Abstract
During endospore formation in Bacillus subtilis, over two dozen polypeptides are assembled into a multilayered structure known as the spore coat, which protects the cortex peptidoglycan (PG) and permits efficient germination. In the initial stages of coat assembly a protein known as CotE forms a ring around the forespore. A second morphogenetic protein, SpoVID, is required for maintenance of the CotE ring during the later stages, when most of proteins are assembled into the coat. Here, we report on a protein that appears to associate with SpoVID during the early stage of coat assembly. This protein, which we call SafA for SpoVID-associated factor A, is encoded by a locus previously known as yrbA. We confirmed the results of a previous study that showed safA mutant spores have defective coats which are missing several proteins. We have extended these studies with the finding that SafA and SpoVID were coimmunoprecipitated by anti-SafA or anti-SpoVID antiserum from whole-cell extracts 3 and 4 h after the onset of sporulation. Therefore, SafA may associate with SpoVID during the early stage of coat assembly. We used immunogold electron microscopy to localize SafA and found it in the cortex, near the interface with the coat in mature spores. SafA appears to have a modular design. The C-terminal region of SafA is similar to those of several inner spore coat proteins. The N-terminal region contains a sequence that is conserved among proteins that associate with the cell wall. This motif in the N-terminal region may target SafA to the PG-containing regions of the developing spore.
In response to nutrient depletion, Bacillus subtilis can undergo a complex differentiation process culminating in the formation of a dormant endospore (34). Sporulation begins with an asymmetric cell division that partitions the sporangium into a large mother cell and a smaller prespore, each of which contains one copy of the bacterial chromosome. Hydrolysis of the septal peptidoglycan (PG) allows the mother cell to engulf the prespore, a process that yields a cellular compartment, the forespore, completely surrounded by the mother cell cytoplasm. Maturation of the spore proceeds by the coordinated synthesis of the necessary gene products from both the forespore and the mother cell sporangial compartments. At the end of the developmental process, the spore is released into the environment upon lysis of the mother cell (20, 34).
The endospore is highly resistant to a variety of insults, including dehydration, organic solvents, lysozyme treatment, and extreme temperatures. However, in spite of its dormant nature, the spore can sense its environment and initiate germination within minutes of exposure to germinants. These spore properties are attributable to the physical and chemical composition of the structures which encase the spore (6, 12, 20). Thin-section electron microscopy reveals two main layers surrounding the core of mature spores: a thick electron-transparent layer of a modified PG known as the cortex (reviewed in reference 4) and a multilayered electron-dense protein structure known as the spore coat (reviewed in references 6 and 12). The coat is composed of more than two dozen proteins organized into three layers: an amorphous undercoat, a thin lamellar inner coat, and a thick striated outer coat (reviewed in references 6 and 12).
Synthesis of most proteins required for coat assembly relies on the action of two mother cell-specific RNA polymerase sigma factors (reviewed in references 6 and 12). Thus, most of the coat proteins are synthesized in the mother cell, where they assemble around the forespore membrane. ςE functions in the preengulfment stages of mother cell development, and following engulfment it is replaced by ςK (34). The earliest events of coat assembly require ςE to drive the production of three morphogenetic proteins, called SpoIVA, SpoVID, and CotE (3, 20, 24, 32, 36). The ςE-dependent period is marked by the organization of SpoIVA in a layer closely apposed to the outer forespore membrane (7, 16, 22) and the subsequent concentric positioning of a ring of CotE 75 nm from SpoIVA (7, 21). The gap between SpoIVA and CotE defines a compartment referred to as the matrix, which becomes the site of assembly of the inner coat proteins. The CotE ring is implicated as the nucleation site for the assembly of outer coat proteins (7, 36). Activation of ςK triggers the synthesis of the cortex layer (between the forespore inner and outer membranes) and of most of the coat structural proteins (6, 12, 20, 34). SpoVID function is required at about this stage for the maintenance of the CotE ring around the forespore. In spoVID-null mutants the CotE ring forms but does not persist, and the coat misassembles as swirls of semiorganized material in the mother cell cytoplasm (3, 7). As a consequence, spoVID spores have an exposed cortex and are extremely sensitive to lysozyme treatment (3). Thus, SpoVID is an important determinant of protein localization to the nascent coat. It is not known how SpoVID is itself targeted to the matrix or how it influences the fate of the CotE ring. To begin analyzing these issues, we sought to identify proteins that interact with SpoVID. Here we report that the product of the yrbA gene appears to associate with SpoVID during the early stage of coat assembly.
MATERIALS AND METHODS
Isolation of safA mutants.
The B. subtilis strains used in this study are listed in Table 1. Escherichia coli strain DH5α (Bethesda Research Laboratories) was used for transformation and amplification of all plasmid constructs. E. coli strain BL21(DE3)(pLysS) (Novagen) was used to overproduce His-tagged SpoVID and SafA.
TABLE 1.
