Abstract
The cell wall provides an attractive target for antibiotics against Mycobacterium tuberculosis. Agents such as isoniazid and ethambutol that work by inhibiting cell wall biosynthesis are among the most highly effective antibiotics against this pathogen. Although considerable progress has been made identifying the targets for cell wall active antibiotics, little is known about the intracellular mechanisms that are activated as a consequence of cell wall injury. These mechanisms are likely to have an important role in growth regulation and in the induction of cell death by antibiotics. We previously discovered three isoniazid-induced genes (iniB, iniA, and iniC) organized in tandem on the M. tuberculosis genome. Here, we investigate the unique features of the putative iniBAC promoter. This promoter was specifically induced by a broad range of inhibitors of cell wall biosynthesis but was not inducible by other conditions that are toxic to mycobacteria via other mechanisms. Induction required inhibitory concentrations of antibiotics and could be detected only in actively growing cells. Analysis of the iniBAC promoter sequence revealed both a regulatory element upstream and a potential repressor binding region downstream of the transcriptional start site. The induction phenotype and structure of the iniBAC promoter suggest that a complex intracellular response occurs when cell wall biosynthesis is inhibited in M. tuberculosis and other mycobacteria.
New therapies are urgently needed to treat Mycobacterium tuberculosis, one of the leading causes of death from a single infectious disease worldwide (15). Although this pathogen can be treated effectively with multidrug therapy (1), incomplete treatment has led to the development of drug-resistant strains (17, 30). In general, antibiotics that inhibit cell wall biosynthesis are among the most effective agents against bacterial pathogens. In the case of M. tuberculosis, a number of highly effective antibiotics including isoniazid (INH) and ethambutol work by inhibiting biosynthesis of the mycobacterial cell wall. Because the cell wall is an attractive target for antibiotic development, considerable effort has been focused on discovering the metabolic steps that are essential for biosynthesis of cell wall components (10, 11, 23, 28). The enzymatic targets of many cell wall active antibiotics also have been discovered recently (7, 12, 25, 38).
Despite progress in characterizing the mycobacterial cell wall, very little is known about the intracellular mechanisms that are activated as a consequence of cell wall injury. Antibiotic-induced cell death may involve events that are not directly related to inhibition of the primary antibiotic target. For example, autolysins are activated when cell wall biosynthesis is inhibited by β-lactam antibiotics in many gram-positive bacteria (13, 19, 39). Microarray technology has revealed that diverse sets of genes are induced following treatment of cells with drugs (14, 42). In the case of M. tuberculosis, novel antibiotics that directly target these pathways may be highly effective in the treatment of disease due to this organism, either alone or synergistically with other cell wall-active agents. The identification of promoters that are specifically induced by cell wall damage could also be valuable as part of a screen that used reporter assays to discover novel cell wall-active compounds (25).
In the course of studying the differential gene expression of M. tuberculosis in response to INH, we discovered three INH-induced genes organized in tandem on the M. tuberculosis genome that we termed iniB, iniA, and iniC (3). We postulated that all three ini genes comprised a single operon, iniBAC. Northern blot and reverse transcription-PCR experiments demonstrated that the iniA gene was induced by both INH and ethambutol (3), two antibiotics that act on the cell wall by different mechanisms (7, 12, 25, 38). The kasA gene has been shown to be induced by INH and the related compound ethionamide (25, 42). However, no mycobacterial promoters that are induced by unrelated antibiotics targeting different pathways of cell wall biosynthesis have been identified. Little is known about the structure of regulated promoters in M. tuberculosis. The inducible promoters that have been characterized in M. tuberculosis include promoters for the sigma factors sigB (20) and sigF (27), the DNA repair protein recA (26), and the response regulator mtrA (41). In the case of recA, a putative upstream activation sequence has been identified, and a Cheo-like box that binds to the transcriptional repressor LexA has been found (26). Here, we demonstrate that the promoter of the putative iniBAC operon is specifically induced by a broad range of inhibitors to cell wall biosynthesis including antibiotics that inhibit the synthesis of (i) peptidoglycan (ampicillin and ampicillin/sulbactam [24]), (ii) arabinogalactam (ethambutol [12, 38]), (iii) mycolic acids (INH, ethionamide, and 2-alkynoic acid [KOAs]), and (iv) fatty acids (5-chloropyrazinamide [5-chloro-PZA]) (7, 25; O. Zimhony et al., unpublished data). We characterize the nature of the induction and demonstrate the suitability of the promoter to screen for cell wall-active antibiotics using luciferase reporter plasmids. The structure of the promoter is investigated and likely regulatory sequences are identified.
MATERIALS AND METHODS
Bacteria and culture methodology.
Escherichia coli DH5α (33) was used for all plasmid constructions. E. coli cultures were grown at 37°C in Luria-Bertani medium (33) with the addition of hygromycin B (200 μg/ml; Sigma, St. Louis, Mo.) or kanamycin (40 μg/ml; Sigma) where appropriate. Mycobacterium bovis BCG Pasteur and BCG Montreal strains ATCC 35735 and ATCC 35747, and M. smegmatis strain mc2155 (34) were grown at 37°C on a rotary shaker in Middlebrook 7H9 medium (Difco, Detroit, Mich.) containing 0.05% Tween 80, 0.02% glycerol, and 10% oleic-albumin dextrose complex (Becton Dickinson, Cockeysville, Md.), with the addition of hygromycin B (50 μg/ml) for strains containing pYUB509-based constructs or kanamycin (24 μg/ml) for mc2155 strains containing pCV125-based constructs. For induction experiments, additional antibiotics were added at the indicated final concentrations. BCG and mc2155 strains were cultured in 150-cm2 tissue culture flasks (Corning, Cambridge, Mass.) at 100 ml per flask, starting from 1:100 dilutions of strain stocks. Cultures were grown to an optical density at 590 nm (OD590) of approximately 0.4, except for experiments specifically designed to examine promoter induction at various ODs. The cultures were then split into 30-ml square medium bottles (Nalgene, Rochester, N.Y.) at 5 ml per bottle, antibiotics and other reagents were added to the growing cultures as indicated, and the cultures were then returned to the incubator. Culture aliquots were removed at specified time points for luciferase or β-galactosidase assays.
Sequence positions.
The sequence numbering used in this study corresponds to the M. tuberculosis genomic sequence position (16; National Center for Biotechnology Information [NCBI] database [http://www.ncbi.nlm.nih.gov/cgi-bin/Entrez/framik?db=Genome&gi=135]). By this convention, the iniB gene (Rv0341 or MTCY13E10.01) starts at 409354, the iniA gene (Rv0342 or MTCY13E10.02) starts at 410824, and the iniC gene (Rv0343 or MTCY13E10.03) starts at 412755. The translational start sites designated in this investigation vary slightly from previous annotations.
Plasmids and strains.
