Abstract
Cyclic AMP (cAMP) is a ubiquitous second messenger synthesized by most living organisms. In bacteria, it plays highly diverse roles in metabolism, host colonization, motility, and many other processes important for optimal fitness. The main route of cAMP perception is through transcription factors from the diverse and versatile CRP–FNR protein superfamily. Since the discovery of the very first CRP protein CAP in Escherichia coli more than four decades ago, its homologs have been characterized in both closely related and distant bacterial species. The cAMP-mediated gene activation for carbon catabolism by a CRP protein in the absence of glucose seems to be restricted to E. coli and its close relatives. In other phyla, the regulatory targets are more diverse. In addition to cAMP, cGMP has recently been identified as a ligand of certain CRP proteins. In a CRP dimer, each of the two cyclic nucleotide molecules makes contacts with both protein subunits and effectuates a conformational change that favors DNA binding. Here, we summarize the current knowledge on structural and physiological aspects of E. coli CAP compared with other cAMP- and cGMP-activated transcription factors, and point to emerging trends in metabolic regulation related to lysine modification and membrane association of CRP proteins.
Keywords: nucleotide second messenger, cAMP, cGMP, CRP transcription factors, allosteric control, CRP regulon
This review summarizes the current knowledge on CRP transcription factors allosterically activated by cAMP or cGMP to control highly diverse regulons in a broad range of bacteria.
Adenosine 3′, 5′-cyclic monophosphate (cAMP) signaling is widespread in the domain of bacteria. It represents a key regulatory mechanism in bacterial physiology, allowing bacteria to respond to their environment. cAMP is a second messenger produced in response to a variety of environmental stimuli, including nutrient availability, stress, and quorum sensing, and its cellular concentration controls a regulatory switch between different bacterial lifestyles, such as motility and virulence. Perception of cyclic mononucleotides (cNMPs), including cAMP and guanosine 3′, 5′-cyclic monophosphate (cGMP), is mostly mediated by cAMP receptor proteins, which act as transcription regulators. These regulators are members of the CRP–FNR protein superfamily. We will first provide an overview of this superfamily and then focus on the diversity of cNMP receptor CRP proteins in the bacterial domain to reveal structural and functional similarities and differences among these regulators.
CRP–FNR protein superfamily
Early studies of diauxic growth of Escherichia coli led to the discovery of the phenomenon of catabolite repression involving the cyclic AMP-responsive transcription factor CAP (for catabolite activator protein, also later named CRP for cyclic AMP receptor protein). Together with the structurally similar fumarate and nitrate reduction regulator FNR from E. coli, CAP (CRP) was a founder of the CRP–FNR superfamily of transcription regulators present in most of the bacterial phyla. The common trait of CRP–FNR superfamily members is an allosteric regulation by binding of a small molecule in the regulatory domain. This domain shares structural similarities with a nucleotide-binding domain (NBD), which translates the ligand binding into structural adjustments in the helix-turn-helix DNA binding domain (DBD), promoting or inhibiting binding to specific DNA sites. The CRP–FNR proteins include both global regulators with large regulons and more specialized transcription factors. Their functions are highly versatile. They are indispensable for adaptation to changing environmental conditions since they have crucial roles in governing small molecule metabolism and respiration, cell envelope biosynthesis, motility, stress response, and many other traits, important for bacterial fitness and, in case of pathogens, also virulence. CRP–FNR proteins are commonly working as transcription activators with DNA binding sites upstream of the promoter, but they can also act as repressors if bound further downstream.
The CRP–FNR proteins were categorized into 21 subclasses based on protein sequence similarity and named after regulators with known functions (Körner et al. 2003). A total of 13 of these subclasses include proteins similar to the anaerobic regulator FNR from E. coli. These proteins respond to inorganic molecules, mostly with the help of a cofactor. Escherichia coli FNR, e.g. senses microaerobic conditions via an iron–sulfur cluster, which has to be in the reduced state to promote FNR dimerization and DNA binding (Mettert and Kiley 2018), and DnrR from Pseudomonas aeruginosa was suggested to sense nitric oxide via heme (Giardina et al. 2008). A direct interaction with an inorganic molecule can also serve as a regulatory cue. An example is the Bradyrhizobium japonicum microaerobic regulator FixK2, which is able to bind the target DNA in its apo-form, whereas oxidation of a single cysteine residue in the DBD interferes with DNA binding (Bonnet et al. 2013).
The remaining eight subclasses contain proteins similar to E. coli CAP. These are generally referred to as CRP proteins. The arsenal of regulatory cues to which these proteins respond includes both organic and inorganic molecules. The founding member CAP is activated by cAMP (Emmer et al. 1970) and cyanobacterial NtcA is activated by 2-oxoglutarate and the accessory protein PipX (Llácer et al. 2010). A prominent example of a combined regulation by organic and inorganic factors is Desulfitobacterium spp. CprK, which is activated by binding of o-chlorophenol and inhibited by oxidation (Levy et al. 2008). CooA subtype proteins, such as CooA in Rhodospirillum rubrum or Carboxydothermus hydrogenoformans, sense carbon monoxide via heme located in the regulatory domain (Lanzilotta et al. 2000, Borjigin et al. 2007).
The term CRP-like is usually applied to proteins similar to CAP, but being active in their apo-form. Their activity as transcription regulators is controlled by other factors. Xanthomonas and Lysobacter Clp proteins are inhibited by binding of c-di-GMP (Dong and Ebright 1992, Leduc and Robert 2009, Xu et al. 2018, 2021), Myxococcus xanthus MrpC activity is regulated at the level of protein biosynthesis and phosphorylation (Lux and Shi 2005, Robinson et al. 2014), and in case of Thermus thermophilus TAP, the mechanism is still unknown (Feng et al. 2016). Many bacterial species contain multiple copies of CRP–FNR family proteins, specialized for particular physiological needs (Körner et al. 2003). Most of the CRP–FNR protein genes described so far are nonessential. In some phylogenetic branches, they might have been acquired via horizontal gene transfer. This is suggested by a lack of correlation between sequence similarities of CRP proteins and global genome sequence similarities of the respective bacterial species (Soberon-Chavez et al. 2017). In the following, we describe the structural and functional diversity of bacterial cNMP receptor CRP proteins and compare this diversity to the CAP paradigm of E. coli, which we first describe in detail.
The CAP model
Catabolite repression in E. coli is a regulatory feature of carbon metabolism that involves preferential utilization of glucose over other carbon sources. Expression of genes for uptake and metabolism of nonpreferred carbon sources depends on transcriptional activation by CAP–cAMP, which is reduced in presence of glucose in the medium (Notley-McRobb et al. 1997, Saier 1998). Escherichia coli CAP was the first transcription factor with known crystal structure (McKay and Steitz 1981) and remains one of the best-studied bacterial proteins.
Protein structure
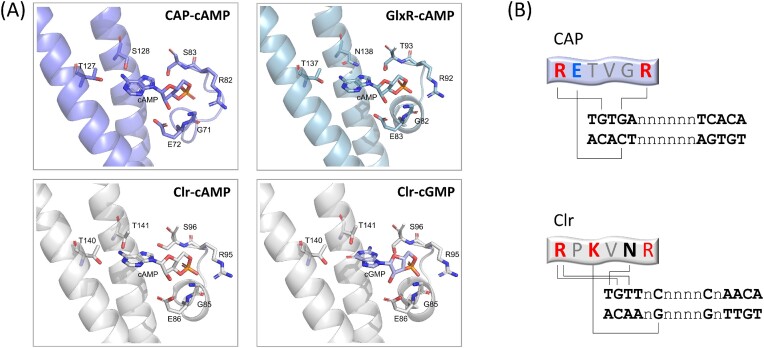
The X-ray structural analysis of CAP revealed a dimer in which each protomer consists of an N-terminal NBD and C-terminal DBD, connected by a C-helix adopting coiled–coil structure and serving as a dimer interface (Passner and Steitz 1997, Passner et al. 2000, Popovych et al. 2009, Seok et al. 2014). The NBD contains a hydrophobic pocket for nucleotide binding and can bind one cAMP molecule in anti-conformation or one cGMP molecule in syn-conformation (Passner and Steitz 1997, Passner et al. 2000, Seok et al. 2014). In addition, a low-affinity binding site for cAMP in syn-conformation was detected at the interface between the NBD and DBD, although biological significance of this binding site was not confirmed (Passner and Steitz 1997). The nucleotide syn- and anti-conformations are defined by the rotation of the nucleobase around the glycosidic bond, which results in the nucleobase pointing in the same (syn-) or in the opposite (anti-) direction as the phosphate group. The anti-conformation is a stretched form, i.e. common in nucleic acids, whereas the syn-conformation is a compact, rare form, occurring, e.g. in active sites of ribozymes (Sokoloski et al. 2011). The phosphoribose of the cAMP molecule is accommodated in the CAP nucleotide binding pocket by bonding to R82 and S83 (RS signature motif) and E72, whereas the nucleobase interacts with the C-helix residues T127 of the same protomer and S128 of the adjacent protomer (TS signature motif) (Figs 1 and 2A; Table 1) (Passner et al. 2000).
Figure 1.
Selected features of CAP–cAMP and GlxR–cAMP structures. The cAMP-binding signature residues are boxed and highlighted in pink, the DNA-binding F-helix signature is boxed and highlighted in yellow. The C- and D-helices in apo-CAP and apo-GlxR are indicated by dashed lines. The residues constituting AR1, AR2, and AR3 in CAP are underlined. PDB structures: CAP, 4N9H; CAP–cAMP, 1G6N; GlxR, 4BYY; and GlxR-cAMP, 4CYD.
Figure 2.
Structural features of cyclic nucleotide- and DNA-binding signature motifs. (A), cAMP/cGMP binding to selected Crp-like proteins. (B) CAP–DNA and Clr–DNA interaction interfaces.
Table 1.
Overview of CRP proteins experimentally shown to bind cAMP or cGMP.

