Abstract
Partition cassettes, essential for the segregational stability of low-copy-number bacterial plasmids, typically encode two autoregulated proteins and an adjacent cis-acting centromere analog to which one or perhaps both proteins bind. The diminutive partition region of pTAR of Agrobacterium spp. was reported to be exceptional, encoding only a single protein, ParA (D. R. Gallie and C. I. Kado, J. Mol. Biol. 193:465–478, 1987). However, resequencing of the region revealed two small downstream genes, parB and orf-84, of which only parB was found to be essential for partitioning in A. tumefaciens. Purified ParA exhibited a weak ATPase activity that was modestly increased by nonspecific DNA. ParB bound in vitro to repeated sequences present in a region, parS, that possesses centromere and operator functions and within which we identified the primary transcription start site by primer extension. In certain respects the Par proteins behave normally in the foreign host Escherichia coli. In E. coli, as in A. tumefaciens, ParB repressed the partition operon; ParA, inactive alone, augmented this repression. Functional similarities between the partition system of pTAR and those of other plasmids and bacteria are prominent, despite differences in size, organization, and amino acid sequence.
Partition (par) operons are a characteristic feature of low-copy-number bacterial plasmids. They confer segregational stability and may do so by a factor of 100 or more. The most thoroughly studied par operons, those of plasmids P1, F, R1, and NR1, are simple in structure and exhibit a number of similarities. Each consists of an autogenously regulated gene pair (10, 19, 22, 32, 39) and a centromere analog that is either upstream (for R1 [7] and NR1 [38]) or downstream (for P1 [1] and F [34]). The first gene encodes an ATPase with recognizable motifs (4, 25, 33). The second gene encodes a protein that can bind tightly to plasmid-specific iterated sequences within the cognate centromere analog (3, 5, 11, 12, 32, 39). Homologs of plasmid partition genes with apparently analogous function have been reported in Bacillus (17, 21, 26, 37) and in Caulobacter (31) spp., as well as in members of an increasing number of bacterial genera.
The par region of Agrobacterium tumefaciens plasmid pTAR (a 44-kb plasmid that confers the ability to catabolize tartaric acid [16]) is relatively small. It has been reported to be contained within a 1,259-bp segment of pTAR DNA and to encode only a single partition protein, ParA (14). Features of the amino acid sequence suggested that this ParA belongs to the family of ATPases to which most other partition ATPases (demonstrated or putative) have been assigned (25, 33).
The supposed simplicity of the pTAR partition system is inconsistent with evidence obtained by Gallie and Kado that insertions distal to parA reduced expression of the par operon and could reduce the efficiency of plasmid partition (14). As suspected, pTAR does encode a second partition protein, which we show here to be unusually small but otherwise unexceptional in its characteristics.
MATERIALS AND METHODS
Bacteria and plasmids.
Bacteria are listed in Table 1. Escherichia coli K-12 strains MC1061 and DH5α served as hosts for cloning and plasmid propagation. To allow our results to be interpreted in relation to those of earlier studies, we confined our experiments to the LBA4301 strain of A. tumefaciens, obtained from the Kado laboratory. As received, the strain was found to be sensitive to rifampin rather than rifampin resistant as originally described (15).
TABLE 1.
Bacterial strains
| Strain | Genotype and/or characteristics | Source or reference |
|---|---|---|
| A. tumefaciens LBA4301 (BR6394) | rec (UVs) rpoB (Rifs) | 15 |
| E. coli BL21(DE3) (BR7591) | F−ompT hsdSB(rB− mB−) gal dcm | Novagen |
| E. coli BR5806 (M510) | pcnB380 zad::Tn10 (destabilizes pBR322) | J. Beckwith via D. Sledjeski (27) |
| E. coli BR6326 | MC1061 (parSpTAR-lacZ)attλ (transcriptional fusion to lacZ) | This work |
| E. coli DH5α (BR2966) | F−endA1 hsdR17(rK− mK+) supE44 thy1 recA1 gyrA (Nalr) relA1 Δ(lacIZYA-argF) U169 deoR [φ80dlacΔ(lacZ)M15] | New England Biolabs |
| E. coli MC1061 (BR6545) | F−araD139 Δ(ara-leu)7696 galE15 galK16 Δ(lac)X74 rpsL (Str) hsdR2(rK− mK−) mcrA mcrB1 | Lab collection |
Parental plasmids are listed in Table 2. Restriction enzymes were purchased from New England Biolabs or Boehringer Mannheim. The shuttle vector pUCD2000 and its par+ derivative, pUCD2001, carrying the partition region of pTAR were kindly supplied by Clarence Kado. The in-frame deletion within parA (designated ΔparA1) was made as an NcoI-generated deletion in pUCD550 (see Fig. 1 for the locations of NcoI sites) to generate pUCD550ΔNcoI. In pUCD55 the parA and parB genes are flanked by an EcoRV site (as in Fig. 1) and a BamHI site artificially introduced at position 1259 of the sequence in Fig. 1. The in-frame deletion within parA was introduced into pUCD2001 by substitution of the EcoRV-BamHI fragment (containing wild-type parA and parB genes) by the corresponding fragment from pUCD550ΔNcoI. The resulting plasmid does not confer tetracycline resistance. pUCD2000parS+ parA+ was constructed by excision of the BglII-BamHI fragment from the parental plasmid pUCD2001. Deletion of the par structural genes from pUCD2001 to yield pUCD2000parS+ was accomplished similarly, by the excision of a ClaI fragment (from the site at position 247 in Fig. 1 to the ClaI site in the promoter of the pBR322 tetracycline resistance gene).
