS, CCRP
Mayo Clinic, Rochester:
Ronald Petersen, MD, PhD
Sara S. Mason, RN
Colleen S. Albers, RN
David Knopman, MD
Kris Johnson, RN
Baylor College of Medicine:
Javier Villanueva‐Meyer, MD
Valory Pavlik, PhD
Nathaniel Pacini, MA
Ashley Lamb, MA
Joseph S. Kass, MD, LD, FAAN
Rachelle S. Doody, MD, PhD – Past Investigator
Victoria Shibley, MS – Past Investigator
Munir Chowdhury, MBBS, MS – Past Investigator
Susan Rountree, MD – Past Investigator
Mimi Dang, MD – Past Investigator
Columbia University Medical Center:
Yaakov Stern, PhD
Lawrence S. Honig, MD, PhD
Akiva Mintz, MD, PhD
Washington University, St. Louis:
Beau Ances, MD, PhD, MSc
John C. Morris, MD
David Winkfield, BS
Maria Carroll, RN, MSN, GCNS‐BC
Georgia Stobbs‐Cucchi, RN, CCRP—Past
Investigator
Angela Oliver, RN, BSN, MSG – Past Investigator
Mary L. Creech, RN, MSW – Past Investigator
Mark A. Mintun, MD – Past Investigator
Stacy Schneider, APRN, BC, GNP – Past
Investigator
University of Alabama ‐ Birmingham:
David Geldmacher, MD
Marissa Natelson Love, MD
Randall Griffith, PhD, ABPP – Past Investigator
David Clark, MD – Past Investigator
John Brockington, MD – Past Investigator
Daniel Marson, JD, PhD – Past Investigator
Mount Sinai School of Medicine:
Hillel Grossman, MD
Martin A. Goldstein, MD
Jonathan Greenberg, BA
Effie Mitsis, PhD – Past Investigator
Rush University Medical Center:
Raj C. Shah, MD
Melissa Lamar, PhD
Patricia Samuels
Wien Center:
Ranjan Duara, MD
Maria T. Greig‐Custo, MD
Rosemarie Rodriguez, PhD
Johns Hopkins University:
Marilyn Albert, PhD
Chiadi Onyike, MD
Leonie Farrington, RN
Scott Rudow, BS
Rottislav Brichko, BS
Stephanie Kielb, BS – Past Investigator
University of South Florida: USF Health Byrd Alzheimer's Institute:
Amanda Smith, MD
Balebail Ashok Raj, MD – Past Investigator
Kristin Fargher, MD – Past Investigator
New York University:
Martin Sadowski, MD, PhD
Thomas Wisniewski, MD
Melanie Shulman, MD
Arline Faustin, MD
Julia Rao, PhD
Karen M. Castro, BA
Anaztasia Ulysse, BA
Shannon Chen, BA
Mohammed O. Sheikh, MD – Past Investigator
Jamika Singleton‐Garvin, CCRP – Past Investigator
Duke University Medical Center:
P. Murali Doraiswamy, MBBS, FRCP
Jeffrey R. Petrella, MD
Olga James, MD
Terence Z. Wong, MD
Salvador Borges‐Neto, MD – Past Investigator
University of Pennsylvania:
Jason H. Karlawish, MD
David A. Wolk, MD
Sanjeev Vaishnavi, MD
Christopher M. Clark, MD – Past Investigator
Steven E. Arnold, MD – Past Investigator
University of Kentucky:
Charles D. Smith, MD
Gregory A. Jicha, MD, PhD
Riham El Khouli, MD
Flavius D. Raslau, MD
University of Pittsburgh:
Oscar L. Lopez, MD
MaryAnn Oakley, MA
Donna M. Simpson, CRNP, MPH
University of Rochester Medical Center:
Anton P. Porsteinsson, MD
Kim Martin, RN
Nancy Kowalski, MS, RNC
Melanie Keltz, RN
Bonnie S. Goldstein, MS, NP – Past Investigator
Kelly M. Makino, BS – Past Investigator
M. Saleem Ismail, MD – Past Investigator
Connie Brand, RN – Past Investigator
University of California Irvine IMIND:
Gaby Thai, MD
Aimee Pierce, MD
Beatriz Yanez, RN
Elizabeth Sosa, PhD
Megan Witbracht, PhD
University of Texas Southwestern Medical School:
Brendan Kelley, MD
Trung Nguyen, MD
Kyle Womack, MD
Dana Mathews, MD, PhD– Past Investigator
Mary Quiceno, MD– Past Investigator
Emory University:
Allan I. Levey, MD, PhD
James J. Lah, MD, PhD
Ihab Hajjar, MD
Janet S. Cellar, DNP, PMHCNS‐BC – Past
Investigator
University of Kansas, Medical Center:
Jeffrey M. Burns, MD
Russell H. Swerdlow, MD
William M. Brooks, PhD
University of California, Los Angeles:
Daniel H.S. Silverman, MD, PhD
Sarah Kremen, MD
Liana Apostolova, MD – Past Investigator
Kathleen Tingus, PhD – Past Investigator
Po H. Lu, PsyD – Past Investigator
George Bartzokis, MD – Past Investigator
Ellen Woo, PhD – Past Investigator
Edmond Teng, MD, PhD – Past Investigator
Mayo Clinic, Jacksonville:
Neill R Graff‐Radford, MBBCH, FRCP (London)
Francine Parfitt, MSH, CCRC Kim Poki‐Walker, BA
Indiana University:
Martin R. Farlow, MD
Ann Marie Hake, MD – Past Investigator
Brandy R. Matthews, MD – Past Investigator
Jared R. Brosch, MD