The plasmids and strains used in this study are listed in Table 1. Plasmid pYUB509, containing lacZ and fflux reporter genes, was used to construct pG4697-6 and pG1697-3 for testing iniBAC promoter activity in luciferase and β-galactosidase assays (Table 1). Plasmid pG4697-6, which contained a sequence beginning 211 bp upstream of the first iniBAC open reading frame (iniB) extending to the translational start site, was transformed into the antibiotic-susceptible BCG Montreal strain ATCC 35735 (BCGS) to create strain BCGS(pG4697-6). Plasmid pG4697-6 was also transformed into the INH-resistant BCG Montreal strain ATCC 35747 (BCGR), which contains a deletion in the katG gene, to create strain BCGR(pG4697-6). Plasmid pG1697-3, which contained the same 211-bp region in the reverse orientation, was transformed into BCGS to create strain BCGS(pG1697-3). Plasmid pCV125, an integrating vector containing kanamycin resistance and a promoterless lacZ gene, was obtained from MedImmune (Gaithersburg, Md.). This plasmid was used as the basis for constructs aimed at testing the activity of partial promoter deletions. Fourteen plasmids were constructed by ligating successive 5′ or 3′ deletions of the 211-bp sequence upstream of iniB into pCV125 (Table 1). These 14 plasmids were transformed into M. smegmatis strain mc2155 to create strains mc2155(pG4799-1) through mc2155(pG4799-12), mc2155(pG15499-1), and mc2155(pG15499-2). The finished plasmid constructs were subjected to automated DNA sequencing in order to exclude mutations that could occur during the PCR or cloning process. The unpublished plasmid pKB15 was a gift of Graham Hatfull, this integrating plasmid carries the fflux gene under control of the L5 phage pL promoter, resulting in constitutive expression of luciferase. Plasmid pKB15 also contains hygromycin and ampicillin resistance genes and L5 attP, int, and oriE. pKB15 was transformed into BCG Pasteur, resulting in strain BCG(pKB15).
TABLE 1.
Plasmids used in this study
| Plasmid | Description or source |
|---|---|
| pYUB509 | The complete coding region for leuD and leuC was generated by PCR of pYUB 516 (8) using primers B1A and B4B. The PCR fragment was digested with SnaB1 and NsiI and cloned into SnaB1/NsiI-digested pYUB469 (see below) to generate pLCD1. Plasmid pLCD1 was digested with SnaB1 and HindIII (partial); the resulting 5.2-kb fragment was treated with Klenow enzyme and cloned into SnaB1/PmlI-digested pYUB503 (see below) to generate pYUB506. The kanamycin resistance gene present in pYUB506 was removed by SpeI and HindIII (partial) digestion and replaced with the hygromycin gene generated by PCR of pMV261-H (a hygromycin-resistant derivative of pMV261 (36), using primers HYG1 and HYG2 to generate pYUB509. |
| pYUB503 | Plasmid pYUB178 (31) containing an EcoRI polylinker with PacI, SnaB1, KpnI, PmlI, and PacI restriction sites cloned into the EcoRI site. The KpnI site of pYUB178 was also destroyed with Klenow enzyme. |
| pYUB469 | A general laboratory plasmid containing the full-length, promoterless E. coli lacZ and firefly fflux genes, cloned in tandem into pBluescript KS (Stratagene, La Jolla, Calif.). |
| pG4697-6 | The integrating plasmid pYUB509 with a 211-bp insertion containing the iniBAC promoter region. The insert consisted of the sequence beginning 211 bp upstream of the first iniBAC open reading frame (iniB) extending to the translational start site (409142–409353). The sequence was generated from M. tuberculosis strain H37Rv genomic DNA by a seminested PCR using primers iigBPT and iigBPIIB followed by a second PCR using primers iigBproxhoT and iigBpro2xhoB, which contain 5′ XhoI sites. The PCR fragment was then inserted into the unique XhoI site of pYUB509 in the correct orientation. |
| pG1697-3 | Same as pG4697-6 but inserted into pYUB509 in the reverse orientation |
| pCV125 | Described in Materials and Methods |
| pG21898-12 | 211-bp iniBAC promoter sequence generated by PCR of pG4697-6 plasmid DNA using primers iniBproEcoT and iniBproSalB, inserted into the integrating plasmid pCV125 between the EcoRI and SalI restriction sites |
| pG21298-1 | pG21898-12 with 18-bp 5′ deletion of the 211-bp fragment, generated by PCR using primers iniBproEcoT20 and iniBproSalB |
| pG20298-2 | pG21898-1 with 42-bp 5′ deletion, generated using primers iniBproEcoT43 and iniBproSalB |
| pG20298-3 | pG21898-1 with 64-bp 5′ deletion, generated using primers iniBproEcoT65 and iniBproSalB |
| pG21298-4 | As in pG21898-1 with 89-bp 5′ deletion, generated using primers iniBproEcoT90 and iniBproSalB |
| pG20298-5 | pG21898-1 with 112-bp 5′ deletion, generated using primers iniBproEcoT113 and iniBproSalB |
| pG1199-6 | pG21898-1 with 133-bp 5′ deletion, generated using primers iniBproEcoT134 and iniBproSalB |
| pG1199-7 | pG21898-1 with 153-bp 5′ deletion, generated using primers iniBproEcoT154 and iniBproSalB |
| pG20298-8 | pG21898-1 with 173-bp 5′ deletion, generated using primers iniBproEcoT175 and iniBproSalB |
| pG20298-9 | pG21898-1 with 188-bp 5′ deletion, generated using primers iniBproEcoT189 and iniBproSalB |
| pG20298-10 | pG21898-1 with 19-bp 3′ deletion, generated using primers iniBproEcoT and iniBproSalB193 |
| pG599-11 | pG21898-1 with 39-bp 3′ deletion, generated using primers iniBproEcoT and iniBproSalB173 |
| pG15499-2 | pG20298-10 with 19-bp 3′ deletion and 19-bp 3′ spacer sequence, generated using primers iniBproEcoT and iniBproSalB193spa |
| pG15499-1 | pG21898-1 with 65-bp 3′ deletion, generated using primers iniBproEcoT and iniBproSalB148 |
| pG7897-4 | XmnI-PvuII fragment of the M. tuberculosis genome containing the iniB and iniA gene, cloned into the PvuII site of pMV261 (36) |
| pKB115 | Described in Materials and Methods |
Chromosomal DNA and RNA extraction.
Chromosomal DNA from different mycobacterial species was extracted using a sodium dodecyl sulfate (SDS)-hexadecyltrimethylammonium bromide (Fisher, Pittsburgh, Pa.) protocol as described previously (2). For RNA preparation, BCG strain ATCC 35735 was grown to an OD590 of 0.5. INH was then added to the culture for a final concentration of 1.0 μg/ml, or the culture was allowed to continue without added INH. After an additional 18-h incubation, RNA was extracted using a TRIzol (Life Technologies, Gaithersburg, Md.) based protocol as described previously (3).
PCR generation of amplicons.
PCRs were performed in 50-μl volumes containing either 10 ng of chromosomal DNA or 1 ng of plasmid DNA with 2.5 mM each deoxynucleosides triphosphate, 20 pmol each of upstream and downstream primers, 1.25 U of Taq polymerase with a final concentration of 1× PCR buffer (Gibco BRL, Grand Island, N.Y.), and 2 mM MgCl2. DNA was amplified in an Applied Biosystems Geneamp 9700 thermal cycler (Perkin-Elmer, Foster City, Calif.) for 30 cycles of 94°C for 1 min, annealing at 55°C for 1 min, and 72°C for 1 min, followed by 72°C for 10 min.
Luciferase assays.
