Abstract
Acral dermatoses, including hyperkeratotic palmoplantar eczema (HPE), palmoplantar psoriasis (PP), and mycosis fungoides palmaris et plantaris (MFPP), can be challenging to diagnose clinically and histopathologically. In this setting, cytokine biomarkers may be able to help provide diagnostic clarity. Therefore, we evaluated IL-17A, IFN-γ, and IL-13 expression in PP, HPE, and MFPP and compared their expression profiles with nonacral sites. We used biopsy specimens from the Yale Dermatopathology database, selecting cases of HPE (n = 12), PP (n = 8), MFPP (n = 8), normal acral skin (n = 9), nonacral eczema (n = 10), and nonacral psoriasis (n = 10) with classic clinical and histopathologic features. IL17A mRNA expression by RNA in situ hybridization differentiated PP (median score 63.1 [interquartile range 9.4–104.1]) from HPE (0.8 [0–6.0]; P = 0.003), MFPP (0.6 [0–2.6]; P = 0.003), and normal acral skin (0 [0–0]; P < 0.001). Unexpectedly, both PP and HPE showed co-expression of IFNG and IL13 mRNA. In contrast, nonacral psoriasis and eczema showed divergent patterns of IFNG and IL13 mRNA expression. Taken together, we show that IL17A mRNA expression may be a useful biomarker of PP, and we further show that acral dermatoses exhibit distinct immunology compared to nonacral sites, with implications for clinical management.
Introduction
Hyperkeratotic palmoplantar eczema (HPE) and palmoplantar psoriasis (PP) are both characterized by inflammation and hyperkeratosis on acral surfaces (Kim et al., 2022). In contrast to dyshidrotic eczema and pustular PP, which also affect palmoplantar skin and are usually relatively straightforward to diagnose, HPE and PP can sometimes be difficult to differentiate from each other because of significant clinical overlap. As therapies in dermatology become increasingly targeted—and selection of the wrong therapy can have consequences ranging from neutral to detrimental (Espinosa et al., 2020)—accurate differentiation of HPE and PP becomes essential. Furthermore, these diseases may have overlapping features with mycosis fungoides palmaris et plantaris (MFPP). Although biopsy is often helpful, some cases remain difficult to definitively classify (Kim et al., 2022; Park et al., 2017).
In this setting, biomarkers reflecting the underlying immunology of these disorders have the potential to provide both diagnostic clarity and therapeutic guidance. In canonical eczema/atopic dermatitis and psoriasis, well-established divergent patterns of T helper 2 (Th2) and Th1/Th17 inflammation are seen, respectively (Wang et al., 2021), informing the rational selection of therapies targeting these inflammatory pathways. In contrast, the immunologic changes of these dermatoses at acral sites have not been rigorously investigated, and cytokine-directed therapies have varied efficacy when applied to acral sites. For instance, some patients with HPE show an incomplete response to dupilumab, which blocks Th2 cytokine activity (Oosterhaven et al., 2019). Patients with PP may also experience an inferior response to the IL-17A inhibitor secukinumab compared to patients with plaque psoriasis (Gottlieb et al., 2017; Langley et al., 2014; Reolid et al., 2022). These acral dermatoses may therefore involve inflammatory pathways overlapping with but not identical to their nonacral counterparts. Furthermore, canonical mycosis fungoides exhibits a Th2-predominant response but can also express cytokines associated with other Th subsets (Liu et al., 2022; Rindler et al., 2021); whether MFPP shows similar diversity remains unknown.
In this study, we used RNA in situ hybridization (RISH) to evaluate classic cases of HPE, PP, and MFPP biopsy specimens for markers of Th1 (IFN-γ), Th2 (IL-13), Th17 (IL-17A), and innate immune (IL-36α, IL-36γ) inflammation, as well as expression of inducible nitric oxide synthase (NOS2), an established psoriasis biomarker (Wang et al., 2021). Our goals were two-fold: (i) to identify markers specific for each acral dermatosis and (ii) to compare the immunologic profile of these acral conditions to their nonacral counterparts.
Results
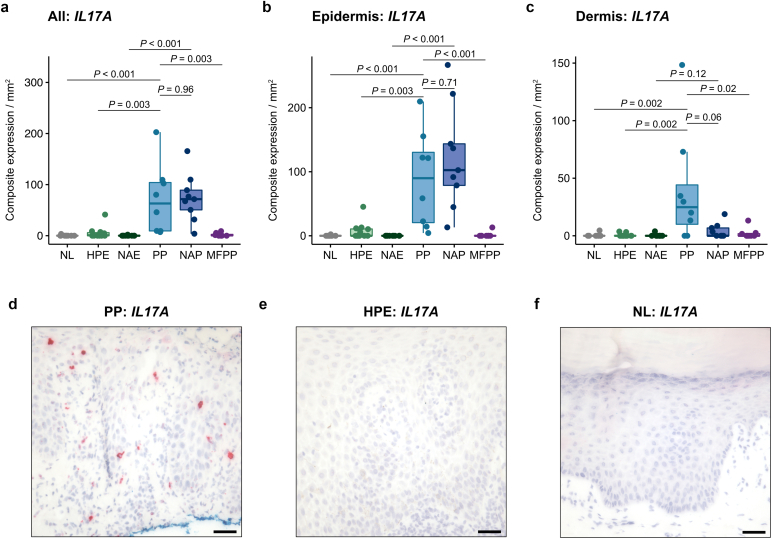
In total, 57 cases from 53 patients (mean [SD] age, 51.3 [18.1] years; 23 [43.4%] female) were evaluated (Table 1). Consistent with previous work showing IL17A to be a specific marker of nonacral psoriasis (Wang et al., 2021), we found that IL17A mRNA expression was also significantly increased in PP (median score 63.1 [IQR 9.4–104.1]) compared with HPE (0.8 [0–6.0]; P = 0.003), MFPP (0.6 [0–2.6]; P = 0.003), and normal acral skin (0 [0–0]; P < 0.001) (Figure 1a–f). Expression of IL17A was highest in the epidermis (Figure 1b).
Table 1.