Scott Herring, RN, CCRC
Yale University School of Medicine:
Christopher H. van Dyck, MD
Adam P. Mecca, MD, PhD
Adam P. Mecca, MD, PhD
Susan P. Good, APRN
Martha G. MacAvoy, PhD
Richard E. Carson, PhD
Pradeep Varma, MD
McGill Univ., Montreal‐Jewish General Hospital:
Howard Chertkow, MD
Susan Vaitekunis, MD
Chris Hosein, MEd
Sunnybrook Health Sciences, Ontario:
Sandra Black, MD, FRCPC
Bojana Stefanovic, PhD
Chris (Chinthaka) Heyn, BSC, PhD, MD, FRCPC
U.B.C. Clinic for AD & Related Disorders:
Ging‐Yuek Robin Hsiung, MD, MHSc, FRCPC
Ellen Kim, BA
Benita Mudge, BS
Vesna Sossi, PhD
Howard Feldman, MD, FRCPC – Past Investigator
Michele Assaly, MA – Past Investigator
St. Joseph's Health Care: Elizabeth Finger, MD
Stephen Pasternak, MD
Irina Rachinsky, MD
Andrew Kertesz, MD – Past Investigator
Dick Drost, MD – Past Investigator
John Rogers, MD – Past Investigator
Northwestern University:
Ian Grant, MD
Brittanie Muse, MSPH
Emily Rogalski, PhD
Jordan Robson
M.‐Marsel Mesulam, MD – Past Investigator
Diana Kerwin, MD – Past Investigator
Chuang‐Kuo Wu, MD, PhD – Past Investigator
Nancy Johnson, PhD – Past Investigator
Kristine Lipowski, MA– Past Investigator
Sandra Weintraub, PhD – Past Investigator
Borna Bonakdarpour, MD– Past Investigator
Nathan Kline Institute: Nunzio Pomara, MD
Raymundo Hernando, MD
Antero Sarrael, MD
University of California, San Francisco:
Howard J. Rosen, MD
Bruce L. Miller, MD
David Perry, MD
Georgetown University Medical Center:
Raymond Scott Turner, MD, PhD
Kathleen Johnson, NP
Brigid Reynolds, NP
Kelly MCCann, BA Jessica Poe, BS
Brigham and Women's Hospital:
Reisa A. Sperling, MD
Keith A. Johnson, MD
Gad A. Marshall, MD
Stanford University:
Jerome Yesavage, MD
Joy L. Taylor, PhD
Steven Chao, MD, PhD
Jaila Coleman, BA
Jessica D. White, BA – Past Investigator
Barton Lane, MD – Past Investigator
Allyson Rosen, PhD – Past Investigator
Jared Tinklenberg, MD – Past Investigator
Banner Sun Health Research Institute:
Christine M. Belden, PsyD Alireza Atri, MD, PhD
Bryan M. Spann, DO, PhD
Kelly A. Clark
Edward Zamrini, MD – Past Investigator
Marwan Sabbagh, MD – Past Investigator
Boston University:
Ronald Killiany, PhD
Robert Stern, PhD
Jesse Mez, MD, MS
Neil Kowall, MD – Past Investigator
Andrew E. Budson, MD – Past Investigator
Howard University:
Thomas O. Obisesan, MD, MPH
Oyonumo E. Ntekim, MD, PhD
Saba Wolday, MSc
Javed I. Khan, MD
Evaristus Nwulia, MD
Sheeba Nadarajah, PhD
Case Western Reserve University:
Alan Lerner, MD
Paula Ogrocki, PhD
Curtis Tatsuoka, PhD
Parianne Fatica, BA, CCRC
University of California, Davis – Sacramento:
Evan Fletcher, PhD
Pauline Maillard, PhD
John Olichney, MD
Charles DeCarli, MD
Owen Carmichael, PhD – Past Investigator
Dent Neurologic Institute:
Vernice Bates, MD
Horacio Capote, MD
Michelle Rainka, PharmD, CCRP
Parkwood Institute:
Michael Borrie, MB ChB
T‐Y Lee, PhD
Dr Rob Bartha, PhD
University of Wisconsin:
Sterling Johnson, PhD
Sanjay Asthana, MD
Cynthia M. Carlsson, MD, MS
Banner Alzheimer's Institute:
Allison Perrin, PhD
Anna Burke, PhD – Past Investigator
Ohio State University:
Douglas W. Scharre, MD
Maria Kataki, MD, PhD
Rawan Tarawneh, MD
Brendan Kelley, MD – Past Investigator
Albany Medical College: David Hart, MD
Earl A. Zimmerman, MD
Dzintra Celmins, MD
University of Iowa College of Medicine
Delwyn D. Miller, PharmD, MD
Laura L. Boles Ponto, PhD
Karen Ekstam Smith, RN
Hristina Koleva, MD
Hyungsub Shim, MD
Ki Won Nam, MD – Past Investigator
Susan K. Schultz, MD – Past Investigator
Wake Forest University Health Sciences:
Jeff D. Williamson, MD, MHS
Suzanne Craft, PhD
Jo Cleveland, MD
Mia Yang, MD– Past Investigator
Kaycee M. Sink, MD, MAS – Past Investigator
Rhode Island Hospital:
Brian R. Ott, MD
Jonathan Drake, MD
Geoffrey Tremont, PhD
Lori A. Daiello, Pharm.D, ScM
Jonathan D. Drake, MD
Cleveland Clinic Lou Ruvo Center for Brain Health:
Marwan Sabbagh, MD
Aaron Ritter, MD
Charles Bernick, MD, MPH – Past Investigator
Donna Munic, PhD – Past Investigator
Akiva Mintz, MD, PhD – Past Investigator
Roper St. Francis Healthcare:
Abigail O'Connelll, MS, APRN, FNP‐C
Jacobo Mintzer, MD, MBA
Arthur Wiliams, BS
Houston Methodist Neurological Institute:
Joseph Masdeu, PhD
Barrow Neurological Institute:
Jiong Shi, MD, PhD
Angelica Garcia, BS
Marwan Sabbagh – Past Investigator
Vanderbilt University Medical Center:
Paul Newhouse, PhD
Long Beach VA Neuropsychiatric Research Program:
Steven Potkin, PhD
Butler Hospital Memory and Aging Program:
Stephen Salloway, MD, MS
Paul Malloy, PhD
Stephen Correia, PhD
Neurological Care of CNY:
Smita Kittur, MD – Past Investigator
Hartford Hospital, Olin Neuropsychiatry Research Center:
Godfrey D. Pearlson, MD – Past Investigator
Karen Blank, MD – Past Investigator
Karen Anderson, RN – Past Investigator
Dartmouth‐Hitchcock Medical Center:
Laura A. Flashman, PhD – Past Investigator
Marc Seltzer, MD – Past Investigator
Mary L. Hynes, RN, MPH – Past Investigator
Robert B. Santulli, MD – Past Investigator
Cornell University
Norman Relkin, MD, PhD – Past Investigator
Gloria Chiang, MD – Past Investigator
Athena Lee, PhD
Michael Lin, MD – Past Investigator
Lisa Ravdin, PhD – Past Investigator
II. DOD ADNI
Part A: Leadership and Infrastructure
Principal Investigator
| Michael W. Weiner, MD | University of California, San Francisco |
ATRI PI and Director of Coordinating Center Clinical Core
| Paul Aisen, MD | University of Southern California |
| Co Director Clinical Core Ron Petersen | Mayo Clinic |
Executive Committee
| Michael W. Weiner, MD | University of California, San Francisco |
| Paul Aisen, MD | University of Southern California |
| Ronald Petersen, MD, PhD | Mayo Clinic, Rochester |
| Robert C. Green, MD, MPH | Brigham and Women's Hospital/ Harvard Medical School |
| Danielle Harvey, PhD | University of California, Davis |
| Clifford R. Jack, Jr., MD | Mayo Clinic, Rochester |
| William Jagust, MD | University of California, Berkeley |
| John C. Morris, MD | Washington University St. Louis |
| Andrew J. Saykin, PsyD | Indiana University |
| Leslie M. Shaw, PhD | Perelman School of Medicine, University of Pennsylvania |
| Arthur W. Toga, PhD | University of Southern California |
| John Q. Trojanowki, MD, PhD | Perelman School of Medicine, University of Pennsylvania |
Psychological Evaluation/PTSD Core
| Thomas Neylan, MD | University of California, San Francisco |
Traumatic Brain Injury/TBI Core
| Jordan Grafman, PhD | Rehabilitation Institute of Chicago, Feinberg School of Medicine, Northwestern University |
Data and Publication Committee (DPC)
| Robert C. Green, MD, MPH | BWH/HMS (Chair) |
Resource Allocation Review Committee
| Tom Montine, MD, PhD | University of Washington (Chair) |
Clinical Core Leaders and Key Personnel
| Michael W. Weiner MD | Core PI |
| Ronald Petersen, MD, PhD | Mayo Clinic, Rochester (Core PI) |
| Paul Aisen, MD | University of Southern California (Core PI) |
| Gustavo Jimenez, MBS | University of Southern California |
| Michael Donohue, PhD | University of Southern California |
| Devon Gessert , BS | University of Southern California |
| Jennifer Salazar, MBS | University of Southern California |
| Caileigh Zimmerman, MS | University of Southern California |
| Sarah Walter, MSc | University of Southern California |
| Olusegun Adegoke, MSc | University of Southern California |
| Payam Mahboubi, MPH | University of Southern California |
| Lindsey Hergesheimer, BS | University of Southern California |
| Sarah Danowski, MA | University of Southern California |
| Godfrey Coker, MBA, MPH | University of Southern California |
| Taylor Clanton, MPH | University of Southern California |
| Jeremy Pizzola, BA | University of Southern California |
| Elizabeth Shaffer, BS | University of Southern California |
| Catherine Nguyen‐Barrera, MS | University of Southern California |
San Francisco Veterans Affairs Medical Center
| Thomas Neylan, MD | University of California, San Francisco |
| Jacqueline Hayes | University of California, San Francisco |
| Shannon Finley | University of California, San Francisco |
Biostatistics Core Leaders and Key Personnel