At specified time points, 25 μl of each culture was removed and added to 75 μl of 7H9 medium in a glass cuvette (Lumacuvette-P; Celsis-Lumac, Landgraaf, The Netherlands). Luciferase activity was measured using 40 mM luciferin (Sigma) in 1 M C6H5Na3O7 · 2H2O (pH 4.5) in a Lumac 2010A luminometer (Celsis-Lumac) according to the manufacturer's recommendations. Induction was calculated as follows: relative light units (RLU) for sample culture/RLU for medium-only control culture, if RLU for sample > RLU for control. A decrease in luciferase activity of the sample culture compared to the control culture (repression) was calculated as RLU for control/RLU for sample.
β-Galactosidase assays.
At specified time points, 500 μl of each culture was set aside on ice for subsequent measurement of the OD590. An additional 500 μl of each culture was simultaneously removed and added to 500 μl of Z buffer (60 mM Na2HPO4 · 7H2O, 40 mM NaH2PO4 · H2O, 10 mM KCl, 1 mM MgSO4 · 7H2O, 50 mM β-mercaptoethanol, adjusted to pH 7.0) in a 2-ml microcentrifuge tube. Two drops of chloroform and one drop of 0.1% SDS were then added, and the tubes were vortexed for 30 s. The tubes were incubated at 28°C for 5 min, 200 μl of fresh o-nitrophenyl-β-d-galactopyranoside (ONPG) (4 mg/ml; Sigma) in Z buffer was added to each tube, and the tubes were shaken well and incubated at 28°C. When a faint yellow color appeared in the control tube (5 to 20 min), the reaction was stopped with 0.5 ml of 1 M Na2CO3 and spun in a microcentrifuge at top speed for 5 min, and the OD420 of the supernatant was measured. β-Galactosidase units were calculated using the formula 1,000 × OD420/time (minutes) × 0.5 × OD590.
Primer extension.
Oligonucleotide primers were end labeled with [γ32P]ATP at their 5′ ends, using T4 polynucleotide kinase as described in the primer extension kit (Promega, Madison, Wis.); 0.1 pmol of labeled primer was annealed to 6.5 μg of total RNA at 75°C for 0.5 h. Extension was carried out with avian myeloblastosis virus reverse transcriptase (Promega) according to the manufacturer's instructions at 42°C for 0.5 h. The reactions were added to 20 μl of loading dye (98% [vol/vol] formamide, 10 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue) and denatured at 90°C for 10 min. The reaction products were then run on an 8% polyacrylamide-urea sequencing gel. Bands were visualized by autoradiography. Sequencing reactions were carried out by cycle sequencing (Perkin-Elmer) according to the manufacturer's instructions.
Southern blots.
Genomic DNA from different mycobacterial species was digested with PvuII, subjected to electrophoresis in a 0.7% agarose gel, and transferred by capillary action to Biotrans Plus nylon membranes (ICN Pharmaceuticals, Costa Mesa, Calif.). The blots were prehybridized at 50°C in Rapid-Hyb buffer (Amersham, Arlington Heights, Ill.) and then hybridized overnight with [α32P]dCTP-radiolabeled (Megaprime labeling kit; Amersham) probes. The probe complementary to a 400-bp segment of the iniA gene was generated by PCR using the primers iniART-T and iniART-B. The probe complementary to the entire iniB gene was generated by a BamHI-NruI digestion of pG7897-4 (Table 1). The blots were washed in progressively more stringent conditions until autoradiography revealed clear bands and minimal background hybridization.
Statistical analysis.
Mean induction or repression and 95% confidence internals were calculated using Microsoft Excel 97 software.
RESULTS
Induction of the iniBAC promoter.
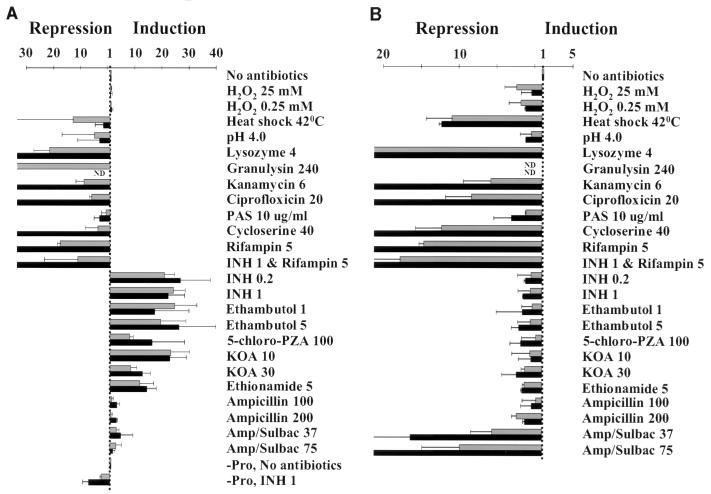
The discovery that the iniA gene was induced by ethambutol as well as INH (3) suggested that induction was not specific to inhibition of mycolic acid biosynthesis. Integrating reporter plasmids containing the iniBAC promoter fused to the genes encoding luciferase and β-galactosidase were constructed to further investigate the induction characteristics. The iniA gene appeared to be the second gene in a three-gene operon consisting of iniB, iniA, and iniC. We chose to investigate the promoter activity of the sequence extending 211 bp upstream of iniB to the translational start site of iniB (positions 409142 to 409353). BCGS(pG4697-6), which contained the full-length 211-bp sequence, was cultured to log phase and then split into untreated portions or portions that were treated with antibiotics and other reagents. Induction was assessed by comparing luciferase activity in the treated portions to that in the untreated control. Significant induction occurred in the presence of many different cell wall-active agents despite the divergence of their known mechanisms of action (Fig. 1A). After 24 and 48 h of incubation with antibiotics, induction by INH, ethambutol, ethionamide, 5-chloro-PZA, and KOA was 10- to 30-fold greater than control cultures. Induction by ampicillin, and by the combination β-lactam–β-lactamase inhibitor Unasyn (ampicillin/sulbactam), was consistently three- to fivefold greater than in control cultures. Induction by INH was reversed by coincubation with rifampin, indicating that the induction was due to increased rates of transcription. Among the cell wall biosynthesis inhibitors tested, only cycloserine did not result in induction of the reporter.
FIG. 1.
Effects of different compounds on iniBAC promoter activity as measured by luciferase assays of integrated transcriptional fusion plasmids. Induction is shown after incubation with compounds for 24 h (grey bars) or 48 h (black bars). Final concentrations are indicated in micrograms per milliliter. Error bars represent 95% confidence intervals. (A) INH-susceptible BCGS(pG4697-6) containing the iniBAC promoter fused to lacZ and fflux genes [-Pro indicates assays performed with BCGS(pG1697-3), which contains the same construct, except that the transcriptional fusion was performed by inserting the promoter in the opposite orientation from the coding region]. (B) INH-susceptible BCG(pMKB15) containing the L5 phage pL promoter fused to the fflux gene. (C) INH-resistant BCGR(pG4697-6) containing the iniBAC promoter fused to lacZ and fflux genes.
