Abstract
CelE, one of the three major proteins of the cellulosome of Clostridium cellulolyticum, was characterized. The amino acid sequence of the protein deduced from celE DNA sequence led us to the supposition that CelE is a three-domain protein. Recombinant CelE and a truncated form deleted of the putative cellulose binding domain (CBD) were obtained. Deletion of the CBD induces a total loss of activity. Exhibiting rather low levels of activity on soluble, amorphous, and crystalline celluloses, CelE is more active on p-nitrophenyl–cellobiose than the other cellulases from this organism characterized to date. The main product of its action on Avicel is cellobiose (more than 90% of the soluble sugars released), and its attack on carboxymethyl cellulose is accompanied by a relatively small decrease in viscosity. All of these features suggest that CelE is a cellobiohydrolase which has retained a certain capacity for random attack mode. We measured saccharification of Avicel and bacterial microcrystalline cellulose by associations of CelE with four other cellulases from C. cellulolyticum and found that CelE acts synergistically with all tested enzymes. The positive influence of CelE activity on the activities of other cellulosomal enzymes may explain its relative abundance in the cellulosome.
Clostridium cellulolyticum is a mesophilic anaerobic bacterium that can grow on cellulose as its sole carbon and energy sources. Like some other clostridia (2–4), the bacterium degrades cellulose by secreting several enzymes organized in a tight complex termed a cellulosome (2–5, 13). Over the past few years, efforts have been made to develop a complete description of this complex and its in vitro reconstitution. Prior to this study, five cellulase-encoding genes (celA, celC, celD, celF, and celG) had been entirely sequenced and another (celE) had been partially sequenced (1, 9, 27, 31). They have been overexpressed in Escherichia coli, and the biochemical properties of the corresponding recombinant proteins have been investigated (10–12, 28, 32). CelA, CelC, CelD, CelF, and CelG contain a catalytic core belonging to the glycosyl hydrolase families 5, 8, 5, 48, and 9, respectively (P. M. Coutinho and B. Henrissat, online [http://afmb.cnrs-mrs.fr/∼pedro/CAZY/db.html]) and a dockerin domain which is involved in the binding to the scaffolding protein CipC (22–24). These enzymes are actually subunits of the cellulosome complex in which CelF and CelE are the most abundant enzymatic components (13). In the present study, we describe the sequencing of celE and the purification and characterization of the celE gene product (which has been completely sequenced). CelE is a multidomain enzyme which, from the N to C terminus, is made up of a family IV cellulose binding domain (CBD) (Coutinho and Henrissat, online), an immunoglobulin-like (Ig) domain (17), a glycosyl hydrolase family 9 (GH9) catalytic domain (Coutinho and Henrissat, online), and a dockerin domain (3). Two genetic constructions were generated, one leading to the complete enzyme without the signal sequence and the other leading to a truncated form lacking the N-terminus CBD. We purified the two forms to homogeneity and studied their catalytic properties. Moreover, the ability of CelE to act synergistically with other cellulases from C. cellulolyticum was explored.
MATERIALS AND METHODS
Bacterial strains and plasmids.
C. cellulolyticum ATCC 35319 was used as the source of genomic DNA. E. coli DH5α (Bethesda Research Laboratories) was used as the host for pET-22b(+) (Novagen) derivatives. E. coli BL21(DE3) (Novagen) was used as the host for pET-22b(+) derivative expression vectors. C. cellulolyticum was grown anaerobically at 32°C on basal medium supplemented with cellobiose as the carbon and energy source, and chromosomal DNA was obtained as previously described (24). E. coli was grown at 37°C in Luria-Bertani (LB) medium supplemented with ampicillin (100 or 200 μg/ml) when required.
DNA sequencing of celE.
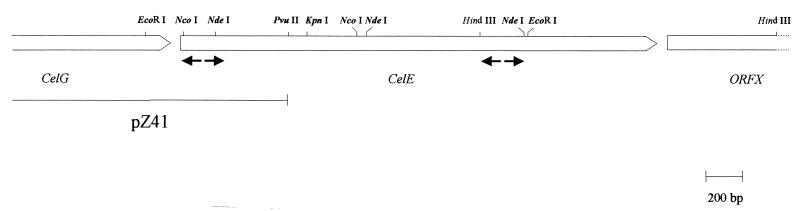
Plasmid pZ41, containing the 5′ end of the celE gene, has already been sequenced (1). To obtain the sequence of the entire gene, two successive inverse PCR experiments from C. cellulolyticum genomic DNA were performed (Fig. 1). In a first step, two divergent primers derived from the previously determined sequence of pZ41 allowed us to obtain a 2.3-kb EcoRI fragment which was sequenced. A second step, using two other divergent primers derived from the newly sequenced fragment, allowed us to obtain an overlapping 1.3-kb HindIII fragment. The DNA was sequenced by Genome Express Society using a Perkin-Elmer 383 fluorescent sequencing apparatus (Applied Biosystem dye terminator method).
FIG. 1.
Gene map showing the restriction sites used for construction of plasmid pET-E-1.4 containing the entire celE gene. Divergent arrows in bold correspond to positions of divergent primers used.