|
In apo-CAP, the DBD is dimerized via coiled–coil interactions between the D-helices, which locks the DNA recognition F-helix in a position, incompatible with DNA binding. cAMP binding results in an extension of the C-helix and shortening of the D-helix by one turn (Fig. 1). This abolishes the D-helix coiled–coil dimerization and causes rotation of the DBDs, ensuring correct orientation of the F-helices (Popovych et al. 2009, Sharma et al. 2009). Binding of two cAMP molecules in a CAP dimer elicited the same allosteric effect upon either subunit, but proceeded with negative cooperativity, with only the first one being energetically favorable (Popovych et al. 2009). In contrast to cAMP, cGMP binding did not induce the same conformational changes. It was suggested that it either has no appreciable effect on the CAP structure (Popovych et al. 2009) or is able to stabilize the inactive conformation due to guanosine bonding with S83 and K130, thereby playing an active negative role (Seok et al. 2014). The properly positioned F-helix can be inserted into the major grove of the target DNA, where positively charged R180 and R185 of the RETVGR motif bond with G2 and G4 of the recognition motif TGTGA, and negatively charged E181 interacts with the G4 cytosine base pair mate (Fig. 2B) (Parkinson et al. 1996). Binding of the CAP dimer to the palindrome consensus binding sequence TGTGA N6 TCACA results in significant DNA kinks and twists at both DNA–protein interfaces, exemplified by roll angles of 52 and 35 degrees and twist angles of 17 and 12 degrees observed in an X-ray crystallography study (Parkinson et al. 1996). This DNA deformation is additionally promoted by contacts between phosphates of the DNA outside the core recognition motif and other mostly charged amino acid residues located at the surface of the DBD and NBD (Parkinson et al. 1996).
Transcription regulation
The transcription activation by CAP can proceed via different molecular mechanisms, depending on the promoter structure. CAP can interact with the RNA polymerase (RNAP) at three distinctive surface patches termed activating regions AR1, AR2, and AR3 (Fig. 1). AR1 is located within the DBD, and AR2 and AR3 within the NBD (Feng et al. 2016, Liu et al. 2017). In Class I promoters, the CAP dimer is bound at varying distances upstream of the −35 promoter element and interacts with an α-CTD of the RNAP via the AR1. This interaction is proposed to stabilize RNAP–DNA binding, facilitating the RNAP–promoter closed complex formation (Benoff et al. 2002). In Class II promoters, the CAP dimer is bound in the −35 promoter region and interacts with both RNAP α-CTDs via AR1, with RNAP α-NTD and the β-subunit via AR2 and with σR4 region via AR3 (reviewed in Busby and Ebright 1999, Lawson et al. 2004). Class III represents a combination of Class I and Class II promoters. In Class II promoters, additional contacts between the AR2 and AR3 and the RNAP contribute to isomerization of the RNAP–promoter closed complex to the RNAP–promoter open complex, as suggested by crystal structure analysis of CAP in complex with DNA and RNAP, as well as T. thermophilus CAP homolog TAP in complex with RNAP, promoter DNA and a ribotetranucleotide primer (reviewed in Lawson et al. 2004, Feng et al. 2016). A recent cryo-electron microscopy study of an intact Class-II CAP-dependent transcription activation complex revealed transient conformational changes in the RNAP upon CAP binding, favoring transcription initiation (Shi et al. 2020). These changes were reversed during further RNA transcript synthesis, suggesting that CAP may activate transcription by inducing intermediate state (Shi et al. 2020). In a recent high-resolution ChIP-exonuclease (ChIP-exo) study, CAP generated binding patterns closely resembling those generated by σ70, suggesting that during transcription initiation CAP might dissociate from its DNA binding sites and remain bound to RNAP (Latif et al. 2018). CAP binding downstream of the promoter region can mediate transcriptional repression, e.g. of the pck gene, which is repressed by CAP at very high CAP–cAMP levels (Nakano et al. 2014).
The actual CAP binding sites in the E. coli genome deviate from the perfect palindrome TGTGA N6 TCACA in single bases within the core motif and in the length of the spacer (Barber et al. 1993, Pyles and Lee 1996). The variable sequences of the spacer and the immediate surrounding of the binding site can influence bending of the DNA upon CAP contact and thereby the overall binding affinity. These DNA-based properties can potentially generate a wide range of CAP binding sites with different affinities that for a specific promoter can be evolutionarily adapted to the physiological need. Low CAP affinity can be compensated, on the other hand, by interactions with other transcription factors or proteins. Examples include CAP activation of natural competence genes via a noncanonical low-affinity binding site TGCGA, supported by the regulator Sxy (Cameron and Redfield 2006). Binding of CAP together with other transcription factors in the same promoter region provides the possibility of competition, synergistic interactions, or antiactivation (reviewed in Busby and Ebright 1999).
A chromatin immunoprecipitation study showed that besides the strong binding in promoter regions of CAP target genes, CAP also interacts with thousands of weaker sites across the chromosome (Grainger et al. 2005). This led to the hypothesis that CAP may represent an intermediate state in the evolution of a transcription factor from a nucleoid-associated protein (NAP) (Heyde et al. 2021). Of the known proteins recognized as NAPs, CAP most closely resembles the integration host factor IHF in terms of DNA bending and the pronounced binding site specificity (reviewed in Dorman et al. 2020).
CAP physiological effects
The most comprehensive analysis of the CAP regulon using a genomic SELEX (Systematic Evolution of Ligands by EXponential Enrichment) approach identified a minimum of 378 promoters as regulation targets of cAMP–CAP (Shimada et al. 2011). These findings pointed towards a key regulatory role of E. coli CAP in carbohydrate metabolism, including the selective transport of carbon sources, glycolysis, gluconeogenesis and tricarboxylic acid cycle, pyruvate dehydrogenase pathway, and aerobic respiration (Shimada et al. 2011). Most of the CAP binding sites were situated within or upstream of the −35 promoter element, suggesting gene activation, whereas a minor part was found further downstream, indicative of gene repression. A total of 70 transcription factor genes were described as CAP targets, including regulators for carbon or nitrogen metabolism, stress–response, nucleoid proteins, and uncharacterized transcription factors (Shimada et al. 2011). Recent large-scale functional analysis identified 202 feed-forward regulatory loops, which were directly affected by CAP (Yang et al. 2018).
Interestingly, although CAP–cAMP-mediated gene regulation promotes normal growth of E. coli in optimal laboratory conditions, it interferes with survival and persister cell formation in unfavorable conditions, such as prolonged starvation, high hydrostatic pressure used in food sterilization, and exposure to antibiotics and bactericidal agents like hypochlorite, hydrogen peroxide, and low pH (Mok et al. 2015, Gayán et al. 2017, Molina-Quiroz et al. 2018, Nosho et al. 2018, Zeng et al. 2022). Moreover, killing of E. coli by the type VI secretion system-delivered toxin of Vibrio cholerae was reduced in a crp-deficient strain (Crisan et al. 2021). A recent study found that polynucleotide phosphorylase (PNPase) represses CAP gene expression at the transcriptional level, as a key regulator of persister cell formation (Wu et al. 2022). This supports an important negative role of CAP regulation in stress survival. Furthermore, persistence in presence of antibiotics was partially attributed to CAP-dependent regulation of the toxin–antitoxin gene pair mqsR–mqsA (Uppal and Jawali 2016). Targeted engineering of CAP to increase bacterial stress tolerance generated variants, which had lower binding affinity for all three classes of CAP-dependent promoters compared to the native protein, supporting an antipersistence role of CAP (reviewed in Geng and Jiang 2015).
Additional pathways of CAP protein function regulation
Consistent with the global regulatory role of CAP, a mechanism of direct coupling of its functionality to the bacteria’s metabolic status was recently reported. Under growth conditions that result in accumulation of acetyl phosphate, CAP can be acetylated at the lysine residue K100. This modification reduces the CAP steady-state levels and interferes with its DNA binding ability, particularly strongly reducing transcription activation at Class II promoters (Davis et al. 2018, Ro et al. 2021). The status of the K100 residue exerts the largest impact on transcription activation by CAP when the spacing between its DNA binding site and the second position of the −10 promoter element is 22 bp, which is an exclusive feature of Class II promoters (Écija-Conesa et al. 2020).
Another impressive regulatory mechanism involves the dual-function RNA Spot 42, which acts as a base-pairing RNA to directly repress genes involved in central and secondary metabolism, redox balancing, and utilization of nonpreferred carbon sources (Beisel and Storz 2011). Its targets are largely CAP-activated genes, whereas transcription of Spot 42 itself is repressed by CAP, providing a feedforward regulatory loop (Beisel and Storz 2011). Additionally, Spot 42 encodes the 15 amino acid peptide SpfP, which inhibits Class II promoter activation. This inhibition relies on binding of SpfP at the AR3 region of CAP, presumably blocking the CAP–RNAP interaction (Aoyama et al. 2022).
Other γ-proteobacteria
Yersinia and Vibrio
The two human pathogens, V. cholerae and Yersinia pestis, are close relatives of E. coli and contain very similar CAP homologues designated Crp (Table 1). The structure of the Y. pestis CRP–cAMP complex was determined and found to be virtually identical to the CAP–cAMP complex, despite three divergent amino acids, all within the C-helix (Ritzert et al. 2019). Inactivation of the crp gene in pathogenic Y. pseudotuberculosis and Y. pestis strains affected expression of at least 6% of all the genes. Besides genes for carbon catabolism, also quorum sensing- and virulence-related genes were affected, including Type III secretion system-encoding genes and the plasminogen activator protease gene pla (reviewed in Heroven and Dersch 2014, Ritzert et al. 2019).