To ensure that the observed induction was not simply due to luciferase release into the media by cell wall-active antibiotics, the experiments were repeated with BCG(pKB15), a BCG strain which expressed luciferase constitutively. In contrast to BCGS(pG4697-6), luciferase expression in the BCG(pKB15) cultures decreased or remained unchanged under all experimental conditions (Fig. 1B). The results demonstrate that the 211-bp sequence immediately upstream of the iniB gene contains the promoter for the putative iniBAC operon. Induction of this promoter was specific to antibiotics that inhibit cell wall biosynthesis. The promoter was either not induced or repressed by other biological stresses, including hydrogen peroxide, heat shock, acidosis, and the antibiotics kanamycin, ciprofloxicin, and rifampin, that do not directly inhibit cell wall biosynthesis (Fig. 1A). Induction also was not observed after incubation with paraminosalicylic acid, an antituberculosis drug with an unknown mechanism of action. Importantly, disruption of the cell wall by lysozyme or granulysin also led to repression rather than induction of luciferase activity. These findings demonstrate that induction of the iniBAC promoter was due to inhibition of cell wall biosynthesis and could not be caused simply by lysis or disruption of the cell wall.
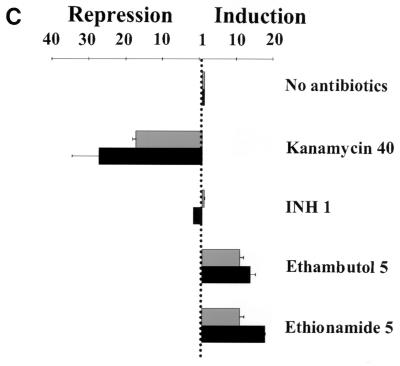
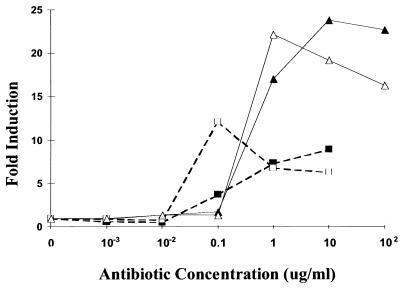
Next, induction was assessed using the INH-resistant strain BCGR(pG4697-6), which contains the same luciferase reporter construct as BCGS(pG4697-6) but is INH resistant due to a deletion in the katG gene. The katG gene encodes catalase-peroxidase, which is required to convert INH into its active form (43). Induction by INH was not observed with this strain; however, it remained inducible in the presence of other antibiotics (Fig. 1C). This demonstrated that induction was due to the biological activity of INH and was not due to nonspecific effects of the unactivated compound. This conclusion is supported by the observation that induction was observed only at antibiotic concentrations at or above their MICs (Fig. 2). Induction did not increase further at concentrations above the MIC, although maximum induction occurred more rapidly at the higher concentrations.
FIG. 2.
Induction of the iniBAC promoter in BCGS(pG4697-6) after incubation with INH for 24 (■) and 48 (□) h and with ethambutol for 24 (▴) and 48 (▵) h.
One inhibitor of cell wall biosynthesis, cycloserine, appeared to result in repression of luciferase expression. However, treatment with cycloserine resulted in rapid cell lysis, a phenomenon that was not observed with the other antibiotics. Induction experiments were repeated with diminishing doses of cycloserine, but induction was not detected at any concentration of this drug. A lacZ reporter system is less dependent on full viability of the cells at the time of the induction assay (40). Induction experiments using ONPG to measure β-galactosidase activity also failed to consistently detect induction by cycloserine (data not shown). In contrast, all of the other inhibitors of cell wall biosynthesis resulted in induction when measured by β-galactosidase assay.
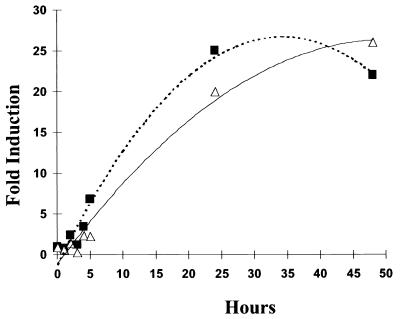
Induction kinetics.
The time course of iniBAC promoter induction after incubation with antibiotics was tested by incubating BCGS(pG4697-6) with INH (1 μg/ml) or ethambutol (5 μg/ml) and assessing serial aliquots for luciferase activity. Induction was apparent as early as 4 h after incubation with either antibiotic, reaching a maximum between 25 and 48 h (Fig. 3).
FIG. 3.
Induction of the iniBAC promoter in BCG strain BCGS(pG4697-6) after incubation with INH (1 μg/ml; ■) and ethambutol (5 μg/ml; ▵) as a function of incubation time.
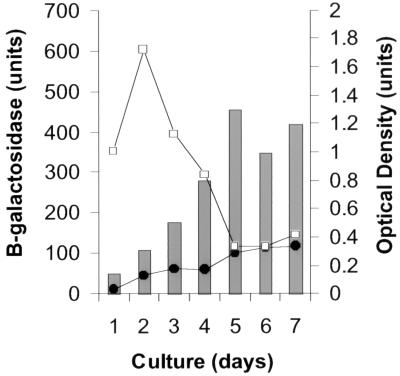
The effect of the growth phase of the culture on induction was also studied. We speculated that ATP levels might be dependent on growth phase; therefore, β-galactosidase assays were used to measure induction in these experiments. BCGS(pG4697-6) was grown from a highly diluted culture until it reached stationary phase. Two aliquots were removed from the culture every 24 h starting at an OD590 of 0.1. Ethambutol was added to one of the paired aliquots, then both were reincubated for an additional 24 h, and β-galactosidase activity was measured. These experiments demonstrated that iniBAC promoter activity increased slowly as the OD590 of the culture increased (Fig. 4). However, induction by ethambutol occurred only during log-phase growth and disappeared when the cultures reached stationary phase. Similar induction characteristics were observed after incubation with INH (data not shown).
FIG. 4.
Induction in different phases of growth. BCGS(pG4697-6) was subcultured by performing a 1:100 dilution of an actively growing culture. Serial aliquots were removed at increasing OD590 and split into paired subcultures. One subculture of each pair was not treated with antibiotics (●); the other subculture of each pair was treated with ethambutol at a final concentration of 5 μg/ml (□). After an additional 24 h, β-galactosidase activity was measured. β-Galactosidase units are shown as a function of the OD590 of the paired subcultures at the time that they were removed from the parent culture.
Species distribution of the iniBAC operon.
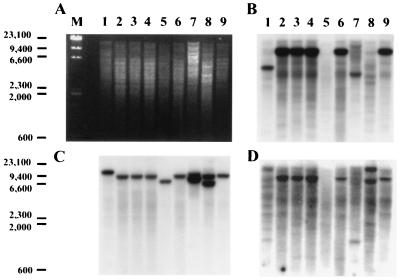
We previously determined that INH induced the iniA genes from both M. tuberculosis H37Rv and BCG (3). The 211-bp promoter region was sequenced in BCG strain ATCC 35735. Alignment with the corresponding sequence in M. tuberculosis strain H37Rv (16; NCBI database) revealed that the two sequences were 100% identical. Because mycobacterial species have similar cell wall structures, the presence of the iniBAC operon in different species of mycobacteria was assessed. Two probes were used, one complementary to a 400-bp region of the iniA gene and the other complementary to the entire iniB gene. The radiolabeled probes were hybridized separately to Southern blots containing genomic digests of three M. tuberculosis strains (Erdman, H37Rv, and H37Ra), of mycobacterial strain BCG, and of M. smegmatis (strain mc2155), M. avium, M. marinum, M. microti, and M. nonchromogenicum. The iniA probe hybridized strongly to single bands in all of the species except M. avium and M. nonchromogenicum. Hybridization to the M. nonchromogenicum genomic DNA resulted in two weak bands (Fig. 5). The iniB probe also failed to hybridize to M. avium, although it hybridized strongly to all of the other species except M. marinum. A control probe complementary to a region of the M. tuberculosis 16S rRNA gene hybridized to all of the species tested.
