Abstract
Background
Long noncoding RNAs (lncRNAs) and microRNAs (miRNAs) have been reported to play regulatory roles in ulcerative colitis (UC). In this study, we aimed to determine the specific roles and action mechanism of the nuclear paraspeckle assembly transcript 1 (NEAT1) in UC.
Methods
Reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR) was used to determine the lncRNA NEAT1 and miR‐493‐5p expression levels in patients with UC and healthy volunteers. We determine the forecast linkage points of NEAT1 and miR‐493‐5p using Starbase and those of miR‐493‐5p and Rab27A using TargetScan, and further verified them using a double luciferase gene reporter kit. RT‐qPCR and Western blot analysis were used to determine the lncRNA NEAT1, miR‐493‐5p, and Rab27A expression levels in lipopolysaccharide (LPS)‐induced Caco‐2 cells. Flow cytometry and cell counting kit‐8 were used to assess Caco‐2 cell viability. Tumor necrosis factor‐α, interleukin (IL)‐6, IL‐8, and IL‐1β levels were determined via an enzyme‐linked immunosorbent assay.
Results
Expression levels of NEAT1 were upregulated and those of miR‐493‐5p were downregualted in 10 ng/mL LPS‐treated Caco‐2 cells and patients with UC. Dual‐luciferase gene reporter assay revealed that miR‐493‐5p is linked to NEAT1, and Rab27A is a downstream target of miR‐493‐5p. Overexpression of miR‐493‐5p inhibited the apoptosis and inflammation in LPS‐treated Caco‐2 cells. Moreover, downregulation of lncRNA NEAT1 expression also inhibited the apoptosis and inflammation in LPS‐treated Caco‐2 cells, which was reversed by Rab27A plasmid cotransfection.
Conclusion
Our results revealed that NEAT1 participates in UC progression by inhibiting miR‐493‐5p expression.
Keywords: long noncoding RNA, miRNA‐493‐5p, NEAT1, Rab27A, ulcerative colitis
The present study aimed to explore the potential mechanism of long noncoding RNAs (LncRNA) nuclear paraspeckle assembly transcript 1 (NEAT1) in ulcerative colitis (UC). Results revealed that LncRNA NEAT1 is involved in UC procession by dampening miR‐493‐5p and may thus be a novel target for treating patients with UC.
1. INTRODUCTION
As a chronic inflammatory bowel disease (IBD), ulcerative colitis (UC) generally occurs at a young age and lasts for the lifetime of the individual. 1 UC is characterized by abdominal pain and diarrhea with mucus and blood. Despite extensive research, the etiology and pathogenesis of UC remain unclear. 2 , 3 Some scholars suggest that UC is caused by immune dysfunction and environmental factors affecting the genetically susceptible populations, while others believe that it is caused by nitrification stress. 4 Owing to its complex pathology and varying characteristics, treatment of UC is serious public health problem.
Long noncoding RNAs (lncRNAs) are epigenetic regulatory genes involved in genomic regulation. LncRNAs are a type of RNA molecule over 200 nt in length that regulate tumor processes, such as epithelial–mesenchymal transition, cell proliferation, and apoptosis. 5 , 6 , 7 , 8 Some lncRNAs related to the pathogenesis of UC have been reported. For instance, lncRNA taurine upregulated 1 has been reported to regulate UC via the microRNA (miR)‐142‐5p/suppressor of cytokine signaling 1 axis. 9 LncRNA nuclear paraspeckle assembly transcript 1 (NEAT1) has been implicated in various human diseases, such as diabetes, 10 acute pancreatitis, 11 and cancer. 12 However, the specific role of NEAT1 in UC remains unknown.
miRNAs are conserved noncoding small RNAs that are 18−20 bp long. 13 miRNAs have been reported to participate in many diseases. 14 , 15 Recently, some studies have reported the critical roles of miRNAs, such as miR‐182‐5p 16 and miR‐26b, 17 in the development of UC. However, the role of miR‐493‐5p and its underlying mechanism in UC remain unknown.
Rab27A is an oncogene that belongs to the RAB protein family. 18 Rab27A has been reported to be involved in several human diseases, such as hepatocellular carcinoma. 19 It has also been repored to be associated with inflammatory pain. 20 However, the specific function of Rab27A in UC remains unknown.
Elucidation of the pathogenesis mechanism is necessary for the development of novel treatment strategies for UC. Therefore, in this study, we aimed to determine whether NEAT1 regulates UC progression via miR‐493‐5p. Our findings reveal a novel biomarker for UC.