Expression of recombinant CelE protein.
The vector used for the genetic construction was the plasmid pET-22b(+). The celE gene was ligated to the NdeI and XhoI sites of the vector to obtain a mature protein located in the E. coli cytoplasm.
Figure 1 shows a partial restriction map of the celE gene. As two NdeI sites are located inside this sequence, the construction was achieved in successive steps. As the N terminus of the mature protein was previously determined by Gal et al. (13), a synthetic linker restoring the first 58 bp of the DNA encoding the mature protein (from the 5′ end to the first NcoI site) was obtained using oligonucleotides celE 215119 (5′ tatgCTTGTTGGGGCAGGAGATTTGATTCGAAACCATACCTTTGACAACAGAGTAGGTCTTC 3′) and celE306956 (5′ CATGGAAGACCTACTCTGTTGTCAAAGGTATGGTTTCGAATCAATCTCCTGCCCCAACAAG ca 3′). These oligonucleotides create an NdeI site upstream of the coding sequence (in bold). This linker was inserted into the pET-22b(+) vector linearized with NdeI and NcoI restriction sites. The resulting plasmid was called pCelE-2A.
The celE gene was produced by PCR using the primers celE 4045 (5′ TTGTTGGGGCAGGAGA 3′) and celE 315895 (5′ ccgctcgagCTGTGTGATTTTTCC 3′). This last primer creates an XhoI site at the 3′ extremity of the gene (in bold). The purified PCR fragment was first digested by KpnI, leading to two fragments (766 and 1,870 bp). The smallest fragment was then digested by NcoI, and the largest was digested by XhoI. These two fragments were purified using a Prepa A Gene purification kit (Bio-Rad) and ligated into NcoI- and XhoI-digested pCelE-2A. The coding sequence of celE was fused in frame with a downstream sequence encoding six histidine residues (His tag). The resulting plasmid, named pET-E-1.4, was used to transform E. coli BL 21(DE3) cells for expression of the recombinant protein.
Expression of a truncated form of CelE.
To obtain a truncated form of the protein containing the catalytic domain and the dockerin domain, two synthetic primers, celE 315895 (see above) and celE 613735 (5′ cttaaggcatatgGCAGGATATACTGAAGA 3′; partially homologous with the 5′ extremity of the DNA coding for the catalytic domain of CelE and creating an NdeI site [in bold] upstream of the coding sequence), were used to obtain a truncated celE gene. The purified PCR fragment was digested by NdeI and NcoI. The 570-bp NdeI-NcoI fragment (including the KpnI site) was purified, and its identity was confirmed by sequencing. The 1,500-bp NcoI-XhoI fragment was obtained by digestion of plasmid pET-E-1.4 with the same enzymes and purified after separation on a 0.7% agarose gel. The two fragments (570-bp NdeI-NcoI and 1,500-bp NcoI-XhoI) were ligated into NdeI- and XhoI-digested pET22b(+). The resulting plasmid, named pET-Etr, was used to transform E. coli BL21(DE3) cells for expression of the truncated form of CelE.
Purification of entire and truncated forms of celE.
The soluble proteins were produced as follows. Cells were grown at 37°C with shaking in LB medium (3 liters) supplemented with glycerol (12 g/liter) and ampicillin (200 μg/ml) until the optical density at 600 nm reached 1.8. They were cooled at 16°C without shaking. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 50 μM, and the culture was incubated for 16 h at 16°C with shaking. At this stage, the optical density at 600 nm was about 6 U. The cells were cooled at 4°C, harvested by centrifugation, and resuspended in 80 ml of ice-cold 0.5 mM CaCl2–30 mM Tris-HCl (pH 8) buffer and broken twice in a French pressure cell. The crude extract was centrifuged at 10,000 × g for 15 min, and the supernatant was loaded onto a 5-ml Ni-nitrilotriacetic acid column previously equilibrated with the same buffer. After two washes with the same buffer supplemented with 5 and 10 mM imidazole, CelE was eluted with a 50 mM imidazole solution. The eluate was immediately dialyzed and concentrated in an Amicon concentrator. Small aliquots of the protein solution were kept frozen (−20°C) until utilization.
Protein quantification.
The protein concentrations were determined as described by Lowry et al. (21), with fraction V bovine serum albumin (Merck) as the standard.
Substrates used.
Avicel (Merck), carboxymethyl cellulose, medium viscosity (CMC) (Sigma), barley glucan (Megazyme), laminarin (Sigma), xylan (Sigma), and lichenan (Sigma) were prepared as 1% (wt/vol) solutions in 25 mM potassium phosphate buffer, pH 7.0 (PPB). Phosphoric acid swollen cellulose (PASC) was prepared from Avicel as described by Walseth (34); its concentration was estimated by the phenol-sulfuric acid method (20) and suitably adjusted to obtain a final concentration of 1% (wt/vol) in PPB. Bacterial microcrystalline cellulose (BMCC) was a generous gift from H. Chanzy (CERMAV, Grenoble, France); its concentration was adjusted to 0.4% (wt/vol) in PPB. p-Nitrophenyl (pNP)-cellobiose was purchased from Sigma.