| Danielle Harvey, PhD | University of California, Davis (Core PI) |
| Michael Donohue, PhD | University of California, San Diego |
MRI Core Leaders and Key Personnel
| Clifford R. Jack, Jr., MD | Mayo Clinic, Rochester (Core PI) |
| Matthew Bernstein, PhD | Mayo Clinic, Rochester |
| Bret Borowski, RT | Mayo Clinic |
| Jeff Gunter, PhD | Mayo Clinic |
| Matt Senjem, MS | Mayo Clinic |
| Kejal Kantarci | Mayo Clinic |
| Chad Ward | Mayo Clinic |
| Duygu Tosun‐Turgut, PhD | University of California, San Francisco |
| Stephanie Rossi Chen, BA | NCIRE / The Vererans Health Research Institute |
PET Core Leaders and Key Personnel
| Susan Landau, PhD | University of California, Berkeley Core PI |
| Robert A. Koeppe, PhD | University of Michigan |
| Norm Foster, MD | University of Utah |
| Eric M. Reiman, MD | Banner Alzheimer's Institute |
| Kewei Chen, PhD | Banner Alzheimer's Institute |
| Neuropathology Core Leaders | |
| John C. Morris, MD | Washington University St. Louis |
| Richard J. Perrin, MD, PhD | Washington University St. Louis |
| Erin Franklin, MS | Washington University St. Louis |
Biomarkers Core Leaders and Key Personnel
| Leslie M. Shaw, PhD | Perelman School of Medicine, University of Pennsylvania |
| John Q. Trojanowki, MD, PhD | Perelman School of Medicine, University of Pennsylvania |
| Magdalena Korecka, PhD | Perelman School of Medicine, University of Pennsylvania |
| Michal Figurski, PhD | Perelman School of Medicine, University of Pennsylvania |
Informatics Core Leaders and Key Personnel
| Arthur W. Toga, PhD | University of Southern California (Core PI) |
| Karen Crawford | University of Southern California |
| Scott Neu, PhD | University of Southern California |
Genetics Core Leaders and Key Personnel
Andrew J. Saykin, PsyD University
| Tatiana M. Foroud, PhD | Indiana University |
| Steven Potkin, MD UC | UC Irvine |
| Li Shen, PhD | Indiana University |
| Kelley Faber, MS, CCRC | Indiana University |
| Sungeun Kim, PhD | Indiana University |
| Kwangsik Nho, PhD | Indiana University |
| Kristi Wilmes, MS, CCRP | NCRAD |
Part B: Investigators By Site
University of Southern California:
Lon S. Schneider, MD
Sonia Pawluczyk, MD
Mauricio Becerra, MD
Liberty Teodoro, RN
Karen Dagerman, MS
Bryan M. Spann, DO, PhD – Past Investigator
University of California, San Diego:
James Brewer, MD, PhD
Helen Vanderswag, RN
Adam Fleisher, MD – Past Investigator
Columbia University Medical Center:
Yaakov Stern, PhD
Lawrence S. Honig, MD, PhD
Akiva Mintz, MD, PhD
Rush University Medical Center:
Raj C. Shah, MD
Ajay Sood, MD, PhD
Kimberly S. Blanchard, DNP, APRN, NP‐C
Debra Fleischman, PhD – Past Investigator
Konstantinos Arfanakis, PhD – Past Investigator
Wien Center:
Dr. Ranjan Duara MD PI
Dr. Daniel Varon MD Co‐PI
Maria T Greig HP Coordinator
Duke University Medical Center:
P. Murali Doraiswamy, MBBS, FRCP
Jeffrey R. Petrella, MD
Olga James, MD– Past Investigator
Salvador Borges‐Neto, MD
Terence Z. Wong, MD
University of Rochester Medical Center:
Anton P. Porsteinsson, MD Bonnie Goldstein, MS, NP
Kimberly S. Martin, RN
University of California, Irvine:
Gaby Thai, MD
Aimee Pierce, MD
Christopher Reist, MD
Beatriz Yanez, RN
Elizabeth Sosa, PhD
Megan Witbracht, PhD
Premiere Research Inst (Palm Beach Neurology):
Carl Sadowsky, MD
Walter Martinez, MD
Teresa Villena, MD
University of California, San Francisco:
Howard Rosen, MD David Perry
Georgetown University Medical Center:
Raymond Scott Turner, MD, PhD
Kathleen Johnson, NP
Brigid Reynolds, NP
Kelly MCCann, BA Jessica Poe, BS
Brigham and Women's Hospital:
Reisa A. Sperling, MD
Keith A. Johnson, MD
Gad Marshall, MD
Banner Sun Health Research Institute:
Christine M. Belden, PsyD Alireza Atri, MD, PhD
Bryan M. Spann, DO, PhD
Kelly A. Clark
Edward Zamrini, MD – Past Investigator
Marwan Sabbagh, MD – Past Investigator
Howard University:
Thomas O. Obisesan, MD, MPH
Oyonumo E. Ntekim, MD, PhD
Saba Wolday, MSc
Evaristus Nwulia, MD
Sheeba Nadarajah, PhD, RN
University of Wisconsin:
Sterling Johnson, PhD
Sanjay Asthana, MD
Cynthia M. Carlsson, MD, MS
University of Washington:
Elaine R. Peskind, MD
Eric C. Petrie, MD, MS
Gail Li, MD, PhD
Stanford University:
Jerome Yesavage, MD
Joy L. Taylor, PhD
Steven Chao, MD, PhD
Jaila Coleman, BA
Jessica D. White, BA – Past Investigator
Barton Lane, MD – Past Investigator
Allyson Rosen, PhD – Past Investigator
Jared Tinklenberg, MD – Past Investigator
Cornell University:
Michael Lin, PhD
Gloria Chiang, MD
Lisa Ravdin, PhD
Norman Relkin, MD, PhD – Past Investigator
Roper St. Francis Healthcare:
Abigail O'Connelll, MS, APRN, FNP‐C
Jacobo Mintzer, MD, MBA
Arthur Wiliams, BS
III. ADNI Depression
Part A: Leadership and Infrastructure
Principal Investigator
| Scott Mackin, PhD | University of California, San Francisco |
ATRI Coordinating Center Clinical Core
| Paul Aisen, MD | University of Southern California |
| Rema Raman, PhD | University of Southern California |
| Gustavo Jimenez‐Maggiora, MBS | University of Southern California |
| Michael Donohue, PhD | University of Southern California |
| Devon Gessert, BS | University of Southern California |
| Jennifer Salazar, MBS | University of Southern California |
| Caileigh Zimmerman, MS | University of Southern California |
| Sarah Walter, MSc | University of Southern California |
| Olusegun Adegoke, MSc | University of Southern California |
| Payam Mahboubi, MPH | University of Southern California |
Executive Committee
| Scott Mackin, PhD | University of California, San Francisco |
| Michael W. Weiner, MD | University of California, San Francisco |
| Paul Aisen, MD | University of Southern California |
| Rema Raman, PhD | University of Southern California |
| Clifford R. Jack, Jr., MD | Mayo Clinic, Rochester |
| Susan Landau, PhD | University of California, Berkeley |
| Andrew J. Saykin, PsyD | Indiana University |
| Arthur W. Toga, PhD | University of Southern California |
| Charles DeCarli, MD | University of California, Davis |
| Robert A. Koeppe, PhD | University of Michigan |
Data and Publication Committee (DPC)
| Robert C. Green, MD, MPH | BWH/HMS (Chair) |
| Erin Drake, MA | BWM/HMS (Director) |
Clinical Core Leaders
| Michael W. Weiner MD | Core PI |
| Paul Aisen, MD | University of Southern California |
| Rema Raman, PhD | University of Southern California |
| Mike Donohue, PhD | University of Southern California |
Psychiatry Site Leaders and Key Personnel
| Scott Mackin, PhD | University of California, San Francisco |
| Craig Nelson, MD | University of California, San Francisco |
| David Bickford, BA | University of California, San Francisco |
| Meryl Butters, PhD | University of Pittsburgh |
| Michelle Zmuda, MA | University of Pittsburgh |
MRI Core Leaders and Key Personnel
| Clifford R. Jack, Jr., MD | Mayo Clinic, Rochester (Core PI) |
| Matthew Bernstein, PhD | Mayo Clinic, Rochester |
| Bret Borowski, RT | Mayo Clinic, Rochester |
| Jeff Gunter, PhD | Mayo Clinic, Rochester |
| Matt Senjem, MS | Mayo Clinic, Rochester |
| Kejal Kantarci, MD | Mayo Clinic, Rochester |
| Chad Ward, BA | Mayo Clinic, Rochester |
| Denise Reyes, BS | Mayo Clinic, Rochester |
PET Core Leaders and Key Personnel
| Robert A. Koeppe, PhD | University of Michigan |
| Susan Landau, PhD | University of California, Berkeley |
Informatics Core Leaders and Key Personnel
| Arthur W. Toga, PhD | University of Southern California (Core PI) |
| Karen Crawford | University of Southern California |
| Scott Neu, PhD | University of Southern California |
Genetics Core Leaders and Key Personnel
| Andrew J. Saykin, PsyD | Indiana University |
| Tatiana M. Foroud, PhD | Indiana University |
| Kelley M. Faber, MS, CCRC | Indiana University |
| Kwangsik Nho, PhD | Indiana University |
| Kelly N. Nudelman | Indiana University |
Part B: Investigators By Site
University of California, San Francisco:
Scott Mackin, PhD
Howard Rosen, MD
Craig Nelson, MD
David Bickford, BA
Yiu Ho Au, BA
Kelly Scherer, BS
Daniel Catalinotto, BA
Samuel Stark, BA
Elise Ong, BA
Dariella Fernandez, BA
University of Pittsburgh:
Meryl Butters, PhD
Michelle Zmuda, BS
Oscar L. Lopez, MD
MaryAnn Oakley, MA
Donna M. Simpson, CRNP, MPH