2. MATERIALS AND METHODS
2.1. Clinical sample collection
A total of 20 colonic mucosa samples from patients with UC and healthy individuals were used in this study. The colonic mucosa samples were obtained under the endoscope. Inclusion criteria were as follows: (i) None of the patients received anti‐UC therapy before surgery and (ii) final diagnosis was via pathological determination. Patients who underwent treatment were excluded from the study. This study was approved by the Ethics Committee of the Third Affiliated Hospital of Qiqihar Medical University. Written informed consent was obtained from all patients before their inclusion in this study. All tissues were stored at –80°C before use.
2.2. Cell culture
Human colorectal adenocarcinoma cells (Caco‐2) and 293T cells were obtained from the American Type Culture Collection for the dual‐luciferase assay. Cells were cultured on Ham's F‐12 K medium (Gibco) supplemented with 1% penicillin and streptomycin (Thermo) and 20% fetal bovine serum (Gibco) in an atmosphere of 5% CO2 at 37°C.
Caco‐2 cells were treated with lipopolysaccharide (LPS; L8880; Solarbio) to establish the UC model in vitro. Briefly, Caco‐2 cells were incubated with 10 ng/mL LPS for 24 h and harvested for subsequent experiments.
2.3. Cell transfection
To control the expression of miR‐493‐5p, miR‐493‐5p and NC inhibitors (miR‐493‐5p inhibitor: 5′‐CACAUUCGGUGAAGGUCUA‐3′; NC inhibitor: 5′‐CAGUACUUUUGUGUAGUACAA‐3′), miR‐493‐5p mimic (5′‐UUGUACAUGGUAGGCUUUCAUU‐3′), and NC mimic (5′‐UACUGAGAGACAUAAGUUGGUC‐3′) were purchased from Genscript. Small interfering RNA (siRNA) for NEAT1 (si‐NEAT1: 5′‐GGTGTGTGTTGTGGAATCTGT‐3′) and nonspecific control (si‐NC: 5′‐CACGATAAGACAATGTATTT‐3′; Thermo Fisher; Fermantas) were used to knockdown NEAT1. Rab27A CDS sequence was inserted into the pcDNA3.1 vector. All sequences were transfected into cells grown to 60% confluency with JETprime (Polyplus). After 48 h of culture at 37°C and 5% CO2, cells were collected fro subsequent analysis.
2.4. Reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR)
Total RNA was extracted from cells by using TRIzol (Aladdin) and reverse‐transcribed into cDNA using the Titan One Tube RT‐PCR Kit (Merck), according to the manufacturer's instructions. Expression levels of miRNAs were determined using the TransScript II Multiplex Probe One‐Step qRT‐PCR SuperMix UDG (Transgene), and RT‐qPCR was performed using the PerfectStart Green qPCR SuperMix (Transgene). Relative expression levels were calculated using the method. Primers used in the PCR assay were as follows: U6, 5′‐CGCCCTCTTCAGCAGTTATACTA‐3′ (F) and 5′‐CTTCACGCCTTTGCGGCTCAT‐3′ (R); β‐actin, 5′‐CCATCGCCAGTTGCCGATCC‐3′ (F) and 5′‐GCGAGAGGAGCACAGATACCACCAA‐3′ (R); NEAT1, 5′‐AGGCAGGGAGAGGTAGAAGG‐3′ (F) and 5′‐TGGCATGGACAAGTTGAAGA‐3′ (R); and miR‐493‐5p, 5′‐TCCTACGGAGAGGCTCAG‐3′ (F) and 5′‐TCCTCGTAGTCCAACACG‐3′ (R).
2.5. Cell counting kit (CCK)‐8 assay
Cell proliferation was assessed using the CCK‐8 assay (Solarbio). After transfection and LPS stimulation, Caco‐2 cells were resuspended and seeded into a 96‐well plate (5 × 103 cells/well) and incubated with 10 μL of CCK‐8 reagent for 2–3 h at 37°C and 5% CO2 in the dark. Optical density values were measured at 490 nm using an ultraviolet spectrophotometer (Bio‐Rad).
2.6. Cell apoptosis assay
LPS‐treated Caco‐2 cells (2 × 105) were collected and cultured with 500 μL of buffering agent containing 5 μL annexin V‐Alexa Fluor488 and 5 μL propidium iodide (Fcmacs) at room temperature in the dark for 30 min. Cell apoptosis was analyzed via flow cytometry (Beckman Coulter).