Enzyme assays.
The carboxymethylcellulase (CMCase) activity was assayed by mixing 1 ml of enzyme solution at appropriate concentrations with 4 ml of CMC solution at 37°C (final concentration of CMC, 0.8%). Aliquots of 1 ml were collected at specific intervals and stored on ice, and the reducing sugar contents were determined by the Park and Johnson ferricyanide method (25). One international unit (IU) corresponds to 1 μmol of d-glucose equivalent released per min. Since laminarin solutions strongly react with the Park and Johnson ferricyanide reagents, the enzymatic activities were monitored using 10-μl aliquots made up to 1 ml with PPB before measuring the reducing sugars.
Insoluble sugars xylan, Avicel, and lichenan were used at a final concentration of 0.8% (wt/vol), and BMCC was used at 0.36%, in PPB. Aliquots of 1.5 ml were collected at specific intervals and centrifuged at 5,000 × g for 15 min at 4°C. The reducing sugar content of 1 ml of supernatant and the IU were determined as described above.
pNP-cellobiose was used at 0.1% (wt/vol) in PPB. The enzymatic activities were determined at 37°C by monitoring the pNP released at 400 nm; 1 IU corresponds to 1 μmol of pNP (using pNP from Sigma as the standard) released per min.
Binding assays.
The experiments were performed as described previously (13). Various quantities of protein were incubated with various quantities of Avicel for 30 min in PPB, with slow shaking at 4°C in 1 ml (final volume), and then centrifuged at 5,000 × g for 15 min. The protein concentration in the supernatant was determined by the Lowry method. In all experiments, the bound fraction was estimated by subtracting the protein concentration of the free fraction in the supernatant from the initial protein concentration.
Viscosimetric assays.
Viscosimetric assays were performed by monitoring the flow time of 0.8% CMC solution incubated with various quantities of enzyme at different times. The relative fluidity ΔF was determined as [T0/(T0′ − T0)] − [T0/(T − T0)] where T0 is the flow time measured for the buffer, T0′ the flow time of the CMC solution without enzyme, and T is the flow time of the CMC solution with enzyme.
Synergistic assays.
In all the assays, a final Avicel concentration of 0.8% and a final BMCC concentration of 0.36% were used.
Cellodextrin analysis.
Cellodextrins were analyzed using a Dionex DX 500 chromatographic system (Dionex Co., Sunnyvale, Calif.) by high-performance anion-exchange chromatography on a pellicular ionic column (Carbopac PA 100; Dionex) coupled with pulsed amperometric detection. System control and data acquisition were performed using a PC Dell G1 computer and the Peaknet 5.1 software (Dionex). Chromatography solvents were NaOH (Fisher Scientific), anhydrous sodium acetate (Merck), and ultrapure water (Elga Maxima). Eluant A was NaOH (100 mM)-CH3COONa (5 mM); eluant B was NaOH (100 mM)-CH3COONa (500 mM). A linear gradient was applied from 0 to 50% of eluant B over 20 min before returning to initial conditions. Samples (25 μl) were automatically injected every 30 min.
Nucleotide sequence accession number.
The updated nucleotide sequence of the entire celE gene was submitted to GenBank (accession no. M87018).
RESULTS
Sequence analysis of the celE gene.
In a previous study (1), the 5′ region of the celE gene, located downstream of the celG gene, was cloned and sequenced. The missing part of celE was obtained based on a two-step procedure using inverse PCR. The entire celE gene and the 5′ region of the following open reading frame (ORF) (orfX [A. Bélaich, personal communication]) were obtained. The 2,658-nucleotide celE coding sequence was determined. The ORF encodes a 885-amino-acid polypeptide. It begins with a peptide signal sequence. The N-terminal sequence of the mature protein was previously determined by Gal et al. (13). The mature protein contains 857 aa with a calculated molecular mass of 93,800 Da, which is in good agreement with the value of 94,000 Da determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (13). The introduction of a XhoI site as well as six His codons downstream of the celE gene results in the entire recombinant protein containing 866 aa and possessing a slightly higher molecular mass (95,025 Da).
Sequence analysis of CelE protein.
The catalytic domain of CelE clearly belongs to the GH9 subfamily 1; i.e., it harbors an Ig domain in the N-terminal position of the catalytic core (17). At the N terminus, as previously observed (1), we find a domain homologous to family IV CBDs (now termed CBM4, for carbohydrate binding module [Coutinho and Henrissat, online]), the function of which was determined by Coutinho et al. (7). At the C terminus, a dockerin domain, characteristic of C. cellulolyticum cellulases (5), is found.