Patient Characteristics
| Sample | Age | Sex | Anatomic Site | Specific Acral Site | Race | Hispanic or Latina/Latino/Latinx | Phototype1 | Chronicity |
|---|---|---|---|---|---|---|---|---|
| HPE-1 | 61 | M | L hand | Palmar | Black or African American | No | VI | N/A |
| HPE-22 | 65 | F | R foot | Unknown | White | Yes | N/A | N/A |
| HPE-32 | 65 | F | L hand | Unknown | White | Yes | N/A | N/A |
| HPE-4 | 62 | M | L hand | Palmar | White | No | I | N/A |
| HPE-5 | 58 | M | R foot | Unknown | White | No | I | N/A |
| HPE-6 | 52 | F | R foot | Unknown | White | No | II | 8 mo |
| HPE-7 | 52 | M | L foot | Lateral | White | No | II | 1 y |
| HPE-8 | 70 | M | R hand | Palmar | White | No | II | 3 y |
| HPE-9 | 33 | F | R hand | Unknown | Black or African American | No | VI | N/A |
| HPE-10 | 11 | M | L foot | Plantar | White | Yes | III | 2 y |
| HPE-11 | 51 | F | L foot | Lateral | White | No | II | 3 y |
| HPE-12 | 58 | M | L hand | Palmar | White | No | II | 10 mo |
| MFPP-1 | 84 | M | L hand | Unknown | White | No | N/A | N/A |
| MFPP-2 | 86 | M | L foot | Plantar | White | No | I | N/A |
| MFPP-3 | 46 | M | L foot | Lateral | White | No | II | 1 mo |
| MFPP-4 | 62 | M | L foot | Lateral | White | No | N/A | N/A |
| MFPP-5 3 | 72 | M | L hand | Palmar | N/A | No | I | N/A |
| MFPP-6 3 | 72 | M | R hand | Palmar | N/A | No | I | N/A |
| MFPP-7 3 | 72 | M | R ankle | Dorsal | N/A | No | I | N/A |
| MFPP-8 3 | 72 | M | R ankle | Dorsal | N/A | No | I | N/A |
| NL-1 | 35 | F | L foot | Dorsal | White | N/A | N/A | Normal skin |
| NL-2 | 68 | F | L foot | Dorsal | White | No | II | Normal skin |
| NL-3 | 59 | F | R foot | Lateral | N/A | N/A | II | Normal skin |
| NL-4 | 65 | F | L hand | Palmar | White | No | II | Normal skin |
| NL-5 | 54 | M | L foot | Unknown | White | No | II | Normal skin |
| NL-6 | 31 | F | L foot | Plantar | N/A | N/A | N/A | Normal skin |
| NL-7 | 74 | F | R hand | Unknown | White | No | I | Normal skin |
| NL-8 | 63 | M | R hand | Dorsal | N/A | N/A | N/A | Normal skin |
| NL-9 | 69 | F | R foot | Plantar | White | No | II | Normal skin |
| PP-1 | 71 | F | R foot | Unknown | White | No | N/A | N/A |
| PP-2 | 57 | F | R foot | Lateral | White | No | I | 1 wk |
| PP-3 | 63 | M | L hand | Dorsal | White | No | II | 3–4 y |
| PP-4 | 48 | F | R foot | Plantar | White | No | II | N/A |
| PP-5 | 82 | F | R hand | Unknown | White | Yes | N/A | N/A |
| PP-6 | 78 | M | L hand | Dorsal | White | No | II | 5–6 wk |
| PP-7 | 56 | M | L hand | Dorsal | Black or African American | No | V | 2 y |
| PP-8 | 38 | M | L hand | Dorsal | White | No | III | >10 y |
| NAE-1 | 28 | F | L upper back | Nonacral | Asian | No | III | 1.5 y |
| NAE-2 | 60 | M | R neck | Nonacral | White | No | III | N/A |
| NAE-3 | 26 | F | R axilla | Nonacral | White | No | II | 1.5 y |
| NAE-4 | 24 | M | L arm | Nonacral | Black or African American | No | VI | 5 mo |
| NAE-5 | 35 | F | R hip | Nonacral | Black or African American | No | VI | >1 y |
| NAE-6 | 40 | M | L neck | Nonacral | Black or African American | No | VI | 20 y |
| NAE-7 | 51 | M | R calf | Nonacral | White | No | II | N/A |
| NAE-8 | 33 | F | R hip | Nonacral | N/A | N/A | N/A | N/A |
| NAE-9 | 50 | M | L lower back | Nonacral | White | No | I | N/A |
| NAE-10 | 15 | M | R antecubital fossa | Nonacral | Asian | No | III | N/A |
| NAP-1 | 53 | M | L flank | Nonacral | White | No | I | N/A |
| NAP-2 | 40 | M | R retroauricular neck | Nonacral | N/A | Yes | III | 1 y |
| NAP-3 | 60 | F | R mid back | Nonacral | N/A | N/A | N/A | N/A |
| NAP-4 | 29 | M | L elbow | Nonacral | White | N/A | N/A | N/A |
| NAP-5 | 49 | F | L thigh | Nonacral | White | No | II | N/A |
| NAP-6 | 40 | F | L upper back | Nonacral | Black or African American | No | IV | 3 wk |
| NAP-7 | 62 | M | R thigh | Nonacral | White | No | II | N/A |
| NAP-8 | 18 | M | R arm | Nonacral | White | No | N/A | N/A |
| NAP-9 | 48 | M | R arm | Nonacral | White | No | II | 4 y |
| NAP-10 | 25 | F | R lower back | Nonacral | Black or African American | No | V | 5 y |
Abbreviations: HPE, hyperkeratotic palmoplantar eczema; MFPP, mycosis fungoides palmaris et plantaris; NL, normal acral skin; PP, palmoplantar psoriasis; NAE, nonacral eczema; NAP, nonacral psoriasis; F, female; M, male; R, right; L, left; N/A, data not available.
According to the Fitzpatrick phototype scale.
These samples came from the same patient.
These samples came from the same patient.
Figure 1.
IL17A mRNA expression by RISH in acral and nonacral samples. (a–c) IL17A composite expression in the entire tissue specimen (a), epidermis (b), and dermis (c) of normal acral skin (n = 9), HPE (n = 12), nonacral eczema (n = 10), PP (n = 8), nonacral psoriasis (n = 9), and MFPP (n = 8). Composite expression was normalized to tissue area in mm2. (d–f) Representative IL17A RISH (red) in PP (d), HPE (e), and normal acral (f) tissue. Middle lines represent medians; upper and lower ends of the boxes represent IQRs; whiskers represent values within 1.5 × IQR. Each point represents an individual case. Bar = 100 μm. HPE, hyperkeratotic palmoplantar eczema; IQR, interquartile range; MFPP, mycosis fungoides palmaris et plantaris; NAE, nonacral eczema; NAP, nonacral psoriasis; NL, normal acral skin; PP, palmoplantar psoriasis; RISH, RNA in situ hybridization.