Bacterial strains and plasmids used in this studya
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| 168ΩJMCS | spoVID::kan trpC2 | 3 |
| MB24 | Wild type; trpC2 metC3 Spo+ | Laboratory stock |
| AH45 | trpC2 metC3 spoIIIGΔ1 | 13 |
| AH64 | trpC2 metC3 cotEΔ::cat Cmr | 29 |
| AH74 | trpC2 metC3 spoIIGBΔ::erm Ermr | 29 |
| AH77 | trpC2 metC3 spoIVCBΔ::erm Ermr | 29 |
| AOB24 | trpC2 metC3 spoVID::kan | This work |
| AOB38 | trpC2 metC3 MB24Ω pOZ52 | This work |
| AOB44 | trpC2 metC3 safAΩ pOZ54 | This work |
| AOB48 | trpC2 metC3 safA yrbB yrbC::pOZ53 | This work |
| AOB65 | AOB48Ω pOZ78 | This work |
| AOB68 | trpC2 metC3 safA::pOZ83 | This work |
| AOB88 | AOB68 amyE::pOZ134 | This work |
| Plasmids | ||
| pAH250 | pBluescript II KS(−)b + Spr gene | A. O. Henriques |
| pDH32 | amyE integrational vector | 19 |
| pOZ33 | safA in pet30a+ | This work |
| pAH333 | spoVID in pet30a+ | This work |
| pOZ52 | yrbC in pAH250 | This work |
| pOZ53 | safA internal fragment in pOZ52 | This work |
| pOZ54 | safA in pAH250 | This work |
| pOZ78 | safA in pLITMUS 29c Kanr | This work |
| pOZ79 | yrbBC in pCR2.1 TOPOd | This work |
| pOZ83 | safA deletion vector | This work |
| pOZ134 | pDH32::safA | This work |
The omega symbol indicates that the plasmid was integrated into the B. subtilis chromosome by a Campbell integration event (single crossover in the region of homology).
From Stratagene.
From New England Biolabs.
From the Invitrogen TA-TOPO cloning kit.
Primers were designed to amplify internal fragments of safA (5′GGCGATTAGATCTGGAAAATAGC3 and 5′GCATATGATACATAAGATCTATAT3′) and yrbC (5′CAGGCCATTACTAGTGGAAAAAC3′ and 5′CAAGCATGAGCTCATCTTCTTC3′) from the wild-type (MB24) chromosome. The safA construct (pOZ54) was made by cloning the 430-bp PCR fragment into the BamHI sites of the integrational plasmid pAH250 (8) (Table 1). Similarly, the yrbC construct (pOZ52) was made using a 460-bp PCR fragment cloned into the SacI and SpeI sites of pAH250 (Table 1). Competent cells of wild-type B. subtilis were transformed with pOZ54 and pOZ52, with selection for spectinomycin resistance. Transformants were expected to arise as the result of a single reciprocal crossover (Campbell-type) event, which was confirmed by PCR analysis of the recombinant chromosomes.
A partial deletion of the genes contained in the safA-to-yrbC region was created by allele replacement using pOZ53. pOZ53 was constructed by inserting a 472-bp PCR fragment internal to the safA locus (generated with primers 5′CTCAATACAAAGCTTAGCAATCCA3′ and 5′GTGTGATGAAGCTTTTCCTCTTCT3′) into the HindIII site of pOZ52. This plasmid carries a fragment internal to yrbC cloned between the SacI and SpeI sites of the pAH250 vector (see above). The orientation of the safA gene was confirmed to be the same as that of the yrbC fragment, and thus the resulting plasmid (pOZ53) contained the spectinomycin resistance gene flanked by internal fragments of the safA and yrbC loci (Table 1). Marker replacement recombination with this plasmid resulted in a partial deletion of safA and yrbC and a complete deletion of yrbB. A plasmid (pOZ83) designed to delete the complete safA coding region while leaving the downstream yrbB and yrbC genes intact was constructed in several steps. First, the yrbB-yrbC region was PCR amplified with the primer pair 5′CGTTCGGAACGATGTAAATCG3′ and 5′GAATACACCCATGGAAGTGAACG3′, which yielded a 1.64-kb DNA fragment. Second, the yrbB-yrbC fragment was purified and cloned directly into the pCR2.1 TOPO (from the TA-TOPO cloning system; Invitrogen) to produce pOZ79. Third, a region of 617 bp upstream of the safA start codon was PCR amplified with primers 5′ATTGCGCTTGAAGCTTTGGCTGTGTGGGATCCG3′ and 5′GGATTTTCAAGCTTTTCCCCTCCTATGCAAAACG3′. The PCR product was purified, cleaved with HindIII, and cloned into the corresponding site of pOZ79, in the same orientation as the first insert. The resulting plasmid is referred to as pOZ81. Lastly, a spectinomycin acetyltransferase gene (spc), obtained from AvrII-SpeI digestion of pAH256 (8), was cloned into the SpeI site of pOZ81. This created pOZ83, in which the spc gene is in the same orientation as it is relative to the safA and yrbB-yrbC pieces (Table 1).