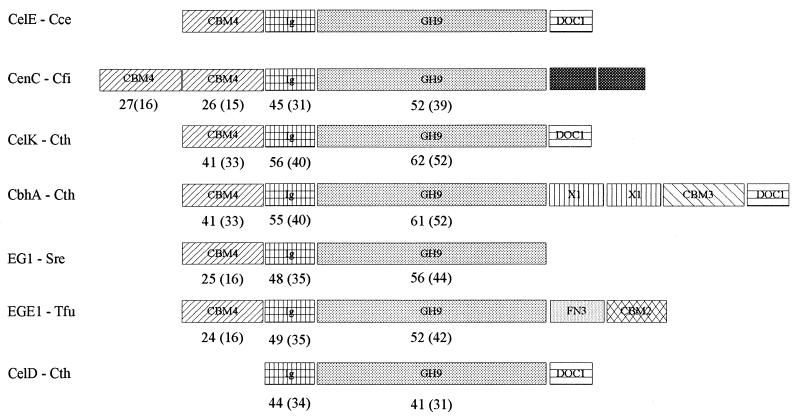
The domain organization of CelE is compared in Fig. 2 with those of closely related cellulases (i.e., harboring CBM4, Ig, and GH9 domains, successively) and with that of CelD from C. thermocellum, for which the structure is known (17). Comparison of the sequences reveals that conservation among catalytic cores is greater than conservation among CBM4, indicating either a lower level of evolutionary pressure on these last domains or changes in function and specificities.
FIG. 2.
Comparison of the subfamily 9-1 cellulase domain structures harboring one or two family IV CBDs (now named CBM4) with CelD (CelD-Cth) from C. thermocellum (14) for which the three-dimensional structure of Ig and GH9 has been established (17). Other abbreviations: CelE-Cce, C. cellulolyticum CelE; CenC-Cfi, Cellulomonas fimi CenC (8); CelK-Cth, C. thermocellum CelK (19); CbhA-Cth, C. thermocellum CbhA (35); EG1-Sre, Streptomyces reticuli Cel1 (30); EGE1-Tfu, Thermonospora fusca EGE1 (16). Numbers indicate percent similarity (percent identity) relative to the corresponding domains in CelE.
Production and purification of CelE.
CelE was purified by a one-step process using properties of the His tag. Expression of celE in E. coli encountered the same problems as those observed with celG (12) and celF (27), in particular the formation of inclusion bodies. Again, as in the case of celG and celF, the culture conditions were optimized so that the protein could be produced in a soluble form (see Materials and Methods). Under these conditions, about 30 mg of pure protein were obtained from 3 liters of growth culture following a one-step process on a Ni-nitrilotriacetic acid column. The recombinant protein had an apparent mass of 94 kDa on SDS-PAGE, which is in good agreement with its theoretical mass (95,025 Da). A 24-fold purification was achieved with a yield of 25%, and the final CMCase specific activity was 0.22 IU/mg.
Catalytic properties of CelE.
Although CelE, like CelG, is a multidomain family 9 cellulase, its activity pattern (Table 1) diverges significantly from that observed for CelG, or for any other cellulases from C. cellulolyticum characterized so far (10–12, 28). CelE appears to have a low activity on CMC. It exhibits levels of activity similar to that observed for CelA on PASC and to those reported for CelA and CelG on Avicel. The most remarkable feature is its high activity toward pNP-cellobiose, since CelE is approximately 20-fold more efficient than CelA, which was the only enzyme of this system characterized so far exhibiting activity on this chromophoric substrate.
TABLE 1.
Activities of CelE on various substrates and comparison with the activities of previously characterized cellulases (proteins with dockerins)
| Substrate | Activity (U/min) (reference)
|
||||
|---|---|---|---|---|---|
| CelE | CelA (11) | CelC (10) | CelF (28) | CelG (12) | |
| CMC | 13.5 | 2,500.0 | 2,160.0 | 0.0 | 1,170.0 |
| Avicel | 5.8 | 5.4 | 0.8 | 13.4 | 5.0 |
| PASC | 71.5a | 100.0 | 336.0 | 42.5 | 38.0 |
| BMCC | 5.5 | NDb | ND | 3.9 | 9.0 |
| Xylan | 2.3 | 280.0 | 6.2 | ND | 0.0 |
| Laminarin | 0.0 | 0.0 | 0.0 | ND | 0.0 |
| Lichenan | 5.8 | 2,090.0 | 528.0 | ND | 140.0 |
| pNP-cellobiose | 34.8a | 1.5 | 0.0 | 0.0 | 0.0 |
Vmax established using Lineweaver-Burk plots.
ND, not determined.
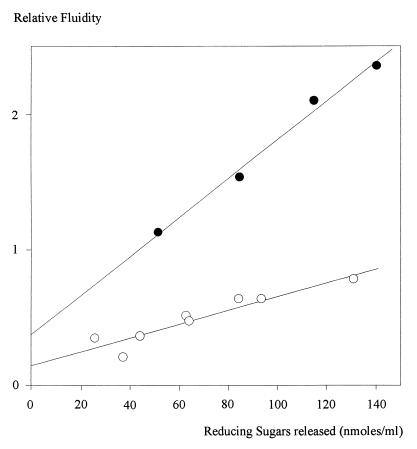
The apparent Km and Vmax of CelE were determined from initial velocities of PASC (Km = 2.8 g/liter and Vmax = 3.4 μM/min with 0.048 μM CelE) and pNp-cellobiose (Km = 0.4 mM and Vmax = 4.2 μM/min with 0.12 μM CelE) degradations, as these compounds represent the best substrates for the enzyme. Compared to CelA, CelE exhibits a 10-times-higher affinity for pNP-cellobiose. Conversely, the degradation of CMC is not concomitant with a decrease in viscosity as large as that observed with CelA (Fig. 3). The optimum pH and temperature measured using PASC as a substrate were 6.0 and 45°C, respectively. These values are very similar to those obtained with the other characterized cellulases from C. cellulolyticum (10–12, 28).