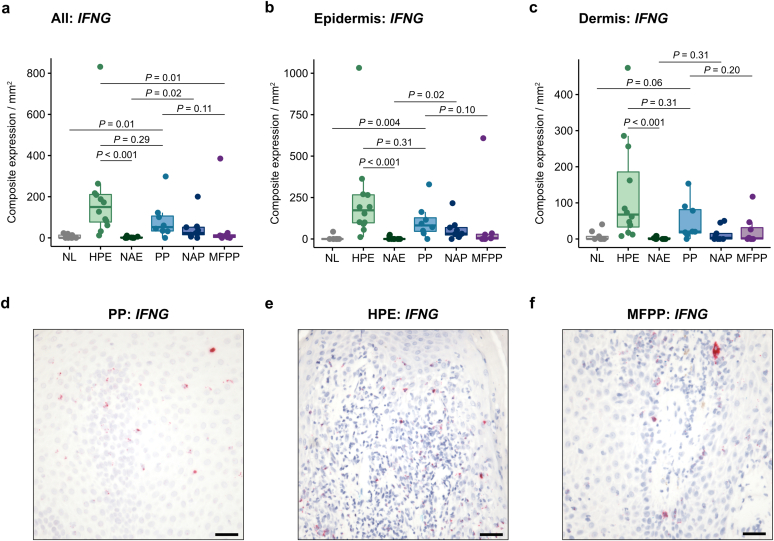
We found that IFNG mRNA expression was not significantly different between PP and HPE (Figure 2a–e). In fact, levels in HPE trended higher (149.9 [76.9–210.9]) than PP (52.4 [33.6–106.1]), although this trend was not significant (P = 0.29). This was unexpected as minimal IFNG expression was present in nonacral eczema (0.8 [0–3.5]; P < 0.001). Earlier studies have reported increased IFNG expression in nonacral eczema in older patients compared to in younger patients (Zhou et al., 2019). It has also been proposed that cytokine profiles in atopic dermatitis may vary across different ethnic backgrounds (Czarnowicki et al., 2019). Therefore, in a sensitivity analysis to account for potential demographic differences between patients with HPE and patients with nonacral eczema in our cohort, we used a multivariable regression model to adjust for age, sex, race, and ethnicity. This analysis showed that IFNG expression was significantly increased in HPE compared with nonacral eczema (regression β = 117.1 [26.0–273.7]; P < 0.001), after adjustment for demographic covariates. IFNG expression was generally low or absent in MFPP (9.4 [2.1–14.8]) except for one case that had very high expression of IFNG (and IL13) in the absence of IL17A (Figure 2f).
Figure 2.
IFNG mRNA expression by RISH in acral and nonacral samples. (a–c) IFNG composite expression in the entire tissue specimen (a), epidermis (b), and dermis (c) of normal acral skin (n = 9), HPE (n = 12), nonacral eczema (n = 10), PP (n = 8), nonacral psoriasis (n = 9), and MFPP (n = 8). Composite expression was normalized to tissue area in mm2. (d–f) Representative IFNG RISH (red) in PP (d), HPE (e), and a high-expressing MFPP (f) tissue. Middle lines represent medians; upper and lower ends of the boxes represent IQRs; whiskers represent values within 1.5 × IQR. Each point represents an individual case. Bar = 100 μm. HPE, hyperkeratotic palmoplantar eczema; IQR, interquartile range; MFPP, mycosis fungoides palmaris et plantaris; NAE, nonacral eczema; NAP, nonacral psoriasis; NL, normal acral skin; PP, palmoplantar psoriasis; RISH, RNA in situ hybridization.
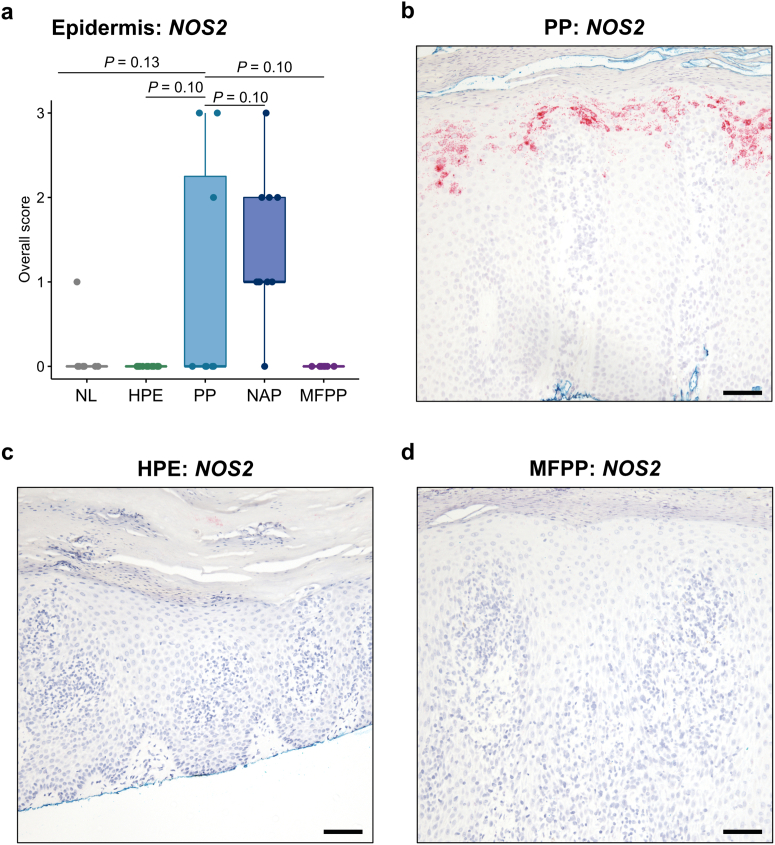
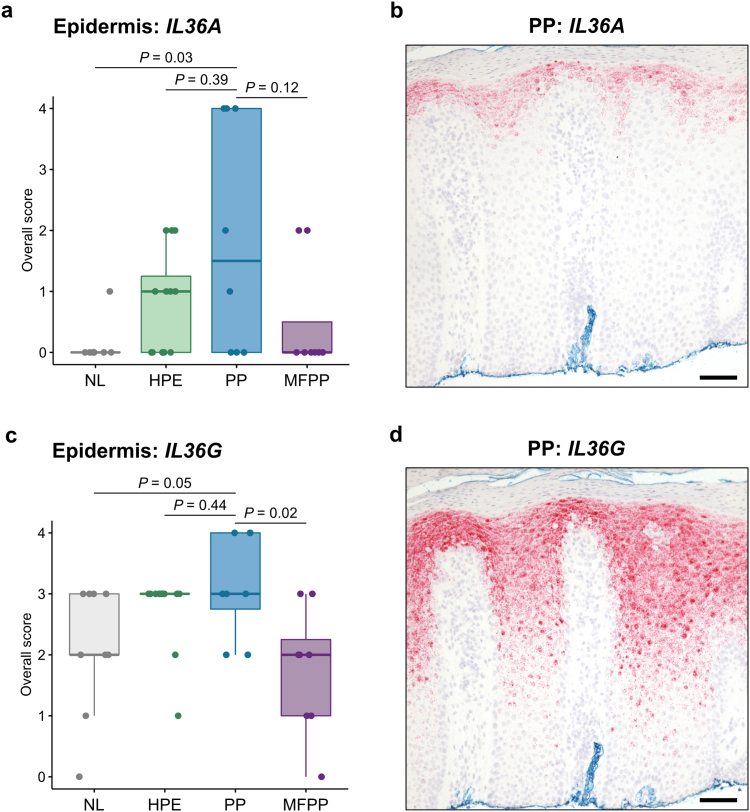
We and others have previously shown that NOS2 mRNA is another useful biomarker in psoriasis, whereas NOS2 expression is absent in eczema (Wang et al., 2021). In addition, immunohistochemistry (IHC) for IL-36α and IL-36γ protein has recently been shown to have utility when examining PP and HPE (Erdem et al., 2022). We therefore interrogated our cohort using these additional RISH markers. While 9 of 10 nonacral psoriasis cases were positive for NOS2, only 3 of 8 PP cases were positive (Figure 3a–d). No staining was observed in HPE or MFPP. For IL36A, there was a slight trend toward increased expression in PP relative to HPE and MFPP, but it was not statistically significant (Figure 4a and b). IL36G staining did not provide additional clarity (Figure 4c and d).
