ABSTRACT
Acinetobacter baumannii causes hospital-acquired infections and is responsible for high mortality and morbidity. The interaction of this bacterium with the host is critical in bacterial pathogenesis and infection. Here, we report the interaction of peptidoglycan-associated lipoprotein (PAL) of A. baumannii with host fibronectin (FN) to find its therapeutic potential. The proteome of A. baumannii was explored in the host-pathogen interaction database to filter out the PAL of the bacterial outer membrane that interacts with the host’s FN protein. This interaction was confirmed experimentally using purified recombinant PAL and pure FN protein. To investigate the pleiotropic role of PAL protein, different biochemical assays using wild-type PAL and PAL mutants were performed. The result showed that PAL mediates bacterial pathogenesis, adherence, and invasion in host pulmonary epithelial cells and has a role in the biofilm formation, bacterial motility, and membrane integrity of bacteria. All of the results suggest that PAL’s interaction with FN plays a vital role in host-cell interaction. In addition, the PAL protein also interacts with Toll-like receptor 2 and MARCO receptor, which suggests the role of PAL protein in innate immune responses. We have also investigated the therapeutic potential of this protein for vaccine and therapeutic design. Using reverse vaccinology, PAL’s potential epitopes were filtered out that exhibit binding potential with host major histocompatibility complex class I (MHC-I), MHC-II, and B cells, suggesting that PAL protein is a potential vaccine target. The immune simulation showed that PAL protein could elevate innate and adaptive immune response with the generation of memory cells and would have subsequent potential to eliminate bacterial infection. Therefore, the present study highlights the interaction ability of a novel host-pathogen interacting partner (PAL-FN) and uncovers its therapeutic potential to combat infection caused by A. baumannii.
KEYWORDS: Acinetobacter baumannii, bacterial resistance, peptidoglycan-associated lipoprotein, fibronectin, far-Western blot, protein-protein interaction, vaccine target, virulence
INTRODUCTION
Acinetobacter baumannii, an ESKAPE pathogen, is a primary global concern to the human health (1). The World Health Organization (WHO) designated this pathogen as a critically prioritized pathogen. A. baumannii evolved different resistant mechanisms, such as the efflux pump (2, 3), beta-lactamases (4), and biofilm formation (5, 6), etc., as well as diverse tolerance mechanisms in the adverse hospital environment (7). The coinfections and transmission of resistant A. baumannii are also seen in Covid-19 patients (8–10). The prime steps of a bacterial infection involve the interaction between the bacterial surface components and the extracellular matrix (ECM) molecules of the host. However, in A. baumannii-mediated pathogenesis, the mode of adhesion and the partners involved in host cell invasion is lagging. The role of fibronectin (FN) during infection of A. baumannii has been investigated, and it was found that FN interacts with OmpA, FBPs, and TonB-dependent copper protein (11). The Tol-Pal protein system has three inner membrane proteins (TolA, TolQ, and TolR) and two outer membrane proteins (TolB and Pal) (12). PAL is anchored to the bacterium’s outer membrane and interacts with Tol proteins, as well as other cell envelope proteins such as OmpA and Lpp proteins (12). The role of PAL in the virulence of Burkholderia mallei has been investigated and found to be involved in resistance to complement-mediated killing and helps in replication inside host cells (13). No studies have been performed to investigate the role of PAL-FN interaction in the pathogenesis of A. baumannii.
Hence, the present study focuses on finding the A. baumannii-host interacting partners and the role of interacting bacterial proteins in bacterial pathogenesis. Therefore, to explore the mechanism, an in silico analysis was performed to screen the interacting partners. The outer membrane-associated interacting partners were filtered, and the PAL-FN interaction complex was shortlisted, which might have a role in bacterial adhesion on the host cell. For further investigation, the pal gene was cloned, expressed, purified, and confirmed by Western blotting and liquid chromatography-mass spectrometry-based sequencing. Various experiments such as bacterial growth kinetics, adherence, invasion, biofilm formation, motility, cell permeability, cell viability, and respiratory bust were performed to show the PAL protein’s role in bacterial pathogenesis. Reverse vaccinology and immune simulation have been implemented to investigate the PAL protein for its therapeutic potential.
RESULTS
Identification of interacting proteins of A. baumannii and human host.
The whole proteome of A. baumannii AYE was downloaded from UniProt, containing 3,652 proteins. Using the Host-Pathogen Interaction Database (HPIDB), protein-protein interactions were analyzed based on homologous pairs of protein interactions across different organisms. A total of 985 interspecies interactions were obtained between Acinetobacter baumannii and the host Homo sapiens. In the manual screening of 985 interactions, a total of 276 unique bacterial proteins have been filtered that interact with different host proteins (see Fig. S1 posted at https://doi.org/10.6084/m9.figshare.22248886.v1). To target bacterial initial adherence capability, outer membrane-associated proteins partners of host and bacteria were screened with online server CELLO and PSORTb. Of 276 proteins, only 9 were located in the outer membrane and extracellular region (see Table S1 posted at https://doi.org/10.6084/m9.figshare.22248886.v1). Cellular localization of bacterial proteins interacting with human proteins was also predicted. Of the 9 bacterial proteins, only 4 were located in the human plasma membrane (see Table S2). A literature survey of these all proteins confirmed that all are found on the surface and play a vital role in the host-pathogen interaction (11, 12, 14–18). Transmembrane helicity and virulence analysis of these four proteins revealed PAL protein as the best target for further study due to its 0 transmembrane helicity score with the highest virulence score (see Table S2).
Interaction analysis of PAL and FN protein using in silico approach.
HPIDB analysis-filtered PAL-FN interaction was evaluated by molecular docking, i.e., PatchDock (19). In the PatchDock server, a docking score of 11303 was found for the PAL-FN complex and further validated by the FireDock server that exhibited binding free energy of −48.73 kcal/mol (see Fig. S2). The PAL-FN complex showed a dissociation constant (Kd) of 1.9 × 10−10 M. The data represented that bacterial PAL protein possibly interacts with the host cell fibronectin (FN) to initiate the host cell adhesion in the infection cycle.
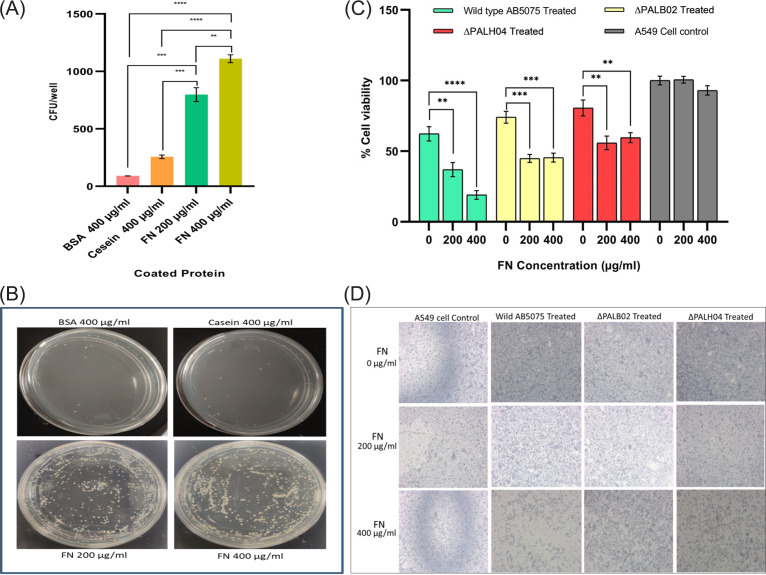
Interaction of Acinetobacter is enhanced by immobilized FN.
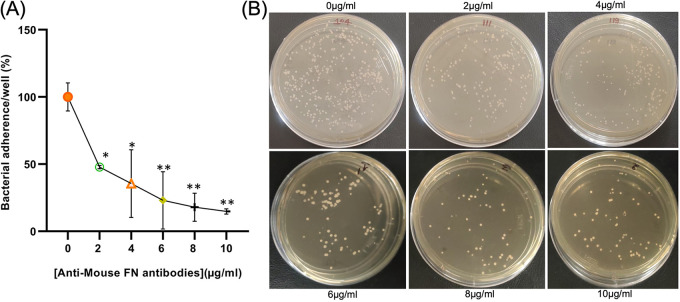
The lung epithelial cells produce FN and release the same on the cell surface, and their role in bacterial adhesion and internalization was identified (20). In the analysis of PAL-FN-mediated interaction, a preliminary experiment was performed to determine the FN-mediated interaction and its role in bacterial adhesion capability. In this experimental setup, the A. baumannii strain significantly (P < 0.0001) bound to an FN-coated (400 μg/mL) plate, and the increased bacterial CFU was counted with increasing concentration of coated FN (Fig. 1A) compared to the control protein. This study showed that fibronectin-mediated interaction facilitated bacterial interaction. The lung epithelial cells produce the FN and release it outside (20). To identify the role of this cell-derived FN in the cell adhesion and invasion process, polyclonal antibodies of FN were incubated with A549 cell lines before the invasion assay. The level of adhesion of wild-type strain of A. baumannii to A549 cell lines was reduced by anti-FN antibody in a dose-dependent manner (Fig. 2). Adhesion was decreased significantly by 85.25% in the presence of 10 μg/mL anti-FN antibodies compared to the absence of antibody (P < 0.0043). This study revealed that the host FN protein has a central role in bacterial infection.
FIG 1.
PAL-FN-mediated host-pathogen interaction. (A) Plot showing CFU/well after incubation of A. baumannii in a well coated with FN protein or control protein (casein and BSA). (B) Images of bacterial CFU counting plates showing the interaction of A. baumannii with the differently coated proteins. (C) Bar showing the percent cell viabilities of A549 epithelial cell lines after infection with wild-type and pal mutant strains of A. baumannii in the presence of exogenous FN. (D) Image showing A549 epithelial cells under an inverted microscope with 5× magnification after infection with A. baumannii. Cell viability in noninfected A549 cell lines (control) was defined as 100%. All reported values in panels A and C are mean values of three experiments ± the SD, and the statistical significance was identified using GraphPad Prism software.
FIG 2.
Effect of cell-derived FN on bacterial adherence of A. baumannii. (A) Percentage of bacterial adherence to A549 cell lines in the presence of different concentrations of anti-FN antibody. The bacterial adhesion is expressed as a percentage of that observed in the absence of anti-FN antibody (control). (B) Image of a petri plate showing CFU count for adhered bacteria of the A549 cell line in the presence of a different concentration of anti-FN antibody. All reported values in panel A are mean values of three experiments ± the SD, and statistical significance was identified using GraphPad Prism software.
Cloning and overexpression of PAL.
The pal encoding gene PCR product of 573 bp was cloned in T/A vector, followed by its subcloning into the pET28a vector and transformation in E. coli DH5α competent cells. The kanamycin resistance selected recombinant colony was used for plasmid isolation. The isolated plasmid was transformed into E. coli C43 BL21(DE3) for overexpression and purification. Sequencing of the pET28a gene construct and its nucleotide BLAST analysis matched (100% similarity) with the deposited PAL sequence of A. baumannii (sequence ID CP040425.1). The translate tool of ExPASy proteomics server (http://www.expasy.ch/tools) was used to learn the amino acid sequence based on the pal gene. The theoretical pI and molecular weight of PAL protein were estimated to be 6.43 and 20.59 kDa, respectively. Overexpression of PAL protein was confirmed by Western blotting. Liquid chromatography-tandem mass spectrometry analysis of purified PAL protein corroborated the presence and purity of the PAL protein.
Purified PAL interacts with pure FN protein.
