Abstract
The trp RNA-binding attenuation protein (TRAP) regulates expression of the Bacillus subtilis trpEDCFBA operon by a novel transcription attenuation mechanism. Tryptophan-activated TRAP binds to the nascent trp leader transcript by interacting with 11 (G/U)AG repeats, 6 of which are present in an antiterminator structure. TRAP binding to these repeats prevents formation of the antiterminator, thereby promoting formation of an overlapping intrinsic terminator. A third stem-loop structure that forms at the extreme 5′ end of the trp leader transcript also plays a role in the transcription attenuation mechanism. The 5′ stem-loop increases the affinity of TRAP for trp leader RNA. Results from RNA structure mapping experiments demonstrate that the 5′ stem-loop consists of a 3-bp lower stem, a 5-by-2 asymmetric internal loop, a 6-bp upper stem, and a hexaloop at the apex of the structure. Footprinting results indicate that TRAP interacts with the 5′ stem-loop and that this interaction differs depending on the number of downstream (G/U)AG repeats present in the transcript. Expression studies with trpE′-′lacZ translational fusions demonstrate that TRAP-5′ stem-loop interaction is required for proper regulation of the trp operon. 3′ RNA boundary experiments indicate that the 5′ structure reduces the number of (G/U)AG repeats required for stable TRAP-trp leader RNA association. Thus, TRAP-5′ stem-loop interaction may increase the likelihood that TRAP will bind to the (G/U)AG repeats in time to block antiterminator formation.
Expression of the Bacillus subtilis tryptophan biosynthetic genes is regulated in response to changes in the intracellular level of tryptophan by the trp RNA-binding attenuation protein (TRAP) (4, 16). The trpEDCFBA operon is regulated by TRAP-mediated transcription attenuation (5, 10, 17, 19, 24, 25) and translational control mechanisms (14, 19, 22). TRAP also regulates expression of the unlinked trpG gene at the translational level (8, 15, 30). TRAP exists as a complex consisting of 11 identical subunits arranged in a single ring termed the β-wheel (1, 3). Tryptophan cooperatively activates TRAP by binding between every adjacent TRAP subunit (3, 6).
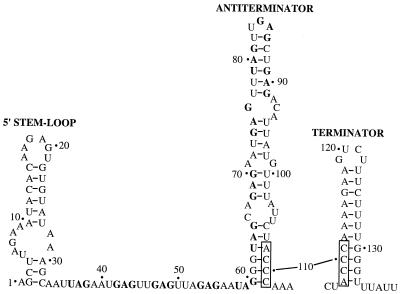
The 203-nucleotide untranslated trp operon leader transcript can fold into three distinct RNA secondary structures that participate in transcription attenuation (Fig. 1). When TRAP is activated by tryptophan, 11 KKR motifs that outline the periphery of the TRAP complex can bind to 11 closely spaced (G/U)AG repeats present in the nascent trp leader transcript, thereby wrapping the RNA around the periphery of the TRAP complex (2, 8, 31). TRAP binding blocks formation of the antiterminator since six of the (G/U)AG repeats are present within this RNA structure (5, 8). Thus, TRAP binding promotes formation of the overlapping intrinsic terminator which results in transcription termination before RNA polymerase can reach the trp operon structural genes. In the absence of TRAP binding, formation of the antiterminator permits transcription of the entire operon (5).
FIG. 1.
Nucleotide sequence of the B. subtilis trp leader transcript showing the 5′ stem-loop and the mutually exclusive antiterminator and terminator structures. Boxed nucleotides mark overlapping segments of the competing secondary structures. The (G/U)AG repeats known to be involved in TRAP-RNA recognition are indicated by boldface type. Numbering is from the start of transcription. RNA secondary structure predictions were performed using MFOLD (29, 32). Note that the 5′ stem-loop is modified from the structure predicted by MFOLD due to the RNA secondary structure mapping data obtained during the course of these studies.
While it is not known how TRAP initially interacts with the nascent trp leader transcript, the interaction must occur quickly to prevent formation of the antiterminator structure. During attenuation regulation of the Escherichia coli trp operon, transcriptional pausing allows the regulatory ribosome to bind to the leader transcript at an appropriate time (20). Since leader peptide synthesis is not involved in transcription attenuation of the B. subtilis trp operon, nor has RNA polymerase pausing been demonstrated to play a role in this regulatory mechanism, we were interested in determining if any factor besides TRAP and the (G/U)AG repeats were involved in TRAP interaction with the nascent trp leader transcript.
We recently demonstrated that, in addition to the antiterminator and terminator, an RNA structure predicted to form at the extreme 5′ end of the nascent trp leader transcript participates in the transcription attenuation mechanism (28). Deletion or disruption of this putative structure resulted in a dramatic increase of trp operon expression in vivo and increased transcriptional readthrough in vitro. This previous study also demonstrated that the 5′ stem-loop functions primarily in TRAP-dependent regulation of the trp operon and that overexpression of TRAP suppressed the defect associated with the 5′ stem-loop deletion. Moreover, we showed that the presumed 5′ structure increased the affinity of TRAP for trp leader RNA (28). Thus, it was possible that the 5′ stem-loop participated in the attenuation mechanism by interacting with TRAP.
In the present study we determined the secondary structure of the 5′ stem-loop and found that TRAP interacts with this structure. We also established that the 5′ stem-loop reduces the number of (G/U)AG repeats required for stable TRAP-trp leader RNA association and that the TRAP-5′ stem-loop interaction differs depending on the number of downstream (G/U)AG repeats that are present in the transcript. Our results suggest that the TRAP-5′ stem-loop interaction increases the probability that TRAP will bind to the (G/U)AG repeats before the antiterminator can form, thereby increasing the likelihood that transcription termination occurs before RNA polymerase can reach the trp operon structural genes.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All of the B. subtilis strains used in this study are listed in Table 1. The plasmids pTZ18U (Stratagene) and pPB77, pPB78, pPB82, and pPB83 (8) have been described. Plasmid pPB310 contains nucleotides 32 to 111 of the B. subtilis trp leader region and was constructed by PCR. The resulting PCR product was digested with EcoRI and BamHI and subcloned into the EcoRI and BamHI sites of the pTZ18U polylinker. Plasmid pHD55, which contains nucleotides 1 to 36 of the B. subtilis trp leader, was also constructed by PCR. In this case the PCR product was digested with EcoRI and KpnI and ligated into the EcoRI and KpnI sites of the pTZ18U polylinker. Plasmid pHD68 was constructed by digesting pHD55 with EcoRI and treating it with mung bean nuclease to remove the cohesive ends followed by self-ligation. Plasmid pHD34 contains the trp promoter and nucleotides 6 to 203 Δ(+1 to +5) of the trp leader. This plasmid was constructed by a two-step process using overlap extension PCR. The final PCR product was digested with EcoRI and HindIII and subcloned into the EcoRI and HindIII sites of PTZ18U. pHD40 carries the trp promoter and a mutant trp leader in which nucleotides 6 to 9 were replaced with a T residue, while pHD46 carries the trp promoter and a leader containing nucleotides 16 to 203 Δ(+1 to +15). Both of these plasmids were constructed in the same manner as pHD34. The B. subtilis integration vector, ptrpBG1-PLK, used for the generation of trpE′-′lacZ translational fusions was described previously (22). The plasmids pHD52, pHD53, and pHD54, which contain trpE′-′lacZ fusions, were constructed by subcloning the trp promoter and leader region from pHD34, pHD40, and pHD46 into the EcoRI and HindIII sites of the ptrpBG1-PLK polylinker, respectively. The three plasmids pHD52, pHD53, and pHD54 were linearized with SalI and separately integrated into the amyE locus of B. subtilis W168. The resulting strains are PLBS138, PLBS139, and PLBS140.