2.7. Relative dual‐luciferase reporter gene activity
Wild‐type (WT) or mutated NEAT1 3′‐UTR sequence and Rab27A were inserted into the pGL3‐U6 vector (Tongyong) for the luciferase activity assay. Subsequently, 293T cells were cotransfected with WT or mutant pGL3‐NEAT1 (or Rab27A)‐3′‐UTR with NC and miR‐493‐5p mimics using JETprime (Polyplus), according to the manufacturer's instructions. Relative luciferase activity was measured using a reporter system 24 h after infection (Promega). The results were normalized to those of Renilla luciferase.
2.8. RNA binding protein immunoprecipitation (RIP) assay
RIP assay was performed using the Magna RIP™ RNA binding protein immunoprecipitation kit (Millipore), according to the manufacturer's protocol. Caco‐2 cells were lysed with RIPA lysis bufer (Beyotime) and then subjected to immunoprecipitation with anti‐Ago2 antibody (Millipore). Normal IgG antibody (Millipore) served as the control. At last, the retrieved RNA was examined by RT‐qPCR. Experiments were carried out in triplicate.
2.9. Western blot analysis (WB) assay
Total protein was isolated from Caco‐2 cells using the radioimmunoprecipitation assay buffer containing protease inhibitors and measured using a BCA kit (Beyotime). Equal concentrations of proteins were separated via sodium dodecyl sulfate‐polyacrylamide gel electrophoresis, transferred onto NC membranes (Whatman), blocked with 5% bovine serum albumin, incubated with primary antibodies at 4°C overnight, washed with PBST, and incubated again with secondary antibodies at 4°C for 4 h. Protein bands were visualized using enhanced chemiluminescence (Fcmacs). Anti‐Rab27A (ab55667; Abcam) and anti‐GAPDH (ab9485; Abcam) primary antibodies were used for WB. GAPDH was used as an internal reference.
2.10. Enzyme‐linked immunosorbent assay (ELISA)
ELISA was used to determine the expression levels of the inflammatory cytokines, tumor necrosis factor (TNF)‐α, interleukin (IL)‐1β, IL‐6, and IL‐8. ELISA kits were obtained from R&D Systems. All operations were performed according to the supplier's instructions.
2.11. Statistical analysis
Data are represented as the average ± standard deviation from triplicate experiments. Student's t‐test was used to compare two groups, and Tukey's multiple‐comparison test was used to compare multiple groups. Statistical significance was set at p < .05.
3. RESULTS
3.1. NEAT1 is highly expressed in UC
We first performed RT‐qPCR to determine the levels of NEAT1 in UC. NEAT1 is highly expressed in UC samples than in the healthy tissue samples (Figure 1A). RT‐qPCR results revealed that NEAT1 expression levels were significantly increased in the UC model established using 10 ng/mL LPS‐treated Caco‐2 cells (Figure 1B).
Figure 1.
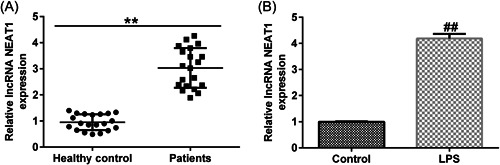
Long noncoding RNA (lncRNA) nuclear paraspeckle assembly transcript 1 (NEAT1) is overexpressed in ulcerative colitis (UC). (A) LncRNA NEAT1 levels in colonic mucosa samples of patients with UC normalized to those in the controls. (B) LncRNA NEAT1 levels in lipopolysaccharide (LPS)‐treated Caco‐2 and control cells. Data are represented as the average ± standard deviation of triplicate experiments.
3.2. miR‐493‐5p binds to NEAT1
We used bioinformatics prediction tools (StarBase) to detemrine the potential binding points of NEAT1 and miR‐493‐5p and found that miR‐493‐5p contained NEAT1 binding sites (Figure 2A). To confirm the link between NEAT1 and miR‐493‐5p, a dual‐luciferase gene assay was performed in 293T cells. As shown in Figure 2B, the relative luciferase levels of the NEAT1 3ʹ‐UTR were notably reduced when the cells were cocultured with a mimic of miR‐493‐5p. When potential connection points were mutated, the miR‐493‐5p mimic had no effect on NEAT expression. These results confirmed that miR‐493‐5p is sponged by NEAT1.
Figure 2.
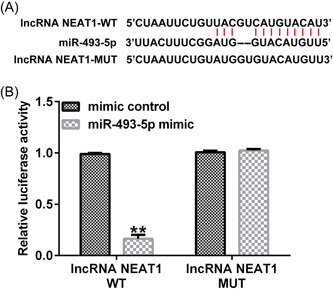
LncRNA NEAT1 targets microRNA (miR)‐493‐5p. (A) Conjunction points of miR‐493‐5p in NEAT1 wild‐type (WT) and mutant (Mut). (B) Luciferase reporter assay of miR‐493‐5p cotransfection with NEAT1 WT and Mut. Data are represented as the average ± SD of triplicate experiments.