To determine the PAL-FN interaction, far-Western blotting was performed. A Western blot containing purified cloned PAL was incubated with soluble human FN. Specific binding of PAL with FN was observed (see Fig. S2). The same experiment without FN/PAL was performed to exclude possible background signals. No binding was observed in the negative control, which confirmed the specific binding of PAL-FN molecules.
PAL regulates the growth kinetics of Acinetobacter.
During bacterial membrane remodeling, PAL protein is recruited via FtsN at the cell pole divisome (21); hence, PAL protein may interfere with the normal cell growth kinetics (22). To analyze this, the growth kinetics pattern of wild-type and mutant cells were examined. The growth kinetics pattern of the mutant strains exhibited a long lag phase and reduced biomass at the stationary phase compared to wild-type strains AB5075 (see Fig. S3 in the supplemental material), which shows the role of PAL in bacterial growth. In the present study, three mutants are used, i.e., wild type (1 to 573), PALB02 (insertion of 73 codons at 138), and PALH04 (insertion of 73 codons at 268).
Reduction in cell viability of A. baumannii-infected A549 cells in the presence of FN.
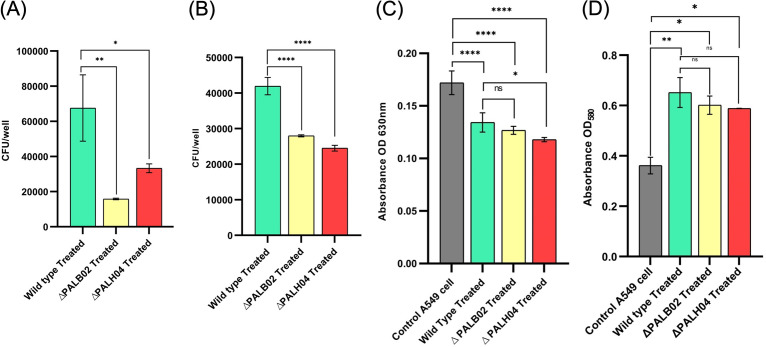
Disrupted respiratory epithelium of hospitalized patients on mechanical ventilation exposes ECM FN, which is recognized by bacterial adhesion proteins (23). To check the role of exposed FN protein in host cell adhesion, a cell viability assay was performed. The results showed that infection of wild-type A. baumannii to A549 cells exhibited a significant (P < 0.0001) decrease (~49%) in A549 cell viability when infection was performed in the presence of FN (200 and 400 μg/mL) compared to its absence (i.e., FN, 0 μg/mL). However, increased viability (approximately 20 to 28%) was witnessed when the A549 epithelial cells were infected with PAL mutant strains (Fig. 1C and D). These results confirmed the important role of PAL-FN-mediated interactions in Acinetobacter adherence on the host cell.
Bacterial adherence and invasion are positively regulated by PAL protein.
Defacement of respiratory epithelial cells facilitates bacterial entry that causes A. baumannii infection. Hence, to assess PAL role in Acinetobacter adherence and invasion, A549 host epithelial cells were infected with wild-type and PAL mutant strains. The adherence of mutant strains (AB5075-PALB02 and AB5075-PALH04) were significantly (P = 0.0021 and P < 0.0155, respectively) impaired by 6 h postinfection (Fig. 3A). However, the adhesion assay consisted of both adhered and invaded bacteria; thus, to examine the internalized bacteria over adherent bacteria, gentamicin protection assay was executed. The invasion potential of PAL mutant strains (AB5075-PALB02 and AB5075-PALH04) was significantly (P < 0.0001) impaired compared to wild-type strain AB5075 (Fig. 3B). These results scrutinized the PAL role in adherence and invasion of Acinetobacter on A549 lung epithelial cells.
FIG 3.
PAL protein role in bacterial pathogenesis. The PAL protein role was evaluated by bacterial interaction analysis in wild-type and PAL mutant strains of A. baumannii using adhesion analysis (A), invasion assay) (B), and respiratory bust analysis using ROS estimation (C) and RNS estimation (D). All reported values are mean values of three experiments ± the SD, and statistical significance was identified using GraphPad Prism software.
PAL protein does not affect the respiratory burst (ROS and RNS) in the host cell.
Previous studies manifested that oxygen-rich environments suppress the production of DAP, and thus the motility of the bacteria (24). Since the PAL protein of A. baumannii possesses higher binding affinity with the DAP protein and facilitates host cell adhesion and Invasion. An NBT assay was performed to scrutinize the role of PAL protein in epithelial cell membrane-associated NADPH oxidase complex-mediated reactive oxygen species (ROS) generation. The results showed that ROS production in the pulmonary cell was significantly (<0.0001) inhibited upon infection of A. baumannii (wild type and PAL mutant strain) compared to the control (A549 epithelial cells only), suggesting that it can persist within the host cells by prevention of oxidative burst. Compared to wild-type AB5075, the mutant strains (AB5075-PALB02 and AB5075-PALH04) showed only a minor (P < 0.0443) decrease in ROS production in the host cell. The decrease was only observed in mutant PALH04 but not in PALB02 (Fig. 3C). ROS production in pulmonary A549 cells was estimated using an EZAssay nitric oxide estimation kit (HiMedia Laboratories, Inc.) using Griess reagent. The results showed that the total nitric oxide concentration of A549 cells were elevated in all three treated samples (wild-type AB5075 [P < 0.0058], AB5075-PALB02 [P < 0.0116], and AB5075-PALH04 [P < 0.0142]) compared to nontreated control cells. With respect to the wild-type strain, the PAL mutant strain (AB5075-PALB02 [P < 0.6095] and AB5075-PALH04 [P < 0.4546]) did not modulate (Fig. 3D) the cell-induced reactive nitrogen species (RNS). These data showed that the PAL protein does not have any role in host cell ROS and RNS generation.
PAL shield bacterial community with strong biofilm formation.
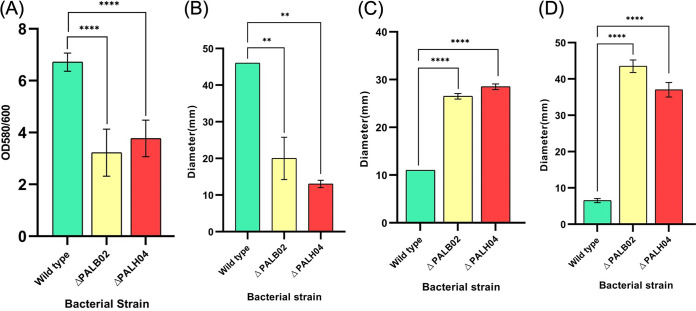
Biofilm formation plays a predominant role in the colonization of A. baumannii on biotic and abiotic surfaces. Here, we investigated the PAL role in biofilm formation ability and found that biofilm was reduced (52.02 and 43.82%, respectively) significantly (P < 0.0001) in PAL mutant strains (AB5075-PALB02 and AB5075-PALH04) compared to wild type AB5075 (Fig. 4A). This suggests the role of PAL in biofilm formation.
FIG 4.
PAL protein pleiotropic role in bacterial cell. In A. baumannii wild-type and PAL mutant strains, bacterial biofilm formation was observed by crystal violet staining (A) and bacterial motility was analyzed by determining bacterial twitching motility (B), swarming motility (C), and swimming motility (D). All reported values in panel A are mean values of three experiments ± the SD, and statistical significance was identified using GraphPad Prism software.
PAL protein downregulates the bacterial twitching motility.
Earlier, nosocomial pathogen A. baumannii was considered nonmotile rod species due to the absence of flagella. However, recent studies showed the motility (twitching and surface-associated) of this ESKAPE pathogen (25). Previous studies showed that bacterial DAP (1,3-diaminopropane) plays a vital role in maintaining bacterial motility and consists of a strong binding affinity with PAL protein (24). To check the PAL role in twitching and/or surface-associated motility, a motility assay was performed. In our study, it was observed that A. baumannii twitching motility had been impaired in mutant AB5075-PALB02 and AB5075-PALH04 strains (P < 0.0046) compared to wild-type AB5075 (Fig. 4B). The previous study has shown that the translucent variant of the AB5075 strain does not exhibit a surface-associated (swarming and swimming) motility (26). However, our study showed an enhanced (P < 0.0001) motile area in PAL mutant strains compared to the wild-type strains (Fig. 4C and D). In accordance with this, switching in the degree of motility helps bacteria maintain their cell locomotion. All of these motility growth areas of the wild-type strain AB5075 and the mutant AB5075-PALB02 and AB5075-PALH04 strains exhibited the role of PAL in bacterial cell motility.
PAL regulates the membrane integrity/permeability.
The outer membrane of A. baumannii plays a crucial role in maintaining its integrity/permeability, and PAL protein is one of its key components. To examine PAL protein’s role in maintaining membrane integrity/permeability, an ethidium bromide (EtBr) accumulation assay was performed (2). The amount of EtBr uptake in the bacterial cell determines the level of impaired outer membrane integrity/permeability. A (1.4 ± 1.7)-fold increase of dye uptake was observed in PAL (AB5075-PALB02 [P < 0.0070] and AB5075-PALH04 [P < 0.0001]) mutants compared to wild-type AB5075 cells (see Fig. S4). Thus, these data determined the potentiality of PAL in the maintenance of outer membrane integrity/permeability, which may be one of the reasons for more EtBr accumulation.
PAL protein has an interaction ability with innate immune cells.
Lipoprotein has properties to induce the host cell immune system with Toll-like receptor 2 (TLR2)-mediated signaling (27). A recent study on A. baumannii moderated infection mechanism revealed that interleukin-10 (IL-10) plays an essential role in the host defense against pulmonary infection of A. baumannii by promoting the antibacterial function of macrophages by regulating MARCO expression through the STAT3-mediated pathway (28). Lipoprotein-mediated interaction with the MARCO receptor is also known (29). As per the literature survey, PAL protein is a lipoprotein; hence, to correlate these two, interaction analysis of PAL protein with TLR2 and MARCO receptor has been performed. In the docking analysis, PAL-TLR2 complex (see Fig. S5A) and PAL-MARCO receptor complex (see Fig. S5B) showed binding free energies of −20.19 and −42.97 kcal/mol, respectively. Molecular dynamics exhibited the stable interaction between the PAL-TLR2 and PAL-MARCO receptors; therefore, it can be stated that PAL protein interacts with TLR2 and MARCO receptor.
Therapeutic potential of PAL protein and its interaction with FN.
(i) Reverse vaccinology and immune simulation confirmed the potentiometric role of PAL as a vaccine target. With the help of reverse vaccinology, the epitopes of PAL protein were screened. In this analysis of PAL protein, 12 major histocompatibility complex class I (MHC-I; see Table S3), 10 MHC-II (see Table S4), and 8 B-cell (see Table S5) interacting epitopes were shortlisted. These shortlisted MHC-I epitopes were further screened by a class I immunogenicity. Eight epitopes with negative immunogenicity scores were filtered out, and the rest of the four proteins were selected (see Table S3). All MHC-I, MHC-II, and B-cell epitopes were screened for their antigenicity, allergenicity, and toxicity potential, in which MHC-I (YLAFPLLSA and RRVEINYEA), MHC-II (AHAQFLMANANSKVA), and B-cell (HFDYDSSDLSTEDYQ, DERGTREYNMA, and ASRKPATTATTGTTNPSTVNTTGLSEDAALNAQNLAGASSKGVTEANKAALAK) interacting epitopes were filtered out. The selected epitopes have the potential to activate the host cell immune response without any allergic and toxicity reactions. Similarly, physicochemical analysis using the ProtParam server exhibited a PAL vaccine instability index of 32.18, which signifies protein stability. The aliphatic index and the grand average of hydropathicity (GRAVY) score of the PAL vaccine were 73.19 and −0.423, respectively, with a half-life of 30 h in mammalian reticulocytes. This in silico screening study showed that PAL could be a potential candidate for vaccine design against A. baumannii.