In Vibrio bacteria, Crp plays a CAP-like role in modulating metabolic pathways; however, it also controls functions involved in natural competence, bioluminescence, pheromone signaling, and colonization of animal hosts (reviewed in Colton and Stabb 2016). In a recent ChIP-seq study, V. cholerae Crp was characterized as an integral component of the regulatory network that controls lifestyle switching, regulating gene expression in response to host colonization (Manneh-Roussel et al. 2018). This study also identified the DNA binding motif TGTGA, identical to the CAP binding motif (Manneh-Roussel et al. 2018). Crp from V. harveyi, deviating from CAP in 10 amino acids, was shown to bind cAMP and cGMP with similar affinity (Chen et al. 1985). However, only cAMP binding resulted in a conformational change in the protein and transcription activation (Chen et al. 1985).
Another level of complexity was recently added with the discovery of membrane association of E. coli CAP and V. cholerae Crp (Gibson et al. 2022). During growth of V. cholerae in nutrient-rich conditions, Crp was found in the membrane fraction in complex with the DNA-binding aminopeptidase PepA, another global transcription regulator (Gibson et al. 2022). In nutrient-poor conditions as well as in stationary growth phase, Crp was mainly found in the cytoplasm. It was shown that lysine succinylation of Crp at K53 and K90 lead to its dissociation from PepA and the membrane, which activated PepA-mediated gene regulation. The membrane association was independent of cAMP. Cross-complementation experiments revealed that CAP can associate with the membrane in a similar manner as V. cholerae Crp, suggesting that this mechanism might be conserved in other species (Gibson et al. 2022).
Pseudomonas
In the human pathogen P. aeruginosa, the CAP homolog was named Vfr because of its primary role as a virulence factor (West et al. 1994). It shares 135 identical amino acids with CAP (Table 1), but it does not serve as catabolite activator (Suh et al. 2002, Wolfgang et al. 2003). Instead, Vfr regulates virulence-related features such as expression of Type IV pili-, Type III and Type II secretion system-, quorum-sensing system-, and exolysin-encoding genes, thereby promoting a planktonic lifestyle and acute invasive infection as opposed to a sessile lifestyle and persistent infection (reviewed in Coggan and Wolfgang 2012, Berry et al. 2018). Furthermore, other important traits such as CRISPR–CAS-related genes are regulated by Vfr (Dela Ahator et al. 2022). In total, the Vfr regulon comprises over 200 genes (Suh et al. 2002, Wolfgang et al. 2003). An X-ray crystallographic analysis of Vfr–cAMP revealed a structure very similar to CAP–cAMP. The only notable alteration is the C-helix threonine-threonine motif of the cAMP binding site, replacing the CAP TS motif. The threonine successfully replaced serine in the function of hydrogen bonding between the CAP C-helices and the cAMP nucleobase (Cordes et al. 2011). The Kd determined for cAMP binding was 1.6 µM, compared with 0.4 µM determined for CAP in the same study (Suh et al. 2002). This suggests a comparable functionality of the NBD in both proteins. Similar to CAP–cAMP, two additional cAMP molecules in syn-conformation were present in the crystal structure at low-affinity binding sites, but their significance was dismissed based on ITC data (Cordes et al. 2011).
Although phylogenetically closely related to P. aeruginosa, P. putida is a soil bacterium with a saprotrophic lifestyle. Inactivation of the cya or crp genes in P. putida affected dipeptide and l-phenylalanine utilization, but not sugar catabolism, and resulted in minor physiological effects mainly related to envelope structures (Milanesio et al. 2011). CrpP. putida (Table 1) shares 82% sequence identity with Vfr. It stood out because of an unusually high affinity to cAMP with a Kd of 45.0 nM. Moreover, strong negative cooperativity of the two binding sites in the dimer was suggested because of a 2:1 stochiometric ratio in CrpP. putida–cAMP complexes, deduced from ITC analysis (Arce-Rodríguez et al. 2021). This high cAMP affinity of CrpP. putida is most probably related to very low cAMP production by P. putida (Milanesio et al. 2011). The latter was attributed to a combination of cAMP degradation by the phosphodiesterase Pde and poor translation of the adenylate cyclase-encoding cya mRNA, resulting in low cAMP synthesis (Arce-Rodríguez et al. 2021). Interestingly, CrpP. putida was found to be able to bind cGMP, however, with approx. 100-fold lower affinity than cAMP showing a Kd of 5.7 µM (Arce-Rodríguez et al. 2021). Yet, the cGMP effect on DNA binding or promoter activation was not investigated. Crystal structure analysis of CrpP. putida could provide insights into the molecular basis of the exceptionally high cAMP affinity or possibly into the molecular consequences of binding only one cAMP molecule per protein dimer.
α-Proteobacteria
The α-class of proteobacteria provided first examples of cGMP-activatable CRP proteins described in Rhodospirillum centenum and Sinorhizobium meliloti. Rhodospirillum centenum is a purple phototrophic bacterium with a complex lifestyle. In addition to planktonic and surface-attached, swarming cells, R. centenum exists as resting encysted forms (Favinger et al. 1989). Encystment is triggered by starvation and is associated with cGMP synthesis by these bacteria. Two guanylate cyclase genes, gcyB and gcyC, were found in an operon with cgrA encoding a CRP protein, which was identified as a regulator of encystment (Marden et al. 2011). A comparison of the transcriptomes of the wild type and a cgrA mutant showed altered expression of 258 genes, including numerous sigma factors and transcription factors, 131 of which were related to cyst development. However, among the 295 CgrA binding sites, identified in a ChIP-seq analysis, only 28 mapped to promoter regions of this differential transcriptome (Dong et al. 2015). The promoter region of the operon, comprising gcyB, gcyC, and cgrA, contains the perfect CAP-binding motif TGTGAactgctTCACA. However, among the CgrA-binding DNA sites identified by ChIP-seq, only a minor part included sequences resembling the CAP-binding motif (Dong et al. 2015, Roychowdhury et al. 2015).
CgrA is the first CRP cGMP receptor discovered in bacteria. Its NBD, although 17 residues longer, contains all the CAP cAMP binding signatures except for the RS motif replaced by RT (Table 1). ITC analysis and fluorescence anisotropy-based DNA-binding assays showed a substantially higher affinity of CgrA to cGMP than to cAMP, as well as a higher affinity of CgrA to the target DNA in presence of cGMP (Kd of 61 nM) than of cAMP (Kd of 1795 nM), and a two-site cGMP binding with negative cooperativity, opposed to one-site cAMP binding (Marden et al. 2011, Roychowdhury et al. 2015). A future structural analysis of CgrA–cGMP may provide insights into the molecular mechanism of preferential activation of a CRP protein by cGMP.
A special case: clr from S. meliloti
Sinorhizobium meliloti is a soil-dwelling bacterium, able to engage in a nitrogen fixing symbiosis with specific host plants. These bacteria invade a root hair via an infection thread, and induce plant cell proliferation in the root cortex, leading to the formation of root nodule, in which the bacteria fix nitrogen to the benefit of the plant (Jones et al. 2007). More than 10 CRP–FNR family transcription factors are encoded in the S. meliloti genome, including FNR-type regulators FixK1, FixK2, and NnrR and the CRP protein Clr (Table 1) (Fischer 1994, Meilhoc et al. 2010, Tian et al. 2012). Clr-mediated gene regulation was required for inhibition of secondary infections on the host plants that already carried nodules originating from primary infections (Tian et al. 2012). Transcriptome analysis identified 72 Clr-regulated genes, mostly related to cell envelope structures, motility, and secondary metabolism, many of which also showed expression changes upon cAMP overproduction (Krol et al. 2016, Zou et al. 2017).
The promoter regions of a fraction of the Clr-regulated genes contained sequences distantly resembling the CAP binding site. Electrophoretic mobility shift assays confirmed that DNA binding by Clr can be induced both by cAMP and cGMP, and promoter–reporter assays verified Clr-dependent transcription regulation by both cyclic nucleotides (Krol et al. 2016, Werel et al. 2023). As determined by ITC analysis, Clr exhibited moderate preference for cAMP over cGMP, reflected by a higher affinity to cAMP (Kd of 6–7 μM) than to cGMP (Kd ∼24 μM) (Werel et al. 2023). The affinity of Clr to its target DNA was similarly stimulated in presence of either cNMP (Kd of 6 µM and 4.5 µM for cAMP and cGMP, respectively) (Werel et al. 2023).
A genome-wide ChIP-seq-based search for Clr–cAMP and Clr–cGMP binding sites identified over 800 genomic regions, largely overlapping between the both cNMPs. Consistent with ChIP-seq studies on CAP and CgrA, showing abundant low-affinity binding sites across the genome (Dong et al. 2015, Latif et al. 2018), only a minor part of the identified Clr-bound DNA contained the Clr binding motif (Werel et al. 2023), arguing for a conservation of this property of CRP proteins.
X-ray crystallography of Clr–cAMP and Clr–cGMP complexes with target DNA revealed remarkably similar structures with both nucleotide ligands (Werel et al. 2023). cAMP in anti- and cGMP in syn-conformation were accommodated by the Clr amino acid residues, corresponding to the signature residues in CAP and other CRPs. The nucleobase of cAMP bonded with both T140 of the same and T141 of the adjacent protomer via the N6 atom of the purine ring and the amine of adenosine, in the same way as in CAP–cAMP (Passner et al. 2000). In contrast to CAP, where cGMP did not fit into the same molecular frame as cAMP (Seok et al. 2014), in Clr, cGMP guanosine successfully bonded with T140 via the carbonyl group whereas both interacting N-atoms were engaged with T141 (Fig. 2A) (Werel et al. 2023). Hydrogen–deuterium exchange analysis suggested rearrangements in the C-helix upon cNMP binding, with cAMP exerting a stronger effect than cGMP (Werel et al. 2023). These effects were similar to the coil-to helix transition in the corresponding region of the CAP C-helix, which resulted in activation of the DBD (Popovych et al. 2009, Sharma et al. 2009).
