Abstract
Human pathogenic yersiniae organisms export and translocate the Yop virulence proteins and V antigen upon contact with a eukaryotic cell. Yersinia pestis mutants defective for production of YscX or YscY were unable to export the Yops and V antigen. YscX and YscY were both present in the Y. pestis cell pellet fraction; however, YscX was also found in the culture supernatant. YscY showed structural and amino acid sequence similarities to the Syc family of proteins. YscY specifically recognized and bound to a region of YscX that included a predicted coiled-coil region. These data suggest that YscY may function as a chaperone for YscX in Y. pestis.
Upon contact with the surface of a eukaryotic cell, Yersinia species pathogenic for humans export a distinct set of plasmid-encoded virulence proteins called Yops (for review, see references 26 and 36). Yop proteins are translocated directly from the cytoplasm of the bacterium into the cytoplasm of the host cell (12, 57, 62). Once translocated into the eukaryotic cell, Yop proteins function to disrupt intracellular signaling pathways (YopH [7, 14, 48, 49] and YpkA [22]), prevent specific cytoskeletal rearrangements (YopE [56]) and YopT [27]), and/or induce apoptotic events (YopJ [38, 39]). This capability enables the yersiniae to avoid phagocytosis and ensures survival of the bacteria within host tissues (14, 15, 17, 54, 55, 65).
Yops are secreted across the bacterial inner and outer membranes by a type III or “contact-dependent” secretion mechanism (36). Maximal expression and secretion of Yops in vitro occurs at 37°C in medium lacking calcium. Genes encoding the Yersinia type III secretion apparatus (11, 25, 47) are clustered within several large transcriptional units which include yscBCDEFGHIJKLM (21, 32, 35, 52), yscNOPQRSTU (6, 16), yscW (2, 32), and yopNtyeAsycNyscXYVlcrR (8, 13, 18, 28, 29, 51). Mutational inactivation of any of the ysc genes (with the exception of yscB and yscH) prevents Yop secretion (2, 3, 6, 28, 35, 36, 51, 52).
Proteins secreted through the Yersinia type III secretion pathway include both effector Yops destined for translocation into the eukaryotic cell and proteins directly required for Yop secretion (YscO [44] and YscP [45]), Yop translocation (YopB [23], YopD [33, 58, 70], YopN, TyeA [29], LcrG [42, 60], and V antigen [42, 50]), and/or the regulation of Yop secretion (YopN [13, 18, 67], TyeA [13, 29] and LcrG [41, 64]). The targeting of proteins for export through the type III secretion apparatus has been shown to involve one or both of two identified secretion targeting signals (4, 9). One secretion signal has been identified within the sequences encoding the initial 15 amino acid residues of YopE, YopN (4), and YopK (YopQ [5]). This signal appears to be encoded in the mRNA sequence rather than being part of the peptide sequence, suggesting a cotranslational mechanism of Yop secretion. A second secretion signal is dependent upon the interaction of the secreted protein with an accessory protein termed a specific Yop chaperone (Syc) (9, 68). The Syc proteins are small cytoplasmic proteins that specifically recognize an amino-terminal region of one or two specific Yops (69, 71). Syc proteins for five Yops have been identified: SycE (68) for YopE, SycH (69, 71) for YopH, SycT (27) for YopT, SycD (40, 69) (LcrH) for YopB and YopD, and SycN (13, 28) and YscB (30) for YopN. The YopE protein contains both an mRNA targeting signal and a Syc-dependent targeting signal (4, 9). Either secretion signal can apparently function independently, although somewhat inefficiently, to target YopE for secretion at 37°C in the absence of calcium in vitro; however, both targeting signals are required for the translocation of YopE into a eukaryotic cell.
Recently, Iriarte and Cornelis described two new genes in Yersinia enterocolitica, yscX and yscY, whose products were required for the secretion of Yop proteins (28, 67). In this study, we confirm a role for the yscX and yscY gene products in Yop export in Yersinia pestis. Polyclonal antisera specific for each of these proteins localized YscX and YscY to the cell pellet fraction; however, YscX, like YscO and YscP, was also found in the culture supernatant at 37°C in the absence of calcium. YscY was demonstrated to possess amino acid sequence and structural characteristics similar to those of the Syc family of proteins. We also demonstrate that YscY directly binds to a region of YscX containing a predicted coiled-coil domain. Together, these data suggest that YscY may function as a chaperone for YscX in Y. pestis.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All Y. pestis strains used in this study are Pgm−, and thus, avirulent from peripheral routes of infection (66). Y. pestis KIM8-3002 (41) [Smr, pCD1, pPCP1− (Pla−), pMT1] served as the parental strain for construction of the yscX and yscY deletion mutants. Plasmid pCD1 is a ca. 70-kb plasmid that encodes the Yops and the components of the type III secretion machinery (47). Plasmid pMT1 is a ca. 100-kb plasmid that encodes the capsular antigen Fra1 (34). Plasmid pPCP1 encodes the outer membrane plasminogen activator protease (Pla), which has been shown to degrade exported Yops (53). Y. pestis KIM8-3002 and derivatives of this strain were routinely grown in heart infusion broth (HIB) or on tryptose blood agar base plates (Difco Laboratories, Detroit, Mich.) at a temperature of 30°C. For growth experiments, Yersinia strains were grown in the presence or absence of 2.5 mM calcium chloride in the defined medium TMH (20) as previously described (51). Overnight TMH cultures (26°C) were used to inoculate 2 ml of fresh TMH (with or without 2.5 mM CaCl2) to an optical density at 620 nm (OD620) of 0.15. Cultures were grown at 26°C with vigorous shaking for 1 h, followed by 5 h at 37°C. Escherichia coli SY327 λpir (37) was used as a host strain for the suicide vector pUK4134 (63) and derivatives of this plasmid. E. coli XL1 blue (Stratagene, La Jolla, Calif.) was used for routine cloning experiments, and E. coli BL21(DE3) (Novagen, Madison, Wis.) was used for overexpression of maltose-binding protein (MBP) fusions and polyhistidine- and FLAG-tagged YscX and YscY.
DNA methods.
Plasmid DNA was isolated by using the Perfect Prep Kit (5-prime, 3-prime, Inc., Boulder, Colo.) or by the method of Kado and Liu (31). DNA fragments were isolated and purified from agarose gels by using the Qiaex DNA purification kit (Qiagen, Inc., Studio City, Calif.). Electroporation of E. coli and Y. pestis cells was done according to the procedure of Perry et al. (46). PCR was performed with the thermostable Pfu polymerase (Stratagene) and 21- to 30-nucleotide primers (Ransom Hill Bioscience, Ramona, Calif.) for 35 cycles according to the manufacturer's directions. Denaturing and annealing temperatures were 94°C for 1 min and 55°C for 1 min, respectively. Pfu polymerase extension times at 72°C varied depending upon the length of the desired PCR product. Double-stranded DNA was sequenced by the University of Miami School of Medicine DNA sequencing core facility by using a DyeDeoxy Terminator Cycle Sequencing kit and an ABI model 373A DNA sequencer (Applied Biosystems, Foster City, Calif.). Nucleotide sequences and amino acid sequences were analyzed with IntelliGenetics computer software (IntelliGenetics, Mountain View, Calif.). The presence and locations of coiled-coil domains were predicted with the MultiCoil program (72).
Construction of yscX and yscY deletion mutants.
Unique BsaBI restriction endonuclease sites flanking the region encoding amino acids 37 to 107 of YscX were used to create an in-frame deletion in yscX of plasmid pGP2 (yopNtyeAsycNyscXYVlcrR) (51). A ca. 4.0-kb XmnI-SmaI fragment (yopNtyeAsycNΔyscXyscYV′) carrying the yscX deletion was inserted into the EcoRV site of pUK4134, creating plasmid pUK4134.P24. An in-frame deletion in yscY eliminating the coding regions for amino acids 13 to 22 was constructed by the PCR-ligation-PCR technique (1) using primers 5′-CCTCTGGGGGTATTTAGCG-3′, 5′-AACTCCTGTTGTCGTTTGG-3′, 5′-TGTGGCCATGCAGAGCGCGC-3′, and 5′-CACGGAGTCGCCGGCGATTAGG-3′. The ca. 2.0-kb amplified product was inserted into the EcoRV site of the suicide vector pUK4134, generating plasmid pUK4134.P25. A second in-frame deletion in yscY eliminating the coding region for amino acid residues 13 to 97 was constructed by the PCR-ligation-PCR technique and by substituting primer 5′-CATGCTTATCAACATTATTTG for primer 5′-TGTGGCCATGCAGAGCGCGC-3′. The resultant PCR product was inserted into pUK4134, generating plasmid pUK4134.P26. Plasmids pUK4134.P24, pUK4134.P25, and pUK4134.P26 were utilized to move the yscX and two separate yscY deletions into Y. pestis KIM8-3002 as previously described (51), generating Y. pestis KIM8-3002.P24 (ΔyscX 37-107) and KIM8-3002.P25 (ΔyscY 13-22) and KIM8-3002.P26 (ΔyscY 13-97).