Competent cells of the wild-type strain, MB24, were transformed with linearized pOZ53 or pOZ83 to force a double-crossover event with the recipient chromosome. The expected integration of pOZ53 (with the concomitant deletion of part of safA, all of yrbB, and part of yrbC) and of pOZ83 (resulting in the deletion of only safA) was confirmed by PCR analysis of the recombinant chromosomes.
safA complementation analysis.
A complete copy of safA was obtained by PCR with primers 5′TAGGAGGGGAAAACCATGGAAAT3′ and 5′CGTTCCGAAAGATCTCTCATTTTC3′ and was cleaved with NcoI and BglII between the NcoI and BamHI sites of pLITMUS 29 (New England Biolabs) to yield pOZ77. A kanamycin resistance gene was released from pKD101 (a gift from W. Haldenwang) with SmaI and introduced into the unique site of pOZ77. The resulting plasmid, pOZ78, was used as a vector for integration of an intact copy of safA into MB24 (wild type) or AOB48 (safA yrbC::spc) to test for complementation. Complementation was also tested by expression of safA at the amyE locus. The safA gene and 598 bp upstream from its start point of translation, including its promoter, were amplified by PCR with primers 5′GATGAATTAGTAGCTGAATCCGGGC3′ and 5′CGTTCCGAAAGATCTCTCATTTTCTTCTTCCGG3′. The resulting 1,788-bp DNA fragment was cloned into PCR2.1 (Invitrogen) to produce pOZ115. In a second step this safA region in pOZ115 was cloned as an EcoRI fragment into the amyE integrational vector pDH32 (19) to produce pOZ134. pOZ134 was linearized with ScaI and used to transform the safA deletion mutant AOB68 to chloramphenicol resistance to produce strain AOB88. PCR analysis was used to confirm that the safA gene had integrated into the chromosome by double crossover in amyE.
Purification of spores for germination assay and electron microscopy.
Spores were harvested 48 h after the onset of sporulation in DS medium (DSM). The spore suspension was washed with 100 ml of sterile water three times in 48 h and either used directly for electron microscopy sample preparation or subjected to further purification on a 50% step gradient of metrizoic acid (Renocal-76 from Squibb Diagnostics), as described previously (8, 9).
Transmission electron microscopy and immunogold localization.
Spores used for electron microscopy were prepared as described in the previous paragraph. Wild-type spores (MB24) and safA mutant spores (AOB48 and AOB68) were fixed for 30 min with 4% paraformaldehyde and 2.5% glutaraldehyde on ice. For immunogold labeling with chemical fixation we reduced the amount of glutaraldehyde to 0.2%. Samples were washed twice with 0.1 M phosphate buffer, pH 7.0. Only samples not to be used for immunogold labeling were treated with 1% osmium tetroxide solution [1.5% K4Fe(CN)6, 1% OsO4, 0.1 M phosphate buffer]. All the samples were dehydrated at room temperature with increasing concentrations of ethanol washes up to 100% and then embedded in the following resins: LR white resin (EM Sciences) at 42°C for the chemical fixation immunogold labeling samples and Epon resin (EM Sciences) for the rest of the samples. Epon sections were processed for observation of the morphology as described by Henriques et al. (11). For immunogold labeling, thin sections were placed on grids and processed with protocols provided by Aurion. Samples were then exposed to blocking agents (acetylated bovine serum albumin and normal goat serum; Aurion), anti-SafA antibodies (1:2,000 dilution), and secondary goat anti-rabbit conjugated to 10-nm-diameter gold or ultrasmall gold beads (Aurion). Transmission electron microscopy was performed on a Hitachi electron microscope operated at 60 keV.
Preparation of B. subtilis whole-cell extracts and immunoblotting.
Samples (10 ml) of DSM cultures of various strains were collected at intervals after the onset of sporulation. Whole-cell lysates were prepared as described by Seyler et al. (29), except that a complete mini-protease inhibitor cocktail (Boehringer Mannheim) was added to the lysis buffer. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting were also done as described by Seyler et al. (29). Anti-SafA or anti-SpoVID antibodies were used overnight at 4°C, at a dilution of 1:10,000 or 1:5,000, respectively. The membranes were washed and incubated for 30 min with anti-rabbit horseradish peroxidase-conjugated antiserum (1:10,000 dilution) in phosphate-buffered saline–Tween (29). After a phosphate-buffered saline–Tween wash, the membranes were developed with enhanced-chemiluminescence reagents, as described by the manufacturer (Amersham). Films were exposed for various times ranging from 30 s to 30 min.
Coimmunoprecipitation of SafA and SpoVID.