Coath W, Modat M, Cardoso MJ, et al. Operationalizing the centiloid scale for [18F]florbetapir PET studies on PET/MRI. Alzheimer's Dement. 2023;15:e12434. 10.1002/dad2.12434
DATA AVAILABILITY STATEMENT
Insight 46 data are available upon request by qualified researchers (https://skylark.ucl.ac.uk/Skylark/). ADNI data are available upon request (https://adni.loni.usc.edu/data‐samples/access‐data/).
REFERENCES
- 1. Jack CR, Knopman DS, Jagust WJ, et al. Hypothetical model of dynamic biomarkers of the Alzheimer's pathological cascade. Lancet Neurol 2010;9:119‐128. 10.1016/S1474-4422(09)70299-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mintun MA, Lo AC, Duggan Evans C, et al. Donanemab in early Alzheimer's disease. N Engl J Med 2021:NEJMoa2100708. 10.1056/NEJMoa2100708 [DOI] [PubMed] [Google Scholar]
- 3. Lane CA, Parker TD, Cash DM, et al. Study protocol: insight 46 – a neuroscience sub‐study of the MRC National Survey of Health and Development. BMC Neurol 2017;17:75. 10.1186/s12883-017-0846-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lopresti BJ, Klunk WE, Mathis CA, et al. Simplified quantification of pittsburgh compound B amyloid imaging PET studies: a comparative analysis. J Nucl Med 2005;46:1959‐1972. [PubMed] [Google Scholar]
- 5. Chiao P, Bedell BJ, Avants B, et al. Impact of reference and target region selection on amyloid PET SUV ratios in the phase 1b PRIME study of aducanumab. J Nucl Med 2019;60:100‐106. 10.2967/jnumed.118.209130 [DOI] [PubMed] [Google Scholar]
- 6. Landau SM, Thomas BA, Thurfjell L, et al. Amyloid PET imaging in Alzheimer's disease: a comparison of three radiotracers. Eur J Nucl Med Mol Imaging 2014;41:1398‐1407. 10.1007/s00259-014-2753-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Landau SM, Breault C, Joshi AD, et al. Amyloid‐β imaging with pittsburgh compound B and Florbetapir: comparing radiotracers and quantification methods. J Nucl Med 2013;54:70‐77. 10.2967/jnumed.112.109009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Properzi MJ, Buckley RF, Chhatwal JP, et al. Nonlinear distributional mapping (NoDiM) for harmonization across amyloid‐PET radiotracers. Neuroimage 2019;186:446‐454. 10.1016/j.neuroimage.2018.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Klunk WE, Koeppe RA, Price JC, et al. The Centiloid Project: standardizing quantitative amyloid plaque estimation by PET. Alzheimers Dement 2015;11:1‐15.e4. 10.1016/j.jalz.2014.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Su Y, Flores S, Hornbeck RC, et al. Utilizing the Centiloid scale in cross‐sectional and longitudinal PiB PET studies. NeuroImage Clin 2018;19:406‐416. 10.1016/j.nicl.2018.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Burgos N, Cardoso MJ, Thielemans K, et al. Attenuation correction synthesis for Hybrid PET‐MR scanners: application to brain studies. IEEE Trans Med Imaging 2014;33:2332‐2341. 10.1109/TMI.2014.2340135 [DOI] [PubMed] [Google Scholar]
- 12. Su Y, Rubin BB, McConathy J, et al. Impact of MR‐based attenuation correction on neurologic PET studies. J Nucl Med 2016;57:913‐917. 10.2967/jnumed.115.164822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ladefoged CN, Law I, Anazodo U, et al. A multi‐centre evaluation of eleven clinically feasible brain PET/MRI attenuation correction techniques using a large cohort of patients. Neuroimage 2017;147:346‐359. 10.1016/j.neuroimage.2016.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bourgeat P, Doré V, Burnham SC, et al. β‐amyloid PET harmonisation across longitudinal studies: application to AIBL, ADNI and OASIS3. Neuroimage 2022;262:119527. 10.1016/j.neuroimage.2022.119527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Navitsky M, Joshi AD, Kennedy I, et al. Standardization of amyloid quantitation with florbetapir standardized uptake value ratios to the Centiloid scale. Alzheimers Dement 2018;14:1565‐1571. 10.1016/j.jalz.2018.06.1353 [DOI] [PubMed] [Google Scholar]