TABLE 2.
Plasmids
| Plasmid | Source of replication origin | Antibiotic resistancea | Other relevant feature(s) | Source or reference |
|---|---|---|---|---|
| pBAD24Km | pBR322 | Km | Vector for cloning under Para | J. Beckwith (18) |
| PCR-Blunt | pUC | Km Ze | Vector for PCR product cloning under Plac | Invitrogen |
| pET-23a (+) | pBR322 | Ap | T7 expression vector for adding C-terminal His6 tag | Novagen |
| pLDR11 | pBR322 | Ap | attλ integration vector | W. Messer (9) |
| pOAR11 | pACYC184 | Cm | lacIq Ptac MCS | O. A. Rodionov |
| pOAR24 | pBR322 | Cm Tc | lacZ under PrepA of P1 | O. A. Rodionov |
| pUCD105 | PBR322 and pTAR | Sp Ap Cm | ori and par of pTAR | 14 |
| pUCD550 | pUC4 | Ap | par of pTAR | 15 |
| pUCD2000 | pBR322 and pTAR | Ap Tc Km | Par− shuttle vector; not stable in A. tumefaciens | 15 |
| pUCD2001 | pBR322 and pTAR | Ap Tc Km | par+ shuttle vector; stable in A. tumefaciens | 15 |
Ap, ampicillin; Cm, chloramphenicol; Km, kanamycin; Sp, spectinomycin; Tc, tetracycline; Ze, zeocin.
FIG. 1.
Nucleotide sequence of the pTAR partition region and deduced sequences of amino acid residues of the encoded proteins. The template for sequencing was pUCD105 (14). The parS repeated sequences are indicated, and putative −35 and −10 sites of RNA polymerase binding are labeled explicitly, as are presumptive ribosome binding sites (rbs) and motifs A, A′, and B in parA, which are characteristic of partition ATPases (25). An asterisk marks the start of the mRNA.
A parS-lacZ fusion was constructed by replacing the parA and parB genes of pUCD2001 (excised by ClaI) with a lacZ gene as a BamHI-SmaI fragment of plasmid pOAR24. The fusion was excised as an EcoRI-BamHI fragment, blunt ended, and inserted into the SmaI site of pLDR11, from which it could be inserted into the λ attachment site (attλ) of E. coli by the method of Diederich et al. (9). The relevant feature of pOAR24 (an intermediate in the construction of indicator strains for gene silencing [36]) is the presence of a lacZ gene flanked by BamHI and SmaI sites which originally derived from pPP112 (35).
For experiments with E. coli, we used the compatible expression vectors pOAR11 (a replicon of p15A) and pBAD24-Km (a replicon of pBR322 [18]). Vector pOAR11, constructed by Oleg Rodionov of this laboratory, was derived from pACYC184 by substitution of its HindIII-SalI fragment by the SspI fragment of pMMB67HE (13) containing lacIq, Ptac, and a multicloning site (MCS). The parAB genes, having been excised from pUCD550 as an EcoRV-BamHI fragment and blunt ended, were inserted into the SalI site of pOAR11 to generate pOAR11parAB. A BglII-KpnI fragment was deleted from pOAR11parAB to generate pOAR11parA; similarly, a PstI-StuI fragment was deleted to generate pOAR11parB. The vector pBAD24-Km is a derivative of pBAD24 (18). It was obtained by insertion of the Km gene of pUC4K (Pharmacia) into the ScaI site (Ap gene) of pBAD24. The DNA sequences of parA, parB, and parA-parB were prepared from plasmid pUCD2001 by PCR with appropriate primers and inserted into the EcoRI and SphI sites of pBAD24-Km.
For the purification of the Par proteins as their His6-tagged derivatives, NdeI-XhoI DNA fragments bearing parA or parB were amplified by PCR from a pUCD2001 template using the high-fidelity Pfu DNA polymerase (Stratagene). PCR primers were custom synthesized (by BioServe Biotechnologies or Genosys Biotechnologies). The fragments were inserted into the expression vector pET-23a(+) (Novagen) for the production of Par proteins with His6 tags at the C termini.
Microbiological methods.