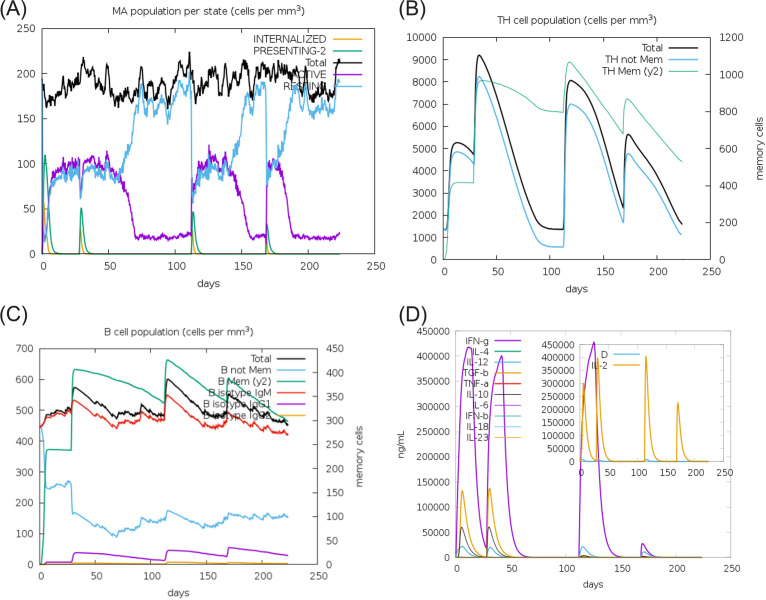
Furthermore, immune simulation analysis using the C-ImmSim model elaborated the role of PAL protein in the activation of the immune response (Fig. 5). In Acinetobacter infection, it has been shown that innate immunity has a significant role in the clearance of infection (30). Injection of PAL protein as a vaccine without lipopolysaccharide (LPS) exhibited elevated innate immune cells (macrophage) (Fig. 5A). The population of adaptive TH cells enhances after injection of the PAL vaccine with the elevated concentration of memory cells (Fig. 5B). Similarly, the total B-cell population with memory cells increases with subsequent injections of PAL protein that show activation of the adaptive immune response. The enhanced B cell remains almost similar with further injections (Fig. 5C). The administration of the PAL vaccine triggers the release of gamma interferon (IFN-γ), transforming growth factor β (TGF-β), IL-10, IL-2, and IL-18 in the host cell (Fig. 5D). In brief, the development of elevated innate and adaptive immune responses with the generation of memory cells would subsequently eliminate bacterial infection.
FIG 5.
Immune simulation of immunization experiment. Injection of PAL protein as a vaccine without LPS was given at day 1, day 28 (4 weeks), day 112 (16 weeks), and day 168 (24 weeks) after initial injection. The innate immune response i.e. macrophage population (A), the T helper (TH) cell population (B), the B-cell population (C), and the cytokine and interleukin population (D) after injection of PAL protein as a vaccine were determined. The result shows that administration of the PAL protein as a vaccine elevated the synthesis of innate immunity, adaptive immune memory cell, and the release of IFN-γ, TGF-β, IL-10, and IL-18 in the host cell.
(ii) Potential drug screening targeting to the pal protein. First, U.S. Food and Drug Administration (FDA) approved library consisting of 2,924 compounds was prepared using the LIGPREP module. The prepared 8,772 stereoisomers were submitted for ADME/T analysis that filtered out 4,542 compounds. During drug target design, interface residues of the PAL-FN docked complex were identified. High-throughput virtual screening (HTVS) docking, followed by standard precision (SP) and extra precision (XP) docking, filtered out three compounds (ZINC00602128, ZINC03794794, and ZINC00000507). Molecular dynamics (MD) simulations of a protein complex (see Fig. S6) with FDA-approved drugs for 10 ns revealed the root mean square deviations (RMSD) for the PAL-FN complex-ZINC00602128 (RMSD = 3.5 Å) and the PAL-FN complex-ZINC03794794 (RMSD = 4.2 Å), whereas the PAL-FN complex-ZINC00000507 interaction was not stable throughout the analysis. In addition, the RMSF values also revealed the lowest fluctuations in the PAL-FN complex with ZINC00602128, whereas the highest fluctuations were observed with ZINC03794794. The PAL complex with all three screened drugs showed different numbers of bond formation, including hydrogen bonds, hydrophobic interactions, and water bridges. Moreover, the interacting fraction exhibited interaction between PAL-drug complex, which means it remains bound to PAL throughout the simulation.
DISCUSSION
A. baumannii, an ESKAPE pathogen, has gained the attention of the medical fraternity worldwide due to its nosocomial infection in hospital setups, mainly in ICUs (4, 6, 31–34). Acinetobacter has developed various resistance mechanisms (MDR, XDR, and PDR) and strategies to evade the host immune system, mobile genetic plasticity, intraspecies heterogeneity, and natural competence (35). To understand the complex disease pathophysiology and shaping into therapeutic diagnosis, substantial studies on the identification of resistant determinants, molecular epidemiology, environmental persistence, and survival strategies employed by A. baumannii have been conducted (30, 36, 37). For host-pathogen interactions over ecological time scales, host susceptibility and pathogen virulence are at the onset of the cross talk. More importantly, understanding the fundamental biology of pathogenesis and their role in host-pathogen interaction is significantly lagging and creating an additional hurdle in designing therapeutics.
In host-pathogen interaction, A. baumannii invade nonphagocytic (epithelial, endothelial, and fibroblast) cells with the help of microfilament and microtubule-dependent zipper-like mechanism for bacterial surface protein interaction with host cell proteins to initiate bacterial entrapment in membrane-bound vacuoles (38). The present study analyzed the bacterium-host interaction and its respective role in bacterial pathogenesis to identify the unique therapeutic targets. A complete screening of the Acinetobacter proteome (3,652 proteins) was conducted with the help of a bioinformatics tool (the HPIDB server). To objectify the initial adhesion targets, virulence-associated outer membrane interacting partners were screened by the CELLO and VFDB servers, respectively. Acinetobacter PAL protein was filtered out as the most virulent associated protein out of all interacting partners. Dabo and his group experimentally validated the Pasteurella multocida OMP16 protein interaction with host fibronectin protein (39), used in this study, and the HPIDB server speculated a possible interaction between Acinetobacter PAL protein and host fibronectin which was further validated by in silico molecular docking.
It has been suggested that the outer membrane proteins of Gram-negative bacteria have key virulence-associated factors exhibiting a role in bacterial adherence, invasion, adaptation, and pathogenesis in the host cell (40). In addition to this, the host ECM is crucial for bacterial colonization, acting as a receptor for the bacterial entry (41). In the field of Acinetobacter adherence, only a few interacting proteins, such as OmpA, EF-Tu, Omp33, TonB-dependent copper receptor, a 34-kDa outer membrane protein, the transporter protein Ata, and ChoP protein, have been known to facilitate bacterial adhesion and invasion (11, 42–47) by interacting with host matrix receptors, including fibronectin, collagen IV, and platelet-derived factor.
To treat A. baumannii infection, polymyxins (polymyxin B and colistin) are last-resort antibiotics (48). In colistin-resistant Acinetobacter, Henry et al. reported that it increases the expression of PAL protein (HMPREF0010_02142) to aid outer membrane stabilization (49). Also, various studies have shown the PAL protein associated immunogenic potency and virulence, thereby executing it as an attractive target to combat widespread epidemic battle (12, 22, 50–52). Moreover, in the host cell, ECM protein acts as a receptor bridge for bacterial (N. meningitidis, S. aureus, and S. suis) entry (41, 53–55). During infection, A. baumannii binds to fibronectin, followed by epithelial and phagocytic cells permitting bacterial invasion and intracellular persistence (56). However, the FN-interacting partners are yet to be explored to discover the interaction machinery and suitable therapeutics. Hence, in vitro validation of PAL-FN interaction was performed to identify their role in bacterial adhesion and pathogenesis.
The proliferation of A. baumannii might be reflected as an essential virulence factor (57). Based on our findings, the PAL mutant strain exhibited a lower growth rate than the wild type, signifying the PAL protein role in the growth kinetics of Acinetobacter. Earlier studies reported that, through matrix binding, bacteria promote their attachment and colonization on the host cell at the beginning of the infection (58). Here, we illustrated that the A. baumannii wild-type strain showed enhanced binding efficiency with FN versus the control casein and BSA proteins, indicating that FN is a potent contributor to A. baumannii host cell interaction. Furthermore, anti-FN antibody treatment reduced bacterial adhesion, indicating that FN binding with A. baumannii plays a key role in efficient binding. PAL binding to FN significantly enhances the adherence of A. baumannii to the lung epithelial A549 cells. Thus, this employs that FN acts as a bridge between the Acinetobacter and host epithelial cell interaction. Further, the bacterial PAL protein was cloned and purified, and FN-mediated interaction was confirmed by far-Western blotting.
In addition, the Acinetobacter PAL mutant strain showed an impaired bacterial internalization (both adhesion and invasion) on host epithelial cells compared to the wild-type strain signifying a PAL-mediated bridge have a profound role in cellular protrusions leading to bacterial entrapment. In the infection process, PAL protein exhibits an active contribution to bacterial adherence and invasion, despite anonymous cascade signaling. PAL protein-associated cellular signaling and dynamic features can be highlighted succinctly by analyzing host immune signaling cascades. One of the initial responses in the host immune system is the reactive species free radical generation (ROS/RNS) to invade bacterial colonization by cellular immune activation. Acinetobacter exploits host cell-induced NADPH oxidase and mitochondrion-derived ROS, which detoxifies H2O2 into water and oxygen to prolong infection (30, 59–61). Our study found that A. baumannii-infected host epithelial cells had diminished ROS production and an elevated RNS production compared to the noninfected control. However, the PAL mutant-infected host cells showed no significant modulation, which indicated that PAL protein does not have a role in reactive species generation.
A. baumannii virulence-associated outer membrane protein A (OmpA) and DAP have been known to possess intricate roles in the regulation of biofilm formation, motility, survival capacity in an oxygen-rich environment, and the maintenance of cell homeostasis (62–64) and also induce host cell apoptosis (42). Bacterial PAL protein containing the OmpA-like domain (65) and exhibiting an interaction with cell peptidoglycan DAP might play a pleiotropic role in bacteria. Upon analysis of the PAL protein, we observed that it maintains bacterial virulence by regulating the bacterial twitching motility, biofilm production, and cellular homeostasis by maintaining membrane integrity.
The past era showed the traditional vaccine design of A. baumannii to be very expensive and time-consuming. In this regard, various antigenic candidates, including live attenuated strains, formalin-inactivated whole cells, bacterial ghosts, K1 capsular polysaccharide, outer membrane complexes [OmpA, trimeric autotransporter protein, biofilm-associated protein, and poly-N-acetyl-β-(1-6)-glucosamine], outer membrane vesicles, and subunit vaccines, have been examined that induce phagocytic and/or antibody-mediated inhibition or killing (66–78). However, these vaccines were not sufficient for a mixed Th1/Th2 or Th1/Th17 response. Therefore, none have passed in clinical trials, which clarifies the challenges behind eradicating this infection. Therefore, there is a critical need for further advancement in understanding the multifarious relationships between Acinetobacter and the human host at every phase of the disease progression (79).