Plasmid pYSCX1, encoding YscX, was constructed by inserting a 0.54-kb PvuII-MscI fragment of pGP2, carrying the entire yscX gene, into SmaI-EcoRV-digested pBluescript SK(−) (Stratagene) such that the yscX gene is transcribed from the vector lac promoter. Plasmid pYSCY1, encoding YscY, was constructed by inserting a 0.46-kb SacII-EcoRV fragment of pGP2 carrying the entire yscY gene into SacII-EcoRV-digested pBluescript SK(−) downstream of the vector lac promoter. Plasmids pYSCX1 and pYSCY1 were used in complementation experiments.
Generation of antisera specific for YscX and YscY.
Overexpression and purification of polyhistidine-tagged YscX and YscY was facilitated by construction of expression plasmids pET-YSCX1 and pET-YSCY1. The entire yscX and yscY genes were amplified by PCR using the oligonucleotide primer pairs YscX-H1 (5′-TCATATGGTGAGTCGCATAATAACTGC-3′) plus YscX-H2 (5′-AGGATCCTCATACTTTGTGCAACAGG-3′) and YscY-H1 (5′-TCATATGAATATTACTTTAACCAA-3′) plus YscY-H2 (5′-AGGATCCTCATGGGGATTCATTATGA-3′), respectively. The oligonucleotides were tailed with NdeI and BamHI restriction endonuclease sites (underlined) to facilitate insertion of the amplified fragments into NdeI-BamHI-digested pET15b (Novagen). Plasmids pET-YSCX1 and pET-YSCY1 were electroporated into E. coli BL21(DE3) for overexpression of polyhistidine-tagged YscX and YscY. The recombinant polyhistidine-tagged YscX and YscY proteins expressed from plasmids pET-YSCX1 and pET-YSCY1 are predicted to be expressed with an additional 20 amino-terminal residues that include the polyhistidine tag and a thrombin cleavage site. Purification of the polyhistidine-tagged proteins was performed according to the manufacturer's protocol by using the His-Bind Resin and Buffer Kit (Novagen). Purified polyclonal antisera specific for YscX and YscY were raised in female New Zealand White rabbits by using the purified YscX and YscY proteins (Animal Pharm Services, Healdsburg, Calif.).
YscX and YscY MBP fusions.
Derivatives of plasmid pMAL-c2 (New England Biolabs, Beverly, Mass.) encoding full-length YscX, YscY, and YopN or truncated versions of YscX fused to the carboxyl terminus of the E. coli MBP were constructed to facilitate stable high-level expression of these proteins. DNA fragments corresponding to the full-length yscX, yscY, or yopN open reading frames or to truncated versions of the yscX gene encoding amino acid residues 30 to 122, 50 to 122, 80 to 122, 1 to 110, or 1 to 90 of YscX were amplified with oligonucleotide primers that introduced an EcoRI site at the 5′ end and a PstI site (NsiI for yopN) at the 3′ end of each fragment. EcoRI-PstI- and EcoRI-NsiI-digested fragments were inserted into EcoRI-PstI-digested pMAL-c2, generating plasmids pMBP-YscX, pMBP-YscY, pMBP-YopN, pMBP-YscXΔ1–29, pMBP-YscXΔ1–49, pMBP-YscXΔ1–79, pMBP-YscXΔ91–122, and pMBP-YscXΔ111–122. Plasmid pMBP-YscXΔ70–90, which contains an internal deletion in yscX eliminating the DNA sequence encoding amino acid residues 70 to 90 of YscX, was constructed by the PCR-ligation-PCR technique.
Expression of YscX- and YscY-FLAG-tagged recombinant proteins.
Plasmids pYSCX-FLAG and pYSCY-FLAG, which express carboxyl-terminal FLAG-tagged YscX and YscY, respectively, were constructed by inserting PCR-amplified fragments of yscX and yscY tailed with HindIII and BglII restriction endonuclease sites into HindIII-BglII-digested pFLAG-CTC (Sigma, St. Louis, Mo.). The recombinant YscX-FLAG and YscY-FLAG proteins expressed from plasmids pYSCX-FLAG and pYSCY-FLAG are predicted to be expressed with an additional three amino-terminal residues (Met-Lys-Leu) and an additional eleven carboxyl-terminal residues (Arg-Ser-Val-Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys), which includes the FLAG epitope (underlined). DNA fragments corresponding to regions of yscX encoding amino acid residues 1 to 60, 1 to 100, and 16 to 122 of YscX were amplified with oligonucleotide primers that introduced a HindIII site at the 5′ end and a BglII site at the 3′ end of each fragment. HindIII- and BglII-digested fragments were inserted into HindIII-BglII-digested pFLAG-CTC, generating plasmids pFLAG-YscXΔ61–122, pFLAG-YscXΔ101–122, and pFLAG-YscXΔ1–15.
SDS-PAGE and immunoblotting.
Cell pellets and culture supernatants were separated by centrifugation at 12,200 × g for 10 min at 4°C. Cell pellets were washed once with ice-cold 100 mM Tris-HCl–1 mM EDTA, pH 8.0, and pelleted by centrifugation. Culture supernatant proteins were precipitated on ice overnight with 10% (vol/vol) trichloroacetic acid and resuspended in 100 mM Tris-HCl–1 mM EDTA, pH 8.0. Volumes of cellular fractions corresponding to equal numbers of bacteria were mixed 1:1 (vol/vol) with 2× electrophoresis sample buffer (20 mM Tris-HCl, 2 mM EDTA, 4% [wt/vol] sodium dodecyl sulfate [SDS], 10% [vol/vol] β-mercaptoethanol) and analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot analysis essentially as previously described (51). Samples to be analyzed with polyclonal antisera specific for YscX or YscY were electrophoresed on 14% (wt/vol) acrylamide gels. Y. pestis proteins were visualized as previously described (13, 30), using polyclonal antisera specific for YopM, YopN, V antigen, YscX, and YscY. FLAG-tagged proteins were detected with the FLAG M2 monoclonal antibody (Sigma).
Yeast two-hybrid system experiment.
The yeast two-hybrid assay was performed by methods recommended by the commercial supplier (Clontech, Palo Alto, Calif.). Vectors pGAD424 and pGBT9 encoding the activation domain and DNA binding domain, respectively, of the yeast GAL4 transcriptional activator were obtained from Clontech laboratories as part of the Matchmaker Two-Hybrid System kit. The complete yscX (5′-CCGGAATTCGTGAGTCGCATAATAACTGCC-3′ and 5′-TCCGCTGCAGTCATACTTTGTGCAACAGGTT-3′) and yscY (5′-CCGGAATTCATGAATATTACTTTAACCAAA-3′ and 5′-TCCGCTGCAGTCATGGGGATTCATTATGATC-3′) coding sequences were obtained by PCR, digested with EcoRI and PstI, and inserted into EcoRI-PstI-digested pGAD424 and pGBT9, generating plasmids pGAD-YscX, pGAD-YscY, pGBT-YscX, and pGBT-YscY. Plasmids pGAD-YopN, pGAD-SycN, pGAD-YscB, pGBT-YopN, pGBT-SycN, and pGBT-YscB were constructed previously (13). Plasmids were transformed into Saccharomyces cerevisiae SFY596 cells according to the manufacturer's instruction (Clontech) and plated on appropriate minimal yeast synthetic dropout medium plates. Colony lift assays and liquid assays for detecting β-galactosidase activity were performed essentially as described by Clontech laboratories.
Protein affinity blotting.
E. coli BL21(DE3) cells transformed with plasmid pYSCY-FLAG were grown at 37°C in 30 ml of HIB containing 100 μg of ampicillin/ml to an OD620 of 1.2. IPTG (isopropyl-β-d-thiogalactopyranoside) was then added to a 1 mM final concentration, and the cells were grown for an additional hour at 30°C. Growth at 30°C increased the percentage of YscY-FLAG protein that was expressed in a soluble form. Cells induced for YscY-FLAG expression were harvested by centrifugation at 8,000 × g for 10 min at 4°C, resuspended in 3 ml of Tris-buffered saline (TBS) (20 mM Tris-HCl, 140 mM NaCl, pH 7.5) and lysed by a single passage through a chilled French pressure cell. Unlysed cells and large debris were removed by centrifugation at 8,000 × g for 5 min at 4°C. The cleared lysate was stored at 4°C. E. coli BL21(DE3) transformed with plasmids pMAL-c2, pMBP-YscX, pMBP-YscY, pMBP-YopN, pMBP-YscXΔ1–29, pMBP-YscXΔ1–49, pMBP-YscXΔ1–79, pMBP-YscXΔ111–122, pMBP-YscXΔ91–122, and pMBP-YscXΔ70–90 were grown at 37°C in 3 ml of HIB containing 100 μg of ampicillin/ml to an OD620 of 1.2. IPTG was then added to a 1 mM final concentration, and the cells were grown for an additional hour at 37°C. Cells were centrifuged at 8,000 × g and resuspended in a volume of 1× electrophoresis sample buffer based on OD measurements of the cell cultures prior to harvest. Cell extracts containing the MBP derivatives were separated on SDS–12% (wt/vol) PAGE gels and transferred to Immobilon membranes as previously described. Membranes were blocked with TBS containing 5% nonfat dry milk for 1 h at room temperature. Blocked membranes were incubated in 50 ml of TBS containing 1% nonfat dry milk and 2 ml of the BL21(DE3) YscY-FLAG cleared lysate for 1 h at room temperature. Membranes were washed with TBS and incubated with the FLAG M2 monoclonal antibody (1:20,000 dilution) for 1 h at room temperature. Membranes were washed with TBS and further incubated with an alkaline phosphatase-conjugated anti-mouse immunoglobulin G secondary antibody (1:5,000 dilution) for 1 h at room temperature. Membranes were washed with TBS and developed as previously described (51).