Unless otherwise specified, bacteria were grown at 30°C in Luria broth (LB) with vigorous aeration or on LB agar plates (29). Media were appropriately supplemented with antibiotics: 100 μg of ampicillin/ml, 50 μg of carbenicillin/ml, 20 μg of chloramphenicol/ml, 30 μg of kanamycin/ml, and 15 μg of tetracycline/ml. Measurements of plasmid retention in A. tumefaciens were performed by replica plating on appropriate antibiotic-containing plates as described previously (14) except that cultures being sampled were maintained continuously in the logarithmic-growth phase by serial dilution. In sampling cultures of A. tumefaciens, we took advantage of the ability of this organism to survive in deionized water at 4°C for several days with no detectable loss of viability. Plasmid retention was measured as described previously (24).
Protein purification.
His6-tagged Par proteins were purified from 1 liter of IPTG (isopropyl-β-d-thiogalactopyranoside)-induced cultures of BL21(DE3) grown at 30°C and carrying the par genes cloned into pET-23a(+). Plasmid amplification and protein purification by elution with imidazole from Ni2+-nitrilotriacetic acid (NTA) His-Bind resin (Qiagen) were performed essentially as recommended by the manufacturer.
Immunochemical assays of the partition proteins.
Rabbit antibodies were raised against C-terminally His6-tagged derivatives of ParA and ParB (BAbCO). Immunoblotting procedures were performed with the ECL protein immunoblot analysis system (Amersham) as recommended by the manufacturer.
Enzyme assays.
Beta-galactosidase was assayed by the method of Miller (29), using sodium dodecyl sulfate (SDS) and chloroform to permeabilize the cells. ATPase was assayed by the release of 32PO4 from [γ-32P]ATP (by a modification of method B of Manne et al. [28]) in reaction mixtures prepared with various salts. The results that we report here were obtained by following the protocol of Davis et al. (8), in which NaCl is present at 150 mM. ParA or ParB (800 ng) was added to 100 μl of a reaction mixture consisting of 1 μl (ca. 10 μCi) of [γ-32P]ATP (3,000 Ci/mmol; Amersham), 30 mM Tris acetate (pH 7.5), 150 mM NaCl, 10 mM Mg acetate, 1 mM dithiothreitol (DTT), 0.1 mg of bovine serum albumin/ml, and 0.1 mM ATP with or without 1 μg of plasmid DNA, as indicated. The mixture was incubated at 30°C, and aliquots (7 μl) were withdrawn at intervals and quickly frozen in dry ice. After all the samples had been collected, 5 μl of each was spotted along the wide edge of a 5-by-10-cm polyethyleneimine (PEI)-cellulose plate (Sigma) and air dried. The plate was placed on its edge in a chromatography chamber containing running buffer (1 M formic acid–0.5 M LiCl) and run until the buffer front reached two-thirds the height of the plate. After air drying, the plate was autoradiographed for 15 to 20 min on Kodak Biomax film. For quantitation, bands were imaged on a Fujix BAS 2000 phosphorimager and analyzed with MacBas computer software (Fuji). The protocol of Jensen and Gerdes (23) in which 50 mM KCl is present, or is replaced by either 50 mM NaCl, 50 mM NH4Cl, or 50 mM K-glutamate, did not further increase the specific activity over background (data not shown).
DNA sequence and protein homology analyses.
The DNA sequence of the GC-rich region of pTAR downstream of parA was obtained with the Thermosequenase radiolabeled terminator cycle sequencing kit of Amersham Life Science and the SequiTherm Excel DNA sequencing kit (Epicenter Technologies). The sequences of cloned genes were verified with the fmol DNA sequencing system of Promega.
DNA binding assays.
Radiolabeled parS double-stranded DNAs, one comprising the sequence shown in Fig. 3B without the ATG-3′ (224 bp) and one comprising that sequence plus an additional 46 bp upstream of the par operon, were prepared, respectively, from pUCD2001 by Lofstrand Laboratories Ltd., Gaithersburg, Md., and from pUCD550 by K.K. as PCR templates. The corresponding 5′-end-labeled forward primers were 5′-GGCATATCCGATTTGATGCG-3′ and 5′-GAATTCCCCGCATTGAAAATTAAC-3′. The corresponding reverse primers were 5′-ATGTCAATTCTCCGGTTAAAT-3′ and 5′-ATGTCATTCTCCGGTTAAATCGAT-3′. Radioactive phosphorus was incorporated using [γ-32P]ATP with T4 polynucleotide kinase (New England Biolabs). Electrophoretic mobility shift assays were performed essentially as described previously (6). Dilutions of the His6-tagged proteins were mixed with 1 nM radiolabeled DNA fragment, incubated for at least 20 min at 25°C in binding buffer (20 mM Tris-HCl [pH 7.6], 150 mM NaCl, 2% glycerol, and 100 μg of bovine serum albumin/ml). Where indicated, ATP was added at a final concentration of 1 mM. Samples were loaded into the wells of a 1- by 150- by 150-mm 5% polyacrylamide gel in 0.5× Tris-borate-EDTA, beginning 10 min after the current was turned on, and were subjected to electrophoresis at 200 V for 1 h. Gels were dried under a vacuum at 80°C on Whatman 3MM paper and autoradiographed 12 to 16 h at 20°C on Kodak Biomax film.