Samples (10 ml) of DSM cultures were collected at h 3 and 4 of sporulation and the cells were harvested by centrifugation. Whole-cell lysates were prepared as described above, except that the cell pellets were resuspended in 1/10 volume of RIPA-d buffer (150 mM NaCl, 100 mM Tris-HCl, 0.1% SDS, 1% NP-40, complete mini-protease inhibitor cocktail [pH 8.0]). A final concentration of 1 mg of lysate per ml was added to protein A-purified anti-SafA and anti-SpoVID antisera at antibody-to-cell lysate ratios of 1:5 and 1:50 (to help optimize the amount of specific antibody to target protein) in a total volume of 500 μl of RIPA-d buffer. Dynabeads (Dynal) conjugated with sheep anti-rabbit antibodies were added, as directed by the manufacturer, to magnetically precipitate immune complexes. The beads were washed with RIPA-d buffer three times and the proteins were eluted by boiling for 2 min in SDS-PAGE loading buffer (Bio-Rad). Proteins were electrophoresed in 12% Tris-glycine SDS-PAGE gels, blotted to nitrocellulose, and immunodetected as described above.
RESULTS
Identification of SafA.
We used phage display to find peptides that may interact with SpoVID. Purified SpoVID was used as the bait protein in successive rounds of selection against a library of random peptide motifs (the Ph.D.12 phage display peptide library kit from New England Biolabs) (27, 31). We determined the sequences of the peptide-encoding regions from 30 phage isolated after four rounds of selection. These 30 sequences were grouped into four consensus sequences, and each consensus sequence was used to search the B. subtilis genome sequence database (15) (www.pasteur.fr/GenoList/Subtilist/) using the BLAST local alignment search method (1). One consensus sequence (WFWPYYHAPSHP) shared 50% identity with a region encoded by a gene previously called yrbA, herein designated safA (for SpoVID-associated factor A). The similarity between the peptide encoded by the phage and the region encoded within safA cannot be considered strong evidence for interaction between the safA-encoded product and SpoVID. However, we examined the effects of mutations in safA, because its function was unknown when we began this study, and the primary structure of its proline-rich C-terminal region is similar over significant regions to the primary structure of the N termini of the inner coat proteins CotJA, CotT, and CotD (2, 5, 9, 29). In addition, this region is also similar to a region in amelogenins (e.g., a human amelogenin [accession number M55418]). These proteins are important components of the developing enamel matrix in vertebrates (23), suggesting a structural role for SafA. During preparation of the manuscript of this report, Takamatsu et al. (35) published the results of a study in which they characterized the yrbA locus. They showed that the safA gene was cotranscribed with yrbB from a ςE-dependent promoter (35). They (35) also found that safA mutant spores were resistant to heat but sensitive to lysozyme and that they exhibited abnormal thin electron-dense outer coats in electron micrographs. We confirmed these results by examining several mutants in which the safA gene had been disrupted as described in Materials and Methods (e.g., strains AOB44, AOB48, and AOB68). As controls we also isolated strains in which the safA mutation was complemented by a wild-type allele of safA located at the amyE locus (strain AOB88) or a copy of the wild-type allele of safA inserted by Campbell-type integration at the safA locus (strain AOB65). We also used SDS-PAGE analysis to examine coat proteins extracted from purified spores as described previously (9), and in confirmation of the results of Takamatsu et al. (35), we found that several coat proteins were extracted in reduced amounts from safA mutant spores (data not shown). One of these is likely to be the 36-kDa CotG protein (25), as suggested by determining the N-terminal sequence of the corresponding species isolated from wild-type spores. We conclude that safA is a morphogenetic protein required for the assembly of CotG, as well as other uncharacterized coat proteins between 6 and 14 kDa, into the spore coat. In contrast to the results of Takamatsu et al. (35), we were unable to identify SafA on a Coomassie blue-stained gel of coat proteins extracted from wild-type spores. Evidently SafA may be less abundant in our wild-type strain.
SafA accumulation begins at h 2 of sporulation and is present in the spore coat.