TABLE 1.
B. subtilis strains used in this study
| Strains | Genotypea | Source or reference |
|---|---|---|
| W168 | Prototroph | BGSCc |
| PLBS44 | amyE::[trpP(−412 to +203) trpE′-′lacZ Cmr] | 28 |
| PLBS104 | amyE::[trpP(−412 to +203) Δ(+3 to +32)trpE′-′lacZ Cmr] | 28 |
| PLBS138 | amyE::[trpP(−412 to +203) Δ(+1 to +5)trpE′-′lacZ Cmr] | This study |
| PLBS139 | amyE::[trpP(−412 to +203) Δ(+6 to +9)Tb trpE′-′lacZ Cmr] | This study |
| PLBS140 | amyE::[trpP(−412 to +203) Δ(+1 to +15)trpE′-′lacZ Cmr] | This study |
trpP denotes the trp promoter. Prime indicates truncation of the gene. −412 to +203 indicates the DNA fragment containing the trp promoter and neighboring regions that was incorporated relative to the transcription start site. “Δ” designates the portion of the leader region that was deleted.
T was substituted for nucleotides 6 to 9.
BGSC, Bacillus Genetic Stock Center, Ohio State University, Columbus, Ohio.
β-Galactosidase assay.
Cells were cultured in minimal Spizizen salts medium (27) containing 0.2% acid-hydrolyzed casein, 0.2% glucose, and 5 μg of chloramphenicol per ml in the presence or absence of 50 μg of tryptophan per ml. Cells were harvested in mid-exponential phase, and cell suspensions were prepared as previously described (28). β-Galactosidase activity was subsequently assayed by the method of Miller (23).
In vitro transcription.
Gel-purified transcripts used in this analysis were synthesized by using the Ambion MEGAscript in vitro transcription kit. Templates consisted of various plasmids that had been linearized with BamHI or HindIII. 5′-End-labeled RNAs were generated by treating in vitro-generated transcripts with calf intestinal phosphatase and subsequently with polynucleotide kinase and [γ-32P]ATP. The unlabeled and labeled RNA was gel purified as previously described (14).
Gel mobility shift assay.
The binding affinity between TRAP and trp leader RNA was estimated by using gel mobility shift assays by modifying a previously published procedure (28). TRAP was purified as described earlier (5). Transcripts used in the analysis were generated from pPB77 (wild type), pPB310 (5′ stem-loop deletion), or pHD68 (5′ stem-loop only) that had been linearized with BamHI. Binding reactions (8 μl) containing 0.2 nM 5′-end-labeled RNA, various concentrations of TRAP (TRAP excess), 1 mM tryptophan in 50 mM Tris-acetate (pH 8.0), 4 mM magnesium acetate, 5 mM dithiothreitol, 10% glycerol, 0.2 mg of E. coli tRNA per ml, 0.1 mg of xylene cyanol per ml, and 400 U of RNasin (Promega) per ml were incubated at 25°C for 20 min. Aliquots of reaction mixtures were fractionated through native polyacrylamide gels containing 375 mM Tris-HCl (pH 8.8), 5% glycerol, and 1 mM EDTA. Electrophoresis was performed at room temperature in running buffer containing 25 mM Tris-glycine (pH 8.3) and 1 mM EDTA. Gels were dried, and the bound and free RNA bands were quantified by using a PhosphorImager (Molecular Dynamics) and the ImageQuant software package. Modifications of the standard reaction are described in the text or the appropriate figure legend. The binding data were fit to the simple binding equation: RNAb = a[TRAP]f/(Kd + [TRAP]f), where a is the maximal fraction of bound RNA (RNAb) that is approximately equal to 1; Kd is defined as the concentration of free protein, [TRAP]f, at which the RNAb reaches 50% saturation; RNAb is the fraction of RNA bound between 0 and 1; and [TRAP]f is the concentration of free TRAP 11-mer which was assumed to be the concentration of total TRAP added since the total TRAP concentration was in at least 12-fold molar excess over RNA.
RNA structure mapping.
RNA structures were predicted by using the MFOLD program (29, 32). RNA structure mapping using unlabeled transcripts followed previously published procedures (14). The unlabeled transcripts used in this analysis were generated from pPB83 linearized with HindIII as template. Titrations of RNases and chemical reagents were routinely performed to determine the amount of each reagent that would prevent multiple cleavages or chemical modifications in any one transcript so that we could minimize the potential of secondary rearrangements in short RNA segments. RNA samples were partially digested with RNase T1 (Gibco-BRL) or RNase V1 (Pharmacia) and recovered as described earlier (14). CMCT and DMS modification reactions, as well as the subsequent recovery of RNA samples, followed a previously published procedure (14). RNA samples were resuspended in primer extension buffer and hybridized to a γ-32P-end-labeled primer, and the primers were extended with Moloney murine leukemia virus (MMLV) reverse transcriptase (U.S. Biochemicals) as described elsewhere (14). After 10 min at 42°C, reactions were terminated by the addition of 3 μl of standard sequencing stop solution. Samples were fractionated through 6% denaturing polyacrylamide gels. Control sequencing reactions were carried out using the same plasmids and end-labeled primer as described above.
5′-end-labeled RNA (see above) generated from various templates (pPB77, pPB78, pPB82, pPB83, or pHD68 digested with BamHI) was renatured by heating at 95°C for 1 min, followed by a 10-min incubation at 37°C. RNA was digested with 0.07 U of RNase V1 per ml for 10 min at 37°C in 40 mM Tris-HCl (pH 8.0)–250 mM KCl–4 mM MgCl2 (TKM buffer). Samples were fractionated through 6% denaturing polyacrylamide gels. The G sequencing ladder was generated by partial RNase T1 digestion under denaturing conditions as described previously (11). Alkali digestion ladders were prepared as described elsewhere (13) from the same end-labeled transcripts.
3′-boundary analysis.
The 3′-boundary analysis followed a published procedure (11). 5′-end-labeled transcripts (see above) generated from pPB77 (wild-type trp leader) or pPB310 (5′ stem-loop deletion trp leader) were treated with alkali to generate an RNA ladder. Then, 100-μl RNA samples (10 pmol) were incubated for 5 min at 95°C in alkaline hydrolysis buffer (100 mM NaHCO3-Na2CO3 [pH 9.0]–2 mM EDTA–0.5 μg of E. coli tRNA per μl) and then recovered by ethanol precipitation. Hydrolyzed RNAs were mixed with 50 μg of TRAP and incubated at 25°C for 20 min in TKM buffer. The reaction mixtures were fractionated through 6% native polyacrylamide gels. Bound and unbound transcripts were visualized by autoradiography, excised from the gel, and subsequently eluted from the gel. RNAs were ethanol precipitated and fractionated through 6% denaturing polyacrylamide gels. RNase T1 and alkali digestion ladders of the same 5′-end-labeled transcripts were used as molecular size standards.
RESULTS
Gel mobility shift analysis of TRAP and trp leader RNA.