3.3. miR‐493‐5p expression is downregulated in UC
We further determined miR‐493‐5p levels in UC. RT‐qPCR results revealed that miR‐493‐5p expression was significantly downregulated in UC (Figure 3A,B).
Figure 3.
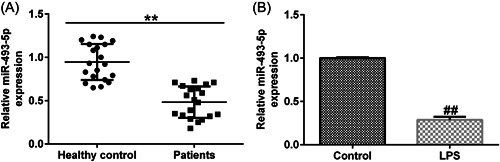
miR‐493‐5p expression is altered in UC. (A) miR‐493‐5p expression levels in colonic mucosa samples of patients with UC were normalized to those in the controls. (B) miR‐493‐5p levels in LPS‐treated Caco‐2 and control cells. Data are represented as the average ± SD of triplicate experiments.
3.4. Inhibition of NEAT1 affects miR‐493‐5p expression in Caco‐2 cells
RT‐qPCR was performed to determine the relationship between miR‐493‐5p and NEAT1 by measuring their expression levels. Compared with the NC siRNA group, NEAT1 was expressed at a lower level by lncRNA NEAT1‐siRNA transfected into Caco‐2 cells (Figure 4A). miR‐493‐5p expression was downregulated in the miR‐493‐5p inhibitor group compared to that in the inhibitor NC control group (Figure 4B). RT‐qPCR results revealed that miR‐493‐5p expression levels were increased in Caco‐2 cells transfected with si‐NEAT1, and cotransfection with the miR‐493‐5p inhibitor reversed this effect (Figure 4C).
Figure 4.
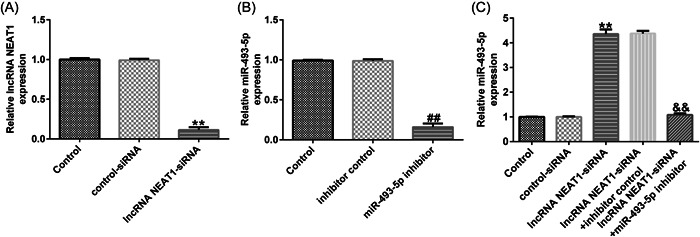
miR‐493‐5p expression is negatively regulated by NEAT1. (A) Efficiency of NEAT1‐small interfering RNA (siRNA) transfection. (B) Efficiency of miR‐493‐5p inhibitor transfection. (C) Levels of miR‐493‐5p with NEAT1‐siRNA and miR‐493‐5p inhibitor cotransfection via reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR). Data are represented as the average ± SD of triplicate experiments.
3.5. Inhibition of NEAT1 suppresses cell apoptosis and inflammation via miR‐493‐5p in LPS‐treated Caco‐2 cells
To determine whether NEAT1 functions via miR‐493‐5p, Caco‐2 cells were transfected with si‐NEAT1 and miR‐493‐5p inhibitor. RT‐qPCR confirmed that NEAT1 was highly expressed and miR‐493‐5p was significantly downregulated in LPS‐treated UC model than in the control group. In the LPS + lncRNA NEAT1‐siRNA group, lncRNA NEAT1‐siRNA knocked down its expression level and enhanced miR‐493‐5p level in contrast with the LPS + control‐siRNA group, but miR‐493‐5p levels were reduced when the cells were cotransfected with the miR‐493‐5p inhibitor (Figure 5A,B). CCK‐8 assay revealed that the capability of LPS‐treated Caco‐2 cells was weaker than that of the negative control group; however, inhibition of NEAT1 rescued this effect, while transfection of miR‐493‐5p inhibitor accelarated this effect (Figure 5C). Flow cytometry revealed that LPS increased apoptosis, si‐NEAT1 transfected into LPS‐treated Caco‐2 cells weakened apoptosis, and transfection of miR‐493‐5p inhibitor increased the percentage of apoptotic cells (Figure 5D,E). Levels of IL‐6, IL‐8, TNF‐α, and IL‐1β were all increased by LPS and decreased by si‐NEAT1 transfection, and these effects were rescued by the miR‐493‐5p inhibitor (Figure 5F–I).
Figure 5.