RESULTS
Construction of Y. pestis yscX and yscY deletion mutants.
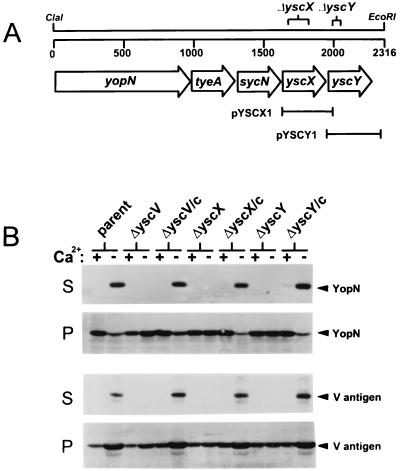
Genes encoding YscX and YscY are located downstream of yopN, tyeA, and sycN in the yopNtyeAsycNyscXYVlcrR operon (Fig. 1A). The yscX gene is predicted to encode a 122-amino-acid residue protein with a predicted molecular weight of 13,756, while yscY is predicted to encode a 114-amino-acid residue protein with a predicted molecular weight of 13,117. Recent data indicates that yscX and yscY of Y. enterocolitica encode essential components of the type III secretion machinery (28). In this study, we identify and localize the Y. pestis YscX and YscY proteins and further define the role of these proteins in Yop export. An in-frame deletion in yscX eliminating the coding region for amino acids 37 to 107 of YscX was constructed by BsaBI digestion. In addition, two separate in-frame deletions were constructed within yscY, the first eliminating the DNA sequences encoding YscY amino acid residues 13 to 22 and the second eliminating the yscY sequences encoding amino acid residues 13 to 97. The yscX and yscY deletions were moved into plasmid pCD1 of Y. pestis KIM8-3002 by allelic exchange, generating the yscX deletion mutant KIM8-3002.P24 and the yscY deletion mutants KIM8-3002.P25 and KIM8-3002.P26. Plasmids pYSCX1 and pYSCY1 were used in complementation studies (Fig. 1). Analysis of the KIM8-3002.P26 strain, which carries a deletion eliminating the coding region for YscY residues 13 to 97, revealed a polar effect on the downstream yscV gene. Immunoblot analysis of YscV expression in this mutant and the mutant complemented with plasmid pYSCY1 revealed no YscV expression (data not shown). KIM8-3002.P24 (ΔyscX) and KIM8-3002.25 (ΔyscY) showed no polar effects on the downstream yscV gene (data not shown) and thus were used for further analysis of these genes and gene products.
FIG. 1.
(A) Genetic organization of the yopNtyeAsycNyscXY region of plasmid pCD1 in Y. pestis KIM. The locations of in-frame deletions within yscX and yscY are shown (see Materials and Methods). Plasmids pYSCX1 and pYSCY1 were used in complementation experiments. (B) Immunoblot analysis of culture supernatant and cell pellet fractions from Y. pestis strains grown at 37°C in the presence (+) or absence (−) of calcium. Antisera specific for YopN and V antigen were used to detect these proteins in the supernatant (S) and cell pellet (P) fractions from Y. pestis KIM8-3002 (parent), a yscV deletion mutant (ΔyscV), the yscX deletion mutant (ΔyscX), and the yscY deletion mutant (ΔyscY). The yscV, yscX, and yscY deletion mutants were complemented (/c) with plasmids pYscV1, pYSCX1, and pYSCY1, respectively.
Secretion of V antigen and YopN by the yscX and yscY deletion mutants.
Secretion of V antigen and YopN by the parent strain Y. pestis KIM8-3002, yscV deletion mutant KIM8-3002.3 (51), the yscX deletion mutant, the yscY deletion mutant, and the yscV, yscX, and yscY deletion mutants complemented with plasmids pYscV1 (pYPD [52]), pYSCX1, and pYSCY1, respectively, was determined by SDS-PAGE and immunoblotting (Fig. 1B). The yscX and yscY deletion mutants, like the previously characterized yscV deletion mutant, failed to secrete the V antigen and YopN at 37°C in the presence or absence of calcium. The parent strain and the yscV, yscX, and yscY deletion mutants complemented with plasmids pYscV1, pYSCX1, and pYSCY1 secreted V antigen and YopN under conditions permissive for Yop secretion. These data indicate that the absence of Yop secretion in the yscX and yscY deletion mutants was due to the lack of YscX or YscY and not to polar effects on downstream genes.
Identification and localization of the yscX and yscY gene products.
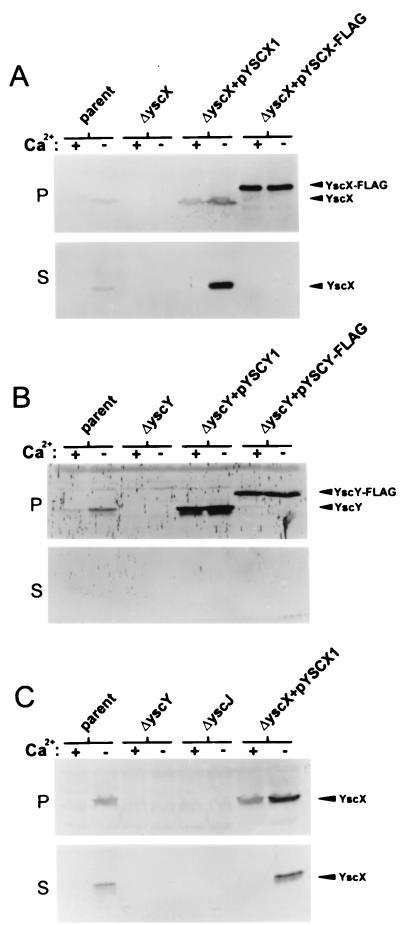
Polyclonal antiserum raised against a polyhistidine-tagged yscX gene product was used to identify YscX in immunoblots from cell pellet and culture supernatant fractions of the parent Y. pestis KIM8-3002, the yscX deletion mutant, and the yscX deletion mutant complemented with pYSCX1 or pYSCX-FLAG (Fig. 2A). An approximately 13-kDa protein was identified as YscX in immunoblots from the parent strain and the yscX deletion mutant complemented with plasmid pYSCX1. An approximately 15-kDa protein was identified in the yscX deletion mutant complemented with plasmid pYSCX-FLAG. As expected, YscX was not detected in any fractions obtained from the yscX deletion mutant. YscX was also detected in the culture supernatant fractions from the parent strain and from the yscX deletion mutant complemented with plasmid pYSCX1 grown at 37°C in the absence of calcium, suggesting that YscX is exported via the type III secretion machinery. Introduction of the pYSCX-FLAG plasmid into the yscX deletion mutant resulted in significant intracellular expression of FLAG-tagged YscX; however, this protein was not secreted in the presence or absence of calcium.
FIG. 2.
Identification and localization of YscX and YscY. Bacterial strains were grown at 37°C in TMH with (+) or without (−) calcium. (A) Proteins from cell pellet (P) fractions and culture supernatant (S) fractions of Y. pestis KIM8-3002 (parent), the yscX deletion mutant (ΔyscX), and the yscX deletion mutant complemented with pYSCX1 or pYSCX-FLAG were subjected to SDS-PAGE and immunoblot analysis with a polyclonal antiserum specific for YscX. (B) Cell pellet (P) and culture supernatant (S) fractions from Y. pestis KIM8-3002 (parent), the yscY deletion mutant (ΔyscY), and the yscY deletion mutant complemented with pYSCY1 or pYSCY-FLAG were subjected to SDS-PAGE and immunoblot analysis with a polyclonal antiserum specific for YscY. (C) Cell pellet (P) and culture supernatant (S) fractions from Y. pestis KIM8-3002 (parent), the yscY deletion mutant (ΔyscY), a yscJ deletion mutant (ΔyscJ), and the yscX deletion mutant complemented with pYSCX1 were subjected to SDS-PAGE and immunoblot analysis with a polyclonal antiserum specific for YscX.
Polyclonal antiserum raised against a polyhistidine-tagged yscY gene product was used to identify YscY in immunoblots from cell pellet and culture supernatant fractions of Y. pestis cultures grown at 37°C in the presence and absence of calcium (Fig. 2B). An approximately 13-kDa protein present in the cell pellet fraction was identified as YscY in immunoblots from the parent strain and from the yscY deletion mutant complemented with plasmid pYSCY1. The YscY protein was absent from immunoblots of samples obtained from the yscY deletion mutant. A ca. 15-kDa protein representing FLAG-tagged YscY was present in the cell pellet fraction obtained from the yscY deletion mutant complemented with pYSCY-FLAG. The YscY protein was not detected in the culture supernatant fraction of any of the strains examined. The predicted YscY amino acid sequence does not contain any stretches of more than four consecutive hydrophobic amino acid residues and is predicted to contain no transmembrane domains (59), suggesting that YscY is a cytoplasmic protein.