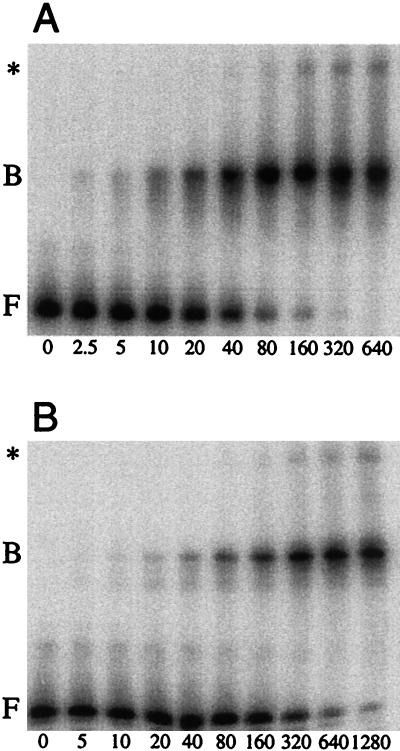
Results from previous in vivo experiments demonstrated that overexpression of mtrB, the gene encoding TRAP (17), suppressed the defect associated with deletion of the 5′ stem-loop (28). Using gel mobility shift assays we further showed that the 5′ stem-loop increases the affinity of TRAP for trp leader RNA approximately fivefold (28). In the previous study (28) we observed a TRAP-dependent band that migrated just behind the free RNA. We assumed that this band resulted from TRAP-trp leader RNA complex dissociation soon after loading the gel. When we repeated the analysis using a modified gel shift procedure (see Materials and Methods) the presence of this band was eliminated, confirming our previous assumption. As was previously observed (28), the presence of the 5′ structure in transcripts that contained all 11 (G/U)AG repeats increased the affinity of TRAP for trp leader RNA (Fig. 2). Binding to the wild-type trp leader transcript was detectable at 2.5 nM TRAP and saturated at approximately 320 nM TRAP (Fig. 2A). With the 5′ stem-loop deletion transcript, comparable binding was detected at 5 nM TRAP but did not reach saturation even at a concentration of 1.28 μM TRAP (Fig. 2B). In each case we observed a prominent shifted complex. Note that we also observed two additional shifted complexes for each of these transcripts. One of these complexes is shown (∗), while the other extremely faint complex is not. Note that these complexes were not observed in our previous study (28). While the most prominent shifted species probably consists of complexes containing one TRAP 11-mer bound to a single trp leader transcript, the composition of the other shifted species is not known. We fit these data to a simple binding equation by using nonlinear least-squares analysis. This method yielded estimated Kd values of 26 ± 5 nM TRAP for the wild-type transcript and 280 ± 50 nM for the 5′ stem-loop deletion transcript. The small difference in these values from those observed previously (28) probably reflects the different binding and gel-running conditions used in the current study.
FIG. 2.
Gel mobility shift analysis of TRAP complexed with wild-type or 5′ stem-loop deletion trp leader transcripts. 5′-end-labeled trp leader transcripts (0.2 nM) were incubated with 1 mM tryptophan and the concentration of TRAP indicated at the bottom of each lane (nanomolar). Each transcript contained the 11 (G/U)AG repeats between nucleotides 36 and 91. Bands corresponding to free (F) and bound (B or ∗) RNA are indicated on the left. (A) Wild-type trp leader transcripts. (B) 5′ stem-loop deletion trp leader transcripts.
The finding that the 5′ stem-loop increased the affinity of TRAP for trp leader RNA approximately 10-fold suggested that the 5′ stem-loop interacted with TRAP. When we performed gel shift experiments with transcripts derived from plasmid pHD68 that only contained the 5′ stem-loop, we did not detect any evidence of TRAP binding (data not shown). We also performed RNA competition experiments with wild-type trp leader transcripts and transcripts that only contained the 5′ stem-loop. While the unlabeled wild-type trp leader transcript was able to compete for TRAP binding to labeled wild-type and 5′ stem-loop deletion trp leader transcripts, the transcript that only contained the 5′ stem-loop only competed away the higher-shifted complexes (∗) (data not shown). Since the RNA that only contained the 5′ stem-loop was an ineffective competitor, these results suggest that TRAP–5′ stem-loop RNA interaction does not involve the KKR motifs known to interact with the (G/U)AG repeats (2, 31).
5′ stem-loop structure mapping.
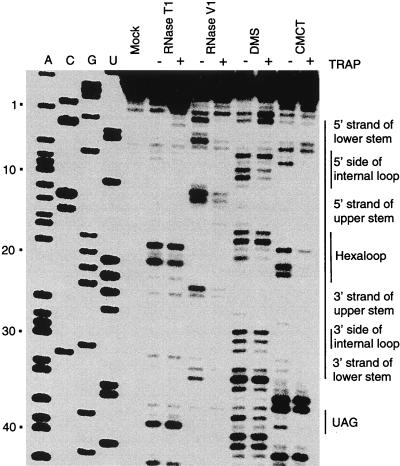
To determine if the predicted 5′ structure actually formed in the trp leader transcript, we probed the structure of a transcript containing the first 68 nucleotides of the trp leader in vitro with structure-specific enzymatic and chemical reagents. This transcript contained the predicted 5′ stem-loop and the first six (G/U)AG repeats known to interact with TRAP, as well as four upstream and downstream nucleotides derived from the vector. Note that computer predictions indicated that these additional residues do not interfere with 5′ stem-loop formation. trp leader transcripts were subjected to partial digestion or chemical modification using RNase T1, RNase V1, DMS, or CMCT. The sites of nuclease cleavage or chemical modification were mapped by primer extension using an end-labeled primer and MMLV reverse transcriptase. Cleavage or chemical modification would give rise to a primer extension product one nucleotide shorter than the corresponding band in the sequencing lane.
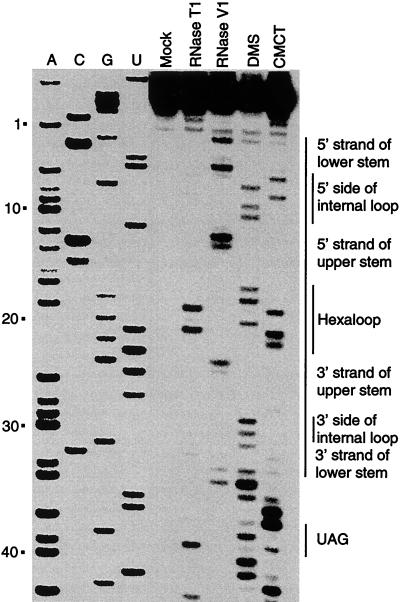
The results of the structure mapping experiments are shown in Fig. 3 and summarized in Fig. 4. The computer-predicted structure of the 5′ stem-loop is identical to the experimentally determined structure except that U5 and A29 are predicted to pair as are A16 and U21. RNase T1 cleaves following unpaired G residues. We observed prominent RNase T1 cleavage following the G residues at positions 18, 20, 38, and 42, indicating that these residues are single stranded. Note that bands corresponding to the G residues at positions 2, 7, 22, 24, and 31 were not detected, suggesting that these residues were base paired (Fig. 3 and 4). With the exception of G7, these results are consistent with the computer predicted secondary structure. RNase V1 is generally specific for base-paired residues; however, this enzyme does not cleave all paired residues, and it sometimes cleaves the first few bases in a single-stranded RNA segment that is adjacent to an RNA duplex (26). We observed prominent RNase V1 cleavage following A1, U4, U11, A12, and U23, as well as weak RNase V1 cleavage following C3, A10, G24, C32, and A33, suggesting that these residues are base paired (Fig. 3 and 4). These results are consistent with the predicted 5′ stem-loop structure. Note that the cleavage of A1 and A33 is likely due to their position immediately adjacent to the lower stem of the structure.