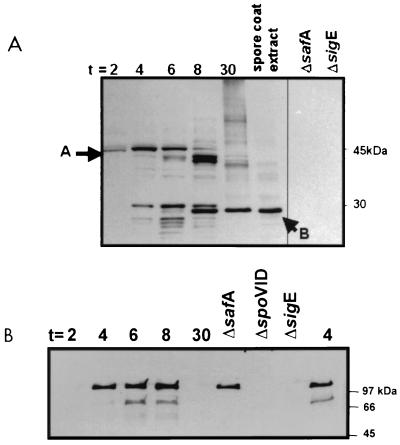
We purified a six-His–SafA fusion protein from E. coli and used it to elicit SafA-specific rabbit antiserum. The antiserum was used in Western blots and was found to react with several species in extracts from wild-type cells (Fig. 1A), as well as sigG and sigK mutants (data not shown). No cross-reacting bands were detected in extracts from safA or sigE mutants (Fig. 1A). This result agrees with the results of Takamatsu et al. (35) that showed that ςE directs safA transcription. The predicted size of SafA is 43 kDa. A product of approximately this size was detected in wild-type cells during the early stages of sporulation (around h 2). However, in later stages (from h 4 of sporulation onwards), the 43-kDa species was progressively replaced with faster-migrating products, including a predominant species of about 30 kDa (Fig. 1A). The 30-kDa species was the major form of the SafA antigen detected during the later stages of development (Fig. 1A, h 30 lane) and in material extracted from purified mature spores (Fig. 1A, spore coat extract lane). Takamatsu et al. (35) also found only the smaller form of SafA in spores. Moreover, Takamatsu et al. (35) determined that the N-terminal sequence of the smaller form begins with a methionine encoded by codon 164 within the safA gene. It is not known whether this or the other smaller forms of SafA detected in whole-cell extracts result from proteolytic processing of the 43-kDa precursor or whether the 30-kDa form arises from translation initiated at codon 164 of safA. In either case, the function and fate of full-length SafA and of its N-terminal region are presently unknown.
FIG. 1.
Accumulation of SafA and SpoVID in sporulating cells and mature spores. Whole-cell extracts were prepared from the wild type or mutant strains at the indicated times (in hours) after the onset of sporulation. Spore coat extracts were prepared from mature spores as described in Materials and Methods. These extracts were subjected to electrophoresis in SDS-polyacrylamide gels, and immunoblots of the gels were stained with anti-SafA (A) or anti-SpoVID (B) antiserum. Extracts from the wild-type strain were harvested 2, 4, 6, 8, and 30 h after the onset of sporulation as indicated above the lanes, but extracts from safA, spoVID, and sigE mutants were harvested 4 h after the onset of sporulation. Arrows A and B indicate the 43- and 30-kDa species of SafA, respectively.
SpoVID accumulation also begins at h 2 of sporulation.
We raised rabbit polyclonal antiserum against purified SpoVID and tested its specificity in the sporulating extracts of a wild-type strain of B. subtilis and in various mutants. Both a 66-kDa form and a 120-kDa form of SpoVID were detected in wild-type extracts between 4 and 8 h after the onset of sporulation but not in extracts from a spoVID deletion mutant (Fig. 1B). Sixty-six kilodaltons is near the predicted size of SpoVID. It is not known whether the 120-kDa band is a SpoVID dimer. The pattern of SpoVID accumulation agrees well with the previously reported transcriptional regulation of the locus (3). As expected, SpoVID was not detected in a sigE mutant (Fig. 1B), confirming its transcriptional regulation by ςE and hence its mother cell-specific accumulation (3, 7). SpoVID accumulation was unaffected by a safA mutation (Fig. 1B). A comparison of the Western blots of SpoVID and SafA shows that the accumulations of these proteins overlap in time (Fig. 1). Moreover, both proteins are likely to accumulate in the mother cell, since we found that SafA production, like that of SpoVID (see above), is dependent on ςE (Fig. 1) and not on ςG or ςK (data not shown). SafA and SpoVID differ in that the SafA was detected in material extracted from purified spores (Fig. 1A), whereas SpoVID was not detected in mature spores (7) (Fig. 1B).
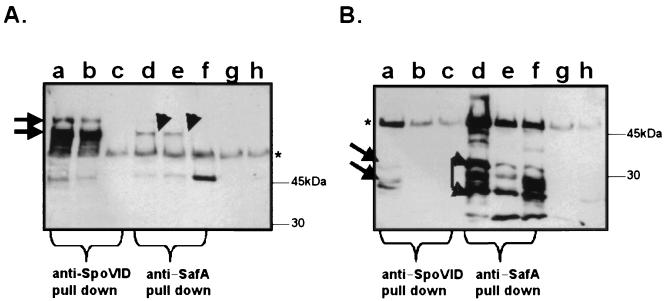
SafA and SpoVID form complexes in sporulating cells.
Our initial phage display results suggested that SafA may interact with SpoVID; therefore, we used coimmunoprecipitation to examine the possibility that SpoVID and SafA form a complex. Lysates of the wild type and a SpoVID mutant (AOB24) were prepared, as described in Materials and Methods, and from samples collected at h 4 of sporulation. Anti-SpoVID or anti-SafA antiserum was used to pull down immune complexes from wild-type and mutant extracts. Preimmune serum was used as a control for nonspecific immunoprecipitation. To detect the immune complexes we used Western blotting with either anti-SpoVID (Fig. 2A) or anti-SafA (Fig. 2B) antibodies. In all lanes of Fig. 2, the presence of the heavy chains of the antibody used in the immunoprecipitation is readily visible at about 50 kDa since all antibodies used in this experiment were raised in rabbits. Anti-SpoVID immunoprecipitated SpoVID (Fig. 2A, lanes a and b) as approximately 66- and 120-kDa forms from wild-type cells but not from a SpoVID mutant (Fig. 2A, lane c). Anti-SpoVID also immunoprecipitated SafA antigen (Fig. 2B, lane a), but not from a SpoVID mutant (lane c). Anti-SafA also precipitated SpoVID from wild-type cells, but only its 66-kDa form (Fig. 2A, lanes d and e). No SpoVID antigen was precipitated from a spoVID mutant (Fig. 2A, lane f). Similarly, anti-SafA precipitated SafA in wild-type extracts (Fig. 2B, lanes d and e) and in spoVID mutant extracts (Fig. 2B, lane f) as three forms between 30 and 21 kDa. We could not clearly distinguish whether the larger, full-length form of SafA was immunoprecipitated, since it migrates at a position that may overlap the region occupied by the heavy chain of the immunoprecipitating antibody. We conclude that SafA associates with SpoVID at the early stages of coat assembly.