- 16. Cardoso MJ, Modat M, Wolz R, et al. Geodesic information flows: spatially‐variant graphs and their application to segmentation and fusion. IEEE Trans Med Imaging 2015;34:1976‐1988. 10.1109/TMI.2015.2418298 [DOI] [PubMed] [Google Scholar]
- 17. Modat M, Cash DM, Daga P, Winston GP, Duncan JS, Ourselin S. A symmetric block‐matching framework for global registration. Med. Imaging 2014 Image Process, vol. 9034, International Society for Optics and Photonics; 2014, p. 90341D. 10.1117/12.2043652 [DOI] [Google Scholar]
- 18. Royse SK, Minhas DS, Lopresti BJ, et al. Validation of amyloid PET positivity thresholds in centiloids: a multisite PET study approach. Alzheimers Res Ther 2021;13:99. 10.1186/s13195-021-00836-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Erlandsson K, Buvat I, Pretorius PH, Thomas BA, Hutton BF. A review of partial volume correction techniques for emission tomography and their applications in neurology, cardiology and oncology. Phys Med Biol 2012;57:R119‐59. 10.1088/0031-9155/57/21/r119 [DOI] [PubMed] [Google Scholar]
- 20. Hutton BF, Thomas BA, Erlandsson K, et al. What approach to brain partial volume correction is best for PET/MRI? Nucl Instrum Methods Phys Res Sect Accel Spectrometers Detect Assoc Equip 2013;702:29‐33. 10.1016/j.nima.2012.07.059 [DOI] [Google Scholar]
- 21. Thomas BA, Cuplov V, Bousse A, et al. PETPVC: a toolbox for performing partial volume correction techniques in positron emission tomography. Phys Med Biol 2016;61:7975‐7993. 10.1088/0031-9155/61/22/7975 [DOI] [PubMed] [Google Scholar]
- 22. Thomas BA, Erlandsson K, Modat M, et al. The importance of appropriate partial volume correction for PET quantification in Alzheimer's disease. Eur J Nucl Med Mol Imaging 2011;38:1104‐1119. 10.1007/s00259-011-1745-9 [DOI] [PubMed] [Google Scholar]
- 23. Farrell ME, Jiang S, Schultz AP, et al. Defining the lowest threshold for amyloid‐PET to predict future cognitive decline and amyloid accumulation. Neurology 2021;96:e619‐31. 10.1212/WNL.0000000000011214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jack CR, Wiste HJ, Weigand SD, et al. Defining imaging biomarker cut points for brain aging and Alzheimer's disease. Alzheimers Dement 2017;13:205‐216. 10.1016/j.jalz.2016.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. La Joie R, Ayakta N, Seeley WW, et al. Multisite study of the relationships between antemortem [11C]PIB‐PET Centiloid values and postmortem measures of Alzheimer's disease neuropathology. Alzheimers Dement 2019;15:205‐216. 10.1016/j.jalz.2018.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Salvadó G, Molinuevo JL, Brugulat‐Serrat A, et al. Centiloid cut‐off values for optimal agreement between PET and CSF core AD biomarkers. Alzheimers Res Ther 2019;11:27. 10.1186/s13195-019-0478-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brendel M, Högenauer M, Delker A, et al. Improved longitudinal [18F]‐AV45 amyloid PET by white matter reference and VOI‐based partial volume effect correction. Neuroimage 2015;108:450‐459. 10.1016/j.neuroimage.2014.11.055 [DOI] [PubMed] [Google Scholar]
- 28. Chen K, Roontiva A, Thiyyagura P, et al. Improved power for characterizing longitudinal amyloid‐β PET changes and evaluating amyloid‐modifying treatments with a cerebral white matter reference region. J Nucl Med 2015;56:560‐566. 10.2967/jnumed.114.149732 [DOI] [PubMed] [Google Scholar]
- 29. Fleisher AS, Joshi AD, Sundell KL, et al. Use of white matter reference regions for detection of change in florbetapir positron emission tomography from completed phase 3 solanezumab trials. Alzheimers Dement 2017;13:1117‐1124. 10.1016/j.jalz.2017.02.009 [DOI] [PubMed] [Google Scholar]
- 30. Landau SM, Fero A, Baker SL, et al. Measurement of longitudinal β‐amyloid change with 18F‐florbetapir PET and standardized uptake value ratios. J Nucl Med 2015;56:567‐574. 10.2967/jnumed.114.148981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schwarz CG, Jones DT, Gunter JL, et al. Contributions of imprecision in PET‐MRI rigid registration to imprecision in amyloid PET SUVR measurements. Hum Brain Mapp 2017;38:3323‐3336. 10.1002/hbm.23622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Delso G, Fürst S, Jakoby B, et al. Performance Measurements of the Siemens mMR Integrated Whole‐Body PET/MR Scanner. J Nucl Med 2011;52:1914‐1922. 10.2967/jnumed.111.092726 [DOI] [PubMed] [Google Scholar]