FIG. 3.
5′ stem-loop structure mapping. RNA containing nucleotides 1 to 68 of the trp leader transcript was used in this analysis (Fig. 1). trp leader RNA was treated with RNase T1, RNase V1, DMS, or CMCT. Residues that were cleaved by RNase T1 or RNase V1 or modified by DMS or CMCT were detected by primer extension by using MMLV reverse transcriptase. The mock-treated control lane without enzymatic or chemical treatment is indicated. Note that the bands observed in the treated lanes are one nucleotide shorter than the corresponding bands in the A, C, G, or U sequencing lanes. The positions of the nucleotides corresponding to the lower stem, the internal loop, the upper stem, the hexaloop, and the first (G/U)AG repeat (UAG) are indicated at the right. Numbering at the left corresponds to the DNA sequencing ladder and is from the start of transcription.
FIG. 4.
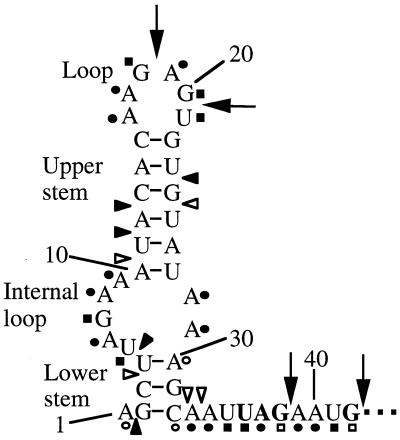
Summary of the 5′ stem-loop structure mapping results. This figure is adapted from the data presented in Fig. 3. Positions of cleavage by the single-stranded probe RNase T1 are indicated by arrows. Positions of cleavage by the double-stranded probe RNase V1 are indicated by arrowheads. Positions of RNA modification using the single-stranded probe DMS (circles) or CMCT (squares) are also indicated. Filled arrowheads, circles, or squares indicate strong modification or cleavage, whereas open arrowheads, circles, or squares indicate weak modification or cleavage. Numbering is from the start of transcription.
To determine the structure of the 5′ stem-loop more precisely, chemical modification experiments with DMS and CMCT were carried out. DMS methylates N1 of adenine and N3 of cytosine when the residues are single stranded, whereas CMCT modifies unpaired G and U residues at the N1 and N3 positions, respectively. These DMS- and CMCT-modified residues are unable to serve as templates for reverse transcriptase. We observed DMS signals at the A and C residues corresponding to positions 1, 6, 8, 9, 16, 17, 19, 28, 29, 30, 32, 33, 34, 37, 39, and 40, suggesting that these residues are single stranded. However, the relatively weak DMS signals at positions 1, 30, and 32 suggest that these residues can be paired or unpaired. The absence of DMS modification of the remaining A and C residues suggests that these residues are base paired. With the exception of A16 and A29, these results are consistent with the predicted structure (Fig. 3 and 4). We observed prominent CMCT signals at the U and G residues corresponding to positions 5, 7, 18, 20, 21, 35, 36, 38, and 41, indicating that these residues are single stranded. The absence of CMCT modification of the remaining U and G residues suggests that these nucleotides are base paired (Fig. 3 and 4). With the exception of G7, which was not cleaved by RNase T1, the CMCT results are consistent with the other reagents tested. Taken together, the results of the structure mapping experiments are consistent with the structure shown in Fig. 4, although it appears that the lower stem is relatively unstable. The structure that we determined differs from the predicted structure by two base pairs. The predicted U5-A29 and the A16-U21 base pairs were not detected in the 5′ stem-loop secondary structure, suggesting the existence of a larger asymmetric internal loop and hairpin loop, respectively (Fig. 4). When taken together, our structure mapping results indicate that the 5′ stem-loop consists of a 3-bp lower stem, a 5-by-2 asymmetric internal loop, a 6-bp upper stem, and a hexaloop at the apex of the structure.
TRAP interacts with the 5′ stem-loop.
Our gel shift analysis indicated that the 5′ stem-loop increases the affinity of TRAP for trp leader RNA but provided little evidence that TRAP interacts with the 5′ structure. We performed TRAP-trp leader RNA footprint experiments to determine if TRAP interacts with the 5′ stem-loop (Fig. 5). We used the same in vitro-generated trp leader transcript (positions 1 to 68), chemical and enzymatic probes, and 5′-end-labeled primer used for the structure-mapping experiments (see above). The cleavage pattern with RNase V1 was dramatically altered when TRAP was bound to the trp leader transcript. Bound TRAP reduced or prevented RNase V1 cleavage at every 5′ stem-loop residue that was cleaved in the absence of TRAP (Fig. 5). Since RNase V1 is generally specific for double-stranded RNA, these results suggested that TRAP bound to the 5′ structure and prevented cleavage. In sharp contrast, the RNase T1 cleavage pattern within the 5′ stem-loop was only slightly altered when TRAP was bound to the transcript (Fig. 5). Interestingly, the RNase T1 cleavage pattern suggests that the GAG sequence in the loop of the 5′ structure does not interact with a TRAP KKR motif (Fig. 4 and 5). Previous results demonstrated that both G residues in GAG repeats are strongly protected from RNase T1 cleavage by bound TRAP (8, 15). This finding is consistent with a previous in vivo study where it was determined that changing this sequence to GUG had little effect on trp operon expression (28). Note that, with the exception of the first UAG repeat, bound TRAP prevented or reduced cleavage of the G residues in the (G/U)AG repeats that were previously shown to interact with TRAP (data not shown) (18).
FIG. 5.
TRAP–5′ stem-loop RNA footprint. RNA containing nucleotides 1 to 68 of the trp leader transcript was used in this analysis (Fig. 1). trp leader RNA was treated with RNase T1, RNase V1, DMS, or CMCT in the presence (+) or absence (−) of bound TRAP. Residues that were cleaved by RNase T1 or RNase V1 or modified by DMS or CMCT were detected by primer extension by using MMLV reverse transcriptase. The mock-treated control lane without enzyme or chemical treatment is indicated. Note that the bands observed in the treated lanes are one nucleotide shorter than the corresponding bands in the A, C, G, or U sequencing lanes. The positions of the nucleotides corresponding to the lower stem, the internal loop, the upper stem, the loop, and the first (G/U)AG repeat (UAG) are indicated on the right. The numbering on the left corresponds to the DNA sequencing ladder and is from the start of transcription.
As was observed for RNase V1 cleavage, the DMS and CMCT RNA modification patterns were significantly altered when TRAP was bound to the trp leader transcript. We found that bound TRAP protected A8, A9, A19, A30, C32, A34, and A37 from DMS methylation, whereas bound TRAP enhanced modification of A1 (Fig. 5). In the case of CMCT, bound TRAP protected G7, G18, G20, U21, and G39 from CMCT modification, whereas modification of U4 was enhanced when TRAP was bound. It should be pointed out that the results with CMCT and RNase T1 are not in agreement. Whereas TRAP protected G18 and G20 from CMCT modification, bound TRAP did not significantly protect either of these residues from RNase T1 cleavage. The reason for this discrepancy is unknown. When taken together, the footprinting results are consistent with a TRAP-5′ stem-loop RNA complex containing the internal loop, the upper stem, the hexaloop, and the 3′ side of the lower stem. Note that in no case did we detect TRAP binding to the trp leader transcript in the absence of tryptophan (data not shown).