FIG. 3.
Identification of the transcription start site of the partition operon by primer extension. (A) Position of runoff transcript. (B) Iterated structure of parS showing the probable transcription start site (underlined “C” with arrow) and an additional site that is not reproducibly observed and that may represent an artifact resulting from RNA degradation or an alternate start site (underlined “C” without arrow).
Determination of transcription start site(s).
RNA was extracted from A. tumefaciens carrying pUCD2000parS+ parA+ and purified with the Qiagen RNA-easy purification kit. Primer extension was performed with the Promega primer extension kit in accordance with the supplier's protocol. Images were obtained with a Fujix BAS2000 phosphorimager and MacBas computer software (Fuji).
Nucleotide sequence accession number.
The region of pTAR sequenced in this study, encompassing the entire par operon (Fig. 1), was deposited in the GenBank database under accession number AF143682.
RESULTS AND DISCUSSION
DNA sequence of the pTAR partition region: revision and extension.
The report by Gallie and Kado (14) that mutations downstream of pTAR parA compromised plasmid partition led us to suspect the existence of a second open reading frame (ORF) downstream of parA. We therefore undertook to resequence the region, including parA (which is relatively AT rich) and the more GC-rich downstream region of the DNA (more typical of Agrobacterium). The relevant bacteria and plasmids were kindly provided by Clarence Kado. We confirmed the published sequence of parA and the upstream region but detected 3 G residues that had been missed in the sequence of the region downstream of parA. Their inclusion reveals an ORF the first ATG start codon of which overlaps the parA TGA stop codon by 2 bp (Fig. 1). Eight base pairs upstream of the ATG is a putative ribosome binding site (GGAG) identical in sequence to the GGAG which precedes the ATG of parA, also by 8 bp. This ORF extends for 94 codons before terminating in a TGA stop codon. Continuation of the sequencing beyond the limits of the fragment that is required for efficient partitioning in A. tumefaciens revealed the presence of an additional ORF, the first ATG start codon of which overlaps the parB TGA stop codon by 2 bp. Eight base pairs upstream of this ATG, a GGAG sequence is again seen. The third ORF of the operon extends for 84 codons before terminating in a TAA stop codon. We give the names parB and orf-84, respectively, to the ORFs downstream of parA.
Analysis of the sequence of the 222 amino acid residues that parA of pTAR could encode led to the prediction that this ParA is a member of the Sop/Par ATPase family (33, 41). The pTAR ParB protein appears to belong to a family entirely different from that of other centromere-binding partition proteins such as ParB of P1 (E. V. Koonin, personal communication). No significant homology between ParB of pTAR and other proteins in the database could be detected. The only feature of the ParB sequence that we consider of possible relevance to its DNA-binding capacity is a central region of 14 residues, 7 of which are basic. As for the orf-84 gene product, it is not needed for partitioning in A. tumefaciens, but it may have a regulatory role under particular conditions or in alternative hosts. We do not further examine its function in this report.
A requirement for both ParA and ParB in plasmid partitioning.
We compared the segregational stability of an Agrobacterium-Escherichia shuttle vector (pUCD2000) carrying, or not, the intact or partially deleted pTAR partition region. As seen from Table 3, a DNA segment containing parS, parA, and parB (and only the first 15 codons of orf-84) can confer an approximately 1,000-fold increase in plasmid stability in A. tumefaciens. This result is consistent with results obtained by Gallie and Kado using similar constructs (14). In their experiments, truncation of the segment from either end (to 62 bp from the initial ATG of parA or to 19 bp from the stop codon that terminates parA) eliminated this stabilizing effect. We attribute the loss of stabilization that occurred upon terminal truncation of the operon to removal of the source of ParB. The alternative, that a cis-acting element required for partitioning is present in this region, is inconsistent with the finding of Gallie and Kado that the only region required in cis for partitioning lies upstream of parA (14). Stabilization experiments were carried out both in A. tumefaciens and in a pcnB strain of E. coli (BR5806). The pcnB mutation reduces the copy number of pBR322 (27), and the shuttle vector is rendered unstable. No statistically significant stabilization was observed in the E. coli host (Table 3).
TABLE 3.
Plasmid stabilization by the pTAR par genes: effect of truncations within the par operon
| pTAR partition gene(s) in pUCD2000 | Loss frequency/generationa
|
|
|---|---|---|
| A. tumefaciens | E. coli pcnB | |
| parS+ parA+ parB+ | 4 × 10−5–6 × 10−5 | 0.4 × 10−2–1 × 10−2 |
| None | 0.7 × 10−1–0.9 × 10−1 | 0.6 × 10−2–4 × 10−2 |
| parS+ | 1.0 × 10−1–1.2 × 10−1 | 1 × 10−2–3 × 10−2 |
| parS+ ΔparA1 parB+b | 1.0 × 10−1–1.2 × 10−1 | 0.2 × 10−2–3 × 10−2 |
| parS+ parA+ | 1.5 × 10−1–2.1 × 10−1 | 0.4 × 10−2–6 × 10−2 |
The ranges of plasmid loss frequencies given are based on the results of three independent experiments with A. tumefaciens and two independent experiments with E. coli BR5806.