In order to confirm that the YscX present in the culture supernatant was exported via the type III secretion apparatus, we measured YscX expression and secretion in both the yscY deletion mutant and in a yscJ deletion mutant (M. W. Jackon, J. B. Day, F. Ferracci, and G. V. Plano, submitted for publication), designated KIM8-3002.P27 (Fig. 2C). YscJ is a bacterial lipoprotein that is required for the secretion of the V antigen and Yops (35). YscX was secreted at 37°C in the absence of calcium from both the parent KIM8-3002 and the yscX deletion strain complemented with pYSCX1; however, no YscX was detected in the supernatant of the yscJ or yscY deletion mutants. Together, these data indicate that YscX is exported to the culture supernatant via the Y. pestis type III secretion machinery.
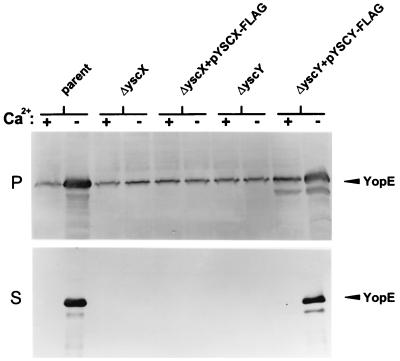
The absence of the FLAG-tagged YscX protein in the culture supernatant of the yscX deletion mutant carrying pYSCX-FLAG (Fig. 2A) could be due to the inability of this protein to complement the general defect in Yop secretion associated with the deletion in yscX, or the addition of the FLAG epitope tag could specifically affect the targeting and secretion of the tagged YscX protein. In order to differentiate between these two possibilities, we evaluated the ability of the pYSCX-FLAG and pYSCY-FLAG plasmids to complement the defect in Yop secretion associated with the yscX and yscY deletions, respectively (Fig. 3). Expression and secretion of YopE by the parent strain, the yscX and yscY deletion strains, and the yscX and yscY deletion strains complemented with pYSCX-FLAG and pYSCY-FLAG, respectively, was determined by SDS-PAGE and immunoblot analysis. The parent strain and the yscY deletion mutant complemented with pYSCY-FLAG secreted YopE at 37°C in the absence of calcium. The yscX deletion mutant, the yscY deletion mutant, and the yscX deletion mutant carrying pYSCX-FLAG failed to secrete YopE in the presence or absence of calcium, indicating that the FLAG-tagged YscX protein was unable to complement the defect in YopE secretion associated with the deletion in yscX.
FIG. 3.
Immunoblot analysis of YopE secretion. Bacterial strains were grown at 37°C in TMH with (+) or without (−) calcium. Proteins from cell pellet (P) fractions and culture supernatant (S) fractions of Y. pestis KIM8-3002 (parent), the yscX deletion mutant (ΔyscX), the yscX deletion mutant complemented with pYSCX-FLAG, the yscY deletion mutant (ΔyscY), and the yscY deletion mutant complemented with pYSCY-FLAG were subjected to SDS-PAGE and immunoblot analysis with a polyclonal antiserum specific for YopE.
Secretion of FLAG-tagged YscX by Y. pestis strains competent for Yop secretion.
FLAG-tagged YscX was unable to complement the defect in Yop secretion associated with the yscX deletion mutant. In order to determine if YscX-FLAG was nonfunctional due to a defect in its ability to be secreted, we moved pYSCX-FLAG into the parent strain Y. pestis KIM8-3002 and monitored secretion of YscX-FLAG by SDS-PAGE and immunoblot analysis with a monoclonal antibody specific for the FLAG epitope (Fig. 4B). Y. pestis KIM8-3002 carrying pYSCX-FLAG secreted YscX-FLAG at 37°C in the absence of calcium, indicating that addition of the FLAG epitope to the carboxyl terminus of YscX disrupted a function of YscX in Yop secretion unrelated to its secretion targeting signal or signals. A significant percentage of the YscX-FLAG protein remained associated with the cell pellet and may represent insoluble YscX-FLAG. In order to confirm that expression of the YscX-FLAG protein did not have an adverse effect upon the Yop secretion pathway, we monitored the secretion of YopE in the parent strain KIM8-3002 and in the parent strain carrying plasmid pYSCX-FLAG (Fig. 4C). YopE was secreted in approximately equal amounts in both the presence and absence of pYSCX-FLAG, indicating that the FLAG-tagged YscX protein did not interfere with the normal functioning of the type III secretion machinery.
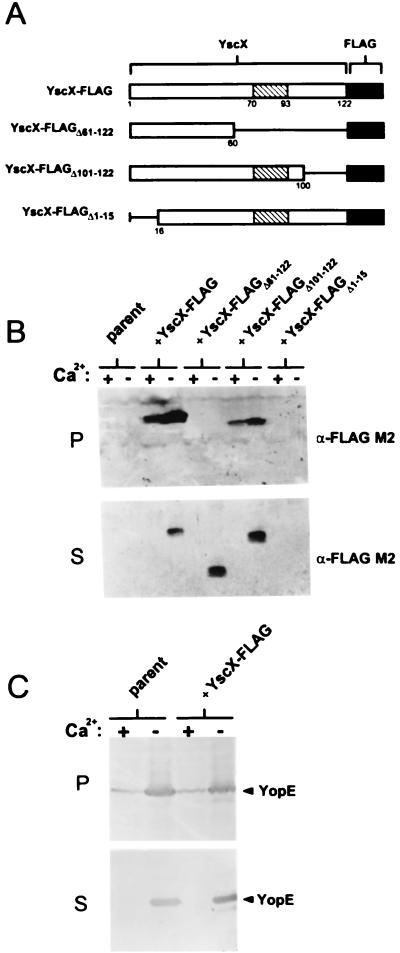
FIG. 4.
Secretion of YscX-FLAG and truncated YscX-FLAG proteins from the parent strain Y. pestis KIM8-3002. (A) Schematic representation of FLAG-tagged YscX (YscX-FLAG), YscX-FLAGΔ61–122, YscX-FLAGΔ101–122, and YscX-FLAGΔ1–15. A predicted coiled-coil domain spanning residues 70 to 93 of YscX is shown (hatched box). (B) Immunoblot analysis of SDS-PAGE-separated culture supernatant (S) and cell pellet (P) fractions from Y. pestis KIM8-3002 (parent) and KIM8-3002 transformed with plasmids pFLAG-YscX, pFLAG-YscXΔ61–122, pFLAG-YscXΔ101–122, and pFLAG-YscXΔ1–15. The FLAG M2 monoclonal antibody was used to detect the FLAG-tagged products. (C) Immunoblot analysis of SDS-PAGE-separated culture supernatant (S) and cell pellet (P) fractions from Y. pestis KIM8-3002 (parent) and KIM8-3002 complemented with plasmid pFLAG-YscX. Antiserum specific for YopE was used to detect this protein in the supernatant and cell pellet fractions.
Secretion of truncated FLAG-tagged yscX gene products by the parent Y. pestis KIM8-3002.
To begin to localize the secretion targeting signal of YscX, we constructed a series of vectors encoding FLAG-tagged amino- or carboxyl-terminal truncated YscX proteins (Fig. 4A). Plasmids carrying deletions in pYSCX-FLAG eliminating DNA sequences encoding YscX amino acid residues 61 to 122, 101 to 122, and 1 to 15 were electroporated into Y. pestis KIM8-3002 cells, and the expression and secretion of the truncated FLAG-tagged YscX proteins was determined by SDS-PAGE and immunoblotting with the FLAG M2 monoclonal antibody (Fig. 4B). FLAG-tagged YscX missing the carboxyl-terminal 22 amino acid residues (YscX-FLAGΔ101–122) was found in both the cell pellet fraction and the culture supernatant fraction. Elimination of the carboxyl-terminal 62 amino acid residues of YscX (YscX-FLAGΔ61–122) resulted in a truncated yscX gene product which was found exclusively in the culture supernatant. This data suggests that a carboxyl-terminal region of YscX is required for the stability of YscX within the cell. YscX-FLAGΔ1–15 was not detected in the cell pellet or culture supernatant fractions, indicating that the gene product was not expressed or was unstable. These data suggest that the carboxyl-terminal 62 residues of YscX are not required for efficient secretion; however, a region of YscX between amino acids 61 and 100 is required for stable expression of YscX within the cell. Interestingly, a predicted coiled-coil region was found between amino acid residues 70 and 93 of YscX (43, 72). A predicted coiled-coil region within the YopN protein sequence has been implicated in the interaction of YopN with its chaperones, YscB and SycN (13, 30).
YscY possesses characteristics similar to those of the Syc proteins.