FIG. 2.
SafA and SpoVID form complexes. Anti-SpoVID (lanes a to c), anti-SafA (lanes d to f), or preimmune serum (lanes g and h) was used to precipitate complexes from cell extracts of wild-type (lanes a, b, d, e, g, and h) or spoVID mutant (lanes c and f) strains harvested 4 h after the onset of sporulation. After separation by SDS-PAGE, immunoblots of the precipitated proteins were probed with anti-SpoVID (A) or anti-SafA (B) antiserum. The arrows indicate the positions of SpoVID (A) and SafA (B). The positions of molecular mass markers run on the same gels are indicated to the right of each panel. The heavy chain of the antibodies used for the immunoprecipitations is visible in every lane, migrating at a position just above the 45-kDa marker (indicated by an asterisk). Tenfold-higher concentrations of cell extract were used in lanes b and e than in lanes a and d, respectively.
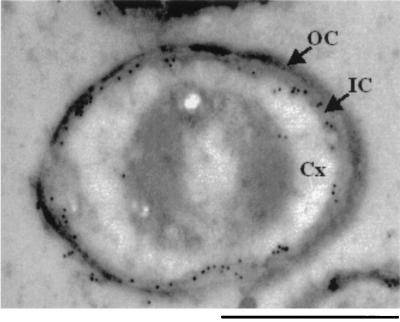
SafA is localized to the cortex-coat interface in mature spores.
We used immunogold electron microscopy to localize SafA in mature spores. In wild-type spores the label localized to the outer rim of the cortex, which was identified by the electron density of the layers (Fig. 3). In some spores the label was also distributed over the cortex region. This labeling was specific, since there were few, or in most spores no, gold particles in the safA mutant spores (data not shown). Thus, SafA is located near cortical PG and the inner coat layers. The localization of SafA is reminiscent of that of the SleB amidase and of the germination protein GerAB, which both localize to the outer edge of the cortex (17, 26).
FIG. 3.
Localization of SafA in wild-type spores. Immunogold electron microscopy was used to localize SafA in spores. Electron-dense gold particles indicate the position of anti-SafA in the electron micrograph of a wild-type spore. Few or no gold particles decorated SafA mutant spores (data not shown). The spore cortex (Cx), outer coat (OC), and inner coat (IC) are indicated. Bar, 500 nm.
DISCUSSION
The mechanisms that target coat proteins to the forespore surface and assemble them into the developing coat are unknown. SpoIVA, CotE, and SpoVID are morphogenetic proteins required for the assembly of many coat structural components or their maintenance around the forespore (3, 7, 20, 24, 32, 36). SafA belongs to this group of early morphogenetic proteins. SafA mutants form coats that appear abnormal by electron microscopy and that lack several polypeptide components. Our results suggest that SafA forms a complex with SpoVID. SafA and SpoVID accumulate during the same developmental period, in the same sporangial compartment, and in close proximity, both located near the forespore membrane (7; also this work). Moreover, SpoVID and SafA were coimmunoprecipitated by either anti-SpoVID or anti-SafA antiserum, which indicates that they exist as a complex in an early stage of coat assembly. Coimmunoprecipitation of SpoVID and SafA does not necessarily indicate that these two proteins directly interact. However, it is unlikely that their association is simply a consequence of their location in a large macromolecular structure such as the cortex or spore coat, because these structures are not present at the stage of development when the SpoVID-SafA complex was coimmunoprecipitated.