FIG. 5.
Testing of the iniBAC operon in different mycobacterial species. Mycobacterial species were tested by low-stringency Southern blot hybridization for the presence of the iniA and iniB genes: M. marinum (lane 1), H37ra (lane 2), M. tuberculosis strains Erdman (lane 3) and H37Rv (lane 4), M. avium (lane 5), BCG (lane 6), M. smegmatis (lane 7), M. nonchromogenicum (lane 8), and M. microti (lane 9). (A) Agarose gel showing PvuII digests of chromosomal DNA. (B) Southern blot of the gel in panel A hybridized with the iniA gene probe. (C) Hybridization with the M. tuberculosis 16S RNA gene probe. (D) Hybridization with the iniB gene.
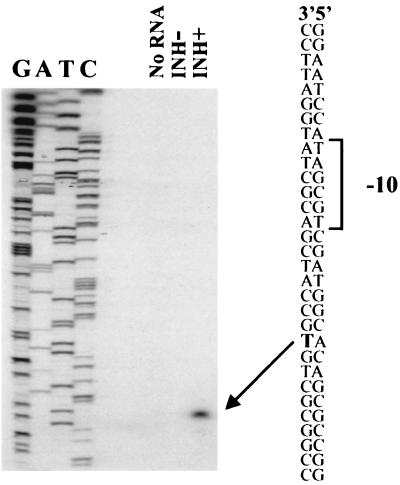
Mapping the transcriptional start site of the BCG iniBAC operon.
The transcriptional start site for the operon was determined by primer extension analysis in order to better understand the structure of the promoter region. Total RNA was extracted from BCGS grown in the presence or absence of INH during the last 18 h of culture. Primer extension was performed with both samples using iniBprimer ext-3, which is complementary to the first 21-bp of the translated M. tuberculosis iniB gene (Table 2). No products were visualized in the primer extensions of BCG RNA that had not been treated with isoniazid. A single product was detected after primer extension of RNA from isoniazid treated BCG. The band was situated 45 bp upstream of the likely translational start site (position 409308) (Fig. 6). Primer extensions performed with two additional primers complementary to other iniB gene sequences (iniBprimer ext-1 and iniBprimer ext-2) yielded identical results (not shown). The complete absence of product from the RNA that was not treated with INH was confirmed by prolonged exposure of the gels. This result is consistent with previously described Northern blot hybridizations of M. tuberculosis in which no iniA RNA was detected unless the samples were treated with INH (3).
TABLE 2.
Primers used in this study
| Name | Sequence, added restriction sites (underlined), spacer sequences (lowercase) | Positiona |
|---|---|---|
| iigBPT | GATCATCACGGCTACGACATCCAC | 409142–409165 |
| iigBPIIB | TGACCGCCGAAACCACCGCTTGAC | 409745–409722 |
| iigBproxhoT | GGAACTCGAGATCATCACGGCTACGACATCCAC | 409143–409165 |
| iigB2proxhoB | TCATCTCGAGTTCCCTTCAATCGAAGAAGC | 409353–409334 |
| iniBproEcoT | CCGTCGAATTCGATCATCACGGCTACGACATCCAC | 409142–409165 |
| iniBproSalB | TCATGTCGACTTCCCTTCAATCGAAGAAGCTGTT | 409353–409330 |
| iniBproEcoT20 | CGGCTGAATTCTCCACGGATAAGTTCCGGACCGGC | 409161–409184 |
| iniBproEcoT43 | TTCCGGAATTCCGTAGGGGTGCCCCATTTCCCCTA | 409184–409207 |
| iniBproEcoT65 | CCCATGAATTCTAATCCCCTAACGCGGCGGCCAGG | 409206–409229 |
| iniBproEcoT90 | GGCGGGAATTCCGATCCCGATAGGTGTTTGGCCGG | 409231–409254 |
| iniBproEcoT113 | TTGTTGAATTCGCTTGCGGATCAGACCCCGATTTC | 409254–409277 |
| iniBproEcoT134 | CAGACGAATTCTTCGGGGTGAGGCGGAATCCATAG | 409275–409298 |
| iniBproEcoT154 | AGGCGGAATTCATAGCGTCGATGGCACAGCGCCGG | 409295–409318 |
| iniBproEcoT175 | ATGGCGAATTCCCGGTCACGCCGGCGAACAGCTTC | 409315–409338 |
| iniBproEcoT189 | TCACGGAATTCAACAGCTTCTTCGATTGAAGGGAA | 409330–409348 |
| iniBproSalB193 | AATCGGTCGACCTGTTCGCCGGCGTGACCGGCGCT | 409334–409311 |
| iniBproSalB173 | GGCTGGTCGACCGCTGTGCCATCGACGCTATGGAT | 409314–409291 |
| iniBproSalB193-spa | TTAAGGTCGACggtctccagtcgctcgactCTGTTCGCCGGCGTGACCGGCGCT | 409334–409311b |
| iniBproSalB148 | TCGGTGTCGACCGCCTCACCCCGAAATCGGGGTCT | 409288–409265 |
| iniBprimer ext-1 | CTGCGGAACAGGCTCAGGATGTAA | 409400–409377 |
| iniBprimer ext-2 | GCCCGTCCCGGAGCGGCAACGAAC | 409442–409419 |
| iniBprimer ext-3 | CGATAAGCGAGGTCATCTTCAT | 409375–409354 |
| iniART-T | GCGCTGGCGGGAGATCGTCAATG | 411552–411574 |
| iniART-B | TGCGCAGTCGGGTCACAGGAGTCG | 412043–412020 |
| B1A | ACATACGTACAACTCGAGAGAGGCACTTCGAGATGGCCTT | NA |
| B4B | ACAATGCATTCAGGGGGCGGGTAGAGTGCGCGGTTTCCA | NA |
| HYG1 | TGCCAACTAGTGCCCGTACCCTGTGAATAGA | NA |
| HYG2 | GAGCAAAGCTTGCGTACGATCGACTGCCAGG | NA |
Position for nucleotides after the added restriction site (if any). NA, not applicable.
Position for nucleotides after the spacer sequence.
FIG. 6.
Mapping the transcriptional start sites of the iniBAC operon. Primer extension experiments were performed with end-labeled iniBprimer ext-3 using either no RNA or RNA isolated from BCG cultured in the absence of INH (INH−) or in the presence of INH at a final concentration of 1 μg/ml (INH+). Sequencing reactions were performed with the identical primer and run alongside the primer extension reactions.
Deletion analysis of the iniBAC promoter.