In recent studies, Pal/OmpA DNA vaccine-associated pVAX1 expression plasmid was vaccinated into mice, which elevated the humoral and mixed Th1/Th2/Th17 response and mouse survival with a lower bacterial burden compared to the unvaccinated group that died within 3 days (80). These results highlighted Pal/OmpA DNA as a potent vaccine target, but further detailed analysis is mandatory to expand delivery, immune response, potency, and DNA vaccine licenses to target the Acinetobacter (80). In our study, in silico interaction of PAL protein with TLR2 and MARCO receptor has been performed, and this identified the role of PAL protein in innate immune responses. PAL protein potential epitopes were filtered out using reverse vaccinology that exhibited high binding potential with host MHC-I, MHC-II, and B cells. Collectively, our results suggested that PAL protein is a potential vaccine target for developing much-required antimicrobials. These immune simulation results also indicated the potential of PAL protein in immune activation to target this emerging infectious threat. In addition, in silico analysis yielded bepotastine as a possible lead to inhibit PAL-FN interaction based on docking, binding free energy scores, and molecular dynamics simulation.
Our study explored the novel interaction between PAL protein and FN. Protein interaction with host FN might expose integrin-binding module (RGD motif), which can further lead to integrin binding on host epithelial cells, followed by bacterial invasion (81–85). A PAL-FN interaction-mediated integrin signaling study might uncover in-depth knowledge of bacterial invasion and inflammation. Most intriguingly, several studies demonstrated that the bacterial twitching motility is associated with high virulence in the infection model compared to surface-associated motility (86, 87), signifying that PAL might play a role in virulence. Moreover, the molecular mechanism behind this phenomenon remains unknown. In addition, a study showed that fibronectin interaction ability with macrophage cells can modulate its functions (88). Based on our findings, we propose that the PAL protein itself or PAL-FN interaction might modulate macrophage cell activation, an interlinking system, a hypothesis that needs to be tested in the future.
Hence, the present study suggests that the PAL-FN complex binding exhibits the foremost role in host cell adhesion. In A. baumannii pathogenesis, the essential role of PAL protein was corroborated that maintains the bacterial cell motility and biofilm formation. Hence, the present result also highlights the potential of the PAL protein as a therapeutic target against A. baumannii infections.
MATERIALS AND METHODS
Materials.
Dulbecco modified Eagle medium (DMEM; HiMedia), HEPES (HiMedia), fetal bovine serum (FBS; HiMedia), amphotericin (HiMedia), vancomycin (HiMedia), gentamicin (HiMedia), the EvaGreen qPCR Mix Plus (Solis BioDyne), the EZAssay nitric oxide estimation kit (HiMedia), the Verso cDNA synthesis kit (Thermo-scientific), mouse polyclonal anti-His antibody (R&D Systems), mouse anti-FN antibodies (R&D Systems), horseradish peroxidase (HRP)-conjugated anti-mouse antibodies (R&D Systems), prestained protein ladder (HiMedia), an Amicon Ultra 3-kDa cutoff filter (Millipore), TMB substrate (HiMedia), human fibronectin (HiMedia, TC618), EEO low melting agarose (SRL), ethidium bromide (CDH), T4 DNA ligase kit (Thermo Scientific), isopropanol (CDH), ethanol (HiMedia), and Trypticase soy broth (HiMedia) were obtained from the indicated suppliers.
Cell lines and culture conditions.
The experiments here involved no animals or humans. The human pulmonary epithelial (A-549) cell line was procured from NCCS Pune and was cultured according to a published protocol (89). Briefly, A549 cells were maintained in DMEM containing 1% HEPES, 10% FBS, 2.5 μg/mL amphotericin, 50 μg/mL vancomycin, and 50 μg/mL gentamicin at 37°C in an atmosphere of 5% CO2. The passage of A549 cell lines was pewrformed after every 2- to 3-day interval using 0.05% trypsin-EDTA. For A. baumannii infection and host gene expression experiments, cells from the confluent flask were seeded in 96- and 6-well plates.
Bacterial strains and culture conditions.
A. baumannii wild-type (AB5075) and PAL mutant (AB5075-B02 and AB5075-H04) strains were taken from the transposon mutant library (90). The mutant strains were prepared by insertion of a tetracycline resistance marker in the pal gene (90). In the AB5075-B02 strain, a tetracycline resistance marker was inserted after 138 bases, whereas in AB5075-H04 a tetracycline resistance marker was inserted after 268 bases of the pal gene. A. baumannii wild-type culture was grown in Luria-Bertani (LB) media, whereas PAL mutant strains were grown in LB medium containing 10 μg/mL tetracycline at 37°C. To perform different assays, the bacterial strains were harvested by centrifugation, washed, and resuspended in sterile phosphate-buffered saline (PBS; 140 mM NaCl, 3 mM KCl, 5 mM Na2HPO47H2O, 2 mM KH2PO4 [pH 7.4]).
In silico analysis of host-pathogen interaction partners.
Identification and analysis of host-pathogen interactions (HPI) is an essential core step to studying A. baumannii infections. The computational prediction of host-pathogen protein interactions was carried out through the HPIDB server 3.0 (https://hpidb.igbb.msstate.edu/). HPIDB predicts HP-PPIs (host-pathogen protein-protein interactions) by serving homologous experimentally derived HP-PPIs as a template. The whole proteome of A. baumannii AYE was downloaded from UniProt (http://www.uniprot.org/) and was given as an input to predict interacting partners that are restricted to Homo sapiens. The cellular localization of interacting partners was determined by the online servers CELLO (91) and PSORTb (92). The bacterial adhesion protein must possess a transmembrane helix of either 0 or 1 and high virulence, so that the in vitro experiment of protein will not be hampered (93). Thus, the interacting partners were further screened based on the number of the transmembrane helix (TMHMM server) (94, 95) and virulence-associated factor (VFDB) (96, 97).
In silico analysis of host fibronectin protein and bacterial PAL protein interaction has been analyzed using molecular docking according to a published method (98). First, the protein PDB IDs, PAL (4G4X) and FN (2CG6), were downloaded from the RCSB protein data bank. Downloaded PDBs were subjected to interaction analysis using the PatchDock server, which provides the best 10 binding score solutions. Using fast interaction refinement in a molecular docking server (FireDock), the global binding energy was calculated from the 10 best scores (99). The protein-protein complex dissociation constant (Kd) was determined with the help of the Prodigy (PROtein binDIng enerGY prediction) server (100) that calculated dissociation constant from ΔG = RT ln(Kd), where R is the ideal gas constant (kcal K−1 mol−1) and T is the temperature in kelvins.
Adherence of Acinetobacter to immobilized FN proteins.
The binding of A. baumannii to immobilized FN protein was studied according to a published protocol (39, 101). Briefly, host FN (200 and 400 μg/mL) protein, casein, or bovine serum albumin (BSA; control, 400 μg/mL) was diluted in coating buffer 0.1 M sodium bicarbonate (0.25 M NaHCO3, 0.25 M Na2CO3 [pH 9.6]) and incubated overnight at 4°C. On the following day, the wells were washed with PBS three times and then blocked with 100 μL of PBST (PBS + 0.1% Tween 20) containing 2% BSA for 2 h at 37°C. For the binding assay, a 100-μL sample (1 × 105 CFU/mL AB5075 culture) was added per well, followed by incubation for 4 h at 37°C. After 4 h, the A549 cell medium was removed carefully, and the plates were rinsed twice with PBS. Adherent bacterial cells were scraped using PBST and plated onto an LA plate. Overnight incubated bacterial cells were counted per well and compared to controls.
For the inhibition assay, before Acinetobacter infection, the A549 cell monolayer was incubated in serum-free media containing mouse anti-human FN antibody (0 to 10 μg/mL) for 30 min. Treated cells were infected with a multiplicity of infection (MOI) of 10 (1 × 105 CFU/mL/well) in serum-free medium. After 6 h of incubation, A549 cells were washed twice with PBS, and cells were scraped with PBST. Then, 100 μL of PBST-scraped lysate was spread onto an LA plate, followed by overnight incubation (102).
Cloning and overexpression of pal gene and purification of the PAL protein.
A. baumannii AB5075 genomic DNA was isolated in accordance with a published protocol (4). The DNA sequence encoding the pal gene was obtained from UNIPORT, and primers were designed. The pal gene-specific primers were PALFw (5′-ACTGGATCCCTAATGGAGATGATGATGAA-3′) and PALRv (5′-GCAAGCTTCGGTTATTTTAATAGAGGAGGAAC-3′) that contained BamHI and HindIII restriction sites. The pal gene PCR product of 573 bp was ligated in the pET28a expression vector, with an N-terminal six-histidine tag to the sequence. The ligated plasmid was transformed in E. coli DH5α competent cells. The kanamycin-resistant recombinant colonies were screened via PCR using vector-specific T7 primer and digestion, followed by gene sequencing.
Transformed pal gene into the expression host C43 BL21(DE3) was grown in LB medium containing 50 μg/mL kanamycin at 37°C until the optical density at 600 nm (OD600) reached 0.5 to 0.6. The expression of the recombinant pal gene was induced at 16°C for 48 h using 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Induced cells were harvested by centrifugation (5,000 rpm, 10 min, 4°C), and pellets were resuspended in sonication buffer (150 mM NaH2PO4, 300 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 1 mg/mL lysozyme; pH 7.6) and disrupted by sonication (SONIC Vibra-cell) with 30-s pulses for eight cycles with a 1-min rest. For purification, the lysate was centrifuged (16,000 rpm, 20 min, 4°C), and the supernatant was loaded onto a Ni-affinity column to purify the His-tagged PAL protein. Protein fractions were checked for the presence of specific PAL protein by Western blotting with mouse polyclonal anti-His antibody, followed by sequencing by mass spectrometry.
PAL-FN binding analysis.
Far-Western blotting was performed to confirm the PAL-FN interaction (103, 104). PAL protein was mixed in a 1:1 ratio with 2× Laemmli buffer (0.125 M Tris-HCl [pH 6.8], 4% sodium dodecyl sulfate [SDS], 20% glycerol, 0.002% bromophenol blue, without β-mercaptoethanol), followed by incubation at room temperature for 10 min before electrophoresis in a 10% SDS (wt/vol) polyacrylamide gel. A prestained protein ladder was used as the molecular weight standard for gel. Samples were run on two separate duplicate gels. Both sets were transferred to a polyvinylidene difluoride membrane using a Bio-Rad Trans-Blot blotting kit. Overnight, the membranes were blocked in blocking buffer (1× PBS buffer [pH 7.2] with 1.5% BSA and 5% skim milk). Afterward, the polyvinylidene difluoride membrane was incubated with human FN (10 μg/mL) in PBS containing 2% NaCl, 5% fetal calf serum, and 0.05% Tween 80 at 4°C for 24 h (105) and then washed three times with PBST buffer. To detect bound FN, the membranes were incubated with mouse anti-FN antibodies in a 1:1,000 dilution in PBST overnight at 4°C, followed by another three washes. Incubation with the HRP-conjugated anti-mouse antibodies at a dilution of 1:1,000 was then performed for 1 h, followed by three washes with PBST buffer. The membrane was finally developed with TMB membrane peroxidase substrate. To confirm the anti-FN antibody’s specificity body, a control blot was exposed to the antibody without the FN binding step (45).
Growth kinetics.