TRAP interaction with the 5′ stem-loop is required for proper regulation of the trp operon.
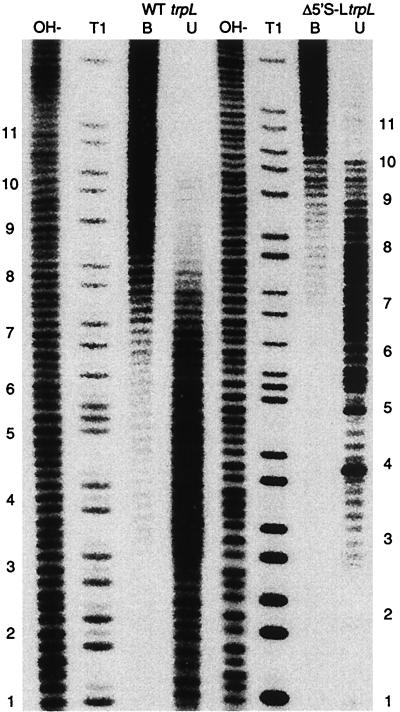
The TRAP–5′ stem-loop footprint results suggested that TRAP does not interact with the 5′ side of the lower stem. To determine if the lower stem is important for 5′ stem-loop function, we deleted the DNA region corresponding to the first five nucleotides of the trp leader transcript. We examined B. subtilis strains containing trpE′-′lacZ translational fusions that were controlled by the wild type (WTtrpL), the 5′ stem-loop deletion Δ(+3 to +32), or the Δ(+1 to +5) trp leader and analyzed β-galactosidase expression when each strain was grown in the presence or absence of exogenous tryptophan. We observed minimal expression in the WTtrpL strain PLBS44 grown in the presence of tryptophan (Table 2). The effect of exogenous tryptophan on the expression of the WTtrpL trpE′-′lacZ fusion can be assessed from the −Trp/+Trp ratio, which was 260. Comparable experiments were performed with the Δ(+3 to +32) strain PLBS104 and the Δ(+1 to +5) strain PLBS138. As was previously observed (28), deletion of the entire 5′ stem-loop resulted in a dramatic increase in expression, especially when cells were grown in the presence of tryptophan. In this case the −Trp/+Trp ratio was only 23, significantly lower than that observed for the WTtrpL strain (Table 2). Interestingly, the expression levels of the Δ(+1 to +5) strain were similar to the wild-type strain (Table 2), indicating that these residues are not required for 5′ stem-loop function (Table 2). This result is consistent with the footprint analysis (Fig. 5).
TABLE 2.
Effect of 5′ stem-loop mutations on trp operon expression
| Strain | 5′ stem-loop mutation | β-Gal activityb (Miller units)
|
β-Gal ratio (−Trp/+Trp) | |
|---|---|---|---|---|
| +Trp | −Trp | |||
| PLBS44 | Wild type | 0.2 ± 0.05 | 52 ± 5 | 260 |
| PLBS104 | Δ(+3 to +32) | 12 ± 0.5 | 274 ± 11 | 23 |
| PLBS138 | Δ(+1 to +5) | 0.4 ± 0.07 | 75 ± 2 | 188 |
| PLBS139 | Δ(+6 to +9) replaced with Ua | 4.2 ± 0.4 | 190 ± 15 | 45 |
| PLBS140 | Δ(+1 to +15) | 3.7 ± 0.2 | 152 ± 12 | 41 |
Nucleotides 6 to 9 (AGAA) were replaced by U in the trp leader transcript.
β-Galactosidase (β-Gal) activity expressed from the trpE′-′lacZ fusion is given in Miller units (23).
The footprint results presented above also indicated that the 5′ side of the asymmetric internal loop is involved in TRAP-5′ stem-loop interaction. We replaced nucleotides 6 to 9 (AGAA) of the internal loop with a single U residue. The predicted structure of this mutant transcript contained a contiguous 12-bp stem without an internal loop (structure not shown). We examined the effect of this trp leader mutation on expression of a trpE′-′lacZ translational fusion (PLBS139). Compared to the wild-type strain, we found that β-galactosidase levels increased 20-fold when this strain was grown in the presence of tryptophan and 4-fold in its absence (Table 2). In this case the −Trp/+Trp ratio was 45, only twofold higher than that observed for the strain in which the entire 5′ stem-loop was deleted. This result indicates that the 5′ side of the asymmetric internal loop is important for 5′ stem-loop function, which again is consistent with the footprint results. We also examined the effect of a deletion that extended from 1 to 15 and found that the expression levels in this strain (PLBS140) were similar to those of the 5′ stem-loop deletion strain (PLBS139) (Table 2).
The 5′ stem-loop reduces the number of (G/U)AG repeats required for tight TRAP-trp leader RNA binding.
Our footprint and gel shift results demonstrated that TRAP interacts with the 5′ stem-loop and that this interaction increases the affinity of TRAP for trp leader RNA. We performed a 3′ boundary analysis using wild type (nucleotides 1 to 111) and 5′ stem-loop deletion (nucleotides 32 to 111) trp leader transcripts to determine if the 5′ stem-loop reduced the number of (G/U)AG repeats that were required for tight TRAP-trp leader RNA binding. Note that these two transcripts were identical to those used in the gel shift analysis (Fig. 2). RNAs were 5′ end labeled, hydrolyzed to obtain a ladder of 5′-end-labeled transcripts, and subsequently mixed with tryptophan-activated TRAP. Bound and unbound RNAs were separated by native gel electrophoresis, gel purified, and separated on a standard denaturing sequencing gel. We observed cutoffs between bound and unbound transcripts with both the wild-type and 5′ stem-loop deletion trp leader transcripts (Fig. 6). The cutoff for the wild-type trp leader transcript was relatively sharp and occurred at between seven and eight (G/U)AG repeats, with bound and unbound lanes showing complementary cutoffs and cutons. Under the binding conditions employed here, this result demonstrated that the first six (G/U)AG repeats were required for stable TRAP-trp leader RNA complex formation when the 5′ stem-loop was present in the transcript. However, a small fraction of the transcripts that contained as few as three repeats was also shifted. Interestingly, the corresponding cutoff for the 5′ stem-loop deletion transcript occurred at between nine and ten (G/U)AG repeats, indicating that the first eight (G/U)AG repeats were required for comparable binding. In this case a small fraction of the transcripts that contained as few as six repeats were also shifted. Note that the short transcripts containing fewer than five (G/U)AG repeats in the unbound 5′ stem-loop deletion sample were not gel purified in this experiment (Fig. 6) since previous experiments indicated that these transcripts were not gel shifted by TRAP. The results of the 3′ boundary analysis demonstrate that the 5′ stem-loop structure reduces the number of (G/U)AG repeats required for stable TRAP association.
FIG. 6.
3′ boundary analysis of wild-type and 5′ stem-loop deletion trp leader transcripts. Limited alkaline hydrolysis ladders of 5′-end-labeled wild-type (WT) or 5′ stem-loop deletion trp leader transcripts were incubated with tryptophan-activated TRAP. TRAP-RNA complexes were separated from unbound RNA on a native gel and subsequently fractionated through a denaturing 6% polyacrylamide gel (shown). Labels for lanes are as follows: OH−, a limited alkaline hydrolysis ladder; T1, partial RNase T1 digest; B and U, bound and unbound are RNA fragments from the limited alkaline hydrolysis that either bound (B) or did not bind (U) TRAP. The numbers on the left (wild-type transcript) or right (5′ stem-loop deletion transcript) indicate the relative positions of the (G/U)AG repeats, with 1 being closest to the 5′ end of the transcript.