FIG. 3.
Variations in the relative fluidity of CMC versus the liberation of reducing extremities by CelE 0.240 μM (○) and CelA 0.015 μM (●).
Properties of truncated CelE.
The truncated celE gene was built to produce a protein deleted of the N-terminal CBD. The boundaries were determined by sequence comparison with CelD from C. thermocellum, which is devoid of CBD (14, 17), and CelK of the same organism, which presents spontaneous cleavage of its CBD (18). The resulting protein contains 692 aa, and its molecular mass is 75,150 Da.
The truncated protein was produced and purified in the same way as was the intact CelE. A soluble protein of the expected size was obtained following purification. Analysis of the N terminus showed that the protein remained intact after its production in E. coli.
Even for protein concentrations 10 times higher than those used with entire CelE, deletion of CBD resulted in total loss of activities on pNP-cellobiose, CMC, and PASC. This observation seems to indicate an interdependence of the catalytic and cellulose binding domains such as is the case for CelG (12).
Binding properties of CelE.
CelE is able to bind to Avicel at 1 g/liter (KD = 0.48 μM and 87 nmol of sites/g) and PASC at 1 g/liter (KD = 1.46 μM and 413 nmol of sites/g), with a higher affinity for Avicel but a greater number of sites on PASC. On Avicel, CelE appears to have a higher affinity than does CelG, although the number of sites remains in the same range (KD = 7.4 μM and 95 nmol of sites/g [12]). These parameters are very different from those obtained with the CBD of mini-CipC1 (KD = 0.13 μM and 280 nmol of sites/g [24]).
Synergistic properties of CelE.
The low activity levels displayed by CelE is an apparent paradox in contrast to its relative abundance in the cellulosome (13). Thus, its possible interactions with the other known enzymes of the system were examined. Surprisingly, on Avicel and BMCC, CelE exhibits strong synergism with all the tested enzymes (Table 2). Synergism appears to be more important between CelE and bona fide endoglucanases, such as CelA and CelC, especially on BMCC, which is the most crystalline substrate tested, but also appears to be very significant with CelG and, unexpectedly, with CelF (Table 2).
TABLE 2.
Maxima of synergism coefficients and percentage of CelE for which they occur
| Substrate | CelA
|
CelC
|
CelF
|
CelG
|
||||
|---|---|---|---|---|---|---|---|---|
| SCa | % CelEb | SC | % CelE | SC | % CelE | SC | % CelE | |
| Avicel | 1.45 | 50 | 1.87 | 17 | 1.81 | 33 | 1.45 | 33 |
| BMCC | 2.70 | 17 | 2.86 | 17 | 1.52 | 16 | 1.48 | 66 |
SC, synergism coefficient, i.e., the ratio measured/theoretical.
Relative quantity of CelE in the mixture with its partners. For all mixtures, the final enzyme concentration was 0.2 μM.
The kinetics of Avicel degradation were examined in the case of the association of CelE and CelG. The released sugars were analyzed and quantified using a Dionex apparatus. The results (Table 3) indicate that combining these two enzymes not only leads to a better efficiency of degradation of crystalline cellulose but also induces a change in the type of released soluble sugars, with a higher production of cellobiose than expected from the quantities liberated by both enzymes alone.
TABLE 3.
Sugar composition of the incubation medium after 2 h of Avicel attack by CelE and/or CelG
| Sugar | CelE (0.2 μM)
|
CelG (0.2 μM)
|
CelE (0.1 μM) + CelG (0.1 μM)
|
SCa | |||||
|---|---|---|---|---|---|---|---|---|---|
| Measured
|
Theoretical
|
||||||||
| nmol/ml | Relative % | nmol/ml | Relative % | nmol/ml | Relative % | nmol/ml | Relative % | ||
| G1 | 2.2 | 3.1 | 14.1 | 27.7 | 5.7 | 6.9 | 8.2 | 13.4 | 0.7 |
| G2 | 65.5 | 92.4 | 18.8 | 36.8 | 68.2 | 83.3 | 42.1 | 69.2 | 1.6 |
| G3 | 3.2 | 4.5 | 16.8 | 33.0 | 8.0 | 9.8 | 10.0 | 16.4 | 0.8 |
| G4 | 0.0 | 0.0 | 1.2 | 2.4 | 0.0 | 0.0 | 0.6 | 1.0 | 0.0 |
| Eq G1b | 142.6 | 103.3 | 166.3 | 123.0 | 1.4 | ||||
SC, synergism coefficient, i.e., the ratio measured/theoretical.
Sum of equivalent glucose from all cellodextrins.