Growth kinetics analysis of the wild-type and mutant strains was performed as described previously (22, 106). Briefly, a single colony of A. baumannii was inoculated and cultured in LB medium (37°C, 200 rpm, 16 h). Overnight-cultured cells were washed, and the bacterial CFU count was measured using serial dilution on the LA plate. Initial inocula of wild-type and mutant strains (1 × 104 CFU/mL) were added to 96-well plates containing 200 μL of fresh LB medium. The culture plate was incubated at 37°C, bacterial growth (OD600) was then measured at an interval of 2h until it reached the stationary phase.
Host cell viability determination in the presence of external FN.
The viability of A549 cell lines in the presence of exogenous FN (0, 200, and 400 μg/mL) was determined after infection (MOI of 10:1) with wild-type and mutant Acinetobacter strains using an MTT assay as previously described (107). Briefly, at 4 h postinfection, 10 μL of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (5 mg/mL) was added, followed by incubation for 3 h at 37°C. Cell medium was removed, and the formed purple formazan crystals were solubilized using dimethyl sulfoxide (DMSO; 100 μL/well). The cell viability of nontreated cells was measured by determining the absorbance at 570 nm, which is directly proportional to living cells. The results are expressed as the percent viability of the A549 cells (108).
Adhesion and invasiveness to A549 human alveolar pulmonary cells.
The role of the PAL protein in bacterial adhesion and invasion was evaluated by following a previously described procedure (89), with some modifications. Briefly, A549 pulmonary epithelial cells were cultured in 5% CO2 at 37°C. Confluent monolayers were washed twice with PBS, coinfected at an MOI of 100 per well, and incubated for 6 h at 37°C. The infected A549 cells were washed twice with PBS and then lysed in 500 μL of PBST (PBS with 0.1% TritonX-100) to examine bacterial adhesion. The cell monolayers were infected, as described above, to determine bacterial invasion. Each well was washed twice with PBS and treated with gentamicin (300 μg/mL) (109) to kill extracellular bacteria, followed by washing and dissolving in 500 μL of PBST. Dilutions of the adhesive and invasive lysates were plated onto an LA plate and incubated at 37°C for 18 h. The CFU were enumerated to determine the attached (adhesion assay) and invaded (invasion assay) bacteria with A549 cells compared to wild type-infected wells. Experiments were conducted in triplicates.
ROS estimation.
ROS production in respiratory epithelial A549 cells was quantified by using a nitroblue tetrazolium assay (110). The pulmonary A549 (1 × 105) cells were incubated overnight in a 96-well plate at 37°C. For bacterial infection, cell monolayers were washed twice with PBS and infected for 2 h with wild-type and mutant strains of A. baumannii at an MOI of 100. In contrast, noninfected cells were used as a control that contained DMEM only. After infection, the cells were washed twice with PBS and incubated with 100 μL of p-nitroblue tetrazolium chloride solution (1.6 mg/mL in PBS) at 37°C and 5% CO2 in the dark for 45 min. Then, A549 cells monolayer were washed twice with Hanks balanced salt solution and once with methanol, followed by the addition of 108 μL of 2 M KOH and 92 μL of DMSO to dissolve the crystals. The absorbance of the solubilized formazan was determined at OD620 using a BioTek synergyH1 microplate reader.
RNS estimation.
RNS were measured in bacterium-infected pulmonary A549 cells using the EZAssay nitric oxide estimation kit. Briefly, bacterial infection was performed similarly to the method used for cellular ROS estimation. After infection with wild-type (AB5075) and mutant strain (AB5075-PALB02 and AB5075-PALH04) bacterial cells, A549 cells were washed twice with PBS and incubated with a reducing agent to reduce nitrate to nitrite. Then, 50 μL each of Griess reagent-I and Griess reagent-II were added, followed by incubation on the plate at 37°C for 4 h. The absorbance was recorded at OD580, which was was proportional to the total nitric oxide produced by the cells.
Quantitative biofilm assay.
To analyze the role of bacterial PAL protein in biofilm formation, A. baumannii wild-type (AB5075) and PAL mutant (AB5075-PALB02 and AB5075-PALH04) strains were streaked onto an LA plate, and single colonies of all strains were inoculated in tryptic soy broth (TSB; 5 mL) containing 0.25% (wt/vol) glucose (37°C, 12 h, 200 rpm) (111). Overnight cultures were pelleted, washed, and resuspended in 5 mL of TSB, and bacterial cells (1 × 105 CFU/well) were incubated (30°C, 48 h) in 6-well ELISA plates under static conditions (112). To measure the biofilm formation, total cell biomasses were first quantified at OD600, and free-floating bacterial cells were washed. Crystal violet (500 μL of a 1% solution) staining of Acinetobacter biofilms was performed at room temperature for 30 min. Stained biofilms were dissolved in 95% ethanol and quantified at OD580 using a BioTek synergyH1 microplate reader. To avoid the bacterial growth differences in various experimental setups, the OD580/OD600 ratio was measured to normalize the amount of biofilm formed to the total cell biomass. Biofilm formation was scored according to a published protocol (113).
Motility assays.
The effects of peptidoglycan-associated lipoprotein on the cell motility and bacterial movement of wild-type and mutant A. baumannii strains were assessed. For swimming motility, A. baumannii wild-type and PAL mutant strains were inoculated onto an LA agar plate and incubated overnight at 37°C. To analyze the swimming motility, bacterial strains were inoculated as 1-μL aliquots of 1 × 108 CFU/mL bacterial cells on a swimming plate in tryptone broth (10 g/L tryptone, 5 g/L NaCl) that contained 0.3% (wt/vol) agarose, followed by incubation at 30°C for 12 to 14 h. For swarming motility, swarming medium plates (0.5% [wt/vol] agar, 8 g/L nutrient broth, 5 g/L glucose) were prepared and allowed to dry at room temperature overnight before being used. The swarming efficiency of bacteria was analyzed by inoculating 1-μL aliquots of the bacterial sample containing 1 × 108 CFU/mL bacteria. The swarming motility was observed on incubated plates after overnight incubation at 30°C. For twitching motility, a twitching motility-associated stab assay was performed on a plate containing LB broth (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl) solidified with 1% (wt/vol) agar. A single-colony inoculum was picked from an overnight-grown LB agar (1.5% [wt/vol]) plate using a toothpick and stabbed to the bottom of the petri dish. After incubation at 37°C for 24 h, the bacterial surface was measured for twitching motility with 0.1% (wt/vol) crystal violet staining of the petri dish surface (114).
Cell permeability assay.
The cell permeability of bacteria was estimated by using an EtBr accumulation assay (115, 116). Briefly, wild-type AB5075 and mutant AB5075-PALB02 and AB5075-PALH04 cells were grown overnight at 37°C. The bacterial cultures were pelleted (5,000 rpm, 5 min), washed, and adjusted to 1 × 105 CFU/mL. The wells containing bacterial suspension without EtBr were kept as controls. For an accumulation assay for the energy source and nonspecific substrate, glucose (0.4%) and EtBr (0.5 μg/mL) were added. The fluorescence emission was examined for 40 min (2-min intervals) using fluorescence spectrophotometer (i.e., a BioTek synergyH1 microplate reader) with excitation (530-nm) and emission (600-nm) wavelengths.
Interaction of PAL protein with TLR2 and MARCO receptor.
To correlate the interaction analysis of PAL protein with TLR2 and MARCO receptor, TLR2 (2Z81) and MARCO receptor (2OY3) PDB IDs were downloaded from the RCSB protein data bank. Interaction analysis was performed using the Patchdock server, and the best 10 complex energy refinements were determined using the FireDock server. Molecular dynamics (MD) simulations of the top-ranked protein-protein complex were performed using GROMACS version 5.4 (3).
Screening of the PAL protein as a vaccine target and immune simulations.
PAL protein is a lipoprotein with host fibronectin interaction properties and virulence-associated factors with potential vaccine targets. To identify this protein’s antigenic and immunogenic property, a reverse vaccinology technique has been applied in accordance with an earlier protocol (117). Briefly, in this approach, PAL protein, MHC-I (NetCTLpan server), MHC-II (IEDB server), and B-cell (Bepipred Linear Epitope Prediction 2.0) interaction epitopes have been analyzed. The analyzed epitopes were further screened based on their class I immunogenicity (IEDB), antigenicity (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html), allergenicity (https://ddg-pharmfac.net/AllergenFP/), and toxicity (https://webs.iiitd.edu.in/raghava/toxinpred/multi_submit.php) scores. Physicochemical property analysis of PAL vaccine was done using the ProtParam server (118). Furthermore, the role of PAL protein-mediated vaccine in immune (humoral and cellular) simulation was examined by using the C-ImmSim online server (http://kraken.iac.rm.cnr.it/C-IMMSIM/), which works based on the Celada-Seiden model (119). The simulation was performed with default parameters (120) with no LPS on day 1, at 4 weeks, at 16 weeks, and at 24 weeks with a multiplication factor 0.2 and an infectivity of 0.6. The different immune cell responses were analyzed in the host after vaccine injection.
Virtual screening of leads via molecular docking and dynamics simulation against the PAL-FN complex.
Virtual screening of drugs and molecular docking was performed using Schrödinger (3). Briefly, the FDA-approved compound library from the ZINC database containing 2,924 compounds was used for the screening. The LIGPREP tool was used for ligand preparation, and ADMET analysis was conducted. Virtual screening was performed by three different methods: high-throughput virtual screening (HTVS) docking, standard precision (SP), and extra precision (XP), and the binding free energies were calculated by MMGBSA (molecular mechanics-generalized Born surface area). MD simulations of the top-ranked PAL-ligand complex were examined using Desmond software using published methods (121).
Statistics.
All reported values are as means ± the standard deviations (SD). One- or two-way analysis of variance was performed to evaluate the randomness of the data and a measure of overall statistical significance between the wild-type and mutant strains. Statistical significance was identified using GraphPad Prism version 5 software. Significance (P) values are indicated by asterisks in the figures (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001).
Data availability.
Figures S1 to S6 and Tables S1 to S5 in the supplemental material are available online (https://doi.org/10.6084/m9.figshare.22248886.v1).
ACKNOWLEDGMENTS
V.T. thanks SERB, India, for funding (SB/YS/LS-07/2014). V.T. thanks RCB, Faridabad, for Mass spectroscopy on a payment basis. V.S. thanks UGC, India, for the JRF fellowship [2061530812/21/06/2015(i)-EU-V] for her Ph.D. fellowship.
V.T. thanks the Department of Biochemistry CURAJ for the use of fluorescence spectrophotometer (BioTek, synergyH1 microplate reader).
V.T. conceived and designed the study. V.S., M.T., and V.T performed the experiments. V.T. analyzed the data. V.T. provided chemicals. V.T. wrote the manuscript. All authors proofread the final version of the manuscript.
We declare that we have no competing interests.
Contributor Information
Vishvanath Tiwari, Email: vishvanath@curaj.ac.in.
Denise Monack, Stanford University.