The first Clr-binding DNA sequence motif, defined as TGTT N8 AACA, was found in one strongly activated promoter (Tian et al. 2012). Later, the Clr binding consensus was redefined as HGTYHC N4 GRWACA, compiled from multiple experimentally verified binding sites (Krol et al. 2016). It overlaps with the CAP binding motif TGTGA N6 TCACA at the conserved G2T3 and A14C15A16 positions, however, the fourth position of the palindrome is not conserved. Instead, the variable spacer of Clr binding sites consists of only four nucleobases and is flanked by conserved palindromic C6 and G11 bases. The structure analysis of Clr–DNA complexes, which included TGTTAC N4 GTAACA as a binding sequence, revealed the importance of the latter two conserved positions. The DNA recognition F-helix motif is represented in CAP by R180ETVGR185 and in Clr by R195PKVNR200. Interaction with G2 and T3 of the DNA motif is mediated by the first arginine in both CAP and Clr (Fig. 2B) (Werel et al. 2023). In CAP, E181 and R185 interact with G4 and its cytosine base pair mate of the CAP binding sequence (Pyles and Lee 1996). In Clr, P196 cannot functionally replace CAP E181 and the conformation of R200 is incompatible with DNA binding (Werel et al. 2023). Instead, K197 bonds with the guanine base pair mate of C6 on the opposite DNA strand (Werel et al. 2023). Thus, the Clr F-helix motif evolved to recognize the DNA sites, in which the sixth position is more important and conserved than the fourth. Among the cNMP-binding CRP proteins studied experimentally to date, the proline at the second and lysine at the third position of the F-helix DNA binding motif are unique to Clr. In contrast, asparagine at the fifth position, which is involved in the interaction of Clr with T1, is also conserved in CRP proteins from Actinomyceta (Table 1).
Clr is well-conserved in the Sinorhizobium genus, including the unusual DNA-binding amino acid motif. Similar proteins are absent from the protein databases of the well-studied, closely related symbiotic nitrogen-fixing rhizobia Rhizobium leguminosarum and Rhizobium etli. However, individual isolates of Rhizobium sp. and Bradyrhizobium diazoefficiens possess Clr homologs. This mosaic phylogenetic distribution of this particular subtype of CRP proteins is consistent with the hypothesized inheritance of these proteins by both vertical and horizontal gene transfer (Soberon-Chavez et al. 2017).
Actinobacteria
Mycobacterium
As a major health hazard pathogen, Mycobacterium tuberculosis and closely related Mycobacterium bovis Bacille Calmette-Guérin (BCG), used in a tuberculosis vaccine, are of special research interest. Mycobacterium tuberculosis is capable of infecting human lung macrophages. It either causes acute lung infection or establishes life-long persistence in a dormant form that can be reactivated under favorable conditions (reviewed in Verma et al. 2022).
Mutation of the crp gene encoding CAP homolog Crp or Rv3676 (Table 1) reduced M. tuberculosis growth and pathogenicity (Rickman et al. 2005). An early transcriptome study comparing cultured wild type and crp-deficient bacteria identified only 16 differentially expressed genes (Rickman et al. 2005). In a more recent ChIP study, 121 Crp binding sites were identified upstream of the coding sequences, and 52 of these genes were found among the differentially expressed genes in the transcriptome analysis (Kahramanoglou et al. 2014). The Crp binding sites mostly clustered in the region between −150 and +50 around the transcription start sites, similar to the E. coli CAP binding sites (Shimada et al. 2011, Kahramanoglou et al. 2014). By combining the SELEX method and computational prediction of Rv3676 binding sites, a total of 73 promoter regions regulating 114 genes were identified as potential Rv3676 targets. Collectively, these genes were related to starvation, hypoxia, and other functions and include rpfA and whiB1, potentially involved in M. tuberculosis persistence and reactivation (Bai et al. 2005).
Rv3676 binding to DNA sequence similar to the CAP consensus could be confirmed, with this binding also resulting in DNA bending (Bai et al. 2005, Rickman et al. 2005). Activation of Rv3676 by cAMP was initially neglected (Bai et al. 2005, Rickman et al. 2005), then confirmed (Agarwal et al. 2006), and later reports suggested enhancement of Rv3676 DNA binding ability by cAMP (Reddy et al. 2009, Stapleton et al. 2010). This controversy may be attributable to differences in experimental design, protein preparation, and DNA sequences, but suggests a rather moderate impact of cAMP on DNA binding by Rv3676.
As determined by ITC, Rv3676 bound two molecules of cAMP per dimer with a Kd of 59 µM, indicating low affinity (Stapleton et al. 2010). Rv3676 was also able to bind cGMP, although the effect on the protein secondary structure estimated by trypsin proteolysis differed from that for cAMP (Stapleton et al. 2010). Intoxication of macrophages with cAMP is one of the important M. tuberculosis virulence traits, as elevated levels of cAMP can suppress innate immune functions, including phagosome maturation (reviewed in Dey and Bishai 2014). Therefore, the mycobacterial Crp might be evolutionary adapted to sense high cAMP levels by lowering the binding affinity for this ligand (Green et al. 2014). A recent study suggests that apo-Rv3637 forms high-order oligomers with DNA, through nonspecific interactions with DNA or through preformed protein–DNA complexes (Gárate et al. 2021). Binding of cAMP binding to Rv3676 reduced oligomerization and nonspecific binding but did not increase the DNA affinity, suggesting an allosteric regulation mechanism distinct from that of CAP–cAMP (Gárate et al. 2021).
The structures of apo- and cAMP-bound Rv3676 were determined by crystallography (Reddy et al. 2009, Kumar et al. 2010). The cAMP binding signatures (RS and TS signatures in CAP) are represented in Rv3676 NBD by arginine-threonine and threonine-asparagine (Table 1), which are functional for cAMP binding. The N-atoms of the cAMP purine ring are able to bond with threonine and asparagine residues, providing molecular contact with the adjacent protein monomer (Reddy et al. 2009, Kumar et al. 2010). In contrast to CAP, undergoing a dramatic conformational change upon cAMP binding, the folding and conformation of the DBD in apo-Rv3676 was differed only slightly from the cAMP-bound protein (Green et al. 2014), consistent with less strict requirement of cAMP for DNA binding.
Similar to E. coli CAP and V. cholera Crp, Rv3676 activity can be modulated by acetylation. This modification at the conserved lysine 193 inhibits DNA binding and transcription activation and occurs less frequently under Crp-activating conditions, such as the presence of cAMP, low pH, high temperature, or oxidative stress (Di et al. 2023). Rv3676 acetylation can be reversed by the action of a NAD+-dependent deacetylase, providing an additional level of controlling Rv3676-mediated regulation of the bacterial pathogenicity (Di et al. 2023). Interestingly, in M. tuberculosis and M. smegmatis, cAMP binds to the NBD of a GCN5-related N-acetyltransferase and promotes its activity in lysine acetylation of the universal stress protein USP (Nambi et al. 2010). This unexpected discovery suggests even broader connections between cAMP-dependent regulation and protein acetylation.
Corynebacterium
Corynebacteria constitute a diverse group of bacteria including soil saprophytes, gut commensals, and opportunistic pathogens. Corynebacterium glutamicum is widely used in biotechnology, in particular for industrial biosynthesis of amino acids (reviewed in Wendisch et al. 2016). The C. glutamicum CAP homolog was initially discovered as regulator of the glyoxylate bypass gene aceB, prompting the name GlxR (Table 1) (Kim et al. 2004). Unlike CAP, which has been reported mainly as transcription activator, GlxR mediates both gene activation and repression. Examples of gene regulation include repression of genes for isocitrate lyase and malate synthase, alcohol dehydrogenase, acetaldehyde dehydrogenase, glucose-specific permease of the sugar phosphotransferase system, and citrate uptake as well as activation of succinate dehydrogenase-encoding gene, anaerobic nitrate reductase operon narKGHJI, and phosphate uptake operon pstSCAB (Bussmann et al. 2009, Park et al. 2010, Nishimura et al. 2011, Panhorst et al. 2011, Toyoda et al. 2011, Subhadra and Lee 2013, Subhadra et al. 2015). In contrast to the E. coli paradigm, cAMP levels in C. glutamicum are higher in cells growing on glucose than in acetate-fed cells (Kim et al. 2004), and therefore, gene regulation by GlxR is triggered by different metabolic circumstances. However, the global regulatory nature of CAP is preserved in GlxR, which exerts a profound effect on carbon metabolism and energy conversion. ChIP-chip, ChIP-seq, and bioinformatic analyses of the GlxR regulon identified over 200 target genes, related to carbon metabolism, respiration, ATP synthesis, nitrogen assimilation, fatty acid biosynthesis as well as cell separation (Kohl et al. 2008, Toyoda et al. 2011, Jungwirth et al. 2013). Although the gene regulation in glxR mutant strains was similar to those of adenylate cyclase-deficient strains, implying GlxR activation by cAMP (Nishimura et al. 2011, Subhadra and Lee 2013), a ChIP-chip study showed that in a cAMP-deficient strain, the ability of GlxR to interact with its target sites was reduced, but not abolished (Toyoda et al. 2011).
GlxR shares 62 identical amino acids with CAP and it is much more similar to Rv3676, with which it shares 178 identical amino acids (Table 1). Crystal structure analysis of apo- and cAMP-bound GlxR revealed a picture similar to the M. tuberculosis homolog Rv3676, with little overall difference between the two structures in terms of folding and conformation of the DBD (Townsend et al. 2014). The length of the α-helices, corresponding to the C-and D-helices in CAP, increased by one residue each upon cAMP binding, in stark contrast to CAP, where the C-helix was prolonged by 11 and the D-helix reduced by six amino acids (Fig. 1). In the apo-form, the DBD showed a higher degree of flexibility and greater differences from Rv3676 than in the holo-form, and cAMP binding stabilized the DBD in a position optimal for DNA binding (Townsend et al. 2014). The cAMP binding signature motif is represented by the same residues as in Rv3676. Replacement of CAP S128 by N138 in GlxR allows for cAMP binding, as the carbonyl group is bonding with the N-atoms of the purine ring (Fig. 2A). The fact that S128 of CAP can form an additional H-bond to the backbone carbonyl oxygen of L124, which is not possible with an asparagine, was considered to be one of the reasons for the differences in the allosteric behavior of the two proteins (Townsend et al. 2014). The Kd in the GlxR–DNA binding assay decreased from 8.3 µM to 87 nM upon cAMP binding, which represents a 100-fold increase in affinity, similar to the cAMP effect on CAP (Townsend et al. 2014).