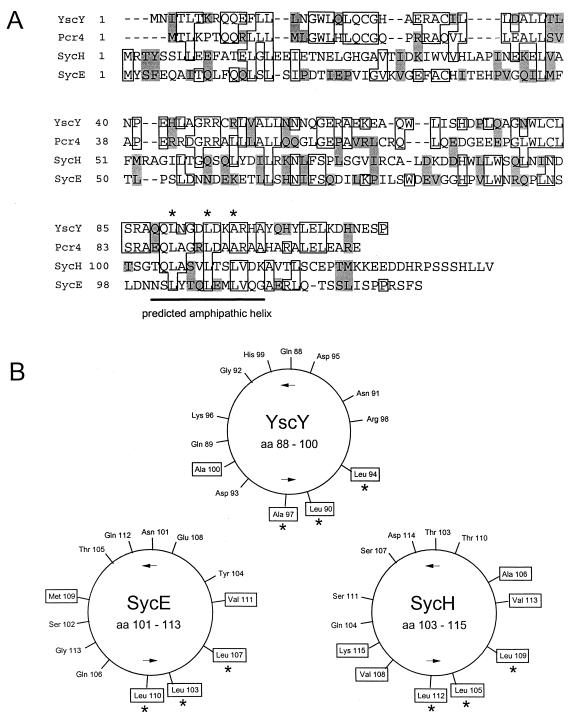
Efficient secretion and translocation of Yop proteins often requires the assistance of an accessory protein termed a specific Yop chaperone. These chaperones are small, acidic cytoplasmic proteins which normally are encoded in close proximity to the gene encoding the protein they interact with. YscY is a small (ca. 13 kDa) acidic (pI = 6.5) cytoplasmic protein that is encoded directly upstream of yscX (28, 67). Alignment (24) of YscY with SycE and SycH, the specific Yop chaperones for YopE and YopH, respectively, revealed similarities between the amino acid sequences (Fig. 5A). YscY also contains a predicted carboxyl-terminal amphipathic alpha-helical region similar in location and arrangement to those found in the other chaperones (69) (Fig. 5B). These data suggest that YscY may function as a chaperone, possibly for the secreted YscX protein.
FIG. 5.
(A) Amino acid alignment of YscY with Pcr4 of P. aeruginosa and with SycE and SycH. Boxed residues, sequence identity; shaded residues, related or similar residues; underlined sequence, region predicted to form an amphipathic helix; dashes within sequences, gaps introduced to optimize alignment. (B) Helical wheel representations (61) of the underlined regions of YscY, SycE, and SycH. Hydrophobic amino acid residues are boxed.
Interaction of YscY with YscX.
The yeast two-hybrid system was utilized to investigate whether YscX and YscY interact with one another or with proteins encoded by the yopNtyeAsycNyscXYVlcrR operon. The interaction of hybrid proteins encoded for by fusions between the entire tyeA, sycN, yscX, yscY, yscB, and yopN genes to sequences of plasmids pGAD424 and pGBT9 encoding the GAL4 activation and DNA binding domains, respectively, were measured by both colony lifts and quantitative liquid β-galactosidase assays (Table 1).
TABLE 1.
Yeast two-hybrid analysis of YscX and YscY interactions
| Plasmids useda
|
Colony lift assayb | β-Gal activityc | |
|---|---|---|---|
| Derivative of pGAD424 | Derivative of pGBT9 | ||
| pGAD424 | pGBT9 | − | <1.0 |
| pGAD-YopE | pGBT-SycE | + | 111.9 ± 3.9 |
| pGAD-YscX | pGBT-YopN | − | <1.0 |
| pGAD-YscX | pGBT-TyeA | − | <1.0 |
| pGAD-YscX | pGBT-SycN | − | <1.0 |
| pGAD-YscX | pGBT-YscB | − | <1.0 |
| pGAD-YscX | pGBT-YscX | − | <1.0 |
| pGAD-YscX | pGBT-YscY | +/− | 5.7 ± 0.8 |
| pGAD-YscY | pGBT-YopN | − | <1.0 |
| pGAD-YscY | pGBT-TyeA | − | <1.0 |
| pGAD-YscY | pGBT-SycN | − | <1.0 |
| pGAD-YscY | pGBT-YscB | − | <1.0 |
| pGAD-YscY | pGBT-YscY | − | <1.0 |
| pGAD-YscY | pGBT-YscX | + | 120.0 ± 3.8 |
The SFY596 reporter strain was transformed with derivatives of the indicated plasmids.
Plus and minus signs indicate the relative intensity of the blue color developed after exposure of permeabilized cells to X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) for 1 h.
Expression of the reporter lacZ gene was measured as described by Clontech laboratories. The given values are averages (± standard deviations) assayed in triplicate. β-Gal, β-galactosidase.
S. cerevisiae SFY596 cells containing pGAD-YscY and pGBT-YscX produced 120 U of β-galactosidase, implying that these proteins directly interact with one another. The reciprocal pairing of pGAD-YscX with pGBT-YscY produced significantly less β-galactosidase activity (5.9 U); however, this level was still significantly greater than levels obtained from SFY596 transformed with control plasmids (pGAD424 and pGBT9) or plasmids containing gene fusions to yopN, tyeA, sycN, or yscB (<1.0 U). These data suggest that YscX and YscY directly interact with one another.
FLAG-tagged YscY recognizes and directly binds an MBP-YscX hybrid protein.
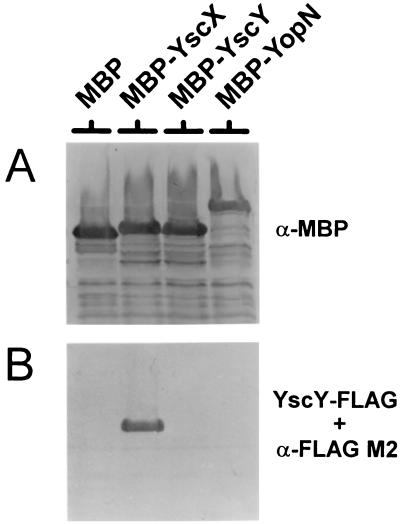
Previous studies have demonstrated that SycE specifically binds to YopE that has been transferred to nitrocellulose membranes (68). In order to confirm the interaction between YscX and its putative chaperone YscY, we performed a similar protein affinity blot experiment using FLAG-tagged YscY to probe Immobilon membranes containing MBP, MBP-YscX, MBP-YscY, and MBP-YopN hybrid proteins (Fig. 6). MBP fusions were detected with a rabbit polyclonal antisera specific for the E. coli MBP (Fig. 6A), while bound YscY-FLAG was detected with the FLAG M2 monoclonal antibody (Fig. 6B). FLAG-tagged YscY directly bound to the MBP-YscX hybrid protein but not to MBP alone or to the MBP-YscY or MBP-YopN hybrid proteins, confirming that YscY specifically recognizes and binds to YscX.
FIG. 6.
Binding of YscY-FLAG to MBP-YscX. Cell pellet fractions from BL21(DE3) expressing MBP, MBP-YscX, MBP-YscY, and MBP-YopN were separated by SDS-PAGE and transferred to Immobilon membranes. MBP migrated slower than expected due to an in-frame fusion between malE and the vector LacZ α-peptide-encoding sequences. (A) Detection of MBP, MBP-YscX, MBP-YscY, and MBP-YopN with antiserum specific for MBP. (B) Immobilon membrane containing SDS-PAGE-separated MBP, MBP-YscX, MBP-YscY, and MBP-YopN probed with a BL21(DE3) extract containing YscY-FLAG. Bound YscY-FLAG was detected with the FLAG M2 monoclonal antibody.
Localization of the region of YscX required for YscY binding.
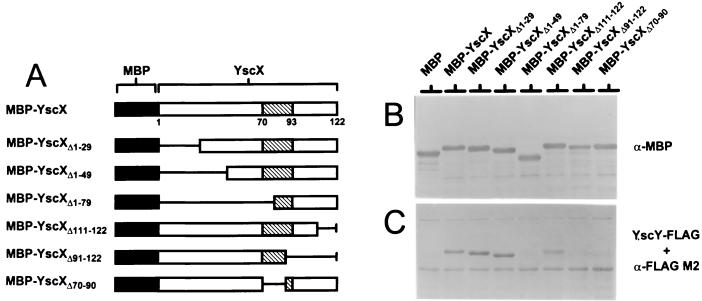
To localize the region of YscX recognized by YscY, a series of in-frame fusions between the malE gene and portions of the yscX gene encoding amino acid residues 1 to 29 (MBP-YscXΔ1–29), 1 to 49 (MBP-YscXΔ1–49), 1 to 79 (MBP-YscXΔ1–79), 111 to 122 (MBP-YscXΔ111–122), and 91 to 122 (MBP-YscXΔ91–122) and an internal deletion eliminating amino acid residues 70 to 90 (MBP-YscXΔ70–90) (Fig. 7A) were constructed and transformed into BL21(DE3). Lysates from BL21(DE3) containing MBP-YscX and the MBP-YscX deletion derivatives were separated by SDS-PAGE, transferred to Immobilon membranes, and probed with antiserum specific for the MBP (Fig. 7B) or with FLAG-tagged YscY (Fig. 7C) as described for Fig. 6.
FIG. 7.
Localization of the YscY binding domain of YscX by using truncated and internally deleted MBP-YscX hybrid proteins. (A) Schematic representation of truncated and internally deleted MBP-YscX hybrid proteins used to detect the binding of YscY-FLAG. A predicted coiled-coil domain spanning residues 70 to 93 of YscX is shown (hatched boxes). (B) Detection of MBP-YscX and derivatives with antiserum specific for MBP. MBP migrated slower than expected due to an in-frame fusion between malE and the vector LacZ α-peptide-encoding sequences. (C) Immobilon membrane containing SDS-PAGE-separated MBP-YscX, MBP-YscXΔ1–29, MBP-YscXΔ1–49, MBP-YscXΔ1–79, MBP-YscXΔ111–122, MBP-YscXΔ91–122, and MBP-YscXΔ70–90 probed with a BL21(DE3) extract containing YscY-FLAG. Bound YscY-FLAG was detected with the FLAG M2 monoclonal antibody.