The ΔparA1 gene carries the ΔNcoI in-frame deletion.
Autogenous regulation of the pTAR Par operon.
The possibility that the parA gene is autoregulated was suggested by Gallie and Kado (14) on the basis of increased transcription from the par promoter when the parA coding region (and the region we now know to include parB) was deleted. In order to determine which member(s) of the operon contributes to autoregulation, we examined the effects of inducible sources of ParA and ParB on the expression of a transcriptional fusion of lacZ to the promoter region of the par operon. This reporter was inserted into the chromosome of E. coli as described in Materials and Methods.
Two main conclusions can be drawn from the data of Table 4. We note first that the ParA and ParB proteins of pTAR can be expressed in E. coli, as well as in A. tumefaciens, such that they are functional for repression. Second, ParB can repress the partition operon. ParA, inactive alone, can modestly augment this repression. These findings are consistent with the generalization that cooperation between the par gene products is a common theme in par operon regulation, but the relation of the operator region to the centromere region may influence the choice of primary repressor. In the case of R1, NR1 and pTAR, the two regions are intimately associated and the second protein of the operon is the primary repressor. In the case of P1 and F, the two regions are at opposite ends of the operon and the first protein of the operon, the ATPase, is the primary repressor.
TABLE 4.
Effects of ParA and ParB on the expression of a parS-lacZ fusion in E. coli
| Plasmidsa | β-Galactosidase sp act (Miller units) with:
|
|||||
|---|---|---|---|---|---|---|
| No arabinose and:
|
0.2% Arabinose and:
|
|||||
| No IPTG | 0.06 mM IPTG | 0.13 mM IPTG | No IPTG | 0.06 mM IPTG | 0.13 mM IPTG | |
| pBAD24-parA + pOAR11 (vector) | 110 ± 7 | 108 ± 4 | 107 ± 3 | 110 ± 7 | 108 ± 5 | 107 ± 2 |
| pBAD24 (vector) + pOAR11-parB | 103 ± 5 | 42 ± 3 | 2 ± 0 | 102 ± 4 | 41 ± 4 | 2 ± 0 |
| pBAD24-parA + pOAR11-parB | 105 ± 1 | 44 ± 1 | 2 ± 1 | 103 ± 6 | 15 ± 2 | 1 ± 0 |
Cultures of BR6326 harboring the indicated plasmids were grown for five generations with or without inducers as shown.
Purification and some features of the products of parA and parB.
In order to characterize the protein products of the two par genes, the genes were cloned separately into expression vectors, the proteins were purified as His6-tagged derivatives, and antibodies were raised to them as detailed in Materials and Methods. Since all the in vitro studies were carried out with the C-terminally His6-tagged derivatives, the possibility that some of the properties reported below are modified by the presence of the added residues cannot be excluded.
The parA gene could encode a protein of 222 residues with a unique cysteine residue at position 38. In the presence of the reducing agent β-mercaptoethanol at 2.5 mM, the electrophoretic mobility of denatured ParA that had been boiled for 10 min in 1× Laemmli sample buffer (Bio-Rad) in a 10% polyacrylamide gel with Tris-glycine-SDS running buffer (Bio-Rad) was that of a monomer, whereas in the absence of β-mercaptoethanol, ParA migrated as a dimer (data not shown). Possibly the intracellular form of ParA is monomeric, and dimerization occurred during isolation. The parB gene encodes a protein without any cysteine residues (as does orf-84). ParB migrated to the position expected of a monomer, whether β-mercaptoethanol was present or not (data not shown).
The results shown in Table 4 and the behavior of other pairs of partition proteins suggest that ParA may interact with ParB directly, possibly without requiring the mediation of parS. Our attempts to demonstrate an interaction in vitro by the hetero-oligomerization assay of Hope and Struhl (20) were not successful. The two purified His6-tagged proteins did not comigrate upon electrophoresis through a nondenaturing gel following incubation together whether ATP was present (1 mM) or absent during incubation and whether they were treated with the cross-linking agent glutaraldehyde (0.02%) or not. An interaction between the proteins might nevertheless be demonstrable under altered preincubation or assay conditions.
Binding of Par proteins to parS DNA.