Gordonia and streptomyces
In Actinobacteria of industrial importance, CRP proteins were found to play a role in regulating biotechnologically important functions, which instigated their molecular characterization. Gordonia polyisoprenivorans from the order Corynebacteriales is a rubber-degrading bacterium in which the CRP protein CRPVH2 (Table 1) was proposed as a key regulator of poly(cis-1,4-isoprene) degradation (de Witt et al. 2020). CAP-like motifs were found in promoter regions of rubber degradation genes. cAMP-dependent CRPVH2 binding in the intergenic region between the gene encoding latex clearing protein and its repressor genes simultaneously activated the first gene and repressed the second (de Witt et al. 2020).
In Streptomyces coelicolor, the CRP protein Crp(Sco) (Table 1) was confirmed as a cAMP-binding protein by cAMP affinity chromatography (Derouaux et al. 2004). DNA-binding ability, however, was not confirmed in vitro. It is known that this protein is required for germination and its overproduction leads to increased production of antibiotics (Derouaux et al. 2004, Gao et al. 2012). In other Streptomyces, Crp homologs were characterized as global regulators of primary and secondary metabolism, able to activate production of antibiotics like monensin in S. cinnamonensis and daptomycin in S. roseosporus (Gao et al. 2012, Lin et al. 2020, Wu et al. 2021). Both CRPVH2 and Crp(Sco) contain conserved cAMP- and DNA-binding signatures, identical to the ones in Rv3676 and GlxR (Derouaux et al. 2004, de Witt et al. 2020).
Cyanobacteria
Among marine cyanobacteria, only a subset of species has CRP homologs (Xu and Su 2009). The most extensively studied CRP protein in this phylum is SYCRP1 from Synechocystis sp. PCC6803 (Table 1) (Ochoa de Alda and Houmard 2000, Yoshimura et al. 2000). Its predicted structure in complex with cAMP and DNA is very similar to that of CAP, and binding of SYCRP1-cAMP to a motif similar to TGTGA N6 TCACA was experimentally verified (Yoshimura et al. 2002, Omagari et al. 2008). Interestingly, the CAP cAMP binding C-helix TS motif is represented by leucine and asparagine in SYCRP1, suggesting a mode of cAMP binding different from CAP. A role in regulation of twitching motility and inorganic carbon metabolism was assigned to SYCRP1 (Song et al. 2018, Bantu et al. 2022).
SYCRP1 was found associated with the membrane under low CO2 conditions, when cAMP levels are low. Upon addition of cAMP to the growth medium or during a shift from low to high CO2 conditions, which promotes cAMP accumulation and SYCRP1-mediated gene regulation, SYCRP1 was released into the cytosol (Bantu et al. 2022). Considering that E. coli CAP and V. cholerae Crp were also found associated with the membrane, this phenomenon appears to be more ubiquitous than previously recognized. Although cGMP synthesis by a guanylate cyclase Cya2 was reported in Synechocystis sp. PCC6803 (Ochoa De Alda et al. 2000), a cGMP receptor protein is awaiting discovery. Since no report of a DNA-SYCRP1 binding assay in presence of cGMP was published, the possibility remains that SYCRP1 might fulfill the role of such a receptor.
Thermus thermophilus
Thermus thermophilus is an extreme thermophile from the Deinococcus–Thermus group, isolated from a Japanese hot spring. A total of four CRP–FNR family proteins are encoded in its genome. One of them, TTHB099 or TAP, was used in a crystallization experiment that provided valuable insight into Class II promoter activation (Feng et al. 2016). However, TAP has a much smaller NBD of only 82 amino acids and binds DNA in its apo-form. Interestingly, TAP did not require DNA binding for interaction with α-CTD, suggesting a “prerecruitment” mechanism, in which TAP first binds to RNAP and then to the DNA (Feng et al. 2016). Another T. thermophilus CRP, TTHA1437 (Table 1), was characterized as a cAMP-dependent transcription factor in an in vitro transcription assay (Shinkai et al. 2007). This protein was able to bind cAMP, although the CAP C-helix TS motif is represented by alanine and aspartate. Transcriptome analysis revealed a relatively small regulon of TTHA1437 comprising 22 genes, including a CRISPR–Cas system, a transcription regulator and other genes with nonrelated functions, and the associated predicted DNA binding sites are only remotely similar to the CAP binding motif (Shinkai et al. 2007).
Concluding remarks
The cAMP-dependent regulation of bacterial physiology by CRP proteins appears to be adapted specifically to the lifestyle of the particular bacterium. Only in closely related phyla, such as Vibrio or Yersinia, the E. coli paradigm of catabolite repression is maintained. In more distant bacterial phyla, the metabolic pathways, virulence and surface structures are often among the CRP–cNMP regulated processes, but the set of target genes can vary substantially, as well as the number of the genes in the regulon. In contrast to the diversity of physiological effects of CRP-dependent regulation and variability of the CRP amino acid sequences, the CAP DNA binding motif TGTGA(N6)TCACA with the highly conserved GT(N10)AC core motif shows a much higher degree of conservation even in phylogenetically distant bacterial species. Genome-wide ChIP analysis of CRP protein binding sites performed in several species yielded similar results, suggesting a broad range of DNA affinities and DNA site conservation. Several studies independently indicated membrane association of CRP proteins and their post-translational modification by lysine acetylation depending on the metabolic status. This emerging research field unveils a new layer of complexity in cAMP-dependent regulation.
The cAMP binding in a CRP protein dimer involves a hydrophobic binding pocket inside the NBD that ensures binding of the cAMP phosphosugar, and the nucleobase interactions with the dimerization C-helices, causing a conformational change in the whole dimer. Whereas the NBD signature is invariantly E(X)nRS or E(X)nRT in all cAMP/cGMP-binding CRP proteins studied so far, the C-helix signature motif is represented by threonine and serine or threonine and threonine in Gram-negatives and by threonine and asparagine, leucine and asparagine, or even alanine and aspartate in the Gram-positives. Although structural data of CRP proteins in their apo-form is limited to a few bacterial species, two distinct types of allosteric activation by cAMP binding can be defined: substantial movements and reorganization of DBD α-helices in CAP, or much smaller rearrangements with subtle conformation changes as observed for Rv3637 and GlxR.
Several CRP proteins from γ-proteobacteria and Gram-positives were reported to bind both cAMP and cGMP. However, cGMP binding to these proteins occurred with much lower affinity and did not induce the same structural rearrangements as cAMP. Clr and CgrA from α-proteobacteria are pioneer members of a new subtype of CRP proteins that can be activated by cGMP. The CAP DNA-binding F-helix site RETVGR is only partially conserved in all cAMP-binding CRP proteins. The first, second, and the last residues that directly interact with the target DNA are highly conserved, although the last position can be occupied by a lysine instead of arginine. In contrast, the glycine at the fifth position is conserved only within γ-proteobacterial CRP proteins, and representative proteins from other bacterial clades have an N, T, or S in this position. A special case is Clr, which contains the RPKVNR F-helix motif and uses the first, the third, and the fifth position to specifically interact with the DNA binding motif.
The largely diverse CRP proteins most likely have a common evolutionary origin and share some basic structural features at the protein level and concerning their DNA binding motifs. They have been exploited for gene regulation in different bacterial clades and their regulons vary widely. Equally diverse are the mechanisms of control of CRP activity. The initial focus on studying the molecular mechanisms underlying the control of E. coli CAP and its regulatory function left in the dark that in the bacterial domain CRPs have exploited almost every option for their condition-dependent regulation. This includes regulation of CRP levels and localization, as well as modulation of CRP activity by ligands, covalent modification, and interaction with other proteins. This has only recently become apparent from an increasing number of CRP studies in a broad variety of bacteria. We are only beginning to recognize the commonalities in this diversity and to understand the specific adaptations both mechanistically at the molecular level and in their biological function.
Acknowledgement
We are also grateful to Regine Hengge who established the priority program SPP 1879 ‘Nucleotide Second Messenger Signaling in Bacteria’ in Germany.
Contributor Information
Elizaveta Krol, Department of Biology, Philipps-Universität Marburg, Karl-von-Frisch-Straße 8, 35043 Marburg, Germany; Center for Synthetic Microbiology (SYNMIKRO), Philipps-Universität Marburg, Karl-von-Frisch-Str. 14, 35043 Marburg, Germany.
Laura Werel, Department of Chemistry, Philipps-Universität Marburg, Hans-Meerwein-Straße 4, 35032 Marburg, Germany.
Lars Oliver Essen, Department of Chemistry, Philipps-Universität Marburg, Hans-Meerwein-Straße 4, 35032 Marburg, Germany.
Anke Becker, Department of Biology, Philipps-Universität Marburg, Karl-von-Frisch-Straße 8, 35043 Marburg, Germany; Center for Synthetic Microbiology (SYNMIKRO), Philipps-Universität Marburg, Karl-von-Frisch-Str. 14, 35043 Marburg, Germany.
Conflict of interest
The authors declare no conflict of interest.
Funding
This work was supported by grants of the German Research Foundation (DFG) to LOE (ES 152/14) and AB (BE 2121/8) in the framework of the priority program SPP 1879.