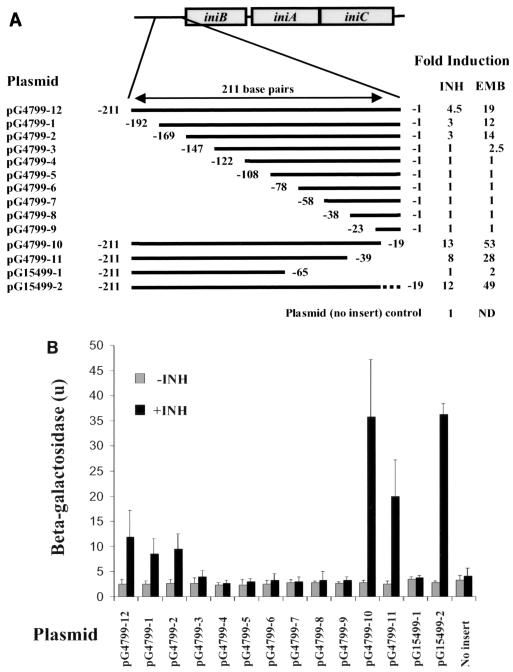
The induction kinetics of the 211-bp M. tuberculosis iniBAC promoter region was investigated in M. smegmatis using strain mc2155(pG4799-12). This strain contained the full-length M. tuberculosis promoter region cloned into the integrating vector pCV125 upstream of lacZ. We observed that M. smegmatis had an induction phenotype similar to that of BCG, although higher concentrations of INH were required because M. smegmatis is relatively INH resistant. Induction by INH (100 μg/ml) and 5 ethambutol (5 μg/ml) was seen as early as 30 min after incubation with antibiotics and was maximal after 4 h of incubation. These conditions were used to investigate the amount of upstream DNA sequence that was required to induce the β-galactosidase reporter. Inserts containing serial 19 to 25-bp 5′ deletions of the 211-bp promoter region were cloned into pCV125 (Fig. 7A), and β-galactosidase activity was measured. Induction by both antibiotics was preserved until a 22-bp region 169 to 147 bp upstream from the translational start site (409184 to 409206) was deleted. After deletion of this sequence, both induction and uninduced expression fell to levels of the promoterless plasmid control (Fig. 7B).
FIG. 7.
Effect of promoter deletions on antibiotic induction. Serial 5′ and 3′ deletions of the 211-bp sequence immediately upstream of the translational start site of iniB were fused to the lacZ gene in the integrating plasmid pCV125 and transformed into M. smegmatis strain mc2155. (A) Schematic of promoter deletions and induction after 4 h of treatment with either INH (100 μg/ml) or ethambutol (EMB; 5 μg/ml). The dotted line indicates the position of a 19-bp spacer that is unrelated to the sequence it replaced; induction represents mean results from at least three experiments. Numbering corresponds to the distance from the translational start site. (B) β-Galactosidase activity of promoter deletions without (grey) or with (black) 4 h of incubation with INH (100 μg/ml). Values represent means of at least three experiments. Error bars represent 95% confidence intervals.
Effects of downstream deletions between the promoter and the translational start site were investigated using serial 3′ deletions (Fig. 7A). Interestingly, a 19-bp deletion 1 to 19 bp from the translational start site (409353 to 409334) increased induction from 4.5- to 13-fold for INH, and from 19- to 53-fold for ethambutol, without changing uninduced expression. The increased induction was not due to changes in the spacing between the promoter and the translational start site of the iniB gene. The higher level of induction continued to be observed after the absolute size of the promoter sequence was restored by replacing the 19-bp 3′ deletion with a 19-bp spacer sequence that was unrelated to the promoter sequence. Further 3′ deletions preserved induction until a 26-bp sequence 39 to 65 bp from the translational start site (409314 to 409288) was removed. When this last sequence was deleted, induction and uninduced expression fell to levels of the promoterless plasmid (Fig. 7B).
DISCUSSION
The iniBAC operon encodes genes that are induced specifically by a broad range of antibiotics that inhibit cell wall biosynthesis. With the exception of cycloserine, the promoter was induced by all clinically relevant antibiotics that act on the M. tuberculosis cell wall. Other toxic stimuli did not induce the promoter, demonstrating the specificity of induction. Significant repression of luciferase activity was observed after treatment with a number of toxic agents that do not act by cell wall inhibition. It is likely that the decreased availability of intracellular ATP levels in the dying cells contributed to this effect.
With the exception of general stress responses, it is extremely uncommon for antibiotics with different mechanisms of action to induce the same set of genes in bacteria. VanA-type vancomycin resistance can be induced by different antibiotics that inhibit cell wall synthesis, possibly through the binding of peptidoglycan precursors in a two-component regulatory system (4, 22, 40). AmpC, the chromosomal β-lactamase in gram-negative bacteria, is induced by different β-lactam antibiotics. Induction is oppositely controlled by cytoplasmic concentrations of biosynthetic and degradative intermediates of murein metabolism (muropeptides) (21). However, VanA-type vancomycin resistance is also induced by cell wall hydrolytic enzymes such as lysozyme (40). We found that the iniBAC promoter was not induced by either lysozyme or granulysin, a molecule released by cytotoxic CD8+ lymphocytes that directly kills extracellular M. tuberculosis by altering the membrane integrity of the bacillus (35). The ampC gene is induced by different β-lactam antibiotics, but these compounds are likely to have similar mechanisms of action. In contrast, the iniBAC promoter is induced by cell wall biosynthesis inhibitors that act on different components of the cell wall. It is possible that the iniBAC operon is induced by osmotic stress; however, we believe this to be unlikely. While antibiotics might lead to alterations in permeability of the bacterial cell wall and subsequent osmotic shock, both lysozyme and granulysin would also be expected to result in permeability changes. Neither of these compounds led to induction of the promoter, suggesting that a different mechanism is involved in induction.
The functions of the genes encoded by the iniBAC operon are unknown. The iniB gene has weak homology to cell wall structural proteins. The iniA and iniC genes are hypothetical proteins with no close homologs (3). Given the induction pattern of this operon, it is possible that these genes participate in the regulation of cell wall growth. It is also possible that the iniBAC operon has either a causal or a protective role in cell death, when killing is initiated by inhibition of cell wall biosynthesis. Induction of the iniBAC operon was not detectable until 4 h of incubation with antibiotics; in contrast, induction of the FAS-II complex and most of the other genes known to be INH induced occurs as early as 20 min after exposure to INH (42). The delay in iniBAC induction corresponds to the time required for INH to decrease the viability of M. tuberculosis in culture (37), suggesting a link between the iniBAC operon and cell death. The rapid induction kinetics of the FAS-II complex and other INH-induced genes closely parallel the repressive effect of INH on mycolate biosynthesis (37). This suggests that unlike the iniBAC operon, these genes are induced by events related to the initial binding of INH to its target. The genes of the iniBAC operon lack close homologs in nonmycobacterial species. However, the iniA and iniB genes were detectable by low-stringency hybridization in the fast-growing and avirulent M. smegmatis and in both virulent and avirulent slow-growing mycobacteria. It is intriguing that only M. avium did not hybridize to either gene probe. It remains possible that these genes are present in M. avium but have insufficient homology to be detected by this method.