REFERENCES
- 1.Kyriakidis I, Vasileiou E, Pana ZD, Tragiannidis A. 2021. Acinetobacter baumannii antibiotic resistance mechanisms. Pathogens 10:373. doi: 10.3390/pathogens10030373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verma P, Tiwari M, Tiwari V. 2022. Potentiate the activity of current antibiotics by naringin dihydrochalcone targeting the AdeABC efflux pump of multidrug-resistant Acinetobacter baumannii. Int J Biol Macromol 217:592–605. doi: 10.1016/j.ijbiomac.2022.07.065. [DOI] [PubMed] [Google Scholar]
- 3.Verma P, Tiwari V. 2018. Targeting outer membrane protein component AdeC for the discovery of efflux pump inhibitor against AdeABC efflux pump of multidrug resistant Acinetobacter baumannii. Cell Biochem Biophys 76:391–400. doi: 10.1007/s12013-018-0846-5. [DOI] [PubMed] [Google Scholar]
- 4.Tiwari V, Moganty RR. 2014. Conformational stability of OXA-51 β-lactamase explains its role in carbapenem resistance of Acinetobacter baumannii. J Biomol Struct Dyn 32:1406–1420. doi: 10.1080/07391102.2013.819789. [DOI] [PubMed] [Google Scholar]
- 5.Tiwari M, Panwar S, Kothidar A, Tiwari V. 2020. Rational targeting of Wzb phosphatase and Wzc kinase interaction inhibits extracellular polysaccharides synthesis and biofilm formation in Acinetobacter baumannii. Carbohydr Res 492:108025. doi: 10.1016/j.carres.2020.108025. [DOI] [PubMed] [Google Scholar]
- 6.Roy R, Tiwari M, Donelli G, Tiwari V. 2018. Strategies for combating bacterial biofilms: a focus on anti-biofilm agents and their mechanisms of action. Virulence 9:522–554. doi: 10.1080/21505594.2017.1313372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asokan GV, Ramadhan T, Ahmed E, Sanad H. 2019. WHO Global Priority Pathogens List: a bibliometric analysis of Medline-PubMed for knowledge mobilization to infection prevention and control practices in Bahrain. Oman Medical J 34:184–193. doi: 10.5001/omj.2019.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knight GM, Glover RE, McQuaid CF, Olaru ID, Gallandat K, Leclerc QJ, Fuller NM, Willcocks SJ, Hasan R, van Kleef E, Chandler CI. 2021. Antimicrobial resistance and COVID-19: intersections and implications. Elife 10:e64139. doi: 10.7554/eLife.64139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu X, Ge Y, Wu T, Zhao K, Chen Y, Wu B, Zhu F, Zhu B, Cui L. 2020. Coinfection with respiratory pathogens among COVID-2019 cases. Virus Res 285:198005. doi: 10.1016/j.virusres.2020.198005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lima WG, Brito JCM, da Cruz Nizer WS. 2020. Ventilator-associated pneumonia (VAP) caused by carbapenem-resistant Acinetobacter baumannii in patients with COVID-19: two problems, one solution? Med Hypotheses 144:110139. doi: 10.1016/j.mehy.2020.110139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smani Y, McConnell MJ, Pachón J. 2012. Role of fibronectin in the adhesion of Acinetobacter baumannii to host cells. PLoS One 7:e33073. doi: 10.1371/journal.pone.0033073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Godlewska R, Wiśniewska K, Pietras Z, Jagusztyn-Krynicka EK. 2009. Peptidoglycan-associated lipoprotein (Pal) of Gram-negative bacteria: function, structure, role in pathogenesis and potential application in immunoprophylaxis. FEMS Microbiol Lett 298:1–11. doi: 10.1111/j.1574-6968.2009.01659.x. [DOI] [PubMed] [Google Scholar]
- 13.Dyke JS, Huertas-Diaz MC, Michel F, Holladay NE, Hogan RJ, He B, Lafontaine ER. 2020. The peptidoglycan-associated lipoprotein Pal contributes to the virulence of Burkholderia mallei and provides protection against lethal aerosol challenge. Virulence 11:1024–1040. doi: 10.1080/21505594.2020.1804275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ceremuga I, Seweryn E, Bednarz-Misa I, Pietkiewicz J, Jermakow K, Banas T, Gamian A. 2014. Enolase-like protein present on the outer membrane of Pseudomonas aeruginosa binds plasminogen. Folia Microbiol (Praha) 59:391–397. doi: 10.1007/s12223-014-0311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi N, Nishino K, Hirata T, Yamaguchi A. 2003. Membrane topology of ABC-type macrolide antibiotic exporter MacB in Escherichia coli. FEBS Lett 546:241–246. doi: 10.1016/s0014-5793(03)00579-9. [DOI] [PubMed] [Google Scholar]
- 16.Peng S, Fitzen M, Jornvall H, Johansson J. 2010. The extracellular domain of Bri2 (ITM2B) binds the ABri peptide (1-23) and amyloid beta-peptide (Abeta1-40): implications for Bri2 effects on processing of amyloid precursor protein and Abeta aggregation. Biochem Biophys Res Commun 393:356–361. doi: 10.1016/j.bbrc.2009.12.122. [DOI] [PubMed] [Google Scholar]
- 17.Koide A, Bailey CW, Huang X, Koide S. 1998. The fibronectin type III domain as a scaffold for novel binding proteins. J Mol Biol 284:1141–1151. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- 18.Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. 1987. Structure of the human class I histocompatibility antigen, HLA-A2. Nature 329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- 19.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. 2005. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res 33:W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metinko A. 2004. Neonatal pulmonary host defense mechanisms, p 1620–1673. In Polin RA, Fox WW, Abman SH (ed), Fetal and neonatal physiology, 3rd ed. WB Saunders, Philadelphia, PA. doi: 10.1016/B978-0-7216-9654-6.50167-3. [DOI] [Google Scholar]
- 21.Bohuszewicz O, Liu J, Low HH. 2016. Membrane remodelling in bacteria. J Struct Biol 196:3–14. doi: 10.1016/j.jsb.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy TF, Kirkham C, Lesse AJ. 2006. Construction of a mutant and characterization of the role of the vaccine antigen P6 in outer membrane integrity of nontypeable Haemophilus influenzae. Infect Immun 74:5169–5176. doi: 10.1128/IAI.00692-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paulsson M, Riesbeck K. 2018. How bacteria hack the matrix and dodge the bullets of immunity. Eur Respir Rev 27:180018. doi: 10.1183/16000617.0018-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vijayakumar S, Rajenderan S, Laishram S, Anandan S, Veeraraghavan B, Biswas I. 2016. Biofilm formation and motility depend on the nature of the Acinetobacter baumannii clinical isolates. Front Public Health 4. doi: 10.3389/fpubh.2016.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harding CM, Hennon SW, Feldman MF. 2018. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol 16:91–102. doi: 10.1038/nrmicro.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tipton KA, Rather PN. 2017. An ompR-envZ two-component system ortholog regulates phase variation, osmotic tolerance, motility, and virulence in Acinetobacter baumannii strain AB5075. J Bacteriol 199:e00705-16. doi: 10.1128/JB.00705-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moffatt JH, Harper M, Mansell A, Crane B, Fitzsimons TC, Nation RL, Li J, Adler B, Boyce JD. 2013. Lipopolysaccharide-deficient Acinetobacter baumannii shows altered signaling through host Toll-like receptors and increased susceptibility to the host antimicrobial peptide LL-37. Infect Immun 81:684–689. doi: 10.1128/IAI.01362-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang M-J, Jang A-R, Park J-Y, Ahn J-H, Lee T-S, Kim D-Y, Lee M-S, Hwang S, Jeong Y-J, Park J-H. 2020. IL-10 protects mice from the lung infection of Acinetobacter baumannii and contributes to bacterial clearance by regulating STAT3-mediated MARCO expression in macrophages. Front Immunol 11:270. doi: 10.3389/fimmu.2020.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukouhara T, Arimoto T, Cho K, Yamamoto M, Igarashi T. 2011. Surface lipoprotein PpiA of Streptococcus mutans suppresses scavenger receptor MARCO-dependent phagocytosis by macrophages. Infect Immun 79:4933–4940. doi: 10.1128/IAI.05693-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris FC, Dexter C, Kostoulias X, Uddin MI, Peleg AY. 2019. The mechanisms of disease caused by Acinetobacter baumannii. Front Microbiol 10:1601–1601. doi: 10.3389/fmicb.2019.01601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tiwari M, Roy R, Tiwari V. 2016. Screening of herbal-based bioactive extract against carbapenem-resistant strain of Acinetobacter baumannii. Microb Drug Resist 22:364–371. doi: 10.1089/mdr.2015.0270. [DOI] [PubMed] [Google Scholar]
- 32.Tiwari V, Kapil A, Moganty RR. 2012. Carbapenem-hydrolyzing oxacillinase in high resistant strains of Acinetobacter baumannii isolated from India. Microb Pathog 53:81–86. doi: 10.1016/j.micpath.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 33.Tiwari V, Tiwari M, Biswas D. 2018. Rationale and design of an inhibitor of RecA protein as an inhibitor of Acinetobacter baumannii. J Antibiot (Tokyo) 71:522–534. doi: 10.1038/s41429-018-0026-2. [DOI] [PubMed] [Google Scholar]
- 34.Tiwari V, Vashistt J, Kapil A, Moganty RR. 2012. Comparative proteomics of inner membrane fraction from carbapenem-resistant Acinetobacter baumannii with a reference strain. PLoS One 7:e39451. doi: 10.1371/journal.pone.0039451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonzalez-Villoria AM, Valverde-Garduno V. 2016. Antibiotic-resistant Acinetobacter baumannii increasing success remains a challenge as a nosocomial pathogen. J Pathog 2016:7318075. doi: 10.1155/2016/7318075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee C-R, Lee JH, Park M, Park KS, Bae IK, Kim YB, Cha C-J, Jeong BC, Lee SH. 2017. Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front Cell Infect Microbiol 7:55. doi: 10.3389/fcimb.2017.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong D, Nielsen TB, Bonomo RA, Pantapalangkoor P, Luna B, Spellberg B. 2017. Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin Microbiol Rev 30:409–447. doi: 10.1128/CMR.00058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi CH, Lee JS, Lee YC, Park TI, Lee JC. 2008. Acinetobacter baumannii invades epithelial cells and outer membrane protein A mediates interactions with epithelial cells. BMC Microbiol 8:216. doi: 10.1186/1471-2180-8-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dabo SM, Confer AW, Hartson SD. 2005. Adherence of Pasteurella multocida to fibronectin. Vet Microbiol 110:265–275. doi: 10.1016/j.vetmic.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 40.Yin H, Flynn AD. 2016. Drugging membrane protein interactions. Annu Rev Biomed Eng 18:51–76. doi: 10.1146/annurev-bioeng-092115-025322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaca DJ, Thibau A, Schütz M, Kraiczy P, Happonen L, Malmström J, Kempf VAJ. 2020. Interaction with the host: the role of fibronectin and extracellular matrix proteins in the adhesion of Gram-negative bacteria. Med Microbiol Immunol 209:277–299. doi: 10.1007/s00430-019-00644-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi CH, Hyun SH, Lee JY, Lee JS, Lee YS, Kim SA, Chae JP, Yoo SM, Lee JC. 2008. Acinetobacter baumannii outer membrane protein A targets the nucleus and induces cytotoxicity. Cell Microbiol 10:309–319. [DOI] [PubMed] [Google Scholar]