The main product of CelE when acting on Avicel is cellobiose (more than 90%), whereas CelG produces glucose, cellobiose, and cellotriose in similar proportions as well as traces of cellotetraose. Mixing the two enzymes clearly deviates hydrolysis toward formation of cellobiose (60% more in 2 h) and increases Avicel attack (40%). Thus, the large cellodextrins (G3 and G4) produced by CelG seem to be substrates for CelE, and the degradation of Avicel is increased.
DISCUSSION
The complete sequence of celE, which encodes CelE, one of the main components of the C. cellulolyticum cellulosome along with CipC and CelF (13), was obtained. CelE is a multidomain protein involving CBM4 associated with an Ig domain, GH9 catalytic core, and dockerin.
With the exception of PASC and pNP-cellobiose, the activities of CelE were much lower than the activities of other endoglucanases from the same organism (CelA, CelC, and CelG [10–12]) and resemble those found for CelF (28). CelE, however, among all the enzymes tested, shows a rather high activity on pNP-cellobiose. Moreover, it attacks CMC with only a slight decrease in viscosity of this substrate, and the main product of its activity on Avicel is cellobiose. All of these data indicate that CelE is a cellobiohydrolase.
To our knowledge, enzymatic properties of only two enzymes presenting associations of CBM4, Ig, and GH9 domains have been described so far: CenC from Cellulomonas fimi, which is considered by Tomme et al. (33) to be a semiprocessive enzyme, and CelK from C. thermocellum (18, 19). The latter, which presents the greatest structural similarity to CelE (Fig. 2), seems to have the closest activity pattern (18), but it does not decrease the viscosity of CMC at all (19). CenC seems to be a better CMCase (equally or more active on this substrate than either CenA or CenB from the same organism [33]), but as was observed for CelE this activity is not correlated with an important decrease in viscosity, and the main product of PASC and BMCC hydrolysis is cellobiose (98%). These data are in agreement with the existence of a class of cellulases (associating CBM4, Ig, and GH9 domains) which acts first by random mode on cellulose and mainly releases cellobiose. The association of these domains seems of great importance for the occurrence of these particular properties because enzymes from the same organisms (CenB in Cellulomonas fimi and CelG in C. cellulolyticum), which exhibit the same GH9 domain, present totally different activity spectra (12, 33). Moreover, CelD from C. thermocellum, which contains only Ig and GH9 domains, is very active on CMC, an activity which is accompanied by a large decrease in viscosity (15). Thus, the characteristic properties of CelE seem to be due to the presence of a CBM4 domain. Moreover, the deletion of CBM4 induces a total inactivation of the enzyme. Cellulose binding properties of CelE are in agreement with those described for the two CBM4 located at the N terminus of CenC from Cellulomonas fimi (7), i.e., a greater number of sites on PASC than on Avicel. On this latter substrate, binding parameters are in the same range as those observed with CelG (12) and are much lower (in affinity and number of sites) to those determined for mini-CipC1 (24). It was previously observed that the CBD (CBM3) and catalytic domain (GH9) of CelG were both required for activity (12). Recently, Sakon et al. (29) solved the three-dimensional structure of cellulase E4 from Thermonospora fusca. This cellulase is structurally similar to CelG from C. cellulolyticum, and a direct interaction and specific relative orientation of the CBM3 and GH9 domains was observed (29), in agreement with our biochemical data on CelG (12) and consistent with the conclusion that this type of CBM domain is directly involved in the catalytic function of the enzyme. It would therefore appear that in the case of CelG and CelE from C. cellulolyticum, the CBMs (less efficient on crystalline cellulose than the CBM domain from Cip or other cellulases) have two other functions in addition to the binding to cellulose (possibly inducing greatest efficiency on amorphous cellulose), involvement in catalysis (possibly by arrangement of the cellulose fibers), and structural implication (perhaps by stabilizing either the catalytic core or the active site). Structural studies on both enzymes are required to elucidate these two last points. Solving the three-dimensional structure of CelG by X-ray crystallography is in progress, and preliminary results of crystallogenetic assays on CelE are promising.
In light of these considerations, the ability of CelE to act in synergism with the four other cellulases from C. cellulolyticum, cellulases which have been characterized in our laboratory (10–12, 28), is of great importance. Synergism between CelE, which appears to be a cellobiohydrolase, and both CelA and CelC, which are true endoglucanases (10, 11), is thus not surprising. These enzymes pairs gave the best scores on BMCC, the most crystalline substrate used. The association of CelE with CelG, which was found to be very efficient on BMCC (12), led to an increase in activity on this substrate. Examination of the soluble products of Avicel hydrolysis by these last two enzymes, both separately and in combination, indicates that the synergism results in an increase in cellulose degradation (enhancement of 40% after 2 h of incubation) and a change in the nature of the products through the action of CelE on large cellodextrins. More surprising is the important synergism observed between CelE and CelF. CelF, which along with CelE is the most abundant enzyme in the cellulosome (13), has been shown to be a processive enzyme (28) and exhibit an overall capacity to act in synergism with other cellulases (26). Moreover, the third main protein of the C. cellulolyticum cellulosome is the CipC, and it has been recently demonstrated that a fragment of this protein, which includes a CBD and a cohesin (mini-CipCC1), stimulates activity of CelA on Avicel (23). Thus, the efficiency of crystalline cellulose hydrolysis by the cellulosome would be due to an association of enzymes possessing various and complementary activities. The present study supports this hypothesis. CelE, through its own action and its positive influence on the activities of other enzymes, appears to be a key enzyme in cellulosome efficiency, in the same way as CelF. To gain further insight into this mechanism, two strategies are planned. First, the resolution of the CelE structure is required to understand the interactions between the different domains and how these interactions induce the peculiar properties of this enzyme. Second, studies of synergism involving CelE, all the available enzymes, and CipC (or miniCipC) will be undertaken to understand the relative role of each component by partial reconstitution of the cellulosome.