References
- Agarwal N, Raghunand TR, Bishai WR. Regulation of the expression of whiB1 in Mycobacterium tuberculosis: role of cAMP receptor protein. Microbiology. 2006;152:2749–56.. 10.1099/mic.0.28924-0. [DOI] [PubMed] [Google Scholar]
- Aoyama JJ, Raina M, Zhong Aet al. Dual-function spot 42 RNA encodes a 15-amino acid protein that regulates the CRP transcription factor. Proc Natl Acad Sci USA. 2022;119:e2119866119. 10.1073/pnas.2119866119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arce-Rodríguez A, Nikel PI, Calles Bet al. Low CyaA expression and anti-cooperative binding of cAMP to CRP frames the scope of the cognate regulon of Pseudomonas putida. Environ Microbiol. 2021;23:1732–49.. 10.1111/1462-2920.15422. [DOI] [PubMed] [Google Scholar]
- Bai G, McCue LA, McDonough KA. Characterization of Mycobacterium tuberculosis Rv3676, a cyclic AMP receptor protein-like DNA binding protein. J Bacteriol. 2005;187:7795–804.. 10.1128/JB.187.22.7795-7804.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantu L, Chauhan S, Srikumar Aet al. A membrane-bound cAMP receptor protein, SyCRP1 mediates inorganic carbon response in Synechocystis sp. PCC 6803. Biochim Biophys Acta Gene Regul Mech. 2022;1865:194803. 10.1016/j.bbagrm.2022.194803. [DOI] [PubMed] [Google Scholar]
- Barber AM, Zhurkin VB, Adhya S. CRP-binding sites: evidence for two structural classes with 6-bp and 8-bp spacers. Gene. 1993;130:1–8.. 10.1016/0378-1119(93)90339-5. [DOI] [PubMed] [Google Scholar]
- Beisel CL, Storz G. The base-pairing RNA spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli. Mol Cell. 2011;41:286–97.. 10.1016/j.molcel.2010.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoff B, Yang H, Lawson CLet al. Structural basis of transcription activation: the CAP-alpha CTD-DNA complex. Science. 2002;297:1562–6.. 10.1126/science.1076376. [DOI] [PubMed] [Google Scholar]
- Berry A, Han K, Trouillon Jet al. cAMP and Vfr control exolysin expression and cytotoxicity of Pseudomonas aeruginosa taxonomic outliers. J Bacteriol. 2018;200:e00135–18.. 10.1128/JB.00135-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet M, Kurz M, Mesa Set al. The structure of Bradyrhizobium japonicum transcription factor FixK2 unveils sites of DNA binding and oxidation. J Biol Chem. 2013;288:14238–46.. 10.1074/jbc.M113.465484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borjigin M, Li H, Lanz NDet al. Structure-based hypothesis on the activation of the CO-sensing transcription factor CooA. Acta Crystallogr D Biol Crystallogr. 2007;63:282–7.. 10.1107/S0907444906051638. [DOI] [PubMed] [Google Scholar]
- Busby S, Ebright RH. Transcription activation by catabolite activator protein. J Mol Biol. 1999;293:199–213.. 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- Bussmann M, Emer D, Hasenbein Set al. Transcriptional control of the succinate dehydrogenase operon sdhCAB of Corynebacterium glutamicum by the cAMP-dependent regulator GlxR and the LuxR-type regulator RamA. J Biotechnol. 2009;143:173–82.. 10.1016/j.jbiotec.2009.06.025. [DOI] [PubMed] [Google Scholar]
- Cameron AD, Redfield RJ. Non-canonical CRP sites control competence regulons in Escherichia coli and many other gamma-proteobacteria. Nucleic Acids Res. 2006;34:6001–14.. 10.1093/nar/gkl734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PF, Tu SC, Hagag Net al. Isolation and characterization of a cyclic AMP receptor protein from luminous Vibrio harveyi cells. Arch Biochem Biophys. 1985;241:425–31.. 10.1016/0003-9861(85)90566-1. [DOI] [PubMed] [Google Scholar]
- Coggan KA, Wolfgang MC. Global regulatory pathways and cross-talk control Pseudomonas aeruginosa environmental lifestyle and virulence phenotype. Curr Issues Mol Biol. 2012;14:47–70. [PubMed] [Google Scholar]
- Colton DM, Stabb EV. Rethinking the roles of CRP, cAMP, and sugar-mediated global regulation in the Vibrionaceae. Curr Genet. 2016;62:39–45.. 10.1007/s00294-015-0508-8. [DOI] [PubMed] [Google Scholar]
- Cordes TJ, Worzalla GA, Ginster AMet al. Crystal structure of the Pseudomonas aeruginosa virulence factor regulator. J Bacteriol. 2011;193:4069–74.. 10.1128/JB.00666-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisan CV, Nichols HL, Wiesenfeld Set al. Glucose confers protection to Escherichia coli against contact killing by Vibrio cholerae. Sci Rep. 2021;11:2935. 10.1038/s41598-021-81813-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R, Écija-Conesa A, Gallego-Jara Jet al. An acetylatable lysine controls CRP function in E. coli. Mol Microbiol. 2018;107:116–31.. 10.1111/mmi.13874. [DOI] [PubMed] [Google Scholar]
- de Witt J, Oetermann S, Parise Met al. Global regulator of rubber degradation in Gordonia polyisoprenivorans VH2: identification and involvement in the regulation network. Appl Environ Microbiol. 2020;86:e00774–20.. 10.1128/AEM.00774-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dela Ahator S, Liu Y, Wang Jet al. The virulence factor regulator and quorum sensing regulate the type I-F CRISPR-cas mediated horizontal gene transfer in Pseudomonas aeruginosa. Front Microbiol. 2022;13:987656. 10.3389/fmicb.2022.987656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derouaux A, Dehareng D, Lecocq Eet al. Crp of Streptomyces coelicolor is the third transcription factor of the large CRP-FNR superfamily able to bind cAMP. Biochem Biophys Res Commun. 2004;325:983–90.. 10.1016/j.bbrc.2004.10.143. [DOI] [PubMed] [Google Scholar]
- Dey B, Bishai WR. Crosstalk between Mycobacterium tuberculosis and the host cell. Semin Immunol. 2014;26:486–96.. 10.1016/j.smim.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Y, Xu S, Chi Met al. Acetylation of cyclic AMP receptor protein by acetyl phosphate modulates mycobacterial virulence. Microbiol Spectr. 2023;11:e0400222. 10.1128/spectrum.04002-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Q, Ebright RH. DNA binding specificity and sequence of Xanthomonas campestris catabolite gene activator protein-like protein. J Bacteriol. 1992;174:5457–61.. 10.1128/jb.174.16.5457-5461.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Q, Fang M, Roychowdhury Set al. Mapping the CgrA regulon of Rhodospirillum centenum reveals a hierarchal network controlling Gram-negative cyst development. BMC Genomics. 2015;16:1066. 10.1186/s12864-015-2248-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman CJ, Schumacher MA, Bush MJet al. When is a transcription factor a NAP?. Curr Opin Microbiol. 2020;55:26–33.. 10.1016/j.mib.2020.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Écija-Conesa A, Gallego-Jara J, Lozano Terol Get al. An ideal spacing is required for the control of class II CRP-dependent promoters by the status of CRP K100. FEMS Microbiol Lett. 2020;367:fnaa164. 10.1093/femsle/fnaa164. [DOI] [PubMed] [Google Scholar]
- Emmer M, deCrombrugghe B, Pastan Iet al. Cyclic AMP receptor protein of E. coli: its role in the synthesis of inducible enzymes. Proc Natl Acad Sci USA. 1970;66:480–7.. 10.1073/pnas.66.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favinger J, Stadtwald R, Gest H. Rhodospirillum centenum, sp. nov., a thermotolerant cyst-forming anoxygenic photosynthetic bacterium. Antonie Van Leeuwenhoek. 1989;55:291–6.. 10.1007/BF00393857. [DOI] [PubMed] [Google Scholar]
- Feng Y, Zhang Y, Ebright RH. Structural basis of transcription activation. Science. 2016;352:1330–3.. 10.1126/science.aaf4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer HM. Genetic regulation of nitrogen fixation in rhizobia. Microbiol Rev. 1994;58:352–86.. 10.1128/mr.58.3.352-386.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Hindra, Mulder Det al. Crp is a global regulator of antibiotic production in Streptomyces. Mbio. 2012;3:e00407–12.. 10.1128/mBio.00407-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gárate F, Dokas S, Lanfranco MFet al. cAMP is an allosteric modulator of DNA-binding specificity in the cAMP receptor protein from Mycobacterium tuberculosis. J Biol Chem. 2021;296:100480. 10.1016/j.jbc.2021.100480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayán E, Cambré A, Michiels CWet al. RpoS-independent evolution reveals the importance of attenuated cAMP/CRP regulation in high hydrostatic pressure resistance acquisition in E. coli. Sci Rep. 2017;7:8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H, Jiang R. cAMP receptor protein -mediated resistance/tolerance in bacteria: mechanism and utilization in biotechnology. Appl Microbiol Biotechnol. 2015;99:4533–43.. 10.1007/s00253-015-6587-0. [DOI] [PubMed] [Google Scholar]
- Giardina G, Rinaldo S, Johnson KAet al. NO sensing in Pseudomonas aeruginosa: structure of the transcriptional regulator DNR. J Mol Biol. 2008;378:1002–15.. 10.1016/j.jmb.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Gibson JA, Gebhardt MJ, Santos RERSet al. Sequestration of a dual function DNA-binding protein by Vibrio cholerae CRP. Proc Natl Acad Sci USA. 2022;119:e2210115119. 10.1073/pnas.2210115119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger DC, Hurd D, Harrison Met al. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc Natl Acad Sci USA. 2005;102:17693–8.. 10.1073/pnas.0506687102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J, Stapleton MR, Smith LJet al. Cyclic-AMP and bacterial cyclic-AMP receptor proteins revisited: adaptation for different ecological niches. Curr Opin Microbiol. 2014;18:1–7.. 10.1016/j.mib.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heroven AK, Dersch P. Coregulation of host-adapted metabolism and virulence by pathogenic yersiniae. Front Cell Infect Microbiol. 2014;4: 146. 10.3389/fcimb.2014.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyde SAH, Frendorf PO, Lauritsen Iet al. Restoring global gene regulation through experimental evolution uncovers a NAP-like behavior of Crp/Cap. Mbio. 2021;12:e0202821. 10.1128/mBio.02028-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KM, Kobayashi H, Davies BWet al. How rhizobial symbionts invade plants: the Sinorhizobium–Medicago model. Nat Rev Microbiol. 2007;5:619–33., 10.1038/nrmicro1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungwirth B, Sala C, Kohl TAet al. High-resolution detection of DNA binding sites of the global transcriptional regulator GlxR in Corynebacterium glutamicum. Microbiology. 2013;159:12–22.. 10.1099/mic.0.062059-0. [DOI] [PubMed] [Google Scholar]