The nature of the TRAP–5′ stem-loop RNA interaction is dependent on the number of downstream (G/U)AG repeats.
Results from a previous study demonstrated that the 5′ stem-loop functions in the transcription attenuation mechanism that controls expression of the trp operon (28). Furthermore, the results described above indicate that TRAP interacts with the 5′ stem-loop and that this interaction increases the affinity of TRAP for trp leader RNA. Moreover, we found that the presence of the 5′ structure reduces the number of (G/U)AG repeats required for stable TRAP-trp leader RNA association. One possible explanation for these results is that the 5′ stem-loop might tether TRAP to the nascent trp leader transcript such that TRAP would be in position to bind to the (G/U)AG repeats as soon as they are transcribed. A multipartite binding mechanism such as this might increase the probability that tryptophan-activated TRAP would bind to the trp leader in time to block antiterminator formation. For this binding mechanism to have the greatest impact on trp operon expression, one would predict that TRAP–5′ stem-loop interaction would occur in the absence of any downstream (G/U)AG repeats.
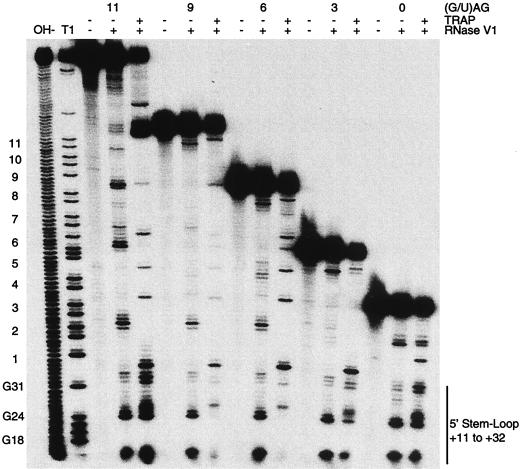
We performed a TRAP-RNA footprint experiment using 5′-end-labeled trp leader transcripts that contained the 5′ stem-loop in the absence of any downstream (G/U)AG repeats (nucleotides 1 to 36) to determine if TRAP could interact with a transcript that only contained the 5′ structure. The RNase V1 cleavage pattern in the absence of TRAP differed from the cleavage pattern when TRAP was present (Fig. 7). We observed appreciable RNase V1 cleavage in the presence or absence of TRAP following U11, A12, U23, and G24. Surprisingly, cleavage following U25, A26, G31, and C32 was only observed in the presence of TRAP. These results indicate that TRAP can interact with the 5′ stem-loop in the absence of the 11 (G/U)AG repeats and that this interaction was transient, resulting in a 5′ stem-loop that is more highly structured. The fact that our gel shift assay was unable to detect a complex between TRAP and a transcript that only contained the 5′ stem-loop (data not shown) is consistent with rapid TRAP-5′ stem-loop RNA complex dissociation.
FIG. 7.
TRAP–5′ stem-loop RNA footprint analysis with transcripts containing various numbers of (G/U)AG repeats. 5′-end-labeled trp leader RNA containing the 5′ stem-loop and either 0, 3, 6, 9, or 11 (G/U)AG repeats was used in this analysis. trp leader RNA was treated with RNase V1 (+) in the presence (+) or absence (−) of tryptophan-activated TRAP. The positions of the nucleotides corresponding to the 5′ stem-loop are indicated at the right. The relative positions of the 11 (G/U)AG repeats, as well as G18, G24, and G31, are shown on the left. The lanes corresponding to partial alkaline hydrolysis (OH−) and partial RNase T1 digestion (T1) ladders generated from the transcript containing all 11 (G/U)AG repeats are indicated.
We then examined the effect TRAP binding had on 5′ stem-loop RNase V1 cleavage patterns when transcripts contained the 5′ structure and 3, 6, 9, or 11 (G/U)AG repeats. As expected, in the absence of TRAP we found that the cleavage pattern within the 5′ stem-loop was essentially identical in all of the transcripts tested (Fig. 7). However, the cleavage pattern of the various transcripts in the presence of bound TRAP differed considerably. When the transcript contained the first three (G/U)AG repeats (nucleotides 1 to 51), we observed a reduction in cleavage following U11, A12, U23, and G24, as well as increased cleavage following U25, A26, G31, C32, and U35 (Fig. 7). Note that the increase in cleavage following U25, A26, G31, and C32 was not as substantial as that observed for the transcript that only contained the 5′ stem-loop. The cleavage pattern in the transcripts containing the first six (1 to 68) or nine (1 to 84) (G/U)AG repeats were similar to one another, although they differed from the other transcripts tested. RNase V1 cleavage was essentially absent following U11, A12, U23, G24, A26, G31, and C32 (Fig. 7). Note that there was no increase in cleavage following U25, A26, G31, and C32 (Fig. 7). Remarkably, the RNase V1 cleavage pattern in the transcript containing all 11 (G/U)AG repeats (1 to 111) was essentially identical to the pattern observed for the transcript that only contained the 5′ stem-loop. When taken together, these results indicate that TRAP can interact with 5′ stem-loop in the absence of any downstream (G/U)AG repeats and that the TRAP-5′ stem-loop complex differs depending on the number of (G/U)AG repeats following the 5′ structure.
DISCUSSION
The transcription attenuation mechanism that controls expression of the B. subtilis trpEDCFBA operon in response to tryptophan relies on TRAP and three RNA secondary structures. When TRAP binds to the 11 (G/U)AG repeats present in the nascent trp leader transcript the antiterminator structure cannot form. Instead, an overlapping intrinsic terminator can form which results in transcription termination upstream of the trp operon structural genes (Fig. 1). A recent genetic study demonstrated that the 5′ stem-loop also participates in the transcription attenuation mechanism (28).
In the current study we examined the molecular basis of 5′ stem-loop function. We determined the secondary structure of the 5′ stem-loop and found that it consists of a relatively unstable 3-bp lower stem, a 5-by-2 asymmetric internal loop, a 6-bp upper stem, and a hexaloop at the apex of the structure (Fig. 3 and 4). It is interesting to note that while both RNase T1 and CMCT are single-stranded specific G probes, only CMCT detected G7 in the structure-mapping experiments. One possible explanation for this difference is that G7 participates in a non-Watson-Crick base-pairing interaction that prevents RNase T1 cleavage but leaves the N1 position available for CMCT modification. Our footprinting results suggest that TRAP interacts with both sides of the asymmetric internal loop, the upper stem, the hexaloop, and the 3′ side of the lower stem (Fig. 5). It is interesting that the hexaloop contains a GAG sequence (nucleotides 18 to 20), while a single AAG sequence is present in the residues comprising the 3′ side of the asymmetric loop and the 3′ side of the lower stem (nucleotides 29 to 31). The TRAP binding target in the trp leader contains four UAG and seven GAG repeats between nucleotides 36 and 91 (Fig. 1) (8), while the TRAP binding site in the unlinked trpG transcript consists of one AAG, one UAG, and seven GAG repeats (15). Since it is known that 11 KKR motifs on TRAP interact with GAG, UAG, and AAG repeats (2, 5, 15, 31), it is possible that KKR motifs contribute to the TRAP–5′ stem-loop complex by interacting with the GAG and/or AAG present within the 5′ structure (Fig. 1). However, as pointed out in Results, substantial evidence suggests that the GAG sequence in the hexaloop interacts with a region of TRAP that is distinct from the KKR motifs. If a KKR motif interacts with the AAG sequence, then the spacing of four nucleotides between the AAG and the first UAG (nucleotides 36 to 38) (Fig. 1) is suboptimal. The optimal spacing between repeats is two nucleotides (7), although it was determined that three-nucleotide spacers are tolerated if present in the appropriate context (9). Moreover, spacers of five and eight nucleotides were identified in the trpG transcript (15); thus, it is possible that a TRAP KKR motif interacts with this AAG sequence. This would bring the number of triplet repeats in the B. subtilis trp leader TRAP target to 12, the same number identified in the Bacillus stearothermophilus trp leader (12). Note that the UAG sequence (nucleotides 5 to 7) is unlikely to interact with a TRAP KKR motif since deletion of the first five residues had virtually no effect on trp operon expression (Table 2).