Figure 3.
NOS2 mRNA expression by RISH in acral and nonacral samples. (a) Quantification of NOS2 RISH in normal acral skin (n = 9), HPE (n = 12), PP (n = 8), nonacral psoriasis (n = 10), and MFPP (n = 8) following the scoring system in Table 3. (b–d) Representative NOS2 RISH (red) in PP (b), HPE (c), and MFPP (d) tissues. Middle lines represent medians; upper and lower ends of the boxes represent IQRs; whiskers represent values within 1.5 × IQR. Each point represents an individual case. Bar = 200 μm. HPE, hyperkeratotic palmoplantar eczema; IQR, interquartile range; MFPP, mycosis fungoides palmaris et plantaris; NAP, nonacral psoriasis; NL, normal acral skin; PP, palmoplantar psoriasis; RISH, RNA in situ hybridization.
Figure 4.
IL36A and IL36G mRNA expression by RISH in acral samples. (a) Quantification of IL36A RISH in normal acral skin (n = 9), HPE (n = 12), PP (n = 8), and MFPP (n = 8) following the scoring system in Table 3. (b) Representative IL36A RISH (red) in PP tissue. (c) Quantification of IL36G RISH in normal acral skin (n = 9), HPE (n = 12), PP (n = 8), and MFPP (n = 8) following the scoring system in Table 4. (d) Representative IL36G RISH (red) in PP tissue. Middle lines represent medians; upper and lower ends of the boxes represent IQRs; whiskers represent values within 1.5 × IQR. Each point represents an individual case. Bar = 200 μm. HPE, hyperkeratotic palmoplantar eczema; IQR, interquartile range; MFPP, mycosis fungoides palmaris et plantaris; NL, normal acral skin; PP, palmoplantar psoriasis; RISH, RNA in situ hybridization.
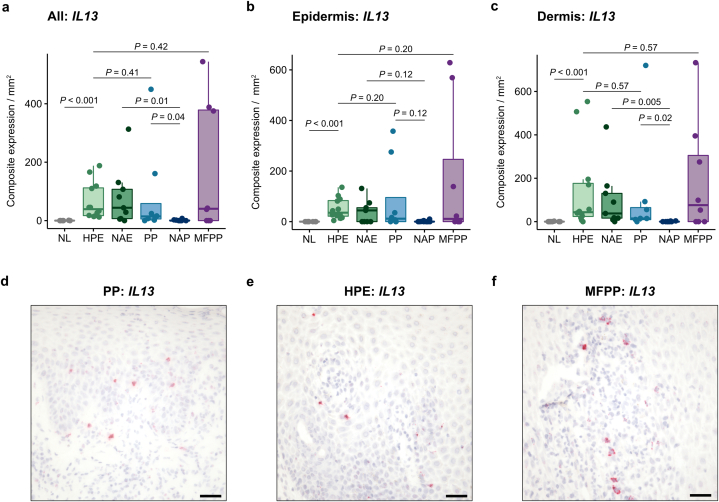
Given the known Th2 bias of nonacral eczema and the lack of Th2 inflammation in nonacral psoriasis (Wang et al., 2021), we evaluated whether IL13 mRNA expression was different when comparing HPE with PP and MFPP. Unexpectedly, PP expressed comparable levels of IL13 (14.4 [5.7–58.4]) to HPE (39.1 [17.1–112.1]; P = 0.41), in contrast to the near complete absence of IL13 expression in nonacral psoriasis (0 [0–1.4]; P = 0.04) (Figure 5a–e). Adjusting for age, sex, race, and ethnicity, we again found that PP demonstrated increased levels of IL13 compared to nonacral psoriasis (regression β = 32.0 [4.6–84.0]; P = 0.004). MFPP exhibited variable IL13 levels (40.6 [0–378.5]), with some cases showing no expression and other cases showing more than 10-fold the median level of IL13 seen in HPE (Figure 5a and f).
Figure 5.