- Kahramanoglou C, Cortes T, Matange Net al. Genomic mapping of cAMP receptor protein (CRP Mt) in Mycobacterium tuberculosis: relation to transcriptional start sites and the role of CRP Mt as a transcription factor. Nucleic Acids Res. 2014;42:8320–9.. 10.1093/nar/gku548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim TH, Kim Yet al. Identification and characterization of glxR, a gene involved in regulation of glyoxylate bypass in Corynebacterium glutamicum. J Bacteriol. 2004;186:3453–60.. 10.1128/JB.186.11.3453-3460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl TA, Baumbach J, Jungwirth Bet al. The GlxR regulon of the amino acid producer Corynebacterium glutamicum: in silico and in vitro detection of DNA binding sites of a global transcription regulator. J Biotechnol. 2008;135:340–50.. 10.1016/j.jbiotec.2008.05.011. [DOI] [PubMed] [Google Scholar]
- Körner S, Sofia HJ, Zumft WG.. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol Rev. 2003;27:559–592. [DOI] [PubMed] [Google Scholar]
- Krol E, Klaner C, Gnau Pet al. Cyclic mononucleotide- and Clr-dependent gene regulation in Sinorhizobium meliloti. Microbiology. 2016;162:1840–56.. 10.1099/mic.0.000356. [DOI] [PubMed] [Google Scholar]
- Kumar P, Joshi DC, Akif Met al. Mapping conformational transitions in cyclic AMP receptor protein: crystal structure and normal-mode analysis of Mycobacterium tuberculosis apo-cAMP receptor protein. Biophys J. 2010;98:305–14.. 10.1016/j.bpj.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzilotta WN, Schuller DJ, Thorsteinsson MVet al. Structure of the CO sensing transcription activator CooA. Nat Struct Biol. 2000;7:876–80. [DOI] [PubMed] [Google Scholar]
- Latif H, Federowicz S, Ebrahim Aet al. ChIP-exo interrogation of crp, DNA, and RNAP holoenzyme interactions. PLoS ONE. 2018;13:e0197272. 10.1371/journal.pone.0197272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson CL, Swigon D, Murakami KSet al. Catabolite activator protein: DNA binding and transcription activation. Curr Opin Struct Biol. 2004;14:10–20.. 10.1016/j.sbi.2004.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leduc JL, Roberts GP. Cyclic di-GMP allosterically inhibits the CRP-like protein of Xanthomonas axonopodis pv. citri. J Bacteriol. 2009;191:7121–2.. 10.1128/JB.00845-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy C, Pike K, Heyes DJet al. Molecular basis of halorespiration control by CprK, a CRP-FNR type transcriptional regulator. Mol Microbiol. 2008;70:151–67.. 10.1111/j.1365-2958.2008.06399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Zhang Y, Wu JHet al. Regulatory patterns of Crp on monensin biosynthesis in Streptomyces cinnamonensis. Microorganisms. 2020;8:271. 10.3390/microorganisms8020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Hong C, Huang RKet al. Structural basis of bacterial transcription activation. Science. 2017;358:947–51.. 10.1126/science.aao1923. [DOI] [PubMed] [Google Scholar]
- Llácer JL, Espinosa J, Castells MAet al. Structural basis for the regulation of NtcA-dependent transcription by proteins PipX and PII. Proc Natl Acad Sci USA. 2010;107:15397–402.. 10.1073/pnas.1007015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux R, Shi W. A novel bacterial signalling system with a combination of a Ser. /Thr kinase cascade and a His/Asp two-component system. Mol Microbiol. 2005;58:345–8.. 10.1111/j.1365-2958.2005.04856.x. [DOI] [PubMed] [Google Scholar]
- Manneh-Roussel J, Haycocks JRJ, Magán Aet al. cAMP receptor protein controls Vibrio cholerae gene expression in response to host colonization. Mbio. 2018;9:e00966–18.. 10.1128/mBio.00966-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marden JN, Dong Q, Roychowdhury Set al. Cyclic GMP controls Rhodospirillum centenum cyst development. Mol Microbiol. 2011;79:600–15.. 10.1111/j.1365-2958.2010.07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay DB, Steitz TA. Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA. Nature. 1981;290:744–9.. 10.1038/290744a0. [DOI] [PubMed] [Google Scholar]
- Meilhoc E, Cam Y, Skapski Aet al. The response to nitric oxide of the nitrogen-fixing symbiont Sinorhizobium meliloti. Mol Plant Microbe Interact. 2010;23:748–59.. 10.1094/MPMI-23-6-0748. [DOI] [PubMed] [Google Scholar]
- Mettert EL, Kiley PJ. Reassessing the structure and function relationship of the O2 sensing transcription factor FNR. Antioxid Redox Signal. 2018;29:1830–40.. 10.1089/ars.2017.7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milanesio P, Arce-Rodríguez A, Muñoz Aet al. Regulatory exaptation of the catabolite repression protein -cAMP system in Pseudomonas putida. Environ Microbiol. 2011;13:324–39.. 10.1111/j.1462-2920.2010.02331.x. [DOI] [PubMed] [Google Scholar]
- Mok WW, Orman MA, Brynildsen MP. Impacts of global transcriptional regulators on persister metabolism. Antimicrob Agents Chemother. 2015;59:2713–9.. 10.1128/AAC.04908-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Quiroz RC, Silva-Valenzuela C, Brewster Jet al. Cyclic AMP regulates bacterial persistence through repression of the oxidative stress response and SOS-dependent DNA repair in uropathogenic Escherichia coli. Mbio. 2018;9:e02144–17.. 10.1128/mBio.02144-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M, Ogasawara H, Shimada Tet al. Involvement of cAMP-CRP in transcription activation and repression of the pck gene encoding PEP carboxykinase, the key enzyme of gluconeogenesis. FEMS Microbiol Lett. 2014;355:93–9.. 10.1111/1574-6968.12466. [DOI] [PubMed] [Google Scholar]
- Nambi S, Basu N, Visweswariah SS. cAMP-regulated protein lysine acetylases in mycobacteria. J Biol Chem. 2010;285:24313–23.. 10.1074/jbc.M110.118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Teramoto H, Toyoda Ket al. Regulation of the nitrate reductase operon narKGHJI by the cAMP-dependent regulator GlxR in Corynebacterium glutamicum. Microbiology. 2011;157:21–28.. 10.1099/mic.0.044552-0. [DOI] [PubMed] [Google Scholar]
- Nosho K, Fukushima H, Asai Tet al. cAMP-CRP acts as a key regulator for the viable but non-culturable state in Escherichia coli. Microbiology. 2018;164:410–9.. 10.1099/mic.0.000618. [DOI] [PubMed] [Google Scholar]
- Notley-McRobb L, Death A, Ferenci T. The relationship between external glucose concentration and cAMP levels inside Escherichia coli: implications for models of phosphotransferase-mediated regulation of adenylate cyclase. Microbiology. 1997;143:1909–18.. 10.1099/00221287-143-6-1909. [DOI] [PubMed] [Google Scholar]
- Ochoa de Alda JAG, Ajlani Get al. Synechocystis strain PCC 6803 cya2, a prokaryotic gene that encodes a guanylyl cyclase. J Bacteriol. 2000;182:3839–42.. 10.1128/JB.182.13.3839-3842.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa de Alda JAG, Houmard J. Genomic survey of cAMP and cGMP signalling components in the cyanobacterium Synechocystis PCC 6803. Microbiology. 2000;146:3183–94.. 10.1099/00221287-146-12-3183. [DOI] [PubMed] [Google Scholar]
- Omagari K, Yoshimura H, Suzuki Tet al. DeltaG-based prediction and experimental confirmation of SYCRP1-binding sites on the Synechocystis genome. FEBS J. 2008;275:4786–95.. 10.1111/j.1742-4658.2008.06618.x. [DOI] [PubMed] [Google Scholar]
- Panhorst M, Sorger-Herrmann U, Wendisch VF. The pstSCAB operon for phosphate uptake is regulated by the global regulator GlxR in Corynebacterium glutamicum. J Biotechnol. 2011;154:149–55.. 10.1016/j.jbiotec.2010.07.015. [DOI] [PubMed] [Google Scholar]
- Park SY, Moon MW, Subhadra Bet al. Functional characterization of the glxR deletion mutant of Corynebacterium glutamicum ATCC 13032: involvement of GlxR in acetate metabolism and carbon catabolite repression. FEMS Microbiol Lett. 2010;304:107–15.. 10.1111/j.1574-6968.2009.01884.x. [DOI] [PubMed] [Google Scholar]
- Parkinson G, Wilson C, Gunasekera Aet al. Structure of the CAP-DNA complex at 2.5 angstroms resolution: a complete picture of the protein-DNA interface. J Mol Biol. 1996;260:395–408.. 10.1006/jmbi.1996.0409. [DOI] [PubMed] [Google Scholar]
- Passner JM, Schultz SC, Steitz TA. Modeling the cAMP-induced allosteric transition using the crystal structure of CAP-cAMP at 2.1 A resolution. J Mol Biol. 2000;304:847–59.. 10.1006/jmbi.2000.4231. [DOI] [PubMed] [Google Scholar]
- Passner JM, Steitz TA. The structure of a CAP-DNA complex having two cAMP molecules bound to each monomer. Proc Natl Acad Sci USA. 1997;94:2843–7.. 10.1073/pnas.94.7.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovych N, Tzeng SR, Tonelli Met al. Structural basis for cAMP-mediated allosteric control of the catabolite activator protein. Proc Natl Acad Sci USA. 2009;106:6927–32.. 10.1073/pnas.0900595106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyles EA, Lee JC. Mode of selectivity in cyclic AMP receptor protein-dependent promoters in Escherichia coli. Biochemistry. 1996;35:1162–72.. 10.1021/bi952187q. [DOI] [PubMed] [Google Scholar]