It is presently unknown whether the localization of either SafA or SpoVID to the matrix requires the other protein. The idea that SafA and SpoVID are independently sorted to the matrix is supported by their primary structures. The first 50 residues of SafA (14) and the last 50 residues of SpoVID (our unpublished results) consist of a motif present in several cell wall binding proteins, including several bacterial or phage murein hydrolases. The cell wall binding motif in SafA and SpoVID may facilitate their deployment to the cortex-coat interface by promoting the interaction of these proteins with cell wall material or with cell wall precursors located in this region. It is tempting to propose that SafA and SpoVID interact with cell wall material between the two membranes of the forespore, which is the site of synthesis of the cortex PG (4) and a thin layer of PG known as the germ cell wall (20). The localization of SafA and SpoVID to the matrix region seems to precede the onset of cortex synthesis (7; also this work), which is a late, ςK-dependent event (4, 20). Moreover, the cortex PG is not required for coat assembly, since mutants (e.g., spoVE mutants) that appear to be deficient in cortex synthesis still assemble an apparently normal coat around the forespore (10, 20). However, these mutant spores may have the germ cell wall PG (20), which potentially provides a target for SafA and SpoVID. If SafA or SpoVID interacts with PG located between the forespore membranes, then SafA must penetrate the membrane. However, unlike most other proteins with a cell wall binding motif (18), neither SafA nor SpoVID (3) has a signal sequence for secretion. Sorting of other proteins to the septal compartment involves, at least in some cases, the intervention of type I signal peptidases. For example, the germination-specific SleB amidase and the CwlD protein, genetically implicated in the hydrolysis of the spore cortex, and the TasA protein, implicated in the assembly of the spore coat, are synthesized as secretory preproteins (17, 28, 33). However, we note that some phage-encoded proteins containing the cell wall binding motif but not containing signal sequences are transported to the PG layer in a process dependent on holins (30). Therefore, SafA may be initially targeted to the thin PG layer located between the two forespore membranes known as the germ cell wall by a holin-dependent mechanism.
ACKNOWLEDGMENTS
We gratefully acknowledge Craig Samford and Lawrence Melsen for their expert technical assistance.
This work was supported by PHS grant GM54395 from the National Institutes of Health to C. P. Moran, Jr. A. O. Henriques was the recipient of a postdoctoral fellowship from Junta Nacional de Investigação Científica e Tecnológica (J.N.I.C.T.).
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Aronson A I, Song H Y, Bourne N. Gene structure and precursor processing of a novel Bacillus subtilis spore coat protein. Mol Microbiol. 1989;3:437–444. doi: 10.1111/j.1365-2958.1989.tb00189.x. [DOI] [PubMed] [Google Scholar]
- 3.Beall B, Driks A, Losick R, Moran C P., Jr Cloning and characterization of a gene required for assembly of the Bacillus subtilis spore coat. J Bacteriol. 1993;175:1705–1716. doi: 10.1128/jb.175.6.1705-1716.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchanan C E, Henriques A O, Piggot P J. Cell wall changes during bacterial endospore formation. In: Ghuysen J-M, Hackenbeck R, editors. Bacterial cell wall. New York, N.Y: Elsevier Science Publishers; 1994. pp. 167–186. [Google Scholar]
- 5.Donovan W, Zheng L B, Sandman K, Losick R. Genes encoding spore coat polypeptides from Bacillus subtilis. J Mol Biol. 1987;196:1–10. doi: 10.1016/0022-2836(87)90506-7. [DOI] [PubMed] [Google Scholar]
- 6.Driks A. Bacillus subtilis spore coat. Microbiol Mol Biol Rev. 1999;63:1–20. doi: 10.1128/mmbr.63.1.1-20.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Driks A, Roels S, Beall B, Moran C P, Jr, Losick R. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 1994;8:234–244. doi: 10.1101/gad.8.2.234. [DOI] [PubMed] [Google Scholar]
- 8.Henriques A O, Beall B W, Moran C P., Jr CotM of Bacillus subtilis, a member of the α-crystallin family of stress proteins, is induced during development and participates in spore outer coat formation. J Bacteriol. 1997;179:1887–1897. doi: 10.1128/jb.179.6.1887-1897.1997. . (Erratum, 179:4455.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henriques A O, Beall B W, Roland K, Moran C P., Jr Characterization of cotJ, a ςE-controlled operon affecting the polypeptide composition of the coat of Bacillus subtilis spores. J Bacteriol. 1995;177:3394–3406. doi: 10.1128/jb.177.12.3394-3406.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henriques A O, de Lencastre H, Piggot P J. A Bacillus subtilis morphogene cluster that includes spoVE is homologous to the mra region of Escherichia coli. Biochimie. 1992;74:735–748. doi: 10.1016/0300-9084(92)90146-6. [DOI] [PubMed] [Google Scholar]
- 11.Henriques A O, Melsen L R, Moran C P., Jr Involvement of superoxide dismutase in spore coat assembly in Bacillus subtilis. J Bacteriol. 1998;180:2285–2291. doi: 10.1128/jb.180.9.2285-2291.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henriques A O, Moran C P., Jr Structure and assembly of the bacterial endospore coat. Methods Companion Methods Enzymol. 2000;20:95–110. doi: 10.1006/meth.1999.0909. [DOI] [PubMed] [Google Scholar]