Our results also indicate that TRAP interacts with the 5′ side of the internal loop and the upper stem (Fig. 5 and Table 2). Moreover, we previously showed that substitution of G7 with A resulted in a 5′ stem-loop defect (28). Thus, it appears that TRAP interaction with the 5′ side of the internal loop and/or non-Watson-Crick base pairing within this RNA segment is crucial for 5′ stem-loop function. Furthermore, we previously demonstrated that disruption of the upper stem by point mutations (C15G or G22C) had similar effects as deleting the entire stem, while a C15G-G22C compensatory change only partially restored expression to wild type-like levels (28). This suggests that both the structure and the sequence of the upper stem are important for TRAP interaction.
While our footprinting and 5′ stem-loop mutation studies demonstrated that TRAP interacts with the 5′ structure and that this interaction is required for proper regulation of the B. subtilis trp operon (Table 2), results from our boundary analysis indicate that TRAP–5′ stem-loop interaction reduces the number of downstream (G/U)AG repeats that are necessary for tight TRAP-trp leader RNA binding (Fig. 6). In addition, our footprinting results demonstrate that TRAP can interact with the 5′ stem-loop without any downstream repeats (Fig. 7). While the nature of the specific interactions are not well understood, it is particularly striking that TRAP interaction with the transcript containing only the 5′ stem-loop resulted in a 5′ hairpin that was more highly structured. A qualitatively identical result occurred when TRAP interacted with the transcript containing the 5′ structure and all 11 downstream (G/U)AG repeats (Fig. 7). Interestingly, the TRAP-dependent RNase V1 cleavage pattern that occurred in the transcripts containing the 5′ stem-loop and six or nine repeats were identical to each other but clearly distinct from the cleavage pattern of the 0- and 11-repeat transcripts. Note that the TRAP-dependent cleavage pattern of the 5′ stem-loop in the transcript that also contained three downstream (G/U)AG repeats is intermediate between the other two RNase V1 cleavage patterns. We believe that this static in vitro experiment captures the essence of the dynamic events taking place during transcription of the trp leader in vivo.
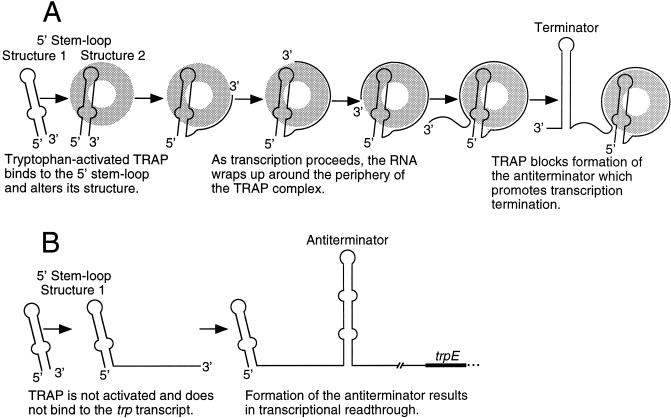
Our current model of the events taking place during transcription attenuation of the B. subtilis trp operon is shown in Fig. 8. Soon after transcription initiates the 5′ stem-loop forms (structure 1) (Fig. 8A). Tryptophan-activated TRAP subsequently binds to structure 1, thereby promoting formation of a more highly structured 5′ hairpin (structure 2). As transcription proceeds, the KKR motifs on the TRAP perimeter interact with the (G/U)AG repeats one at a time as they become available, thereby wrapping the RNA around the periphery of the TRAP complex. Once all of the (G/U)AG repeats are bound, the geometry of this TRAP-trp leader RNA complex is such that the trp leader transcript encircles the entire TRAP 11-mer (2). Once this occurs the 5′ stem-loop can dissociate from TRAP and retain the conformation of stem-loop structure 2 or remain bound. The ability of the 5′ stem-loop to remain bound is supported by the gel shift results, where we observed increased TRAP affinity when the 5′ stem-loop was present in a transcript that contained all 11 (G/U)AG repeats (Fig. 4), while dissociation is supported by the footprint analysis (Fig. 7). As a consequence of TRAP binding, the antiterminator structure cannot form, which promotes formation of the terminator structure and, hence, transcription termination. Since only a relatively short window of opportunity exists for TRAP to block antiterminator formation, it appears that this multipartite binding mechanism increases the probability that TRAP associates with the nascent trp leader transcript in time to promote termination. When the concentration of tryptophan is low, TRAP is not activated and does not bind to the nascent trp leader transcript. In this case, antiterminator formation prevents formation of the intrinsic terminator, resulting in transcription of the entire operon (Fig. 8B). The trp operon leader transcripts of Bacillus pumilus (18), Bacillus caldotenax (31), and B. stearothermophilus (29) also contain 5′ stem-loops and multiple triplet repeats, as well as overlapping antiterminator and terminator structures. Thus, it appears that all four organisms control expression of the trp operon by essentially identical transcription attenuation mechanisms.
FIG. 8.
Transcription attenuation model of the B. subtilis trp operon. (A) Conditions of tryptophan excess. (B) Limiting tryptophan conditions. The 5′ and 3′ ends of the transcript are indicated. TRAP is represented by the gray doughnut structure. See the text for details.
ACKNOWLEDGMENTS
We thank Philip Bevilacqua, Craig Cameron, and Subita Sudershana for discussions throughout the course of this study. We also thank Philip Bevilacqua, Janell Schaak, and Charles Yanofsky for critical reading of the manuscript.
This work was supported by grant GM52840 from the National Institutes of Health.