IL13 mRNA expression by RISH in acral and nonacral samples. (a–c) IL13 composite expression in the entire tissue specimen (a), epidermis (b), and dermis (c) of normal acral skin (n = 9), HPE (n = 12), nonacral eczema (n = 9), PP (n = 8), nonacral psoriasis (n = 10), and MFPP (n = 8). Composite expression was normalized to tissue area in mm2. (d–f) Representative IL13 RISH (red) in PP (d), HPE (e), and a high-expressing MFPP (f) tissue. Middle lines represent medians; upper and lower ends of the boxes represent IQRs; whiskers represent values within 1.5 × IQR. Each point represents an individual case. Bar = 100 μm. HPE, hyperkeratotic palmoplantar eczema; IQR, interquartile range; MFPP, mycosis fungoides palmaris et plantaris; NAE, nonacral eczema; NAP, nonacral psoriasis; NL, normal acral skin; PP, palmoplantar psoriasis; RISH, RNA in situ hybridization.
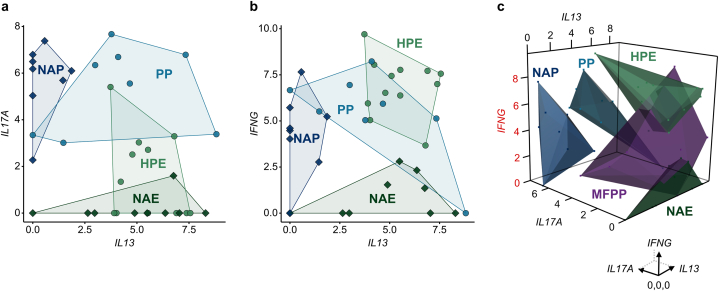
In addition, we analyzed acral and nonacral samples based on their combined expression of IL17A, IFNG, and IL13 mRNA. Nonacral psoriasis and nonacral eczema showed divergent patterns of IL17A/IFNG and IL13 expression, whereas PP showed co-expression of IL17A, IFNG, and IL13 and HPE displayed co-expression of IFNG and IL13 (Figure 6a and b). Combined analysis of all three cytokines was sufficient to distinguish cases of PP (IL17AhiIFNGhiIL13hi), nonacral psoriasis (IL17AhiIFNGhiIL13lo), HPE (IL17AloIFNGhiIL13hi), and nonacral eczema (IL17AloIFNGloIL13hi) (Figure 6c).
Figure 6.
Combined cytokine expression in acral and nonacral samples. (a) Two-dimensional scatter plot of IL17A and IL13 composite expression in the entire tissue specimen of acral and nonacral samples. (b) Two-dimensional scatter plot of IFNG and IL13 composite expression in acral and nonacral samples. (c) Three-dimensional scatter plot of IFNG, IL13, and IL17A composite expression in acral and nonacral samples. HPE (n = 12), NAE (n = 9), PP (n = 8), NAP (n = 8), and MFPP (n = 8). Each point represents an individual case. Circles represent acral samples, and diamonds represent nonacral samples (A and B). Cytokine composite expression was normalized to tissue area in mm2. Axes are log2 (1 + cytokine composite expression/mm2). Polygons indicate the boundaries of cytokine expression for a given group. HPE, hyperkeratotic palmoplantar eczema; MFPP, mycosis fungoides palmaris et plantaris; NAE, nonacral eczema; NAP, nonacral psoriasis; NL, normal acral skin; PP, palmoplantar psoriasis.
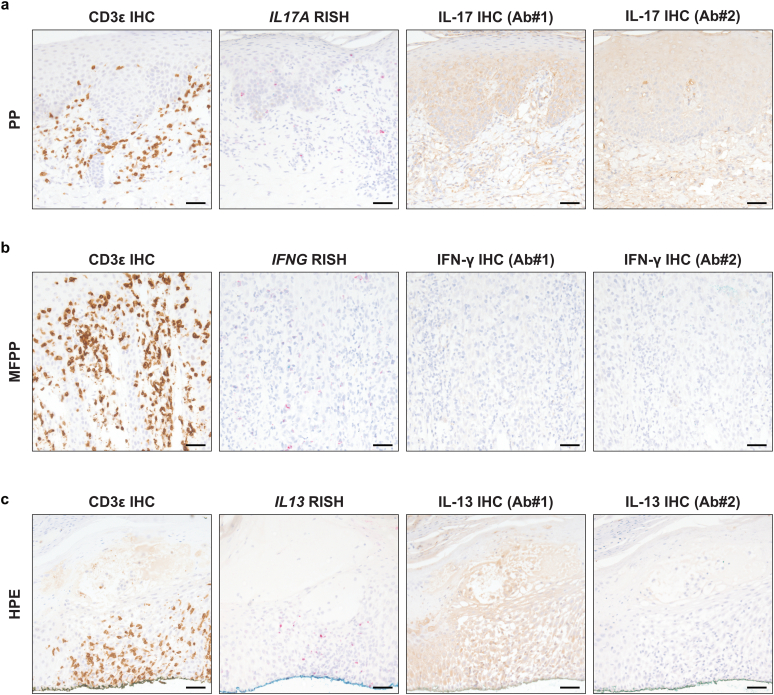
Nonspecific binding is a common problem with commercially available primary antibodies used for IHC, especially for those recognizing secreted signaling proteins (Gautron, 2019). To evaluate this in the context of the current study, we performed IHC to assess whether we could reliably detect cytokine proteins. We performed IHC for IL-17, IFN-γ, IL-13, and CD3ε on three representative samples in which high cytokine mRNA expression had been detected by RISH and three control samples without significant cytokine expression by RISH. Although CD3ε IHC staining, which was used as a technical control, performed well, IHC for IL-17, IFN-γ, and IL-13 showed either poor staining or high background staining and could not be meaningfully quantified (Figure 7a–c).
Figure 7.
Cytokine protein expression by immunohistochemistry in acral samples. (a) CD3ε and IL-17 IHC (brown) and IL17A RISH (red) in serial sections of PP tissue. (b) CD3ε and IFN-γ IHC (brown) and IFNG RISH (red) in serial sections of MFPP tissue. (c) CD3ε and IL-13 IHC (brown) and IL13 RISH (red) in serial sections of HPE tissue. Representative images are shown for each case. Bar = 150 μm. Ab, antibody; HPE, hyperkeratotic palmoplantar eczema; IHC, immunohistochemistry; MFPP, mycosis fungoides palmaris et plantaris; PP, palmoplantar psoriasis; RISH, RNA in situ hybridization.