We assessed the DNA binding activity of the His-tagged ParA and ParB proteins, separately and together, by an electrophoretic mobility shift assay. As a substrate we used an end-labeled fragment of DNA containing the 13 repeated sequences of the centromere-promoter (parS) region (Fig. 3B) and essentially no extraneous DNA or the same region plus an additional 46 bp of DNA upstream of the repeats. Because similar results were obtained with the two preparations, only one is shown. Figure 2 shows a series of smeared bands that are increasingly retarded with increasing concentrations of His6-tagged ParB, suggesting a progressive occupancy of sites within the labeled fragment as the protein concentration was increased. Because the progression of bands is not abrupt, binding to the sites does not appear to be highly cooperative, although some degree of cooperativity is not excluded. ParA protein (also His6 tagged), whether ATP was present or not, caused no further mobility shift under the conditions of our assays, but this failure (data not shown) does not exclude the possibility that at some stage in the partition process the two proteins form a complex with parS in vivo.
FIG. 2.
Electrophoretic mobility shift assay of ParB binding to parS DNA. Serial twofold dilutions of His6-tagged ParB in binding buffer were mixed with an equal volume of 1 nM 32P-labeled parS DNA (including 46 bp upstream of the repeat sequences) in the same buffer, and samples were incubated and electrophoresed as described in Materials and Methods. Similar results (data not shown) were obtained with 32P-labeled DNA that did not include the extra 46-bp region. The amounts of His6-tagged ParB in the lanes containing protein range from 5.4 to 1,400 ng. Identical results were obtained when 1,750 ng of His6-tagged ParA was included in the incubation mixture (data not shown).
Identification of the primary transcription start site.
The observation that ParB binds to iterated sequences immediately upstream of the genes that ParB effectively represses suggested to us that the primary transcription start site may be situated among these repeats. This expectation was confirmed by the primer extension assays, in which we took advantage of the derepression afforded by a ΔparB mutation to increase the signal, otherwise extremely weak in A. tumefaciens (Fig. 3). Similar results were obtained with E. coli (data not shown). Although two bands appeared in some of these experiments, the lower band (not shown) was more variable in intensity and relatively weak; presumably, it represents the product of mRNA degradation or possibly a secondary start site. The upper band locates a start site at a cytosine residue of the template which is appropriately positioned relative to putative −10 and −35 sites of RNA polymerase binding. A central location within the set of iterated sequences is similar to that of the transcription start site in the partition operon of R1 (22).
ATPase activity of ParA.
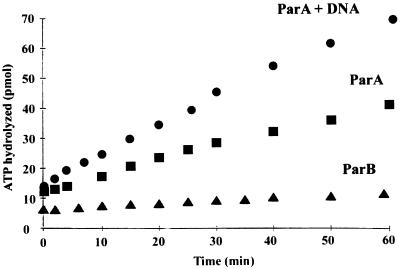
As predicted from its amino acid sequence (33, 41), purified ParA possesses ATPase activity (Fig. 4). The weak activity is comparable to those of other partition ATPases, such as that of P1 (8). Substitution of KCl or K-glutamate for NaCl did not increase the activity, and other nucleotide triphosphates were not significantly hydrolyzed (data not shown). A modest stimulation of ATP hydrolysis by supercoiled plasmid DNA or short linear fragments of double-stranded DNA was observed, and the stimulation did not appear to be any greater if the DNA included the parS region. These results are qualitatively similar to those obtained with the partition ATPases of P1 (8), F (40), and R1 (23).
FIG. 4.
Stimulation of the ATPase activity of ParA by plasmid DNA. ATPase activity was measured as the release of 32PO4 from [γ-32P]ATP by thin-layer chromatography as described in Materials and Methods. Shown are results with pUCD550 (containing parS). Identical results (data not shown) were obtained with the vector pUC4.
Relationships among partition cassettes.
A striking feature of the pTAR partition cassette is the similarity of its organization to that of R1 (or NR1). In each the promoter-operator and centromere regions appear to overlap. In each they are followed by a gene that encodes a weak ATPase whose activity is stimulated by DNA (23, 30). In each a small downstream gene encodes the primary repressor of the operon for which, at least in NR1 (39) as in pTAR, the ATPase serves as a corepressor. In each the binding sites for the primary repressor are a set of tandem repeats within which the transcription start site is centrally situated. Another diminutive putative partition cassette has been described in a linear plasmid of Borrelia burgdorferi (lp 16.9) (2), in which the organization appears to be similar, although the DNA of Borrelia and its numerous plasmids is particularly AT rich, whereas that of Agrobacterium is particularly GC rich. Whether the observed organizational similarities among partition cassettes of diverse provenance are the result of divergent or of parallel evolution is a question that remains to be answered.
ACKNOWLEDGMENTS
We thank our colleagues Roy D. Magnuson, Oleg A. Rodionov, Kyusung Park, and Dhruba K. Chattoraj for advice and constructive criticism; Clarence Kado, Michael Kovacs, and Ann Matthysse for Agrobacterium strains, plasmids, and useful lore; and Lev Sirota for statistical analysis of the data.
K.K. was supported by a Fogarty postdoctoral fellowship. S.S. was a volunteer researcher.