FLAG-tagged YscY bound strongly to MBP-YscX, MBP-YscXΔ1–29, MBP-YscXΔ1–49, and MBP-YscXΔ111–122, indicating that the amino-terminal 49 residues and the carboxyl-terminal 12 residues of YscX are not required for interaction with YscY. FLAG-tagged YscY failed to bind to MBP-YscXΔ1–79 and MBP-YscXΔ91–122, indicating that the region of YscX between residues 50 and 110, which contains the predicted coiled-coil domain (amino acids 70 to 93 of YscX), is essential for interaction with YscY. An in-frame deletion eliminating the coding region for residues 70 to 90 of YscX (MBP-YscXΔ70–90) was not recognized by YscY, suggesting that the predicted coiled-coil domain in YscX is essential for the interaction of YscY with YscX.
DISCUSSION
Many of the individual components of the Yersinia type III secretion system have been shown to consist of membrane-bound, membrane-associated, or cytoplasmic proteins (26). In contrast, our data shows that YscX is not restricted to the intracellular compartments but is able to move into the extracellular environment. The secretion of YscX represents the third example of a component of the Yersinia type III secretion machinery that is secreted by this system (YscO [44] and YscP [45] have also been shown to be secreted). At present, no direct role for the secretion of YscX, or secreted YscX, has been determined.
Secretion of YscX could represent a means of regulating the secretion process through the removal of an essential component of the secretion apparatus. Secretion of YscX could also be a means of eliminating excess YscX from the bacterial cell. These hypotheses infer that cytosolic YscX and YscY perform an essential role in the secretion process; however, secreted YscX is nonfunctional. Alternatively, secreted YscX could be required to complete the formation of a functional type III secretion apparatus. This hypothesis would imply that a hierarchy exists in the type III secretion process, in which YscX secretion is required prior to the secretion of other proteins. A hierarchical type III secretion process has been reported in Salmonella typhimurium, in which the secreted InvJ/SpaN and SpaO proteins are also required for the secretion of the Sip invasion proteins (10).
The cytoplasmic YscY protein shares both structural (size, pI, carboxyl-terminal amphipathic helix) and amino acid sequence similarities with the other members of the Syc family. We demonstrate that YscY, like the other Syc proteins, specifically recognizes and binds to a secreted protein, YscX. Unfortunately, since both YscX and YscY are essential for the function of the Yersinia type III secretion system, we were unable to demonstrate a specific defect in YscX export in the yscY deletion mutant. Interestingly, YscY is the first example of a Syc-like chaperone required for the formation of an export-competent type III secretion system.
The SycE protein has been shown to form a homodimer (68). We have also previously demonstrated that the yeast two-hybrid system can be used to detect both the SycE-SycE interaction and the SycE-YopE interaction. Interestingly, we failed to detect a YscY-YscY interaction by using the yeast two-hybrid system. These data suggest either that YscY functions as a monomer or that the yeast two-hybrid system failed to detect the YscY-YscY interaction. The low β-galactosidase activity (5.7 U) induced by the combination of plasmids pGAD-YscX and pGBT-YscY suggests that the hybrid protein encoded for by one or both of these plasmids is either unstable in yeast or possesses only minimal function. A defective pGAD-YscY gene fusion product could account for the inability to detect a YscY-YscY interaction by using plasmids pGAD-YscY and pGBT-YscY.
YscY was shown to recognize and bind to a region of YscX located between amino acid residues 50 and 110. Similarily, the binding sites for SycE and the SycN-YscB chaperone complex have been localized within amino acid residues 40 to 100 of YopE (68, 71) and YopN (13), respectively. Furthermore, the chaperone binding regions of both YopN (13) and YscX are predicted to contain a coiled-coil region. An MBP-YscX fusion protein deleted for the predicted coiled-coil region of YscX failed to interact with the YscY chaperone, suggesting that the coiled-coil region is involved in the YscY-YscX interaction.
Syc family members have also been shown to be required for the stable soluble cytoplasmic expression of their cognate Yop. For example, intracellular YopE is rapidly degraded in a sycE deletion mutant (9, 19). Our preliminary evidence suggests that YscY may also function to stabilize YscX within the bacterium. YscX-FLAG and the YscX-FLAGΔ101–122-truncated protein, which both possess an intact YscY binding site, were stably expressed in the cytosol and exported from the bacterium. YscX-FLAGΔ61–122, which lacks the coiled-coil region implicated in YscY binding, was also exported at 37°C in the absence of calcium; however, no stable cytosolic FLAG-tagged protein could be detected. These data suggest that YscY is required to maintain a stable cytoplasmic pool of YscX. Furthermore, YscX expressed from the lac promoter of plasmid pYSCX1 exhibited a calcium-dependent expression profile (Fig. 2A), which could be the result of stabilization of YscX by calcium-regulated YscY.
Secretion of YscX occurred only at 37°C in the absence of calcium, suggesting that YscX contains a secretion signal, or signals, similar to those of the exported Yops. YopE has been shown to possess two distinct signals capable of targeting it for export (9). One signal is dependent on the binding of the specific chaperone SycE to an amino-terminal region of YopE, while the second signal is in the mRNA encoding the amino-terminal 15 residues of YopE and is involved in the cotranslational secretion of YopE. Several lines of evidence suggest that YscX may also contain two independent secretion signals. First, as described above, we show that the chaperone-like YscY protein recognizes and directly binds YscX. This is significant because all of the previously identified Syc proteins are directly or indirectly involved in promoting the secretion of their target protein or proteins. Secondly, removal of the YscY binding domain from YscX did not prevent its secretion, which suggests the presence of a second independent secretion signal. Specifically, the parent Y. pestis KIM8-3002 expressing YscX-FLAGΔ61–122, which is deleted for the coiled-coil region implicated in YscY binding, still exported the deleted product to the culture supernatant, although no stable intracellular product was detected. A similar situation has been observed with truncated forms of YopE that lack the SycE binding region (9). Secretion of the truncated YopE proteins deleted for the SycE binding site is thought to occur primarily via a cotranslational mechanism, mediated by the YopE mRNA signal. The secreted YscX-FLAGΔ61–122 may also escape cytosolic degradation by the coupling of its translation and secretion. Interestingly, a yscX construct deleted for the DNA sequences encoding the first 15 amino acid residues of YscX failed to produce a stable YscX product. A similar phenotype has been observed with specific mutations within the first 15 codons of yopE and yopN (4). In this case, the lack of a stable gene product was also explained by an uncoupling of yopE or yopN mRNA translation and protein secretion. Together, these data suggest that YscX may possess both an mRNA signal and a Syc-dependent signal capable of directing its export through the Yersinia type III secretion system.
Messenger RNA secretion signals have been associated with the secretion of the yopE, yopN (4), and yopK (5) gene products. Each of the genes encoding these cotranslationally secreted proteins is the first gene in its transcriptional unit. However, the mRNA from yscX would be predicted to be part of a larger yopNtyeAsycNyscXYVlcrR transcript. Whether or not the secretion of one gene product from the middle of a longer transcript can be translationally coupled is unknown and beyond the scope of this study. Another possibility is that a separate mRNA transcript initiates upstream of yscX. Indeed, in Y. enterocolitica, a putative promoter was identified 62 base pairs upstream of the yscX GTG start codon (67). It is conceivable that only the transcript initiating immediately downstream from this second putative promoter sequence is involved in translational coupling of YscX secretion. Further detailed studies will be necessary to confirm a role for YscY in YscX secretion and to confirm or deny the presence of specific type III secretion targeting signals in the yscX mRNA and gene product.
Interestingly, proteins similar to YscX and YscY, termed Pcr3 and Pcr4, respectively, have been identified in the opportunistic pathogen Pseudomonas aeruginosa (Fig. 5) (73). The genes encoding Pcr3 and Pcr4 lie immediately downstream of genes encoding homologs of YopN, TyeA, and SycN, suggesting that both the operon structure and function of these genes and gene products are conserved between these two bacteria. Interestingly, no proteins with significant amino acid similarity to YscX or YscY have been identified among the components of the type III secretion systems of other bacterial pathogens (26, 28), indicating that these proteins, and their Pseudomonas homologs, perform functions that are unique to the Yersinia and Pseudomonas type III delivery systems.
ACKNOWLEDGMENTS
This study was supported by Public Health Service Grant AI39575.
We thank Susan C. Straley (University of Kentucky) for the kind gift of rabbit anti-V antigen antibody, rabbit anti-YopM antibody, and Y. pestis KIM8-3002.