Discussion
In this study, we evaluated the use of RISH for detecting cytokines in diagnostic biopsies of acral inflammatory dermatoses and cutaneous T-cell lymphoma. We found that IL17A mRNA was the most useful biomarker for distinguishing PP from HPE and MFPP. Unexpectedly, we found that IFNG and IL13 mRNA were expressed at similar levels in PP and HPE, contrasting with their generally divergent expression in nonacral psoriasis and eczema, respectively (Wang et al., 2021). NOS2 mRNA, a robust psoriasis biomarker, was positive in only a minority of PP, suggesting it may have less use on acral sites than on nonacral skin. IHC for cytokine proteins commonly showed nonspecific staining, limiting its use and underscoring the advantage of using RISH to assess cytokine profiles in diagnostic skin biopsies.
Our findings revealed a distinct immunologic profile of psoriasis and eczema/atopic dermatitis at acral sites. In particular, HPE showed a combined Th2 (IL13) and Th1 (IFNG) pattern, whereas nonacral eczema shows a purer Th2 polarization. In PP, there was combined activation of Th17 (IL17A), Th1 (IFNG), and unexpectedly Th2 (IL13) immunity. These immunologic differences between acral and nonacral sites persisted after adjusting for demographic variables, including age, sex, race, and ethnicity. In addition, although an earlier study showed that chronic lesions (>72 hours duration) of nonacral eczema express IFNG in contrast to acute lesions (Gittler et al., 2012), patients with nonacral eczema and those with HPE within our cohort had at least a 3-month history of the disease. Similarly for nonacral psoriasis, one study suggested that differing lesion chronicity may account for the heterogeneous cytokine profiles observed, including an IL-13-strong subset (Swindell et al., 2012). For our study, most biopsies from patients with nonacral psoriasis or PP were taken after at least a month of disease. Therefore, there is not a clear difference in the chronicity of acral and nonacral samples within our cohort that would explain the distinct inflammatory signatures we observed for acral samples. Furthermore, one study reported Th2 activation within a subset of Chinese patients with psoriasis (Chen et al., 2021); however, our cohort of patients with nonacral psoriasis or PP included only patients who identified as White, Black or African American, and Hispanic or Latina/Latino/Latinx.
Our findings raise the question of how psoriasis and eczema on acral skin acquire distinct immunologic profiles compared with these diseases on nonacral skin. Recent work has shown differences in the epidermal lipid profile across anatomically distinct body sites (Merleev et al., 2022). Compared with other sites, palmoplantar skin shows a distinct epidermal lipid profile, including an increased abundance of ceramides comprising an 18-carbon sphingoid base (Merleev et al., 2022). As the lipid composition of keratinocytes may affect their expression of inflammatory cytokines (Merleev et al., 2022), this provides a potential mechanism by which the inflammatory pathways involved in dermatoses such as psoriasis and eczema may differ depending on their anatomical location. Interestingly, a recent report showed that in pustular PP, another acral inflammatory dermatosis in the psoriasis spectrum, there is plasticity in the immune response and overlapping Th2/Th17 polarization may be seen (McCluskey et al., 2022). The simultaneous activation of multiple inflammatory pathways in these conditions on acral sites may explain clinical observations such as the incomplete response to biologics targeting only one cytokine pathway and the often-overlapping morphologic features observed in PP and HPE.
Finally, we note that this RISH-based approach could not specifically identify cases of MFPP, although unusually high cytokine (IFNG or IL13) expression could be a clue to consider this diagnosis in some cases. A Th2-predominant response is well known in nonacral mycosis fungoides (Liu et al., 2022; Rindler et al., 2021). A high index of suspicion should be maintained for MFPP, and molecular analysis of T-cell receptors should be pursued if cutaneous lymphoma is suspected. This study was limited by its relatively small sample size and retrospective design.
Together, these findings demonstrate the utility of cytokine biomarkers for the diagnosis of acral inflammatory dermatoses, highlighting the use of IL17A mRNA expression by RISH to distinguish PP. Our work further suggests a distinct immunology of inflammatory dermatoses on acral sites compared to nonacral sites, with important implications for the selection of cytokine-directed therapies to treat HPE and PP.
Materials and Methods
For this retrospective case series, cases were initially screened by searching the Yale Dermatopathology (New Haven, CT) database for a diagnosis of eczema, psoriasis, HPE, PP, or MFPP. Cases with classic clinical and histopathologic features were selected and were determined by expert consensus among two board-certified dermatologists and dermatopathologists (Erdem et al., 2022; Kim et al., 2022; Park et al., 2017; Wang et al., 2021). A total of 57 cases were selected, comprising HPE (n = 12), PP (n = 8), MFPP (n = 8), nonacral eczema (n = 10), nonacral psoriasis (n = 10), and clinically normal acral skin (n = 9) (Table 1). Clinically normal acral skin was obtained from excision specimens. The Yale University Institutional Review Board reviewed and approved this study. Informed consent was not required per our institutional review board protocol as the study presents no more than minimal risk and identifying material was not collected or included in the study.
RISH was performed on formalin-fixed paraffin-embedded skin biopsies using the chromogenic RNAscope 2.5 HD Reagent Kit-RED (Bio-Techne, catalog [Cat] number #322350; Minneapolis, MN), as previously described (Wang et al., 2021). Probes for IFNG (Cat number #310501), IL13 (Cat number #586241), IL17A (Cat number #310931), IL36A (Cat number #424771), IL36G (Cat number #424791), and NOS2 (Cat number #424991) were purchased from Bio-Techne. RISH scoring was performed by one dermatopathologist blinded to the diagnosis. For IFNG, IL13, and IL17A, positive cells were scored as 1+, 2+, 3+, or 4+ based on the number of dots per cell (Table 2); this is a modification of the manufacturer’s recommended scoring system based on our experience with this approach. 1+ cells were not included in the analysis and likely reflect background staining (Singh et al., 2023). Cells in the epidermis and dermis were quantified separately. Slides were scanned using a NanoZoomer S210 (Hamamatsu, Shizuoka, Japan), and tissue areas of the epidermis and dermis were quantified using HALO software version 3.3.2541.184 (Indica Labs, Albuquerque, New Mexico). The composite cytokine expression for each specimen was calculated using the following formula: ([number of 2+ cells] × 4 + [number of 3+ cells] × 9 + [number of 4+ cells] × 16) / (tissue area in mm2) (Singh et al., 2023). This formula gives more weight to stronger staining cells, which in our experience seem to have more biologic significance (Singh et al., 2023). For IL36A, IL36G, and NOS2, which are primarily expressed by keratinocytes (Erdem et al., 2022; Wang et al., 2021), RISH quantification was performed following the scoring systems in Tables 3 and 4.