ACKNOWLEDGMENTS
We are grateful to H.-P. Fierobe for helpful discussions and to M. Johnson for proofreading the manuscript. We are indebted to C. Villard from the Laboratoire de Biologie et de Biochimie de la Nutrition (Marseille-St-Jérôme) for analysis of cellodextrins using Dionex apparatus.
This research was supported by grants from the Centre National de la Recherche Scientifique, Université de Provence, EEC (BIOTECH contract BIO-CT-94-3018), and Région Provence-Alpes-Côte d'Azur.
REFERENCES
- 1.Bagnara-Tardif C, Gaudin C, Bélaich A, Hoest P, Citard T, Bélaich J-P. Sequence analysis of a gene cluster encoding cellulases from Clostridium cellulolyticum. Gene. 1992;119:17–28. doi: 10.1016/0378-1119(92)90062-t. [DOI] [PubMed] [Google Scholar]
- 2.Bayer E A, Chanzy H, Lamed R, Shoham Y. Cellulose, cellulases and cellulosomes. Curr Opin Struct Biol. 1998;8:548–557. doi: 10.1016/s0959-440x(98)80143-7. [DOI] [PubMed] [Google Scholar]
- 3.Bayer E A, Shimon L J W, Shoham Y, Lamed R. Cellulosomes—structure and ultrastructure. J Struct Biol. 1998;124:221–234. doi: 10.1006/jsbi.1998.4065. [DOI] [PubMed] [Google Scholar]
- 4.Béguin P, Millet J, Aubert J-P. Cellulose degradation by Clostridium thermocellum: from manure to molecular biology. FEMS Microbiol Lett. 1992;79:523–528. doi: 10.1111/j.1574-6968.1992.tb14087.x. [DOI] [PubMed] [Google Scholar]
- 5.Bélaich J-P, Tardif C, Bélaich A, Gaudin C. The cellulolytic system of Clostridium cellulolyticum. J Biotechnol. 1997;57:3–14. doi: 10.1016/s0168-1656(97)00085-0. [DOI] [PubMed] [Google Scholar]
- 6.Chauvaux S, Béguin P, Aubert J-P. Site-directed mutagenesis of essential carboxylic residues in Clostridium thermocellum endoglucanase CelD. J Biol Chem. 1992;267:4472–4478. [PubMed] [Google Scholar]
- 7.Coutinho J B, Gilkes N R, Warren R A J, Kilburn D G, Miller R C., Jr The binding of Cellulomonas fimi endoglucanase C (CenC) to cellulose and Sephadex is mediated by the N-terminal repeats. Mol Microbiol. 1992;6:1243–1252. doi: 10.1111/j.1365-2958.1992.tb01563.x. [DOI] [PubMed] [Google Scholar]
- 8.Coutinho J B, Moser B, Kilburn D G, Warren R A J, Miller R C., Jr Nucleotide sequence of endoglucanase C (CenC) of Cellulomonas fimi, its high-level expression in Escherichia coli, and characterization of its products. Mol Microbiol. 1991;5:1221–1223. doi: 10.1111/j.1365-2958.1991.tb01896.x. [DOI] [PubMed] [Google Scholar]
- 9.Faure E, Bélaich A, Bagnara C, Gaudin C, Bélaich J-P. Sequence analysis of the Clostridium cellulolyticum endoglucanase-A-encoding gene, celCCA. Gene. 1989;84:39–46. doi: 10.1016/0378-1119(89)90137-6. [DOI] [PubMed] [Google Scholar]
- 10.Fierobe H-P, Bagnara-Tardif C, Gaudin C, Guerlesquin F, Sauve P, Bélaich A, Bélaich J-P. Purification and characterization of endoglucanase C from Clostridium cellulolyticum. Catalytic comparison with endoglucanase A. Eur J Biochem. 1993;217:557–565. doi: 10.1111/j.1432-1033.1993.tb18277.x. [DOI] [PubMed] [Google Scholar]
- 11.Fierobe H-P, Gaudin C, Bélaich A, Loutfi M, Faure E, Bagnara C, Baty D, Bélaich J-P. Characterization of endoglucanase A from Clostridium cellulolyticum. J Bacteriol. 1991;173:7956–7962. doi: 10.1128/jb.173.24.7956-7962.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gal L, Gaudin C, Belaich A, Pagès S, Tardif C, Belaich J-P. CelG from Clostridium cellulolyticum: a multidomain endoglucanase acting efficiently on crystalline cellulose. J Bacteriol. 1997;179:6595–6601. doi: 10.1128/jb.179.21.6595-6601.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gal L, Pagès S, Gaudin C, Bélaich A, Reverbel-Leroy C, Tardif C, Bélaich J-P. Characterization of the cellulolytic complex (cellulosome) produced by Clostridium cellulolyticum. Appl Environ Microbiol. 1997;63:903–909. doi: 10.1128/aem.63.3.903-909.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joliff G, Béguin P, Aubert J-P. Nucleotide sequence of the cellulase gene celD encoding endoglucanase D of Clostridium thermocellum overproduced in Escherichia coli. Bio/Technology. 