REFERENCES
- 1.Ali S A, Steinkasserer A. PCR-ligation-PCR mutagenesis: a protocol for creating gene fusions and mutations. BioTechniques. 1995;18:746–750. [PubMed] [Google Scholar]
- 2.Allaoui A, Scheen R, Lambert de Rouvroit C, Cornelis G R. VirG, a Yersinia enterocolitica lipoprotein involved in Ca2+ dependency, is related to exsB of Pseudomonas aeruginosa. J Bacteriol. 1995;177:4230–4237. doi: 10.1128/jb.177.15.4230-4237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allaoui A, Schulte R, Cornelis G R. Mutational analysis of the Yersinia enterocolitica virC operon: characterization of yscE, F, G, I, J, K required for Yop secretion and yscH encoding YopR. Mol Microbiol. 1995;18:343–355. doi: 10.1111/j.1365-2958.1995.mmi_18020343.x. [DOI] [PubMed] [Google Scholar]
- 4.Anderson D M, Schneewind O. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science. 1997;278:1140–1143. doi: 10.1126/science.278.5340.1140. [DOI] [PubMed] [Google Scholar]
- 5.Anderson D M, Schneewind O. Yersinia enterocolitica type III secretion: an mRNA signal that couples translation and secretion of YopQ. Mol Microbiol. 1999;31:1139–1148. doi: 10.1046/j.1365-2958.1999.01254.x. [DOI] [PubMed] [Google Scholar]
- 6.Bergman T, Erickson K, Galyov E, Persson C, Wolf-Watz H. The lcrB (yscN/U) gene cluster of Yersinia pseudotuberculosis is involved in Yop secretion and shows high homology to the spa gene clusters of Shigella flexneri and Salmonella typhimurium. J Bacteriol. 1994;176:2619–2626. doi: 10.1128/jb.176.9.2619-2626.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bliska J B, Guan K, Dixon J E, Falkow S. Tyrosine phosphate hydrolysis of host proteins by an essential Yersinia virulence determinant. Proc Natl Acad Sci USA. 1991;88:1187–1191. doi: 10.1073/pnas.88.4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boland A, Sory M P, Iriarte M, Kerbourch C, Wattiau P, Cornelis G R. Status of YopM and YopN in the Yersinia Yop virulon: YopM of Yersinia enterocolitica is internalized inside the cytosol of PU5-1.8 macrophages by the YopB, D, N delivery apparatus. EMBO J. 1996;15:5191–5201. [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng L W, Anderson D M, Schneewind O. Two independent type III secretion mechanisms for YopE in Yersinia enterocolitica. Mol Microbiol. 1997;24:757–765. doi: 10.1046/j.1365-2958.1997.3831750.x. [DOI] [PubMed] [Google Scholar]
- 10.Collazo C M, Galan J E. Requirement for exported proteins in secretion through the invasion-associated type III system of Salmonella typhimurium. Infect Immun. 1996;64:3524–3531. doi: 10.1128/iai.64.9.3524-3531.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornelis G R, Boland A, Boyd A P, Geuijen C, Iriarte M, Neyt C, Sory M P, Stainier I. The virulence plasmid of Yersinia, an antihost genome. Microbiol Mol Biol Rev. 1998;62:1315–1352. doi: 10.1128/mmbr.62.4.1315-1352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornelis G R, Wolf-Watz H. The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol Microbiol. 1997;23:861–867. doi: 10.1046/j.1365-2958.1997.2731623.x. [DOI] [PubMed] [Google Scholar]
- 13.Day J B, Plano G V. A complex composed of SycN and YscB functions as a specific chaperone for YopN in Yersinia pestis. Mol Microbiol. 1998;30:777–788. doi: 10.1046/j.1365-2958.1998.01110.x. [DOI] [PubMed] [Google Scholar]
- 14.Fallman M, Andersson K, Hakansson S, Magnusson K E, Stendahl O, Wolf-Watz H. Yersinia pseudotuberculosis inhibits Fc receptor-mediated phagocytosis in J774 cells. Infect Immun. 1995;63:3117–3124. doi: 10.1128/iai.63.8.3117-3124.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fallman M, Persson C, Wolf-Watz H. Yersinia proteins that target host cell signaling pathways. J Clin Investig. 1997;99:1153–1157. doi: 10.1172/JCI119270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fields K, Plano G V, Straley S C. A low-Ca2+ response (LCR) secretion (ysc) locus lies within the lcrB region of the LCR plasmid in Yersinia pestis. J Bacteriol. 1994;176:569–579. doi: 10.1128/jb.176.3.569-579.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forsberg Å, Rosqvist R, Wolf-Watz H. Regulation and polarized transfer of the Yersinia outer proteins (Yops) involved in antiphagocytosis. Trends Microbiol. 1994;2:14–19. doi: 10.1016/0966-842x(94)90339-5. [DOI] [PubMed] [Google Scholar]
- 18.Forsberg Å, Viitanen A-M, Skurnik M, Wolf Watz H. The surface-located YopN protein is involved in calcium signal transduction in Yersinia pseudotuberculosis. Mol Microbiol. 1991;5:977–986. doi: 10.1111/j.1365-2958.1991.tb00773.x. [DOI] [PubMed] [Google Scholar]
- 19.Frithz-Lindsten E, Rosqvist R, Johansson L, Forsberg Å. The chaperone-like protein YerA of Yersinia pseudotuberculosis stabilizes YopE in the cytoplasm but is dispensible for targeting to the secretion loci. Mol Microbiol. 1995;16:635–647. doi: 10.1111/j.1365-2958.1995.tb02426.x. [DOI] [PubMed] [Google Scholar]
- 20.Goguen J D, Yother J, Straley S C. Genetic analysis of the low calcium response in Yersinia pestis Mud1 (Ap lac) insertion mutants. J Bacteriol. 1984;160:842–848. doi: 10.1128/jb.160.3.842-848.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haddix P L, Straley S C. Structure and regulation of the Yersinia pestis yscBCDEF operon. J Bacteriol. 1992;174:4820–4828. doi: 10.1128/jb.174.14.4820-4828.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hakansson S, Galyov E E, Rosqvist R, Wolf-Watz H. The Yersinia YpkA Ser/Thr kinase is translocated and subsequently targeted to the inner surface of the HeLa cell plasma membrane. Mol Microbiol. 1996;20:593–603. doi: 10.1046/j.1365-2958.1996.5251051.x. [DOI] [PubMed] [Google Scholar]
- 23.Hakansson S, Schesser K, Persson C, Galyov E E, Rosqvist R, Homble F, Wolf-Watz H. The YopB protein of Yersinia pseudotuberculosis is essential for the translocation of Yop effector proteins across the target cell plasma membrane and displays a contact-dependent membrane disrupting activity. EMBO J. 1996;15:5812–5823. [PMC free article] [PubMed] [Google Scholar]
- 24.Higgins D G, Sharp P M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 25.Hu P, Elliott J, McCready P, Skowronski E, Garnes J, Kobayashi A, Brubaker R R, Garcia E J. Structural organization of virulence-associated plasmids of Yersinia pestis. J Bacteriol. 1998;180:5192–5202. doi: 10.1128/jb.180.19.5192-5202.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hueck C J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iriarte M, Cornelis G R. YopT, a new Yersinia Yop effector protein, affects the cytoskeleton of host cells. Mol Microbiol. 1998;29:915–929. doi: 10.1046/j.1365-2958.1998.00992.x. [DOI] [PubMed] [Google Scholar]
- 28.Iriarte M, Cornelis G R. Identification of SycN, YscX, and YscY, three new elements of the Yersinia Yop virulon. J Bacteriol. 1999;181:675–680. doi: 10.1128/jb.181.2.675-680.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iriarte M, Sory M P, Boland A, Boyd A P, Mills S D, Lambermont I, Cornelis G R. TyeA, a protein involved in control of Yop release and in translocation of Yersinia Yop effectors. EMBO J. 1998;17:1907–1918. doi: 10.1093/emboj/17.7.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackson M W, Day J B, Plano G V. YscB of Yersinia pestis functions as a specific chaperone for YopN. J Bacteriol. 1998;180:4912–4921. doi: 10.1128/jb.180.18.4912-4921.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koster M, Bitter W, de Cock H, Allaoui A, Cornelis G R, Tommassen J. The outer membrane component, YscC, of the Yop secretion machinery of Yersinia enterocolitica forms a ring-shaped multimeric complex. Mol Microbiol. 1997;26:789–797. doi: 10.1046/j.1365-2958.1997.6141981.x. [DOI] [PubMed] [Google Scholar]
- 33.Lee V T, Schneewind O. Type III machines of pathogenic yersiniae secrete virulence factors into the extracellular milieu. Mol Microbiol. 1999;31:1619–1629. doi: 10.1046/j.1365-2958.1999.01270.x. [DOI] [PubMed] [Google Scholar]
- 34.Lindler L E, Plano G V, Mayhew G F, Burland V, Blattner F R. Complete DNA sequence and detailed analysis of the Yersinia pestis KIM5 plasmid encoding murine toxin and capsular antigen. Infect Immun. 1998;66:5731–5742. doi: 10.1128/iai.66.12.5731-5742.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michiels T, Vanooteghem J-C, de Rouvroit C L, China B, Gustin A, Boudry P, Cornelis G R. Analysis of virC, an operon involved in the secretion of Yop proteins by Yersinia enterocolitica. J Bacteriol. 1991;173:4994–5009. doi: 10.1128/jb.173.16.4994-5009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michiels T, Wattiau P, Brasseur R, Ruysschaert J-M, Cornelis G. Secretion of Yop proteins by Yersiniae. Infect Immun. 1990;58:2840–2849. doi: 10.1128/iai.58.9.2840-2849.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mills S D, Boland A, Sory M P, van der Smissen P, Kerbourch C, Finlay B B, Cornelis G R. Yersinia enterocolitica induces apoptosis in macrophages by a process requiring functional type III secretion and translocation mechanisms and involving YopP, presumably acting as an effector protein. Proc Natl Acad Sci USA. 1997;94:12638–12643. doi: 10.1073/pnas.94.23.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monack D M, Mecsas J, Ghori N, Falkow S. Yersinia signals macrophages to undergo apoptosis and YopJ is necessary for this cell death. Proc Natl Acad Sci USA. 1997;94:10385–10390. doi: 10.1073/pnas.94.19.10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neyt C, Cornelis G R. Role of SycD, the chaperone of the Yersinia Yop translocators YopB and YopD. Mol Microbiol. 1999;31:143–156. doi: 10.1046/j.1365-2958.1999.01154.x. [DOI] [PubMed] [Google Scholar]