Table 2.
RISH Scoring System for IFNG, IL13, and IL17A
| RISH Score1 | Scoring Criteria |
|---|---|
| 0+ | No staining |
| 1+ | 1–2 dots per cell2 |
| 2+ | Between 3 and 10 dots per cell, small dot clusters may be present |
| 3+ | >10 dots per cell, medium and/or large dot clusters may be present, cell is not obscured by positive staining |
| 4+ | Large dot clusters predominate, cell is nearly completely or completely obscured by staining |
Abbreviation: RISH, RNA in situ hybridization.
This scoring system was developed based on manufacturer’s recommendations, with some modifications (Singh et al., 2023).
This level of staining is considered background staining and therefore was not included in the quantification.
Table 3.
RISH Scoring System for NOS2 and IL36A
| Score | Scoring Criteria |
|---|---|
| 0 | <1% of epidermis, fuzzy1 |
| 1 | Focal positive, <10% of epidermis |
| 2 | 20–50% of epidermis |
| 3 | >50% of epidermis |
| 4 | Diffuse (∼100% of epidermis) |
Abbreviation: RISH, RNA in situ hybridization.
True staining is cytoplasmic and crisp.
Table 4.
RISH Scoring System for IL36G
| Score | Scoring Criteria |
|---|---|
| 0 | <2% of length of epidermis |
| 1 | <50% of length of epidermis |
| 2 | 50% of length of epidermis to diffuse; if diffuse, there are areas where staining is lost or nearly lost |
| 3 | Diffuse (∼100% of length of epidermis); moderate intensity with areas where staining is limited to 1–2 cell layer thickness in upper epidermis |
| 4 | Diffuse (∼100% of length of epidermis); staining is bright and thick throughout epidermis |
Abbreviation: RISH, RNA in situ hybridization.
IHC was performed on formalin-fixed paraffin-embedded skin biopsies as previously described (Wang et al., 2021). Primary antibodies for IL-17 (Ab number #1: Abcam, Cat number #ab79056, Cambridge, UK) (Ab number #2: R&D Systems, Cat number #AF-317-NA; Minneapolis, MN), IFN-γ (Ab number #1: Abcam, Cat number #ab25101) (Ab number #2: R&D Systems, Cat number #MAB2853), IL-13 (Ab number #1: LSBio, Cat number #LS-B7417; Seattle, WA) (Ab number #2: Thermo Fisher Scientific, clone JES10-5A2; Waltham, MA), and CD3ε (Cell Signaling Technology, clone D7A6E; Danvers, MA) were incubated followed by species-specific secondary antibody staining. Detection was performed using the ImmPRESS horseradish peroxidase reagent (Vector Laboratories; Burlingame, CA) and diaminobenzidine substrate (Vector Laboratories).
Statistical analyses were performed with R version 4.0.2 (R Foundation for Statistical Computing). Data were analyzed using the Kruskal-Wallis test followed by Dunn’s post-test with a Benjamini-Hochberg adjustment for multiple comparisons. Statistical significance was defined as adjusted P < 0.05. For analyses adjusted for demographic covariates, we constructed a multivariable linear regression model with age, sex, and race/ethnicity as independent variables and composite cytokine RISH expression (square root-transformed) as the dependent variable.
Data availability statement
All data are available in the main text or the Supplementary Materials.
ORCIDs
Jennifer S. Chen: http://orcid.org/0000-0002-7808-2670
Michael J. Murphy: http://orcid.org/0000-0001-6494-7250
Katelyn Singh: http://orcid.org/0000-0002-0120-4238
Alice Wang: http://orcid.org/0000-0003-1771-2103
Ryan D. Chow: http://orcid.org/0000-0002-1872-6307
Sa Rang Kim: http://orcid.org/0000-0002-0345-7028
Jeffrey M. Cohen: http://orcid.org/0000-0002-7709-0548
Christine J. Ko: http://orcid.org/0000-0003-2270-2524
William Damsky: http://orcid.org/0000-0003-0975-4071
Conflict of Interest
WD reported consulting fees from Eli Lilly, Pfizer, and TWi Biotech; grants from Pfizer and Advanced Cell Diagnostics/Bio-techne; and licensing fees from EMD/Sigma/Millipore, all outside of the submitted work. No other disclosures were reported.
Acknowledgments
We thank the Yale Dermatopathology Laboratory, especially Robert Criscuolo (Yale School of Medicine), William Sudhoff (Yale School of Medicine), and Dilgash Mekael (Yale School of Medicine). This work was supported by the Colton Center for Autoimmunity at Yale. Advanced Cell Diagnostics/Bio-Techne provided some of the reagents for this work to Yale (WD) through a collaborative research agreement. Yale (WD) has filed a patent application related to this approach. JSC and RDC were supported by National Institutes of Health grants T32GM136651, F30HL149151 (JSC), and F30CA250249 (RDC). WD was supported by a Career Development Award from the Dermatology Foundation.
Author Contributions
Conceptualization: JSC, WD; Data Curation: JSC, WD; Formal Analysis: JSC, RDC, WD; Funding Acquisition: WD; Investigation: JSC, MJM, KS, AW, WD; Methodology: JSC, MJM, KS, AW, SRK, CJK, WD; Resources: SRK, JMC, CJK; Software: JSC, MJM, AW, RDC; Supervision: WD; Visualization: JSC, RDC, WD; Writing - Original Draft Preparation: JSC, WD; Writing - Review and Editing: all authors.
Disclaimer
The funders had no role in the design and conduct of the study; the collection, management, analysis, and interpretation of the data; the preparation, review, or approval of the manuscript; and the decision to submit the manuscript for publication.
accepted manuscript published online XXX; corrected proof published online XXX
Footnotes
Cite this article as: JID Innovations 2023;X:100189
Supplementary material is linked to the online version of the paper at www.jidonline.org, and at https://doi.org/10.1016/j.xjidi.2023.100189.