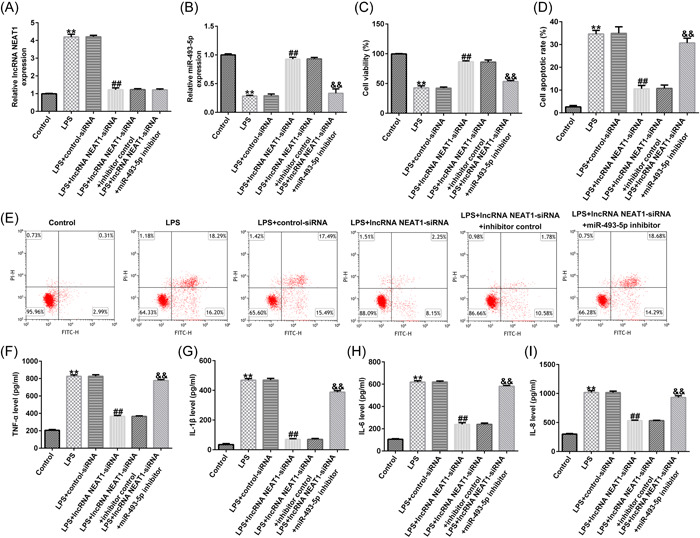
LncRNA NEAT1 targets miR‐493‐5p to regulate the cell damage and inflammatory response in LPS‐treated Caco‐2 cells. (A, B) Levels of NEAT1 and miR‐493‐5p determined via RT‐qPCR. (C) Cell viability determined using the cell counting kit (CCK)‐8 assay. (D, E) Apoptosis ratio of LPS‐treated cells determined via flow cytometry. (F–I) Inflammatory cytokine levels measured using an enzyme‐linked immunosorbent assay (ELISA). Data are represented as the average ± SD of triplicate experiments.
3.6. Rab27A is downstream of miR‐493‐5p
We tested the downstream mRNA of miR‐493‐5p using Targetscan 7.0 and found that Rab27A contained potential miR‐493‐5p binding points (Figure 6A). We used the dual‐relative luciferase method to evaluate this binding. We found that miR‐493‐5p overexpression downregulated the luciferase levels of the Rab27A‐WT reporter gene (Figure 6B). The results of RIP fuether verifed the target relationship between miR‐493‐5p and Rab27A (Figure 6C).
Figure 6.
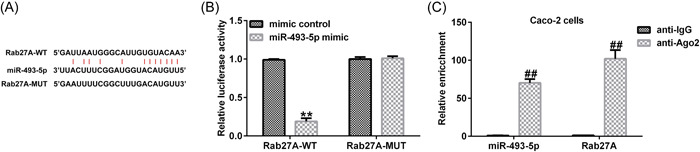
Rab27A is downstream of miR‐493‐5p. (A) Binding points between miR‐493‐5p and Rab27A predicted using TargetScan 7.0. (B) Dual relative luciferase assay to confirm the linkage between miR‐493‐5p and Rab27A. (C) RIP assay was employed to further verify the interaction between miR‐493‐5p and Rab27A. Data are represented as the average ± SD of triplicate experiments.
3.7. miR‐493‐5p modulates Rab27A expression in UC cells
RT‐qPCR was performed to determine the relationship between Rab27A and miR‐493‐5p by measuring their expression levels. miR‐493‐5p expression levels were increased by miR‐493‐5p mimic transfection compared to those in the mimic NC control group (Figure 7A). Rab27A expression was upregulated by transfection with Rab27A‐plasmid compared to its level in the control plasmid group (Figure 7B). Furthermore, overexpression of miR‐493‐5p markedly disturbed the protein and mRNA expression levels of Rab27A in Caco‐2 cells, but Rab27A‐plasmid cotransfection reversed this effect (Figure 7C,D). These results suggest that Rab27A is downstream of miR‐493‐5p and its expression linked to miR‐493‐5p expression in Caco‐2 cells.
Figure 7.
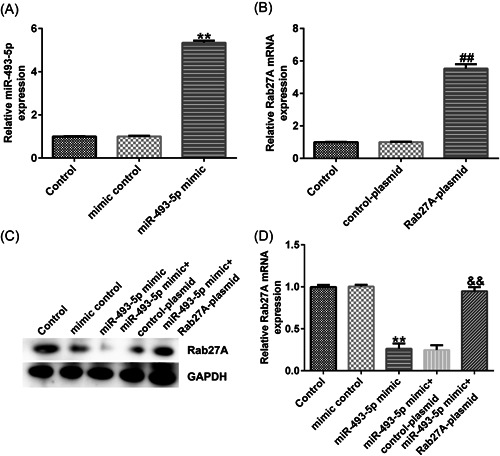
miR‐493‐5p negatively targets Rab27A. (A) Efficiency of miR‐493‐5p mimic transfection. (B) Efficiency of Rab27A‐plasmid transfection. (C and D) Levels of nuclear receptor coactivator 3 (NCOA3) with miR‐142‐5p mimic and Rab27A‐plasmid cotransfection determined via RT‐qPCR and Western blot analysis (WB). Data are represented as the average ± SD of triplicate experiments.