- 13.Kellner E M, Decatur A, Moran C P., Jr Two-stage regulation of an anti-sigma factor determines developmental fate during bacterial endospore formation. Mol Microbiol. 1996;21:913–924. doi: 10.1046/j.1365-2958.1996.461408.x. [DOI] [PubMed] [Google Scholar]
- 14.Kodama T, Takamatsu H, Asai K, Kobayashi K, Ogasawara N, Watabe K. The Bacillus subtilis yaaH gene is transcribed by SigE RNA polymerase during sporulation, and its product is involved in germination of spores. J Bacteriol. 1999;181:4584–4591. doi: 10.1128/jb.181.15.4584-4591.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 16.Lewis P J, Errington J. Use of green fluorescent protein for detection of cell-specific gene expression and subcellular protein localization during sporulation in Bacillus subtilis. Microbiology. 1996;142:733–740. doi: 10.1099/00221287-142-4-733. [DOI] [PubMed] [Google Scholar]
- 17.Moriyama R, Fukuoka H, Miyata S, Kudoh S, Hattori A, Kozuka S, Yasuda Y, Tochikubo K, Makino S. Expression of a germination-specific amidase, SleB, of bacilli in the forespore compartment of sporulating cells and its localization on the exterior side of the cortex in dormant spores. J Bacteriol. 1999;181:2373–2378. doi: 10.1128/jb.181.8.2373-2378.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Navarre W W, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perego M. Integrational vectors for genetic manipulation in Bacillus subtilis. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C.: American Society for Microbiology; 1993. pp. 615–624. [Google Scholar]
- 20.Piggot P J, Coote J G. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976;40:908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pogliano K, Harry E, Losick R. Visualization of the subcellular location of sporulation proteins in Bacillus subtilis using immunofluorescence microscopy. Mol Microbiol. 1995;18:459–470. doi: 10.1111/j.1365-2958.1995.mmi_18030459.x. [DOI] [PubMed] [Google Scholar]
- 22.Price K D, Losick R. A four-dimensional view of assembly of a morphogenetic protein during sporulation in Bacillus subtilis. J Bacteriol. 1999;181:781–790. doi: 10.1128/jb.181.3.781-790.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson C, Brookes S J, Shore R C, Kirkham J. The developing enamel matrix: nature and function. Eur J Oral Sci. 1998;106(Suppl. 1):282–291. doi: 10.1111/j.1600-0722.1998.tb02188.x. [DOI] [PubMed] [Google Scholar]
- 24.Roels S, Driks A, Losick R. Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J Bacteriol. 1992;174:575–585. doi: 10.1128/jb.174.2.575-585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sacco M, Ricca E, Losick R, Cutting S. An additional GerE-controlled gene encoding an abundant spore coat protein from Bacillus subtilis. J Bacteriol. 1995;177:372–377. doi: 10.1128/jb.177.2.372-377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakae Y, Yasuda Y, Tochikubo K. Immunoelectron microscopic localization of one of the spore germination proteins, GerAB, in Bacillus subtilis spores. J Bacteriol. 1995;177:6294–6296. doi: 10.1128/jb.177.21.6294-6296.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scott J K, Smith G P. Searching for peptide ligands with an epitope library. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- 28.Serrano M, Zilhão R, Ricca E, Ozin A J, Moran C P, Jr, Henriques A O. A Bacillus subtilis secreted protein with a role in endospore coat assembly and function. J Bacteriol. 1999;181:3632–3643. doi: 10.1128/jb.181.12.3632-3643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seyler R W, Jr, Henriques A O, Ozin A J, Moran C P., Jr Assembly and interactions of cotJ-encoded proteins, constituents of the inner layers of the Bacillus subtilis spore coat. Mol Microbiol. 1997;25:955–966. doi: 10.1111/j.1365-2958.1997.mmi532.x. [DOI] [PubMed] [Google Scholar]
- 30.Sheehan M M, Stanley E, Fitzgerald G F, van Sinderen D. Identification and characterization of a lysis module present in a large proportion of bacteriophages infecting Streptococcus thermophilus. Appl Environ Microbiol. 1999;65:569–577. doi: 10.1128/aem.65.2.569-577.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith G P. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 32.Stevens C M, Daniel R, Illing N, Errington J. Characterization of a sporulation gene, spoIVA, involved in spore coat morphogenesis in Bacillus subtilis. J Bacteriol. 1992;174:586–594. doi: 10.1128/jb.174.2.586-594.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stöver A G, Driks A. Secretion, localization, and antibacterial activity of TasA, a Bacillus subtilis spore-associated protein. J Bacteriol. 1999;181:1664–1672. doi: 10.1128/jb.181.5.1664-1672.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 35.Takamatsu H, Kodama T, Nakayama T, Watabe K. Characterization of the yrbA gene of Bacillus subtilis, involved in resistance and germination of spores. J Bacteriol. 1999;181:4986–4994. doi: 10.1128/jb.181.16.4986-4994.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng L B, Donovan W P, Fitz-James P C, Losick R. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev. 1988;2:1047–1054. doi: 10.1101/gad.2.8.1047. [DOI] [PubMed] [Google Scholar]