REFERENCES
- 1.Antson A A, Brzozowski A M, Dodson E J, Dauter Z, Wilson K S, Kurecki T, Otridge J, Gollnick P. 11-fold symmetry of the trp RNA-binding attenuation protein (TRAP) from Bacillus subtilis determined by X-ray analysis. J Mol Biol. 1994;244:1–5. doi: 10.1006/jmbi.1994.1698. [DOI] [PubMed] [Google Scholar]
- 2.Antson A A, Dodson E J, Dodson G, Greaves R B, Chen X-P, Gollnick P. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature. 1999;401:235–242. doi: 10.1038/45730. [DOI] [PubMed] [Google Scholar]
- 3.Antson A A, Otridge J, Brzozowski A M, Dodson E J, Dodson G G, Wilson K S, Smith T M, Yang M, Kurecki T, Gollnick P. The structure of trp RNA-binding attenuation protein. Nature. 1995;374:693–700. doi: 10.1038/374693a0. [DOI] [PubMed] [Google Scholar]
- 4.Babitzke P. Regulation of tryptophan biosynthesis: Trp-ing the TRAP or how Bacillus subtilis reinvented the wheel. Mol Microbiol. 1997;26:1–9. doi: 10.1046/j.1365-2958.1997.5541915.x. [DOI] [PubMed] [Google Scholar]
- 5.Babitzke P, Yanofsky C. Reconstitution of Bacillus subtilis trp attenuation in vitro with TRAP, the trp RNA-binding attenuation protein. Proc Natl Acad Sci USA. 1993;90:133–137. doi: 10.1073/pnas.90.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babitzke P, Yanofsky C. Structural features of l-tryptophan required for activation of TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis. J Biol Chem. 1995;270:12452–12456. doi: 10.1074/jbc.270.21.12452. [DOI] [PubMed] [Google Scholar]
- 7.Babitzke P, Bear D G, Yanofsky C. TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a toroid-shaped molecule that binds transcripts containing GAG or UAG repeats separated by two nucleotides. Proc Natl Acad Sci USA. 1995;92:7916–7920. doi: 10.1073/pnas.92.17.7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Babitzke P, Stults J T, Shire S J, Yanofsky C. TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a multisubunit complex that appears to recognize G/UAG repeats in the trpEDCFBA and trpG transcripts. J Biol Chem. 1994;269:16597–16604. [PubMed] [Google Scholar]
- 9.Babitzke P, Yealy J, Campanelli D. Interaction of the trp RNA-binding attenuation protein (TRAP) of Bacillus subtilis with RNA: effects of the number of GAG repeats, the nucleotides separating adjacent repeats, and RNA secondary structure. J Bacteriol. 1996;178:5159–5163. doi: 10.1128/jb.178.17.5159-5163.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babitzke P, Gollnick P, Yanofsky C. The mtrAB operon of Bacillus subtilis encodes GTP cyclohydrolase I (MtrA), an enzyme involved in folic acid biosynthesis, and MtrB, a regulator of l-tryptophan biosynthesis. J Bacteriol. 1992;174:2059–2064. doi: 10.1128/jb.174.7.2059-2064.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bevilacqua P C, George C X, Samuel C E, Cech T R. Binding of the protein kinase PKR to RNAs with secondary structure defects: role of the tandem A-G mismatch and noncontiguous helixes. Biochemistry. 1998;37:6303–6316. doi: 10.1021/bi980113j. [DOI] [PubMed] [Google Scholar]
- 12.Chen X-P, Antson A A, Yang M, Li P, Baumann C, Dodson E J, Dodson G G, Gollnick P. Regulatory features of the trp operon and the crystal structure of the trp RNA-binding attenuation protein from Bacillus stearothermophilus. J Mol Biol. 1999;289:1003–1016. doi: 10.1006/jmbi.1999.2834. [DOI] [PubMed] [Google Scholar]
- 13.Donis-Keller H, Maxam A M, Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977;4:2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du H, Babitzke P. trp-RNA binding attenuation protein-mediated long-distance RNA refolding regulates translation of trpE in Bacillus subtilis. J Biol Chem. 1998;273:20494–20503. doi: 10.1074/jbc.273.32.20494. [DOI] [PubMed] [Google Scholar]
- 15.Du H, Tarpey R, Babitzke P. The trp-RNA binding attenuation protein regulates TrpG synthesis by binding to the trpG ribosome binding site of Bacillus subtilis. J Bacteriol. 1997;179:2582–2586. doi: 10.1128/jb.179.8.2582-2586.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gollnick P. Regulation of the Bacillus subtilis trp operon by an RNA-binding protein. Mol Microbiol. 1994;11:991–997. doi: 10.1111/j.1365-2958.1994.tb00377.x. [DOI] [PubMed] [Google Scholar]
- 17.Gollnick P, Ishino S, Kuroda M I, Henner D J, Yanofsky C. The mtr locus is a two-gene operon required for transcription attenuation in the trp operon of Bacillus subtilis. Proc Natl Acad Sci USA. 1990;87:8726–8730. doi: 10.1073/pnas.87.22.8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffman R J, Gollnick P. The mtrB gene of Bacillus pumilus encodes a protein with sequence and functional homology to the trp RNA-binding attenuation protein (TRAP) of Bacillus subtilis. J Bacteriol. 1995;177:839–842. doi: 10.1128/jb.177.3.839-842.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuroda M I, Henner D, Yanofsky C. cis-acting sites in the transcript of the Bacillus subtilis trp operon regulate expression of the operon. J Bacteriol. 1988;170:3080–3088. doi: 10.1128/jb.170.7.3080-3088.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landick R, Turnbough C L, Yanofsky C. Transcription attenuation. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1996. pp. 1263–1286. [Google Scholar]
- 21.Lowman H B, Draper D E. On the recognition of helical RNA by cobra venom V1 nuclease. J Biol Chem. 1986;261:5396–5403. [PubMed] [Google Scholar]
- 22.Merino E, Babitzke P, Yanofsky C. trp RNA-binding attenuation protein (TRAP)-trp leader RNA interactions mediate translational as well as transcriptional regulation of the Bacillus subtilis trp operon. J Bacteriol. 1995;177:6362–6370. doi: 10.1128/jb.177.22.6362-6370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 24.Otridge J, Gollnick P. MtrB from Bacillus subtilis binds specifically to trp leader RNA in a tryptophan-dependent manner. Proc Natl Acad Sci USA. 1993;90:128–132. doi: 10.1073/pnas.90.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimotsu H, Kuroda M I, Yanofsky C, Henner D J. Novel form of transcription attenuation regulates expression of the Bacillus subtilis tryptophan operon. J Bacteriol. 1986;166:461–471. doi: 10.1128/jb.166.2.461-471.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiratsuchi A, Sato S. Nucleotide sequence of trpE, anthranilate synthase I gene, of Bacillus caldotenax. Biochim Biophys Acta. 1991;1090:348–350. doi: 10.1016/0167-4781(91)90201-v. [DOI] [PubMed] [Google Scholar]
- 27.Spizizen J. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc Natl Acad Sci USA. 1958;44:1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sudershana S, Du H, Mahalanabis M, Babitzke P. A 5′ RNA stem-loop participates in the transcription attenuation mechanism that controls expression of the Bacillus subtilis trpEDCFBA operon. J Bacteriol. 1999;181:5742–5749. doi: 10.1128/jb.181.18.5742-5749.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walter A E, Turner D H, Kim J, Lyttle M H, Mueller P, Mathews D H, Zuker M. Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc Natl Acad Sci USA. 1994;91:9218–9222. doi: 10.1073/pnas.91.20.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang M, de Saizieu A, van Loon A P G M, Gollnick P. Translation of trpG in Bacillus subtilis is regulated by the trp RNA-binding attenuation protein (TRAP) J Bacteriol. 1995;177:4272–4278. doi: 10.1128/jb.177.15.4272-4278.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang M, Chen X-P, Militello K, Hoffman R, Fernandez B, Baumann C, Gollnick P. Alanine-scanning mutagenesis of Bacillus subtilis trp RNA-binding attenuation protein (TRAP) reveals residues involved in tryptophan binding and RNA binding. J Mol Biol. 1997;270:696–710. doi: 10.1006/jmbi.1997.1149. [DOI] [PubMed] [Google Scholar]
- 32.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]