- 43.Gaddy JA, Actis LA. 2009. Regulation of Acinetobacter baumannii biofilm formation. Future Microbiol 4:273–278. doi: 10.2217/fmb.09.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dallo SF, Zhang B, Denno J, Hong S, Tsai A, Haskins W, Ye JY, Weitao T. 2012. Association of Acinetobacter baumannii EF-Tu with cell surface, outer membrane vesicles, and fibronectin. ScientificWorldJournal 2012:128705. doi: 10.1100/2012/128705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smani Y, Dominguez-Herrera J, Pachón J. 2013. Association of the outer membrane protein Omp33 with fitness and virulence of Acinetobacter baumannii. J Infect Dis 208:1561–1570. doi: 10.1093/infdis/jit386. [DOI] [PubMed] [Google Scholar]
- 46.Smani Y, Docobo-Pérez F, López-Rojas R, Domínguez-Herrera J, Ibáñez-Martínez J, Pachón J. 2012. Platelet-activating factor receptor initiates contact of Acinetobacter baumannii expressing phosphorylcholine with host cells. J Biol Chem 287:26901–26910. doi: 10.1074/jbc.M112.344556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bentancor LV, Camacho-Peiro A, Bozkurt-Guzel C, Pier GB, Maira-Litrán T. 2012. Identification of Ata, a multifunctional trimeric autotransporter of Acinetobacter baumannii. J Bacteriol 194:3950–3960. doi: 10.1128/JB.06769-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheah S-E, Li J, Tsuji BT, Forrest A, Bulitta JB, Nation RL. 2016. Colistin and polymyxin B dosage regimens against Acinetobacter baumannii: differences in activity and the emergence of resistance. Antimicrob Agents Chemother 60:3921–3933. doi: 10.1128/AAC.02927-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Henry R, Vithanage N, Harrison P, Seemann T, Coutts S, Moffatt JH, Nation RL, Li J, Harper M, Adler B, Boyce JD. 2012. Colistin-resistant, lipopolysaccharide-deficient Acinetobacter baumannii responds to lipopolysaccharide loss through increased expression of genes involved in the synthesis and transport of lipoproteins, phospholipids, and poly-β-1,6-N-acetylglucosamine. Antimicrob Agents Chemother 56:59–69. doi: 10.1128/AAC.05191-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cordwell SJ, Len AC, Touma RG, Scott NE, Falconer L, Jones D, Connolly A, Crossett B, Djordjevic SP. 2008. Identification of membrane-associated proteins from Campylobacter jejuni strains using complementary proteomics technologies. Proteomics 8:122–139. doi: 10.1002/pmic.200700561. [DOI] [PubMed] [Google Scholar]
- 51.Romero-Saavedra F, Laverde D, Wobser D, Michaux C, Budin-Verneuil A, Bernay B, Benachour A, Hartke A, Huebner J. 2014. Identification of peptidoglycan-associated proteins as vaccine candidates for enterococcal infections. PLoS One 9:e111880. doi: 10.1371/journal.pone.0111880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wessel AK, Liew J, Kwon T, Marcotte EM, Whiteley M. 2013. Role of Pseudomonas aeruginosa peptidoglycan-associated outer membrane proteins in vesicle formation. J Bacteriol 195:213–219. doi: 10.1128/JB.01253-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu F, Li J, Yan K, Li H, Sun C, Zhang S, Yuan F, Wang X, Tan C, Chen H, Bei W. 2017. Binding of fibronectin to SsPepO facilitates the development of Streptococcus suis meningitis. J Infect Dis 217:973–982. doi: 10.1093/infdis/jix523. [DOI] [PubMed] [Google Scholar]
- 54.Unkmeir A, Latsch K, Dietrich G, Wintermeyer E, Schinke B, Schwender S, Kim KS, Eigenthaler M, Frosch M. 2002. Fibronectin mediates Opc-dependent internalization of Neisseria meningitidis in human brain microvascular endothelial cells. Mol Microbiol 46:933–946. doi: 10.1046/j.1365-2958.2002.03222.x. [DOI] [PubMed] [Google Scholar]
- 55.Shinji H, Yosizawa Y, Tajima A, Iwase T, Sugimoto S, Seki K, Mizunoe Y. 2011. Role of fibronectin-binding proteins A and B in in vitro cellular infections and in vivo septic infections by Staphylococcus aureus. Infect Immun 79:2215–2223. doi: 10.1128/IAI.00133-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rubio T, Gagné S, Debruyne C, Dias C, Cluzel C, Mongellaz D, Rousselle P, Göttig S, Seifert H, Higgins PG, Salcedo SP. 2021. Incidence of an intracellular multiplication niche amongst Acinetobacter baumannii. bioRxiv. https://www.biorxiv.org/content/10.1101/2021.04.15.439986v1. [DOI] [PMC free article] [PubMed]
- 57.Eveillard M, Soltner C, Kempf M, Saint-André JP, Lemarié C, Randrianarivelo C, Seifert H, Wolff M, Joly-Guillou ML. 2010. The virulence variability of different Acinetobacter baumannii strains in experimental pneumonia. J Infect 60:154–161. doi: 10.1016/j.jinf.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 58.Li W, Wan Y, Tao Z, Chen H, Zhou R. 2013. A novel fibronectin-binding protein of Streptococcus suis serotype 2 contributes to epithelial cell invasion and in vivo dissemination. Vet Microbiol 162:186–194. doi: 10.1016/j.vetmic.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 59.Cirillo SL, Subbian S, Chen B, Weisbrod TR, Jacobs WR, Jr, Cirillo JD. 2009. Protection of Mycobacterium tuberculosis from reactive oxygen species conferred by the mel2 locus impacts persistence and dissemination. Infect Immun 77:2557–2567. doi: 10.1128/IAI.01481-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun D, Crowell SA, Harding CM, De Silva PM, Harrison A, Fernando DM, Mason KM, Santana E, Loewen PC, Kumar A, Liu Y. 2016. KatG and KatE confer Acinetobacter resistance to hydrogen peroxide but sensitize bacteria to killing by phagocytic respiratory burst. Life Sci 148:31–40. doi: 10.1016/j.lfs.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zeidler S, Müller V. 2019. Coping with low water activities and osmotic stress in Acinetobacter baumannii: significance, current status and perspectives. Environ Microbiol 21:2212–2230. doi: 10.1111/1462-2920.14565. [DOI] [PubMed] [Google Scholar]
- 62.Zeighami H, Valadkhani F, Shapouri R, Samadi E, Haghi F. 2019. Virulence characteristics of multidrug resistant biofilm forming Acinetobacter baumannii isolated from intensive care unit patients. BMC Infect Dis 19:629. doi: 10.1186/s12879-019-4272-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blaschke U, Skiebe E, Wilharm G. 2021. Novel genes required for surface-associated motility in Acinetobacter baumannii. Curr Microbiol 78:1509–1528. doi: 10.1007/s00284-021-02407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Szczepaniak J, Press C, Kleanthous C. 2020. The multifarious roles of Tol-Pal in Gram-negative bacteria. FEMS Microbiol Rev 44:490–506. doi: 10.1093/femsre/fuaa018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Samsudin F, Ortiz-Suarez ML, Piggot TJ, Bond PJ, Khalid S. 2016. OmpA: a flexible clamp for bacterial cell wall attachment. Structure 24:2227–2235. doi: 10.1016/j.str.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 66.Gellings PS, Wilkins AA, Morici LA. 2020. Recent advances in the pursuit of an effective Acinetobacter baumannii vaccine. Pathogens 9:1066. doi: 10.3390/pathogens9121066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ahmad TA, Tawfik DM, Sheweita SA, Haroun M, El-Sayed LH. 2016. Development of immunization trials against Acinetobacter baumannii. Trials Vaccinol 5:53–60. doi: 10.1016/j.trivac.2016.03.001. [DOI] [Google Scholar]
- 68.McConnell MJ, Pachón J. 2010. Active and passive immunization against Acinetobacter baumannii using an inactivated whole cell vaccine. Vaccine 29:1–5. doi: 10.1016/j.vaccine.2010.10.052. [DOI] [PubMed] [Google Scholar]
- 69.Huang W, Yao Y, Long Q, Yang X, Sun W, Liu C, Jin X, Li Y, Chu X, Chen B, Ma Y. 2014. Immunization against multidrug-resistant Acinetobacter baumannii effectively protects mice in both pneumonia and sepsis models. PLoS One 9:e100727. doi: 10.1371/journal.pone.0100727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Russo TA, Beanan JM, Olson R, MacDonald U, Cox AD, St Michael F, Vinogradov EV, Spellberg B, Luke-Marshall NR, Campagnari AA. 2013. The K1 capsular polysaccharide from Acinetobacter baumannii is a potential therapeutic target via passive immunization. Infect Immun 81:915–922. doi: 10.1128/IAI.01184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luo G, Lin L, Ibrahim AS, Baquir B, Pantapalangkoor P, Bonomo RA, Doi Y, Adams MD, Russo TA, Spellberg B. 2012. Active and passive immunization protects against lethal, extreme drug resistant-Acinetobacter baumannii infection. PLoS One 7:e29446. doi: 10.1371/journal.pone.0029446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim SW, Choi CH, Moon DC, Jin JS, Lee JH, Shin J-H, Kim JM, Lee YC, Seol SY, Cho DT, Lee JC. 2009. Serum resistance of Acinetobacter baumannii through the binding of factor H to outer membrane proteins. FEMS Microbiol Lett 301:224–231. doi: 10.1111/j.1574-6968.2009.01820.x. [DOI] [PubMed] [Google Scholar]
- 73.Huang W, Yao Y, Wang S, Xia Y, Yang X, Long Q, Sun W, Liu C, Li Y, Chu XJ, Bai H, Yao Y, Ma Y. 2016. Immunization with a 22-kDa outer membrane protein elicits protective immunity to multidrug-resistant Acinetobacter baumannii. Sci Rep 6:20724. doi: 10.1038/srep20724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang W, Wang S, Yao Y, Xia Y, Yang X, Long Q, Sun W, Liu C, Li Y, Ma Y. 2015. OmpW is a potential target for eliciting protective immunity against Acinetobacter baumannii infections. Vaccine 33:4479–4485. doi: 10.1016/j.vaccine.2015.07.031. [DOI] [PubMed] [Google Scholar]
- 75.McConnell MJ, Domínguez-Herrera J, Smani Y, López-Rojas R, Docobo-Pérez F, Pachón J. 2011. Vaccination with outer membrane complexes elicits rapid protective immunity to multidrug-resistant Acinetobacter baumannii. Infect Immun 79:518–526. doi: 10.1128/IAI.00741-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.García-Quintanilla M, Pulido MR, Pachón J, McConnell M. 2014. Immunization with lipopolysaccharide-deficient whole cells provides protective immunity in an experimental mouse model of Acinetobacter baumannii infection. PLoS One 9:e114410. doi: 10.1371/journal.pone.0114410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chiang M-H, Sung W-C, Lien S-P, Chen Y-Z, Lo AF, Huang J-H, Kuo S-C, Chong P. 2015. Identification of novel vaccine candidates against Acinetobacter baumannii using reverse vaccinology. Hum Vaccin Immunother 11:1065–1073. doi: 10.1080/21645515.2015.1010910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sangroodi YH, Rasooli I, Nazarian S, Ebrahimizadeh W, Sefid F. 2015. Immunogenicity of conserved cork and β-barrel domains of baumannii acinetobactin utilization protein in an animal model. Turk J Med Sci 45:1396–1402. doi: 10.3906/sag-1407-45. [DOI] [PubMed] [Google Scholar]
- 79.Ma C, McClean S. 2021. Mapping global prevalence of Acinetobacter baumannii and recent vaccine development to tackle it. Vaccines 9:570. doi: 10.3390/vaccines9060570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lei L, Yang F, Zou J, Jing H, Zhang J, Xu W, Zou Q, Zhang J, Wang X. 2019. DNA vaccine encoding OmpA and Pal from Acinetobacter baumannii efficiently protects mice against pulmonary infection. Mol Biol Rep 46:5397–5408. doi: 10.1007/s11033-019-04994-2. [DOI] [PubMed] [Google Scholar]