3.8. miR‐493‐5p suppresses LPS‐induced cell damage and inflammation in Caco‐2 cells via Rab27A
Caco‐2 cells were cotransfected with the miR‐493‐5p mimic and Rab27A‐plasmid to determine whether miR‐493‐5p functions by inhibiting Rab27A expression. The findings confirmed that miR‐493‐5p expression was downregulated and Rab27A expression was significantly upregulated in the LPS‐treated model compared to their expression levels in the control group, whereas in the LPS + miR‐493‐5p mimic group, miR‐493‐5p mimic increased miR‐493‐5p expression and downregulated Rab27A, in contrast with the LPS + mimic control, but Rab27A expression was rescued via cortransfection with Rab27A‐plasmid (Figure 8A–C). CCK‐8 assay revealed that the viability of LPS‐treated Caco‐2 cells was lower than that of cells in the negative control group; however, overexpression of miR‐493‐5p reduced the effect of LPS, while cotransfection with Rab27A‐plasmid reversed the miR‐493‐5p overexpression effect (Figure 8D). Flow cytometry revealed that LPS promoted cell apoptosis, miR‐493‐5p mimic transfected into LPS‐treated Caco‐2 cells decreased apoptosis, and cotransfection of Rab27A increased the number of apoptotic cells (Figure 8E,F). Levels of IL‐6, IL‐8, IL‐6, and IL‐1β were significantly increased by LPS treatment and decreased by miR‐493‐5p mimic transfection, and these effects were reversed by the upregulation of Rab27A expression (Figure 8G–J).
Figure 8.

miR‐493‐5p inhibits LPS‐induced cell damage and inflammation in Caco‐2 cells via Rab27A. (A–C) Levels of miR‐493‐5p and Rab27A in cells determined via RT‐qPCR. Rab27A levels in cells determined via WB. (D) Cell viability determined via CCK‐8 assay. (E and F) Apoptosis ratio of LPS‐treated cells determined via flow cytometry. (G–J) Inflammatory cytokine levels measured via ELISA. Data are represented as the average ± SD of triplicate experiments.
4. DISCUSSION
UC is a chronic nonspecific IBD characterized by chronic inflammation and ulcerative changes in the intestinal mucosa. UC lesions are primarily located in the mucosa, colonic submucosa, and rectum. However, UC pathogenesis remains unclear. Some studies have suggested that dysfunction of the immune system is an important factor in UC‐induced intestinal inflammation and tissue damage, while others believe that it may be related to environmental, genetic, infection‐related, and immune factors. Clinical treatment of UC generally involves the use of corticosteroids, immunosuppressants, and aminosalicylates. 21 , 22 , 23 In addition, good psychological care, daily care, and dietary care for UC patients are also crucial for the development of their condition. In this study, we elucidated the mechanism by which NEAT1 accelerates UC in LPS‐treated Caco‐2 cells. We found that the lncRNA NEAT1 inhibits UC via miR‐493‐5p, indicating its potential as a new biomarker for UC.
Development of UC is influenced by various factors. LncRNAs are significant biomarkers of UC. Tian et al. revealed that CDKN2B‐AS1 participates in UC via miR‐16 and miR‐195. 24 Wang et al. reported that MEG3 participates in UC. 25 Ding et al. reported that the lncRNA MIRT2 expression was downregulated in UC. 26 However, only a few studies have investigated the function of NEAT1 in UC. In this study, we first confirmed the abnormal low expression of NEAT1 in UC samples by RT‐qPCR. However, we did not do FISH analysis of NEAT1 in UC samples. This may be a limitation of this study. Then we found that NEAT1 knockdown was effective in treating UC.
Many studies have suggested that lncRNAs regulate the development of various cancers via miRNAs. Zhou reported many miRNAs involved in UC. 27 Qu et al. revealed that the knockdown of miR‐21 and miR‐155 ameliorates UC. 28 Deng et al. reported that miR‐200b participates in UC. 29 In this study, we found that miR‐493‐5p expression was upregulated in UC samples, and miR‐493‐5p expression was disturbed and negatively controlled by the expression of NEAT1 in LPS‐treated Caco‐2 cells.
Rab27A is a member of the Rab family that is involved in tumor cell proliferation and apoptosis in pancreatic, 30 ovarian, 31 and lung cancers. 32 In this study, Rab27A mRNA and protein were found to be highly expressed in the UC model established using the LPS‐treated Caco‐2 cells.