- Reddy MCM, Palaninathan SK, Bruning JBet al. Structural insights into the mechanism of the allosteric transitions of Mycobacterium tuberculosis cAMP receptor protein. J Biol Chem. 2009;284:36581–91.. 10.1074/jbc.M109.041343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickman L, Scott C, Hunt DMet al. A member of the cAMP receptor protein family of transcription regulators in Mycobacterium tuberculosis is required for virulence in mice and controls transcription of the rpfA gene coding for a resuscitation promoting factor. Mol Microbiol. 2005;56:1274–86.. 10.1111/j.1365-2958.2005.04609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritzert JT, Minasov G, Embry Ret al. The cyclic AMP receptor protein regulates quorum sensing and global gene expression in Yersinia pestis during planktonic growth and growth in biofilms. Mbio. 2019;10:e02613–19.. 10.1128/mBio.02613-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro C, Cashel M, Fernández-Coll L. The secondary messenger ppGpp interferes with cAMP-CRP regulon by promoting CRP acetylation in Escherichia coli. PLoS ONE. 2021;16:e0259067. 10.1371/journal.pone.0259067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M, Son B, Kroos Det al. Transcription factor MrpC binds to promoter regions of hundreds of developmentally-regulated genes in Myxococcus xanthus. BMC Genomics. 2014;15:1123. 10.1186/1471-2164-15-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychowdhury S, Dong Q, Bauer CE. DNA-binding properties of a cGMP-binding CRP homologue that controls development of metabolically dormant cysts of Rhodospirillum centenum. Microbiology. 2015;161:2256–64.. 10.1099/mic.0.000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH Jr. Multiple mechanisms controlling carbon metabolism in bacteria. Biotechnol Bioeng. 1998;58:170–4.. . [DOI] [PubMed] [Google Scholar]
- Seok SH, Im H, Won HSet al. Structures of inactive CRP species reveal the atomic details of the allosteric transition that discriminates cyclic nucleotide second messengers. Acta Crystallogr D Biol Crystallogr. 2014;70:1726–42.. 10.1107/S139900471400724X. [DOI] [PubMed] [Google Scholar]
- Sharma H, Yu S, Kong Jet al. Structure of apo-CAP reveals that large conformational changes are necessary for DNA binding. Proc Natl Acad Sci USA. 2009;106:16604–9.. 10.1073/pnas.0908380106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Jiang Y, Deng Yet al. Visualization of two architectures in class-II CAP-dependent transcription activation. PLoS Biol. 2020;18:e3000706. 10.1371/journal.pbio.3000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Fujita N, Yamamoto Ket al. Novel roles of cAMP receptor protein in regulation of transport and metabolism of carbon sources. PLoS ONE. 2011;6:e20081. 10.1371/journal.pone.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai A, Kira S, Nakagawa Net al. Transcription activation mediated by a cyclic AMP receptor protein from Thermus thermophilus HB8. J Bacteriol. 2007;189:3891–901.. 10.1128/JB.01739-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soberón-Chávez G, Alcaraz LD, Morales Eet al. The transcriptional regulators of the CRP family regulate different essential bacterial functions and can be inherited vertically and horizontally. Front Microbiol. 2017;8:959. 10.3389/fmicb.2017.00959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokoloski JE, Godfrey SA, Dombrowski SEet al. Prevalence of syn nucleobases in the active sites of functional RNAs. RNA. 2011;17:1775–87.. 10.1261/rna.2759911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WY, Zang SS, Li ZKet al. Sycrp2 Is essential for twitching motility in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol. 2018;200:e00436–18.. 10.1128/JB.00436-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton M, Haq I, Hunt DMet al. Mycobacterium tuberculosis cAMP receptor protein differs from the Escherichia coli paradigm in its cAMP binding and DNA binding properties and transcription activation properties. J Biol Chem. 2010;285:7016–27.. 10.1074/jbc.M109.047720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subhadra B, Lee JK. Elucidation of the regulation of ethanol catabolic genes and ptsG using a glxR and adenylate cyclase gene deletion mutants of Corynebacterium glutamicum ATCC 13032. J Microbiol Biotechnol. 2013;23:1683–90.. 10.4014/jmb.1310.10031. [DOI] [PubMed] [Google Scholar]
- Subhadra B, Ray D, Han JKet al. Identification of the regulators binding to the upstream region of glxR in Corynebacterium glutamicum. J Microbiol Biotechnol. 2015;25:1216–26.. 10.4014/jmb.1502.02053. [DOI] [PubMed] [Google Scholar]
- Suh SJ, Runyen-Janecky LJ, Maleniak TCet al. Effect of vfr mutation on global gene expression and catabolite repression control of Pseudomonas aeruginosa. Microbiology. 2002;148:1561–9.. 10.1099/00221287-148-5-1561. [DOI] [PubMed] [Google Scholar]
- Tian CF, Garnerone AM, Mathieu-Demazière Cet al. Plant-activated bacterial receptor adenylate cyclases modulate epidermal infection in the Sinorhizobium meliloti–Medicago symbiosis. Proc Natl Acad Sci USA. 2012;109:6751–6.. 10.1073/pnas.1120260109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend PD, Jungwirth B, Pojer Fet al. The crystal structures of apo and cAMP-bound GlxR from Corynebacterium glutamicum reveal structural and dynamic changes upon cAMP binding in CRP/FNR family transcription factors. PLoS ONE. 2014;9:e113265. 10.1371/journal.pone.0113265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda K, Teramoto H, Inui Met al. Genome-wide identification of in vivo binding sites of GlxR, a cyclic AMP receptor protein-type regulator in Corynebacterium glutamicum. J Bacteriol. 2011;193:4123–33.. 10.1128/JB.00384-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppal S, Jawali N. The cyclic AMP receptor protein regulates mqsRA, coding for the bacterial toxin-antitoxin gene pair, in Escherichia coli. Res Microbiol. 2016;167:58–62.. 10.1016/j.resmic.2015.09.001. [DOI] [PubMed] [Google Scholar]
- Verma A, Ghoshal A, Dwivedi VPet al. Tuberculosis: the success tale of less explored dormant Mycobacterium tuberculosis. Front Cell Infect Microbiol. 2022;12:1079569. 10.3389/fcimb.2022.1079569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendisch VF, Jorge JMP, Pérez-García Fet al. Updates on industrial production of amino acids using Corynebacterium glutamicum. World J Microbiol Biotechnol. 2016;32:105. 10.1007/s11274-016-2060-1. [DOI] [PubMed] [Google Scholar]
- Werel L, Farmani N, Krol Eet al. Structural basis of dual specificity of Sinorhizobium meliloti clr, a cAMP and cGMP receptor protein. mBio. 2023;14:e0302822. 10.1128/mbio.03028-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SE, Sample AK, Runyen-Janecky LJ. The vfr gene product, required for Pseudomonas aeruginosa exotoxin A and protease production, belongs to the cyclic AMP receptor protein family. J Bacteriol. 1994;176:7532–42.. 10.1128/jb.176.24.7532-7542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang MC, Lee VT, Gilmore MEet al. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev Cell. 2003;4:253–63.. 10.1016/S1534-5807(03)00019-4. [DOI] [PubMed] [Google Scholar]
- Wu J, Chen D, Wu Jet al. Comparative transcriptome analysis demonstrates the positive effect of the cyclic AMP receptor protein Crp on daptomycin biosynthesis in Streptomyces roseosporus. Front Bioeng Biotechnol. 2021;9:618029. 10.3389/fbioe.2021.618029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N, Zhang Y, Zhang Set al. Polynucleotide phosphorylase mediates a new mechanism of persister formation in Escherichia coli. Microbiol Spectr. 2022;8:e0154622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu GG, Han S, Huo CMet al. Signaling specificity in the c-di-GMP-dependent network regulating antibiotic synthesis in Lysobacter. Nucleic Acids Res. 2018;46:9276–88.. 10.1093/nar/gky803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Lin L, Shen Det al. Clp is a “busy” transcription factor in the bacterial warrior, Lysobacter enzymogenes. Comput Struct Biotechnol J. 2021;19:3564–72.. 10.1016/j.csbj.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Su Z. Computational prediction of cAMP receptor protein binding sites in cyanobacterial genomes. Bmc Genomics. 2009;10:23. 10.1186/1471-2164-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CD, Huang HY, Shrestha Set al. Large-scale functional analysis of CRP-mediated feed-forward loops. Int J Mol Sci. 2018;19:2335. 10.3390/ijms19082335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura H, Hisabori T, Yanagisawa Set al. Identification and characterization of a novel cAMP receptor protein in the cyanobacterium Synechocystis sp. PCC 6803. J Biol Chem. 2000;275:6241–5.. 10.1074/jbc.275.9.6241. [DOI] [PubMed] [Google Scholar]
- Yoshimura H, Yanagisawa S, Kanehisa Met al. Screening for the target gene of cyanobacterial cAMP receptor protein SYCRP1. Mol Microbiol. 2002;43:843–53.. 10.1046/j.1365-2958.2002.02790.x. [DOI] [PubMed] [Google Scholar]
- Zeng J, Hong Y, Zhao Net al. A broadly applicable, stress-mediated bacterial death pathway regulated by the phosphotransferase system and the cAMP-Crp cascade. Proc Natl Acad Sci USA. 2022;119:e2118566119. 10.1073/pnas.2118566119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Gastebois A, Mathieu-Demazière Cet al. Transcriptomic insight in the control of legume root secondary infection by the Sinorhizobium meliloti transcriptional regulator clr. Front Microbiol. 2017;8:1236. 10.3389/fmicb.2017.01236. [DOI] [PMC free article] [PubMed] [Google Scholar]