Deletion studies of the region upstream of the coding sequences demonstrate that regulatory elements essential for gene expression are located in two regions, an upstream region 169 to 147 bp 5′ from the translational start site (409184 to 409206) and a downstream region 65 to 39 bp 5′ from the translational start site (409314 to 409288) (Fig. 8). The upstream region contains a 10-bp sequence flanked by 6-bp inverted repeats and part of two tandem 8-bp direct repeats. Deletion of this region resulted in decreased induction, suggesting the presence of UP elements, curved sequences, or bent sequences that could influence initiation of transcription (32). The downstream region includes both the transcriptional start site and a sequence with strong homology to −10 promoter sequences (9, 29). The decreased induction that resulted from deletion of this region is in agreement with observations in E. coli that changes (or deletions) in the −10 region and the region immediately downstream of the transcriptional start site strongly influence promoter strength (32). The observation that induction can be increased by deleting the 19-bp 3′ end of this promoter region is intriguing and suggests possible binding of a transcriptional repressor downstream of the transcriptional start site. The increased induction is sequence specific and not due to a change in spacing because replacement of the deletion with a spacer sequence does not alter the increased induction that was observed. Database analysis of regulatory sites has shown that the region downstream of −30 binds repressors almost exclusively, whereas activators bind predominantly to positions between −80 and −30 (18). The possibility that a repressor protein binds to this region is currently being investigated. The repressor sequence also appears to include a ribosomal binding site. It is possible that a second, less apparent ribosomal binding site could exist upstream of this sequence.
FIG. 8.
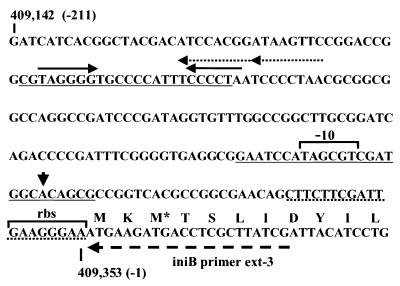
Proposed organization of iniBAC promoter region. Regions found to be essential for induction are underlined with solid lines; the dotted underline indicates the sequence whose deletion leads to increased induction. Also shown are the transcriptional start site (arrowhead), 6-bp inverted repeats (solid arrows), 8-bp direct repeats (dotted arrows), the position of the primer iniBprimer ext-3 used in the primer extension experiments (striped arrow), and possible −10 sequence and ribosomal binding site (rbs). The asterisk marks an alternate translational start site as annotated by Cole et al. (16; NCBI database).
The reporter assays that we describe can be easily adapted to a 96-well plate or solid-phase format. These assays can be used for high-throughput screening of combinatorial libraries with the aim of discovering new classes of compounds that inhibit Mycobacterium cell wall biosynthesis. Assays that are specific for cell wall inhibition may be more useful at finding biologically active compounds than simple screens for inhibition or killing of M. tuberculosis. When testing for inhibitory or cidal compounds through repression of the constitutive luciferase reporter strain BCG(pKB15), highly effective drugs such as INH resulted in little inhibition of luciferase activity. Furthermore, these antibiotics were indistinguishable from relatively ineffective agents such as paraminosalicylic acid using the BCG(pKB15) assay (Fig. 1B).
In conclusion, the iniBAC operon is specifically induced by inhibitors of cell wall biosynthesis. Regulation of transcription is likely to be complex, involving both activator and repressor molecules. Further investigation of the regulatory elements of the iniBAC operon can potentially improve the understanding of the intracellular mechanisms that are activated by cell wall inhibition. Characterization of the proteins encoded by the iniB, iniA, and iniC genes may aid in the development of new antibiotics that may be effective alone or synergistically with other cell wall-active drugs.
ACKNOWLEDGMENTS
We thank Barry R. Bloom and Oren Zimhony for advice and support, and we thank Rosaria Cerny for laboratory assistance. We also thank Graham Hatfull and MedImmune Inc. for use of unpublished vectors, Robert Modlin for his gift of granulysin, and J. P. Welsh for his gift of 5-chloropyrazinamide.
This work was supported by National Institutes of Health grants AI45244 and AI43268 and by the Howard Hughes Medical Research Institute.
REFERENCES
- 1.Ad Hoc Committee of the Scientific Assembly on Microbiology, Tuberculosis, and Pulmonary Infections. Treatment of tuberculosis and tuberculosis infection in adults and children. Clin Infect Dis. 1995;21:9–27. [Google Scholar]
- 2.Alland D, Kalkut G E, Moss A R, McAdam R A, Hahn J A, Bosworth W, Drucker E, Bloom B R. Transmission of tuberculosis in New York City: an analysis by DNA fingerprinting and conventional epidemiologic methods. N Engl J Med. 1994;330:1710–1716. doi: 10.1056/NEJM199406163302403. [DOI] [PubMed] [Google Scholar]
- 3.Alland D, Kramnik I, Weisbrod T R, Otsubo L, Miller L P, Jacobs W R, Jr, Bloom B R. Identification of differentially expressed mRNA in prokaryotic organisms by customized amplification libraries (DECAL): the effect of isoniazid on gene expression in Mycobacterium tuberculosis. Proc Nat Acad Sci USA. 1998;22:13227–13232. doi: 10.1073/pnas.95.22.13227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen N E, Hobbs J N. Induction of vancomycin resistance in Enterococcus faecium by non-glycopeptide antibiotics. FEMS Microbiol Lett. 1995;132:107–114. doi: 10.1111/j.1574-6968.1995.tb07819.x. [DOI] [PubMed] [Google Scholar]
- 5.Arthur M, Molinas C, Courvalin P. The VanK-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1992;174:2582–2591. doi: 10.1128/jb.174.8.2582-2591.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balasubramanian V, Pavelka M S, Jr, Bardarov S S, Martin J, Weisbrod T R, McAdam R A, Bloom B R, Jacobs W R., Jr Allelic exchange in Mycobacterium tuberculosis with long linear recombination substrates. J Bacteriol. 1996;178:273–279. doi: 10.1128/jb.178.1.273-279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um K S, Wilson T, Collins D, de Lisle G, Jacobs W R., Jr inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science. 1994;263:227–230. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 8.Bange F C, Brown A M, Jacobs W R., Jr Leucine auxotrophy restricts growth of Mycobacterium bovis BCG in macrophages. Infect Immun. 1996;64:1794–1799. doi: 10.1128/iai.64.5.1794-1799.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bashym M D, Kaushal D, Sujoy D K, Tyagi A K. A study of the mycobacterial transcriptional apparatus: identification of novel features in promoter elements. J Bacteriol. 1996;178:4847–4853. doi: 10.1128/jb.178.16.4847-4853.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Besra G S, Morehouse C B, Rittner C M, Waechter C J, Brennan P J. Biosynthesis of mycobacterial lipoarabinomannan. J Biol Chem. 1997;272:18460–18466. doi: 10.1074/jbc.272.29.18460. [DOI] [PubMed] [Google Scholar]
- 11.Besra G S, Brennan P J. The mycobacterial cell wall: biosynthesis of arabinogalactan and lipoarabinomannan. Biochem Soc Trans. 1997;25:845–850. doi: 10.1042/bst0250845. [DOI] [PubMed] [Google Scholar]