In conclusion, our study demonstrated that NEAT1 modulates the miR‐493‐5p/Rab27A axis to inhibit UC development. Therefore, targeting the NEAT1/miR‐493‐5p/Rab27A axis is a potential strategy for UC treatment.
AUTHOR CONTRIBUTIONS
Hecheng Wang: Conceptualization, formal analysis, project administration, writing‐original draft, and writing‐review and editing. jiadan teng: supervision, visualization, and writing‐review and editing. Mingtao Wang, Yuhang Zhang, Xiaoshuang Liu, and Zhuya Liu: Investigation, methodology, software, and validation. all authors have read and approved the final manuscript.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ETHICS STATEMENT
This study was sanctioned by Ethics Committee of the Third Affiliated Hospital of Qiqihar Medical University. Written informed content was obtained for each patient. All patients agreed for publication.
ACKNOWLEDGMENTS
The present study was supported by 2022 Qiqihar Science and Technology Plan Joint Guidance Project (grant no. LSFGG‐2022017).
Wang H, Teng J, Wang M, Zhang Y, Liu X, Liu Z. Expression and significant roles of the lncRNA NEAT1/miR‐493‐5p/Rab27A axis in ulcerative colitis. Immun Inflamm Dis. 2023;11:e814. 10.1002/iid3.814
DATA AVAILABILITY STATEMENT
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
REFERENCES
- 1. Ungaro R, Mehandru S, Allen PB, Peyrin‐Biroulet L, Colombel JF. Ulcerative colitis. Lancet. 2017;389:1756‐1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chu H, Khosravi A, Kusumawardhani IP, et al. Gene‐microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science. 2016;352:1116‐1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ming L, Bing W, Xiaotong S, et al. Upregulation of intestinal barrier function in rat with DSS‐Induced colitis by a defined bacterial consortium is associated with expansion of IL‐17A producing gamma delta T cells. Front Immunol. 2017;8:824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pope JL, Bhat AA, Sharma A, et al. Claudin‐1 regulates intestinal epithelial homeostasis through the modulation of notch‐signalling. Gut. 2014;63(4):622‐634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li T, Xie J, Shen C, et al. RETRACTED ARTICLE: upregulation of long noncoding RNA ZEB1‐AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene. 2016;35(12):1575‐1584. [DOI] [PubMed] [Google Scholar]
- 6. Tan DSW, Chong FT, Leong HS, et al. Long noncoding RNA EGFR‐AS1 mediates epidermal growth factor receptor addiction and modulates treatment response in squamous cell carcinoma. Nature Med. 2017;23(10):1167‐1175. [DOI] [PubMed] [Google Scholar]
- 7. Xu Y, Ge Z, Zhang E, et al. The lncRNA TUG1 modulates proliferation in trophoblast cells via epigenetic suppression of RND3. Cell Death Dis. 2017;8(10):3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yuan J, Yang F, Wang F, et al. A long noncoding RNA activated by TGF‐β promotes the invasion‐metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666‐681. [DOI] [PubMed] [Google Scholar]
- 9. Han J, Li Y, Zhang B, Liu H, Wu M, Zhang X. lncRNA TUG1 regulates ulcerative colitis through miR‐142‐5p/SOCS1 axis. Microb Pathog. 2020;143:104139. [DOI] [PubMed] [Google Scholar]
- 10. Shao K, Xi L, Cang Z, Chen C, Huang S. Knockdown of NEAT1 exerts suppressive effects on diabetic retinopathy progression via inactivating TGF‐β1 and VEGF signaling pathways. J Cell Physiol. 2020;235:9361‐9369. [DOI] [PubMed] [Google Scholar]
- 11. Sheng B, Zhao L, Zang X, et al. Quercetin inhibits caerulein‐induced acute pancreatitis through regulating miR‐216b by targeting MAP2K6 and NEAT1. Inflammopharmacology. 2021;29(2):549‐559. [DOI] [PubMed] [Google Scholar]
- 12. Yu X, Li Z, Zheng H, Chan MTV, Wu WKK. NEAT1: a novel cancer‐related long non‐coding RNA. Cell Proliferation. 2017;50:e12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shi Y, Fang N, Li Y, et al. Circular RNA LPAR3 sponges microRNA‐198 to facilitate esophageal cancer migration, invasion, and metastasis. Cancer Sci. 2020;111(8):2824‐2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bandiera S, Pfeffer S, Baumert TF, Zeisel MB. miR‐122 a key factor and therapeutic target in liver disease. J Hepatol. 2015;62(2):448‐457. [DOI] [PubMed] [Google Scholar]