- 81.Schwarz-Linek U, Höök M, Potts JR. 2004. The molecular basis of fibronectin-mediated bacterial adherence to host cells. Mol Microbiol 52:631–641. doi: 10.1111/j.1365-2958.2004.04027.x. [DOI] [PubMed] [Google Scholar]
- 82.Hauck CR, Ohlsen K. 2006. Sticky connections: extracellular matrix protein recognition and integrin-mediated cellular invasion by Staphylococcus aureus. Curr Opin Microbiol 9:5–11. doi: 10.1016/j.mib.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 83.Foster TJ. 2016. The remarkably multifunctional fibronectin binding proteins of Staphylococcus aureus. Eur J Clin Microbiol Infect Dis 35:1923–1931. doi: 10.1007/s10096-016-2763-0. [DOI] [PubMed] [Google Scholar]
- 84.Liang X, Garcia BL, Visai L, Prabhakaran S, Meenan NA, Potts JR, Humphries MJ, Höök M. 2016. Allosteric regulation of fibronectin/α5β1 interaction by fibronectin-binding MSCRAMMs. PLoS One 11:e0159118. doi: 10.1371/journal.pone.0159118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Prystopiuk V, Feuillie C, Herman-Bausier P, Viela F, Alsteens D, Pietrocola G, Speziale P, Dufrêne YF. 2018. Mechanical forces guiding Staphylococcus aureus cellular invasion. ACS Nano 12:3609–3622. doi: 10.1021/acsnano.8b00716. [DOI] [PubMed] [Google Scholar]
- 86.Ayoub MC, Hammoudi HD. 2020. Insights into Acinetobacter baumannii: a review of microbiological, virulence, and resistance traits in a threatening nosocomial pathogen. Antibiotics (Basel) 9:119. doi: 10.3390/antibiotics9030119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Burrows LL. 2012. Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu Rev Microbiol 66:493–520. doi: 10.1146/annurev-micro-092611-150055. [DOI] [PubMed] [Google Scholar]
- 88.Schmidt DR, Kao WJ. 2007. The interrelated role of fibronectin and interleukin-1 in biomaterial-modulated macrophage function. Biomaterials 28:371–382. doi: 10.1016/j.biomaterials.2006.08.041. [DOI] [PubMed] [Google Scholar]
- 89.Tiwari V, Tiwari M, Solanki V. 2017. Polyvinylpyrrolidone-capped silver nanoparticle inhibits infection of carbapenem-resistant strain of Acinetobacter baumannii in the human pulmonary epithelial cell. Front Immunol 8:973. doi: 10.3389/fimmu.2017.00973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gallagher LA, Ramage E, Weiss EJ, Radey M, Hayden HS, Held KG, Huse HK, Zurawski DV, Brittnacher MJ, Manoil C. 2015. Resources for genetic and genomic analysis of emerging pathogen Acinetobacter baumannii. J Bacteriol 197:2027–2035. doi: 10.1128/JB.00131-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yu CS, Chen YC, Lu CH, Hwang JK. 2006. Prediction of protein subcellular localization. Proteins 64:643–651. doi: 10.1002/prot.21018. [DOI] [PubMed] [Google Scholar]
- 92.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FS. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.He Y, Xiang Z, Mobley HLT. 2010. Vaxign: the First web-based vaccine design program for reverse vaccinology and applications for vaccine development. J Biomed Biotechnol 2010:297505. doi: 10.1155/2010/297505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Solanki V, Tiwari V. 2018. Subtractive proteomics to identify novel drug targets and reverse vaccinology for the development of chimeric vaccine against Acinetobacter baumannii. Sci Rep 8:9044. doi: 10.1038/s41598-018-26689-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 96.Solanki V, Tiwari M, Tiwari V. 2019. Prioritization of potential vaccine targets using comparative proteomics and designing of the chimeric multi-epitope vaccine against Pseudomonas aeruginosa. Sci Rep 9:5240–5240. doi: 10.1038/s41598-019-41496-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen L, Yang J, Yu J, Yao Z, Sun L, Shen Y, Jin Q. 2005. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res 33:D325–D328. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Solanki V, Tiwari M, Tiwari V. 2020. Subtractive proteomic analysis of antigenic extracellular proteins and design a multi-epitopes vaccine against Staphylococcus aureus. Microbiol Immunol 65:302–316. doi: 10.1111/1348-0421.12870. [DOI] [PubMed] [Google Scholar]
- 99.Mashiach E, Schneidman-Duhovny D, Peri A, Shavit Y, Nussinov R, Wolfson HJ. 2010. An integrated suite of fast docking algorithms. Proteins 78:3197–3204. doi: 10.1002/prot.22790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xue LC, Rodrigues JP, Kastritis PL, Bonvin AM, Vangone A. 2016. PRODIGY: a web server for predicting the binding affinity of protein-protein complexes. Bioinformatics 32:3676–3678. doi: 10.1093/bioinformatics/btw514. [DOI] [PubMed] [Google Scholar]
- 101.Chia JS, Yeh CY, Chen JY. 2000. Identification of a fibronectin binding protein from Streptococcus mutans. Infect Immun 68:1864–1870. doi: 10.1128/IAI.68.4.1864-1870.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fowler T, Wann ER, Joh D, Johansson S, Foster TJ, Höök M. 2000. Cellular invasion by Staphylococcus aureus involves a fibronectin bridge between the bacterial fibronectin-binding MSCRAMMs and host cell β1 integrins. Eur J Cell Biol 79:672–679. doi: 10.1078/0171-9335-00104. [DOI] [PubMed] [Google Scholar]
- 103.Vazquez V, Liang X, Horndahl JK, Ganesh VK, Smeds E, Foster TJ, Hook M. 2011. Fibrinogen is a ligand for the Staphylococcus aureus microbial surface components recognizing adhesive matrix molecules (MSCRAMM) bone sialoprotein-binding protein (Bbp). J Biol Chem 286:29797–29805. doi: 10.1074/jbc.M110.214981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee JH, Cho NH, Kim SY, Bang SY, Chu H, Choi MS, Kim IS. 2008. Fibronectin facilitates the invasion of Orientia tsutsugamushi into host cells through interaction with a 56-kDa type-specific antigen. J Infect Dis 198:250–257. doi: 10.1086/589284. [DOI] [PubMed] [Google Scholar]
- 105.de Greeff A, Buys H, Verhaar R, Dijkstra J, van Alphen L, Smith HE. 2002. Contribution of fibronectin-binding protein to pathogenesis of Streptococcus suis serotype 2. Infect Immun 70:1319–1325. doi: 10.1128/IAI.70.3.1319-1325.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Horáková K, Greifová M, Seemannová Z, Gondová B, Wyatt GM. 2004. A comparison of the traditional method of counting viable cells and a quick microplate method for monitoring the growth characteristics of Listeria monocytogenes. Lett Appl Microbiol 38:181–184. doi: 10.1111/j.1472-765X.2004.01448.x. [DOI] [PubMed] [Google Scholar]
- 107.Chen H-Y, Lin M-H, Chen C-C, Shu J-C. 2017. The expression of fibronectin is significantly suppressed in macrophages to exert a protective effect against Staphylococcus aureus infection. BMC Microbiol 17:92–92. doi: 10.1186/s12866-017-1003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Seidl K, Zinkernagel AS. 2013. The MTT assay is a rapid and reliable quantitative method to assess Staphylococcus aureus induced endothelial cell damage. J Microbiol Methods 92:307–309. doi: 10.1016/j.mimet.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 109.Schweppe DK, Harding C, Chavez JD, Wu X, Ramage E, Singh PK, Manoil C, Bruce JE. 2015. Host-microbe protein interactions during bacterial infection. Chem Biol 22:1521–1530. doi: 10.1016/j.chembiol.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.García-Pérez BE, Villagómez-Palatto DA, Castañeda-Sánchez JI, Coral-Vázquez RM, Ramírez-Sánchez I, Ordoñez-Razo RM, Luna-Herrera J. 2011. Innate response of human endothelial cells infected with mycobacteria. Immunobiology 216:925–935. doi: 10.1016/j.imbio.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 111.Avila-Novoa M-G, Iñíguez-Moreno M, Solís-Velázquez O-A, González-Gómez J-P, Guerrero-Medina P-J, Gutiérrez-Lomelí M. 2018. Biofilm formation by Staphylococcus aureus isolated from food contact surfaces in the dairy industry of Jalisco, Mexico. J Food Quality 2018:1746139. doi: 10.1155/2018/1746139. [DOI] [Google Scholar]
- 112.Tiwari V, Patel V, Tiwari M. 2018. In-silico screening and experimental validation reveal l-adrenaline as anti-biofilm molecule against biofilm-associated protein (Bap) producing Acinetobacter baumannii. Int J Biol Macromol 107:1242–1252. doi: 10.1016/j.ijbiomac.2017.09.105. [DOI] [PubMed] [Google Scholar]
- 113.Álvarez-Fraga L, Pérez A, Rumbo-Feal S, Merino M, Vallejo JA, Ohneck EJ, Edelmann RE, Beceiro A, Vázquez-Ucha JC, Valle J, Actis LA, Bou G, Poza M. 2016. Analysis of the role of the LH92_11085 gene of a biofilm hyper-producing Acinetobacter baumannii strain on biofilm formation and attachment to eukaryotic cells. Virulence 7:443–455. doi: 10.1080/21505594.2016.1145335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Biswas I, Machen A, Mettlach J. 2019. In vitro motility assays for Acinetobacter species. Methods Mol Biol 1946:177–187. doi: 10.1007/978-1-4939-9118-1_17. [DOI] [PubMed] [Google Scholar]
- 115.Pal S, Misra A, Banerjee S, Dam B. 2020. Adaptation of ethidium bromide fluorescence assay to monitor activity of efflux pumps in bacterial pure cultures or mixed population from environmental samples. J King Saud Univ Sci 32:939–945. doi: 10.1016/j.jksus.2019.06.002. [DOI] [Google Scholar]
- 116.Dennehy R, Romano M, Ruggiero A, Mohamed YF, Dignam SL, Mujica TC, Callaghan M, Valvano MA, Berisio R, McClean S. 2017. The Burkholderia cenocepacia peptidoglycan-associated lipoprotein is involved in epithelial cell attachment and elicitation of inflammation. Cell Microbiol 19. doi: 10.1111/cmi.12691. [DOI] [PubMed] [Google Scholar]
- 117.Solanki V, Sharma S, Tiwari V. 2021. Subtractive proteomics and reverse vaccinology strategies for designing a multiepitope vaccine targeting membrane proteins of Klebsiella pneumoniae. Int J Peptide Res Ther 27:1177–1195. doi: 10.1007/s10989-021-10159-2. [DOI] [Google Scholar]
- 118.Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. 1999. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol 112:531–552. [DOI] [PubMed] [Google Scholar]
- 119.Rapin N, Lund O, Bernaschi M, Castiglione F. 2010. Computational immunology meets bioinformatics: the use of prediction tools for molecular binding in the simulation of the immune system. PLoS One 5:e9862. doi: 10.1371/journal.pone.0009862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Solanki V, Tiwari M, Tiwari V. 2021. Immunoinformatic approach to design a multiepitope vaccine targeting non-mutational hot spot regions of structural and nonstructural proteins of the SARS CoV2. PeerJ 9:e11126. doi: 10.7717/peerj.11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tiwari V. 2021. Denovo designing, retro-combinatorial synthesis, and molecular dynamics analysis identify novel antiviral VTRM1.1 against RNA-dependent RNA polymerase of SARS CoV2 virus. Int J Biol Macromol 171:358–365. doi: 10.1016/j.ijbiomac.2020.12.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Figures S1 to S6 and Tables S1 to S5 in the supplemental material are available online (https://doi.org/10.6084/m9.figshare.22248886.v1).