Supplementary Materials
References
- Chen J., Li C., Li H., Yu H., Zhang X., Yan M., et al. Identification of a TH2-high psoriasis cluster based on skin biomarker analysis in a Chinese psoriasis population. J Eur Acad Dermatol Venereol. 2021;35:150–158. doi: 10.1111/jdv.16563. [DOI] [PubMed] [Google Scholar]
- Czarnowicki T., He H., Krueger J.G., Guttman-Yassky E. Atopic dermatitis endotypes and implications for targeted therapeutics. J Allergy Clin Immunol. 2019;143:1–11. doi: 10.1016/j.jaci.2018.10.032. [DOI] [PubMed] [Google Scholar]
- Erdem O., Leblebici C., Koku Aksu A.E., Erdil D., Kara Polat A., Gürel M.S. IL-36α and IL-36γ expressions in the differential diagnosis of palmoplantar psoriasis and palmoplantar eczema: a retrospective histopathologic and immunohistochemical study. J Cutan Pathol. 2022;49:42–48. doi: 10.1111/cup.14105. [DOI] [PubMed] [Google Scholar]
- Espinosa M.L., Nguyen M.T., Aguirre A.S., Martinez-Escala M.E., Kim J., Walker C.J., et al. Progression of cutaneous T-cell lymphoma after dupilumab: case review of 7 patients. J Am Acad Dermatol. 2020;83:197–199. doi: 10.1016/j.jaad.2020.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautron L. On the necessity of validating antibodies in the immunohistochemistry literature. Front Neuroanat. 2019;13:46. doi: 10.3389/fnana.2019.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittler J.K., Shemer A., Suárez-Fariñas M., Fuentes-Duculan J., Gulewicz K.J., Wang C.Q.F., et al. Progressive activation of T(H)2/T(H)22 cytokines and selective epidermal proteins characterizes acute and chronic atopic dermatitis. J Allergy Clin Immunol. 2012;130:1344–1354. doi: 10.1016/j.jaci.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb A., Sullivan J., van Doorn M., Kubanov A., You R., Parneix A., et al. Secukinumab shows significant efficacy in palmoplantar psoriasis: results from GESTURE, a randomized controlled trial. J Am Acad Dermatol. 2017;76:70–80. doi: 10.1016/j.jaad.2016.07.058. [DOI] [PubMed] [Google Scholar]
- Kim S.R., McNiff J.M., Ko C.J. Clustered intraepidermal lymphocytes and Langerhans cell microgranulomas are consistently observed in hyperkeratotic palmoplantar eczema compared with palmoplantar psoriasis and mycosis fungoides palmaris et plantaris. J Am Acad Dermatol. 2022;87:884–886. doi: 10.1016/j.jaad.2021.11.057. [DOI] [PubMed] [Google Scholar]
- Langley R.G., Elewski B.E., Lebwohl M., Reich K., Griffiths C.E.M., Papp K., et al. Secukinumab in plaque psoriasis--results of two phase 3 trials. N Engl J Med. 2014;371:326–338. doi: 10.1056/NEJMoa1314258. [DOI] [PubMed] [Google Scholar]
- Liu X., Jin S., Hu S., Li R., Pan H., Liu Y., et al. Single-cell transcriptomics links malignant T cells to the tumor immune landscape in cutaneous T cell lymphoma. Nat Commun. 2022;13:1158. doi: 10.1038/s41467-022-28799-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCluskey D., Benzian-Olsson N., Mahil S.K., Hassi N.K., Wohnhaas C.T., APRICOT and PLUM study team, et al. Single-cell analysis implicates TH17-to-TH2 cell plasticity in the pathogenesis of palmoplantar pustulosis. J Allergy Clin Immunol. 2022;150:882–893. doi: 10.1016/j.jaci.2022.04.027. [DOI] [PubMed] [Google Scholar]
- Merleev A.A., Le S.T., Alexanian C., Toussi A., Xie Y., Marusina A.I., et al. Biogeographic and disease-specific alterations in epidermal lipid composition and single-cell analysis of acral keratinocytes. JCI Insight. 2022;7 doi: 10.1172/jci.insight.159762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterhaven J.A.F., Voorberg A.N., Romeijn G.L.E., de Bruin-Weller M.S., Schuttelaar M.L.A. Effect of dupilumab on hand eczema in patients with atopic dermatitis: an observational study. J Dermatol. 2019;46:680–685. doi: 10.1111/1346-8138.14982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.Y., Cho E.B., Park E.J., Park H.R., Kim K.H., Kim K.J. The histopathological differentiation between palmar psoriasis and hand eczema: a retrospective review of 96 cases. J Am Acad Dermatol. 2017;77:130–135. doi: 10.1016/j.jaad.2017.01.005. [DOI] [PubMed] [Google Scholar]
- Reolid A., Armesto S., Sahuquillo-Torralba A., Torres T., Feltes R., Vilarrasa E., et al. Secukinumab is effective and safe in the treatment of recalcitrant palmoplantar psoriasis and palmoplantar pustular psoriasis in a daily practice setting. J Am Acad Dermatol. 2022;87:705–709. doi: 10.1016/j.jaad.2022.05.047. [DOI] [PubMed] [Google Scholar]
- Rindler K., Bauer W.M., Jonak C., Wielscher M., Shaw L.E., Rojahn T.B., et al. Single-cell RNA sequencing reveals tissue compartment-specific plasticity of mycosis fungoides tumor cells. Front Immunol. 2021;12:666935. doi: 10.3389/fimmu.2021.666935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K., Valido K., Swallow M., Wang A., Cohen J., Damsky W. Skin cytokine profiles determined by RNA in situ hybridization correlate with response to dupilumab in patients with eczematous dermatitis. J Am Acad Dermatol. 2023 doi: 10.1016/j.jaad.2022.12.052. [e-pub ahead of print] [DOI] [PubMed] [Google Scholar]
- Swindell W.R., Xing X., Stuart P.E., Chen C.S., Aphale A., Nair R.P., et al. Heterogeneity of inflammatory and cytokine networks in chronic plaque psoriasis. PLoS One. 2012;7 doi: 10.1371/journal.pone.0034594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A., Fogel A.L., Murphy M.J., Panse G., McGeary M.K., McNiff J.M., et al. Cytokine RNA in situ hybridization permits individualized molecular phenotyping in biopsies of psoriasis and atopic dermatitis. JID Innov. 2021;1:100021. doi: 10.1016/j.xjidi.2021.100021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Leonard A., Pavel A.B., Malik K., Raja A., Glickman J., et al. Age-specific changes in the molecular phenotype of patients with moderate-to-severe atopic dermatitis. J Allergy Clin Immunol. 2019;144:144–156. doi: 10.1016/j.jaci.2019.01.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the Supplementary Materials.