- 33. Kameyama M, Ishibash K, Wagatsuma K, Toyohara J, Ishii K. A pitfall of white matter reference regions used in [18F] florbetapir PET: a consideration of kinetics. Ann Nucl Med 2019;33:848‐854. 10.1007/s12149-019-01397-y [DOI] [PubMed] [Google Scholar]
- 34. van Berckel BNM, Ossenkoppele R, Tolboom N, et al. Longitudinal amyloid imaging using 11C‐PiB: methodologic considerations. J Nucl Med 2013;54:1570‐1576. 10.2967/jnumed.112.113654 [DOI] [PubMed] [Google Scholar]
- 35. Lowe VJ, Lundt ES, Senjem ML, et al. White Matter Reference Region in PET Studies of 11C‐Pittsburgh Compound B Uptake: Effects of Age and Amyloid‐β Deposition. J Nucl Med 2018;59:1583‐1589. 10.2967/jnumed.117.204271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brickman AM, Zahra A, Muraskin J, et al. Reduction in cerebral blood flow in areas appearing as white matter hyperintensities on magnetic resonance imaging. Psychiatry Res Neuroimaging 2009;172:117‐120. 10.1016/j.pscychresns.2008.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pietroboni AM, Carandini T, Colombi A, et al. Amyloid PET as a marker of normal‐appearing white matter early damage in multiple sclerosis: correlation with CSF β‐amyloid levels and brain volumes. Eur J Nucl Med Mol Imaging 2019;46:280‐287. 10.1007/s00259-018-4182-1 [DOI] [PubMed] [Google Scholar]
- 38. Auvity S, Tonietto M, Caillé F, et al. Repurposing radiotracers for myelin imaging: a study comparing 18F‐florbetaben, 18F‐florbetapir, 18F‐flutemetamol,11C‐MeDAS, and 11C‐PiB. Eur J Nucl Med Mol Imaging 2020;47:490‐501. 10.1007/s00259-019-04516-z [DOI] [PubMed] [Google Scholar]
- 39. Bodini B, Veronese M, García‐Lorenzo D, et al. Dynamic imaging of individual remyelination profiles in multiple sclerosis. Ann Neurol 2016;79:726‐738. 10.1002/ana.24620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Carotenuto A, Giordano B, Dervenoulas G, et al. [18F]Florbetapir PET/MR imaging to assess demyelination in multiple sclerosis. Eur J Nucl Med Mol Imaging 2020;47:366‐378. 10.1007/s00259-019-04533-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. de Paula Faria D, Copray S, Sijbesma JWA, et al. PET imaging of focal demyelination and remyelination in a rat model of multiple sclerosis: comparison of [11C]MeDAS, [11C]CIC and [11C]PIB. Eur J Nucl Med Mol Imaging 2014;41:995‐1003. 10.1007/s00259-013-2682-6 [DOI] [PubMed] [Google Scholar]
- 42. Stankoff B, Freeman L, Aigrot M‐S, et al. Imaging central nervous system myelin by positron emission tomography in multiple sclerosis using [methyl‐11C]‐2‐(4′‐methylaminophenyl)‐ 6‐hydroxybenzothiazole. Ann Neurol 2011;69:673‐680. 10.1002/ana.22320 [DOI] [PubMed] [Google Scholar]
- 43. Moscoso A, Whitman A, Baker SL, et al. Reduced [18 F] flortaucipir retention in white matter hyperintensities compared to normal‐appearing white matter, ALZ; 2020. [DOI] [PubMed] [Google Scholar]
- 44. Wong DF, Rosenberg PB, Zhou Y, et al. In vivo imaging of amyloid deposition in Alzheimer disease using the radioligand 18F‐AV‐45 (Flobetapir F 18). J Nucl Med 2010;51:913‐920. 10.2967/jnumed.109.069088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Karlberg AM, Sæther O, Eikenes L, Goa PE. Quantitative comparison of PET performance—Siemens Biograph mCT and mMR. EJNMMI Phys 2016;3:5. 10.1186/s40658-016-0142-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moses WW. Fundamental limits of spatial resolution in PET. Nucl Instrum Methods Phys Res Sect Accel Spectrometers Detect Assoc Equip 2011;648:S236‐S240. 10.1016/j.nima.2010.11.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp Information 1
Supp Information 2
Data Availability Statement
Insight 46 data are available upon request by qualified researchers (https://skylark.ucl.ac.uk/Skylark/). ADNI data are available upon request (https://adni.loni.usc.edu/data‐samples/access‐data/).