- 12.Belanger A E, Besra G S, Ford M E, Mikusova K, Belisle J T, Brennan P J, Inamine J M. The embAB genes of Mycobacterium avium encode an arabinosyl transferase involved in cell wall arabinan biosynthesis that is the target for the antimycobacterial drug ethambutol. Proc Natl Acad Sci USA. 1996;93:11919–11924. doi: 10.1073/pnas.93.21.11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Best G K, Best N H, Koval A V. Evidence of participation of autolysins in bactericidal action of oxacillin of Staphylococcus aureus. Antimicrob Agents Chemother. 1974;6:825–830. doi: 10.1128/aac.6.6.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown P O, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999;21:33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- 15.Centers for Disease Control. Estimates of future global tuberculosis morbidity and mortality. Morbid Mortal Weekly Rep. 1993;42:961–964. [PubMed] [Google Scholar]
- 16.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier L, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B G, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 17.Fischl M A, Uttamchandani R B, Daikos G L, Poblete R B, Moreno J N, Reyes R R. An outbreak of tuberculosis caused by multiple-drug-resistant tubercle bacilli among patients with HIV infection. Ann Intern Med. 1992;117:177–183. doi: 10.7326/0003-4819-117-3-177. [DOI] [PubMed] [Google Scholar]
- 18.Gralla J D, Collado-Vides J. Organization and function of transcription regulatory elements. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 1232–1245. [Google Scholar]
- 19.Horne D, Tomasz A. Tolerant response of Streptococcus sanguis to beta-lactams and other cell wall inhibitors. Antimicrob Agents Chemother. 1977;11:888–896. doi: 10.1128/aac.11.5.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu Y, Coates A R M. Transcription of two sigma 70 homologue genes, sigA and sigB, in stationary-phase Mycobacterium tuberculosis. J Bacteriol. 1999;181:469–476. doi: 10.1128/jb.181.2.469-476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobs C, Frere J M, Normark S. Cytosolic intermediates for cell wall biosynthesis and degradation control inducible biosynthesis and degradation control inducible β-lactam resistance in gram-negative bacteria. Cell. 1997;88:823–832. doi: 10.1016/s0092-8674(00)81928-5. [DOI] [PubMed] [Google Scholar]
- 22.Lai M H, Kirsch D R. Induction signals for vancomycin resistance encoded by the vanA gene cluster in Enterococcus faecium. Antimicrob Agents Chemother. 1996;40:1645–1648. doi: 10.1128/aac.40.7.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee R E, Brennan P J, Besra G S. Mycobacterium tuberculosis cell envelope. Curr Top Microbiol Immunol. 1996;215:1–27. doi: 10.1007/978-3-642-80166-2_1. [DOI] [PubMed] [Google Scholar]
- 24.Mandell G E, Douglas G R, Jr, Bennett J E. Principles and practice of infectious diseases. 4th ed. New York, N.Y: Churchill Livingstone; 1995. [Google Scholar]
- 25.Mdluli K, Slayden R A, Zhu Y, Ramaswamy S, Pan X, Mead D, Crane D D, Musser J M, Barry C E., III Inhibition of a Mycobacterium tuberculosis β-ketoacyl ACP synthase by isoniazid. Science. 1998;280:1607–1710. doi: 10.1126/science.280.5369.1607. [DOI] [PubMed] [Google Scholar]
- 26.Movahedzadeh F, Colston J M, Davis E O. Determination of DNA sequences required for regulated Mycobacterium tuberculosis RecA expression in response to DNA-damaging agents suggests that two modes of regulation exist. J Bacteriol. 1997;179:3509–3518. doi: 10.1128/jb.179.11.3509-3518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michele T M, Ko C, Bishai W R. Exposure to antibiotics induces expression of the Mycobacterium tuberculosis sigF gene: implications for chemotherapy against mycobacterial persistors. Antimicrob Agents Chemother. 1999;43:218–225. doi: 10.1128/aac.43.2.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mikusova K, Mikus M, Besra G S, Hancock I, Brennan P J. Biosynthesis of the linkage region of the mycobacterial cell wall. J Biol Chem. 1996;271:7820–7828. doi: 10.1074/jbc.271.13.7820. [DOI] [PubMed] [Google Scholar]
- 29.Mulder M A, Zappe H, Steyn L M. Mycobacterial promoters. Tuberc Lung Dis. 1997;78:211–223. doi: 10.1016/s0962-8479(97)90001-0. [DOI] [PubMed] [Google Scholar]
- 30.Pablos-Mendez A, Raviglione M C, Laszlo A, Binkin N, Rieder H L, Bustreo F, Cohn D L, Lambregts-van Weezenbeek C S, Kim S J, Chaulet P, Nunn P. Global surveillance for antituberculosis-drug resistance, 1994–1997. World Health Organization-International Union against Tuberculosis and Lung Disease Working Group on Anti-Tuberculosis Drug Resistance Surveillance. N Engl J Med. 1998;338:1641–1649. doi: 10.1056/NEJM199806043382301. [DOI] [PubMed] [Google Scholar]
- 31.Pascopella L, Collins F M, Martin J M, Lee M H, Hatfull G F, Stover C K, Bloom B R, Jacobs W R., Jr Use of in vivo complementation in Mycobacterium tuberculosis to identify a genomic fragment associated with virulence. Infect Immun. 1994;62:1313–1319. doi: 10.1128/iai.62.4.1313-1319.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Record M T, Jr, Reznikoff W S, Craig M L, McQuade K L, Schlax P J. Escherichia coli RNA polymerase (Eδ70), promoters, and the kinetics of the steps of transcription initiation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 792–820. [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 34.Snapper S B, Melton R E, Mustafa S, Kieser T, Jacobs W R., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 35.Stenger S, Hanson D A, Teitelbaum R, Dewan P, Niazi K R, Froelich C J, Ganz T, Thoma-Uszynski S, Melian A, Bogdan C, Porcelli S A, Bloom B R, Krensky A M, Modlin R L. An antimicrobial activity of cytolytic T cells mediated by granulysin. Science. 1998;282:121–125. doi: 10.1126/science.282.5386.121. [DOI] [PubMed] [Google Scholar]
- 36.Stover C K, de la Cruz V F, Fuerst T R, Burlein J E, Benson L A, Bennett L T, Bansal G P, Young J F, Lee M H, Hatfull G F, et al. New use of BCG for recombinant vaccines. Nature. 1991;351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 37.Takayama K, Wang L, Hugo D L. Effect of isoniazid on the in vivo mycolic acid synthesis, cell growth, and viability of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1972;2:29–35. doi: 10.1128/aac.2.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Telenti A, Philipp W J, Sreevatsan S, Bernasconi C, Stockbauer K E, Wieles B, et al. The emb operon, a gene cluster of Mycobacterium tuberculosis involved in resistance to ethambutol. Nat Med. 1997;3:567–570. doi: 10.1038/nm0597-567. [DOI] [PubMed] [Google Scholar]
- 39.Tomasz A, Albino A, Zanati E. Multiple antibiotic resistance in a bacterium with suppressed autolytic system. Nature. 1970;227:138–140. doi: 10.1038/227138a0. [DOI] [PubMed] [Google Scholar]
- 40.Ulijasz A T, Grenader A, Weisblum B. A vancomycin-inducible LacZ reporter system in Bacillus subtilis: induction by antibiotics that inhibit cell wall synthesis and by lysozyme. J Bacteriol. 1996;178:6305–6309. doi: 10.1128/jb.178.21.6305-6309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Via L E, Curcic R, Mudd M H, Dhandayuthapani S, Ulmer R J, Deretic V. Elements of signal transduction in Mycobacterium tuberculosis: in vitro phosphorylation and in vivo expression of the response regulator MtrA. J Bacteriol. 1996;178:3314–3321. doi: 10.1128/jb.178.11.3314-3321.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson M, DeRisi J, Kristensen H H, Imboden P, Rane S, Brown P O, Schoolnik G K. Exploring drug-induced alterations in gene expression in mycobacterium tuberculosis by microarray hybridization. Proc Natl Acad Sci USA. 1999;96:12833–12838. doi: 10.1073/pnas.96.22.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Heym B, Allen B, Young D, Cole S. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]