REFERENCES
- 1.Abeles A L, Friedman S A, Austin S J. Partition of unit-copy miniplasmids to daughter cells. III. The DNA sequence and functional organization of the P1 partition region. J Mol Biol. 1985;185:261–272. doi: 10.1016/0022-2836(85)90402-4. . (Erratum, 189:387, 1986). [DOI] [PubMed] [Google Scholar]
- 2.Barbour A G, Carter C J, Bundoc V, Hinnebusch J. The nucleotide sequence of a linear plasmid of Borrelia burgdorferi reveals similarities to those of circular plasmids of other prokaryotes. J Bacteriol. 1996;178:6635–6639. doi: 10.1128/jb.178.22.6635-6639.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biek D P, Shi J. A single 43-bp sopC repeat of plasmid mini-F is sufficient to allow assembly of a functional nucleoprotein partition complex. Proc Natl Acad Sci USA. 1994;91:8027–8031. doi: 10.1073/pnas.91.17.8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bork P, Sander C, Valencia A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc Natl Acad Sci USA. 1992;89:7290–7294. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breuner A, Jensen R B, Dam M, Pedersen S, Gerdes K. The centromere-like parC locus of plasmid R1. Mol Microbiol. 1996;20:581–592. doi: 10.1046/j.1365-2958.1996.5351063.x. [DOI] [PubMed] [Google Scholar]
- 6.Carey J. Gel retardation. Methods Enzymol. 1991;208:103–117. doi: 10.1016/0076-6879(91)08010-f. [DOI] [PubMed] [Google Scholar]
- 7.Dam M, Gerdes K. Partitioning of plasmid R1. Ten direct repeats flanking the parA promoter constitute a centromere-like partition site, parC, that expresses incompatibility. J Mol Biol. 1994;236:1289–1298. doi: 10.1016/0022-2836(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 8.Davis M A, Martin K A, Austin S J. Biochemical activities of the parA partition protein of the P1 plasmid. Mol Microbiol. 1992;6:1141–1147. doi: 10.1111/j.1365-2958.1992.tb01552.x. [DOI] [PubMed] [Google Scholar]
- 9.Diederich L, Rasmussen L J, Messer W. New cloning vectors for integration in the lambda attachment site attB of the Escherichia coli chromosome. Plasmid. 1992;28:14–24. doi: 10.1016/0147-619x(92)90032-6. [DOI] [PubMed] [Google Scholar]
- 10.Friedman S A, Austin S J. The P1 plasmid-partition system synthesizes two essential proteins from an autoregulated operon. Plasmid. 1988;19:103–112. doi: 10.1016/0147-619x(88)90049-2. [DOI] [PubMed] [Google Scholar]
- 11.Funnell B E. Participation of Escherichia coli integration host factor in the P1 plasmid partition system. Proc Natl Acad Sci USA. 1988;85:6657–6661. doi: 10.1073/pnas.85.18.6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Funnell B E, Gagnier L. The P1 plasmid partition complex at parS. II. Analysis of ParB protein binding activity and specificity. J Biol Chem. 1993;268:3616–3624. [PubMed] [Google Scholar]
- 13.Furste J P, Pansegrau W, Frank R, Blocker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 14.Gallie D R, Kado C I. Agrobacterium tumefaciens pTAR parA promoter region involved in autoregulation, incompatibility and plasmid partitioning. J Mol Biol. 1987;193:465–478. doi: 10.1016/0022-2836(87)90260-9. [DOI] [PubMed] [Google Scholar]
- 15.Gallie D R, Novak S, Kado C I. Novel high- and low-copy stable cosmids for use in Agrobacterium and Rhizobium. Plasmid. 1985;14:171–175. doi: 10.1016/0147-619x(85)90078-2. [DOI] [PubMed] [Google Scholar]
- 16.Gallie D R, Zaitlin D, Perry K L, Kado C I. Characterization of the replication and stability regions of Agrobacterium tumefaciens plasmid pTAR. J Bacteriol. 1984;157:739–745. doi: 10.1128/jb.157.3.739-745.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glaser P, Sharpe M E, Raether B, Perego M, Ohlsen K, Errington J. Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes Dev. 1997;11:1160–1168. doi: 10.1101/gad.11.9.1160. [DOI] [PubMed] [Google Scholar]
- 18.Guzman L M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayes F, Radnedge L, Davis M A, Austin S J. The homologous operons for P1 and P7 plasmid partition are autoregulated from dissimilar operator sites. Mol Microbiol. 1994;11:249–260. doi: 10.1111/j.1365-2958.1994.tb00305.x. [DOI] [PubMed] [Google Scholar]