- 15. Elton TS, Selemon H, Elton SM, Parinandi NL. Regulation of the MIR155 host gene in physiological and pathological processes. Gene. 2013;532(1):1‐12. [DOI] [PubMed] [Google Scholar]
- 16. Tang S, Guo W, Kang L, Liang J. MiRNA‐182‐5p aggravates experimental ulcerative colitis via sponging Claudin‐2. J Mol Histol. 2021;52(6):1215‐1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Benderska N, Dittrich AL, Knaup S, et al. miRNA‐26b overexpression in ulcerative colitis‐associated carcinogenesis. Inflamm Bowel Dis. 2015;21(9):2039‐2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee JH, Kim MS, Yoo NJ, Lee SH. BPTF, a chromatin remodeling‐related gene, exhibits frameshift mutations in gastric and colorectal cancers. APMIS. 2016;124:425‐427. [DOI] [PubMed] [Google Scholar]
- 19. Zhu B, Cao X, Zhang W, et al. MicroRNA‐31‐5p enhances the warburg effect via targeting FIH. FASEB J. 2018;33:545‐556. 10.1096/fj.201800803R [DOI] [PubMed] [Google Scholar]
- 20. Gross T, Wack G, Syhr KMJ, et al. Rab27A contributes to the processing of inflammatory pain in mice. Cells. 2020;9(2):1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lord JD. Promises and paradoxes of regulatory T cells in inflammatory bowel disease. World J Gastroenterol. 2015;21(40):11236‐11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu Y, Zhang X, Yang C, Fan H. Oxymatrine protects rat brains against permanent focal ischemia and downregulates NF‐κB expression. Brain Res. 2009;1268:174‐180. [DOI] [PubMed] [Google Scholar]
- 23. Hong‐Li S, Lei L, Lei S, et al. Cardioprotective effects and underlying mechanisms of oxymatrine against ischemic myocardial injuries of rats. Phytother Res. 2008;22(7):985‐989. [DOI] [PubMed] [Google Scholar]
- 24. Tian Y, Cui L, Lin C, Wang Y, Liu Z, Miao X. LncRNA CDKN2B‐AS1 relieved inflammation of ulcerative colitis via sponging miR‐16 and miR‐195. Int Immunopharmacol. 2020;88:106970. [DOI] [PubMed] [Google Scholar]
- 25. Wang Y, Wang N, Cui L, et al. Long non‐coding RNA MEG3 alleviated ulcerative colitis through upregulating miR‐98‐5p‐Sponged IL‐10. Inflammation. 2021;44(3):1049‐1059. [DOI] [PubMed] [Google Scholar]
- 26. Ding G, Ming Y, Zhang Y. lncRNA Mirt2 is downregulated in ulcerative colitis and regulates IL‐22 expression and apoptosis in colonic epithelial cells. Gastroenterol Res Pract. 2019;2019:1‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou J, Liu J, Gao Y, Shen L, Li S, Chen S. miRNA‐Based potential biomarkers and new molecular insights in ulcerative colitis. Front Pharmacol. 2021;12:707776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qu S, Shen Y, Wang M, Wang X, Yang Y. Suppression of miR‐21 and miR‐155 of macrophage by cinnamaldehyde ameliorates ulcerative colitis. Int Immunopharmacol. 2019;67:22‐34. [DOI] [PubMed] [Google Scholar]
- 29. Deng S, Wang H, Fan H, et al. Over‐expressed miRNA‐200b ameliorates ulcerative colitis‐related colorectal cancer in mice through orchestrating epithelial‐mesenchymal transition and inflammatory responses by channel of AKT2. Int Immunopharmacol. 2018;61:346‐354. [DOI] [PubMed] [Google Scholar]
- 30. Kren N, Michaud D, Bagchi S, Greene K, Pylayeva‐Gupta Y. Rab27A plays a dual role in metastatic propensity of pancreatic cancer. Sci Rep. 2020;10(1):7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dorayappan KDP, Wanner R, Wallbillich JJ, et al. Hypoxia‐induced exosomes contribute to a more aggressive and chemoresistant ovarian cancer phenotype: a novel mechanism linking STAT3/Rab proteins. Oncogene. 2018;37(28):3806‐3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mohamed Gamal El‐Din G, Ibrahim FK, Shehata HH, Osman NM, Abdel‐Rahman OM, Ali M. Exosomal expression of Rab27A and its related lncRNA Lnc‐RNA‐RP11‐510M2 in lung cancer. Arch Physiol Biochem. 2020;13:1‐7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