- 41.Nilles M L, Williams A W, Skrzypek E, Straley S C. Yersinia pestis LcrV forms a stable complex with LcrG and may have a secretion-related regulatory role in the low-Ca2+ response. J Bacteriol. 1997;179:1307–1316. doi: 10.1128/jb.179.4.1307-1316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nilles M L, Fields K A, Straley S C. The V antigen of Yersinia pestis regulates Yop vectorial targeting as well as Yop secretion through effects on YopB and LcrG. J Bacteriol. 1998;180:3410–3420. doi: 10.1128/jb.180.13.3410-3420.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pallen M J, Dougan G, Frankel G. Coiled-coil domains in proteins secreted by type III secretion systems. Mol Microbiol. 1997;25:423–425. doi: 10.1046/j.1365-2958.1997.4901850.x. [DOI] [PubMed] [Google Scholar]
- 44.Payne P L, Straley S C. YscO of Yersinia pestis is a mobile core component of the Yop secretion system. J Bacteriol. 1998;180:3882–3890. doi: 10.1128/jb.180.15.3882-3890.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Payne P L, Straley S C. YscP of Yersinia pestis is a secreted component of the Yop secretion system. J Bacteriol. 1999;181:2852–2862. doi: 10.1128/jb.181.9.2852-2862.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perry R D, Pendrak M, Schuetze P. Identification and cloning of a hemin storage locus involved in the pigmentation phenotype of Yersinia pestis. J Bacteriol. 1990;172:5929–5937. doi: 10.1128/jb.172.10.5929-5937.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perry R D, Straley S C, Fetherston J D, Rose D J, Gregor J, Blattner F R. DNA sequencing and analysis of the low-Ca2+-response plasmid pCD1 of Yersinia pestis KIM5. Infect Immun. 1998;66:4611–4623. doi: 10.1128/iai.66.10.4611-4623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Persson C, Carballeira N, Wolf-Watz H, Fallman M. The PTPase YopH inhibits uptake of Yersinia, tyrosine phosphorylation of p130Cas and FAK, and the associated accumulation of these proteins in peripheral focal adhesions. EMBO J. 1997;16:2307–2318. doi: 10.1093/emboj/16.9.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Persson C, Nordfelth R, Holmstrom A, Hakansson S, Rosqvist R, Wolf-Watz H. Cell-surface-bound Yersinia translocate the protein tyrosine phosphatase YopH by a polarized mechanism into the target cell. Mol Microbiol. 1995;18:135–150. doi: 10.1111/j.1365-2958.1995.mmi_18010135.x. [DOI] [PubMed] [Google Scholar]
- 50.Pettersson J, Holmstrom A, Hill J, Leary S, Frithz-Lindsten E, Von Euler-Matell A, Carlsson E, Titball R, Forsberg A, Wolf-Watz H. The V-antigen of Yersinia is surface exposed before target cell contact and involved in virulence protein translocation. Mol Microbiol. 1999;32:9619–9676. doi: 10.1046/j.1365-2958.1999.01408.x. [DOI] [PubMed] [Google Scholar]
- 51.Plano G V, Straley S C. Multiple effects of lcrD mutations in Yersinia pestis. J Bacteriol. 1993;175:3536–3545. doi: 10.1128/jb.175.11.3536-3545.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Plano G V, Straley S C. Mutations in yscC, yscD, and yscG prevent high-level expression and secretion of V antigen and Yops in Yersinia pestis. J Bacteriol. 1995;177:3843–3854. doi: 10.1128/jb.177.13.3843-3854.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Protsenko O A, Anisimov P L, Mozharov O T, Konnov N P, Popov Y A, Kokooshkin A M. Detection and characterization of Yersinia pestis plasmids determining pesticin I, fraction I antigen, and “mouse” toxin synthesis. Genetica. 1983;19:1081–1090. [PubMed] [Google Scholar]
- 54.Rosqvist R, Bölin I, Wolf-Watz H. Inhibition of phagocytosis in Yersinia pseudotuberculosis: a virulence plasmid-encoded ability involving the Yop2b protein. J Bacteriol. 1988;174:3355–3363. doi: 10.1128/iai.56.8.2139-2143.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosqvist R, Forsberg Å, Rimpiläinen M, Bergman T, Wolf-Watz H. The cytotoxic protein YopE of Yersinia obstructs the primary host defence. Mol Microbiol. 1990;4:657–667. doi: 10.1111/j.1365-2958.1990.tb00635.x. [DOI] [PubMed] [Google Scholar]
- 56.Rosqvist R, Forsberg Å, Wolf-Watz H. Intracellular targeting of the Yersinia YopE cytotoxin in mammalian cells induces actin microfilament disruption. Infect Immun. 1990;59:4562–4569. doi: 10.1128/iai.59.12.4562-4569.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosqvist R, Magnusson K, Wolf-Watz H. Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J. 1994;13:964–972. doi: 10.1002/j.1460-2075.1994.tb06341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosqvist R, Persson C, Hakansson S, Nordfeldt R, Wolf-Watz H. Translocation of the Yersinia YopE and YopH virulence proteins into target cells is mediated by YopB and YopD. Contrib Microbiol Immunol. 1995;13:230–234. [PubMed] [Google Scholar]
- 59.Rost B, Casadio R, Fariselli P, Sander C. Prediction of helical transmembrane segments at 95% accuracy. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sarker M R, Sory M P, Boyd A P, Iriarte M, Cornelis G R. LcrG is required for efficient translocation of Yersinia Yop effector proteins into eukaryotic cells. Infect Immun. 1998;66:2976–2979. doi: 10.1128/iai.66.6.2976-2979.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schiffer M, Edmundson A B. Use of helical wheels to represent the structures of proteins and to identify segments with helical potential. Biophys J. 1967;7:121–135. doi: 10.1016/S0006-3495(67)86579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Skrzypek E, Cowan C, Straley S C. Targeting of the Yersinia pestis YopM protein into HeLa cells and intracellular trafficking to the nucleus. Mol Microbiol. 1998;30:1051–1065. doi: 10.1046/j.1365-2958.1998.01135.x. [DOI] [PubMed] [Google Scholar]
- 63.Skrzypek E, Haddix P L, Plano G V, Straley S C. New suicide vector for gene replacement in yersiniae and other gram-negative bacteria. Plasmid. 1993;29:160–163. doi: 10.1006/plas.1993.1019. [DOI] [PubMed] [Google Scholar]
- 64.Skrzypek E, Straley S C. LcrG—a secreted protein involved in negative regulation of the low calcium response in Yersinia pestis. J Bacteriol. 1993;175:3520–3528. doi: 10.1128/jb.175.11.3520-3528.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Straley S C, Cibull M L. Differential clearance and host-pathogen interactions of YopE− and YopK− YopL−Yersinia pestis in BALB/c mice. Infect Immun. 1989;57:1200–1210. doi: 10.1128/iai.57.4.1200-1210.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Une T, Brubaker R R. In-vivo comparison of avirulent Vwa− and Pgm− or Pstr phenotypes of Yersiniae. Infect Immun. 1984;43:895–900. doi: 10.1128/iai.43.3.895-900.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Viitanen A-M, Toivanen P, Skurnik M. The lcrE gene is part of an operon in the lcr region of Yersinia enterocolitica 0:3. J Bacteriol. 1990;172:3152–3162. doi: 10.1128/jb.172.6.3152-3162.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wattiau P, Cornelis G R. SycE, a chaperone-like protein of Yersinia enterocolitica involved in the secretion of YopE. Mol Microbiol. 1993;8:123–131. doi: 10.1111/j.1365-2958.1993.tb01209.x. [DOI] [PubMed] [Google Scholar]
- 69.Wattiau P, Woestyn S, Cornelis G R. Customized secretion chaperones in pathogenic bacteria. Mol Microbiol. 1996;20:255–262. doi: 10.1111/j.1365-2958.1996.tb02614.x. [DOI] [PubMed] [Google Scholar]
- 70.Williams A W, Straley S C. YopD of Yersinia pestis plays a role in negative regulation of the low-calcium response in addition to its role in translocation of Yops. J Bacteriol. 1998;180:350–358. doi: 10.1128/jb.180.2.350-358.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Woestyn S, Sory M P, Boland A, Lequenne O, Cornelis G R. The cytosolic SycE and SycH chaperones of Yersinia protect the region of YopE and YopH involved in translocation across eukaryotic cell membranes. Mol Microbiol. 1996;6:1261–1271. doi: 10.1111/j.1365-2958.1996.tb02645.x. [DOI] [PubMed] [Google Scholar]
- 72.Wolf E, Kim P S, Berger B. MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yahr T L, Goranson J, Frank D W. Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III pathway. Mol Microbiol. 1996;22:991–1003. doi: 10.1046/j.1365-2958.1996.01554.x. [DOI] [PubMed] [Google Scholar]