- 20.Hope I A, Struhl K. GCN4, a eukaryotic transcriptional activator protein, binds as a dimer to target DNA. EMBO J. 1987;6:2781–2784. doi: 10.1002/j.1460-2075.1987.tb02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ireton K, Gunther IV N W, Grossman A D. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1994;176:5320–5329. doi: 10.1128/jb.176.17.5320-5329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen R B, Dam M, Gerdes K. Partitioning of plasmid R1. The parA operon is autoregulated by ParR and its transcription is highly stimulated by a downstream activating element. J Mol Biol. 1994;236:1299–1309. doi: 10.1016/0022-2836(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 23.Jensen R B, Gerdes K. Partitioning of plasmid R1. The ParM protein exhibits ATPase activity and interacts with the centromere-like ParR-parC complex. J Mol Biol. 1997;269:505–513. doi: 10.1006/jmbi.1997.1061. [DOI] [PubMed] [Google Scholar]
- 24.Jensen R B, Gerdes K. Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol Microbiol. 1995;17:205–210. doi: 10.1111/j.1365-2958.1995.mmi_17020205.x. [DOI] [PubMed] [Google Scholar]
- 25.Koonin E V. A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic replication. Nucleic Acids Res. 1993;21:2541–2547. doi: 10.1093/nar/21.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin D C, Grossman A D. Identification and characterization of a bacterial chromosome partitioning site. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- 27.Lopilato J, Bortner S, Beckwith J. Mutations in a new chromosomal gene of Escherichia coli K-12, pcnB, reduce plasmid copy number of pBR322 and its derivatives. Mol Gen Genet. 1986;205:285–290. doi: 10.1007/BF00430440. [DOI] [PubMed] [Google Scholar]
- 28.Manne V, Bekesi E, Kung H F. Ha-ras proteins exhibit GTPase activity: point mutations that activate Ha-ras gene products result in decreased GTPase activity. Proc Natl Acad Sci USA. 1985;82:376–380. doi: 10.1073/pnas.82.2.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 30.Min Y N, Tabuchi A, Womble D D, Rownd R H. Transcription of the stability operon of IncFII plasmid NR1. J Bacteriol. 1991;173:2378–2384. doi: 10.1128/jb.173.7.2378-2384.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohl D A, Gober J W. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell. 1997;88:675–684. doi: 10.1016/s0092-8674(00)81910-8. [DOI] [PubMed] [Google Scholar]
- 32.Mori H, Mori Y, Ichinose C, Niki H, Ogura T, Kato A, Hiraga S. Purification and characterization of SopA and SopB proteins essential for F plasmid partitioning. J Biol Chem. 1989;264:15535–15541. [PubMed] [Google Scholar]
- 33.Motallebi-Veshareh M, Rouch D A, Thomas C M. A family of ATPases involved in active partitioning of diverse bacterial plasmids. Mol Microbiol. 1990;4:1455–1463. doi: 10.1111/j.1365-2958.1990.tb02056.x. [DOI] [PubMed] [Google Scholar]
- 34.Ogura T, Hiraga S. Partition mechanism of F plasmid: two plasmid gene-encoded products and a cis-acting region are involved in partition. Cell. 1983;32:351–360. doi: 10.1016/0092-8674(83)90454-3. [DOI] [PubMed] [Google Scholar]
- 35.Papp P P, Mukhopadhyay G, Chattoraj D K. Negative control of plasmid DNA replication by iterons. Correlation with initiator binding affinity. J Biol Chem. 1994;269:23563–23568. [PubMed] [Google Scholar]
- 36.Rodionov O, Lobocka M, Yarmolinsky M. Silencing of genes flanking the P1 plasmid centromere. Science. 1999;283:546–549. doi: 10.1126/science.283.5401.546. [DOI] [PubMed] [Google Scholar]
- 37.Sharpe M E, Errington J. The Bacillus subtilis soj-spo0J locus is required for a centromere-like function involved in prespore chromosome partitioning. Mol Microbiol. 1996;21:501–509. doi: 10.1111/j.1365-2958.1996.tb02559.x. [DOI] [PubMed] [Google Scholar]
- 38.Tabuchi A, Min Y N, Kim C K, Fan Y L, Womble D D, Rownd R H. Genetic organization and nucleotide sequence of the stability locus of incFII plasmid NR1. J Mol Biol. 1988;202:511–525. doi: 10.1016/0022-2836(88)90282-3. [DOI] [PubMed] [Google Scholar]
- 39.Tabuchi A, Min Y N, Womble D D, Rownd R H. Autoregulation of the stability operon of IncFII plasmid NR1. J Bacteriol. 1992;174:7629–7634. doi: 10.1128/jb.174.23.7629-7634.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe E, Wachi M, Yamasaki M, Nagai K. ATPase activity of SopA, a protein essential for active partitioning of F plasmid. Mol Gen Genet. 1992;234:346–352. doi: 10.1007/BF00538693. [DOI] [PubMed] [Google Scholar]
- 41.Williams D R, Thomas C M. Active partitioning of bacterial plasmids. J Gen Microbiol. 1992;138:1–16. doi: 10.1099/00221287-138-1-1. [DOI] [PubMed] [Google Scholar]