1986;4:896–900. doi: 10.1093/nar/14.21.8605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joliff G, Béguin P, Juy M, Millet J, Ryter A, Poljak R, Aubert J-P. Isolation, crystallisation and properties of a new cellulase of Clostridium thermocellum. Nucleic Acids Res. 1986;14:8605–8613. doi: 10.1093/nar/14.21.8605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung E D, Lao G, Irwin D, Barr B K, Benjamin A, Wilson D B. DNA sequences and expression in Streptomyces lividans of an exoglucanase gene and an endoglucanase gene from Thermonospora fusca. Appl Environ Microbiol. 1993;59:3032–3043. doi: 10.1128/aem.59.9.3032-3043.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juy M, Amit A G, Alzari P M, Poljak R J, Claeyssens M, Béguin P, Aubert J-P. Three-dimensional structure of a thermostable bacterial cellulase. Nature. 1992;357:89–91. [Google Scholar]
- 18.Kataeva I, Li X-L, Chen H, Ljungdahl L G. CelK—a new cellobiohydrolase from Clostridium thermocellum cellulosome: role of N-terminal cellulose binding domain. In: Ohmiya K, Sakka K, Karita S, Hayashi K, Kobayashi Y, Kimura T, editors. Genetics, biochemistry and ecology of cellulose degradation. Tokyo, Japan: Unipublishers Co.; 1998. pp. 454–460. [Google Scholar]
- 19.Kataeva I, Li X-L, Chen H, Ljungdahl L G. Cloning and sequence analysis of a new cellulase gene encoding CelK, a major cellulosome component of Clostridium thermocellum: evidence for gene duplication and recombination. J Bacteriol. 1999;181:5288–5295. doi: 10.1128/jb.181.17.5288-5295.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leray C, Nicoli J, Audiffren P. Microdoasge colorimètrique des oses neutres par l'acide sulfurique et le phénol. Influence des sels et des protéines. Med Trop. 1966;26:382. [PubMed] [Google Scholar]
- 21.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 22.Pagès S, Belaich A, Fierobe H-P, Tardif C, Gaudin C, Belaich J-P. Sequence analysis of scaffolding protein CipC and ORFXp, a new cohesin-containing protein in Clostridium cellulolyticum: comparison of various cohesin domains and subcellular localisation of ORFXp. J Bacteriol. 1999;181:1801–1810. doi: 10.1128/jb.181.6.1801-1810.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pagès S, Bélaich A, Tardif C, Reverbel-Leroy C, Gaudin C, Bélaich J-P. Interaction between the endoglucanase CelA and the scaffolding protein CipC of the Clostridium cellulolyticum cellulosome. J Bacteriol. 1996;178:2279–2286. doi: 10.1128/jb.178.8.2279-2286.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pagès S, Gal L, Bélaich A, Gaudin C, Tardif C, Bélaich J-P. Role of scaffolding protein CipC of Clostridium cellulolyticum in cellulose degradation. J Bacteriol. 1997;179:2810–2816. doi: 10.1128/jb.179.9.2810-2816.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park J T, Johnson M J. A submicrodetermination of glucose. J Biol Chem. 1949;181:149–151. [PubMed] [Google Scholar]
- 26.Reverbel-Leroy C. La cellulase CelF: un composant majoritaire du cellulosome de Clostridium cellulolyticum. Thèse de Doctorat. d'Université. Marseille, France: Université de Provence; 1996. [Google Scholar]
- 27.Reverbel-Leroy C, Bélaich A, Bernadac A, Gaudin C, Bélaich J-P, Tardif C. Molecular study and overexpression of the Clostridium cellulolyticum celF cellulase gene in Escherichia coli. Microbiology. 1996;142:1013–1023. doi: 10.1099/00221287-142-4-1013. [DOI] [PubMed] [Google Scholar]
- 28.Reverbel-Leroy C, Pagès S, Bélaich A, Bélaich J-P, Tardif C. The processive endocellulase CelF, a major component of the Clostridium cellulolyticum cellulosome—purification and characterization of the recombinant form. J Bacteriol. 1997;179:46–52. doi: 10.1128/jb.179.1.46-52.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakon J, Irwin D, Wilson D B, Karplus P A. Structure and mechanism of endo/exocellulase E4 from Thermonospora fusca. Nat Struct Biol. 1997;4:810–818. doi: 10.1038/nsb1097-810. [DOI] [PubMed] [Google Scholar]
- 30.Schlochtermeier A, Walter S, Schröder J, Moorman M, Schrempf H. The gene encoding the cellulase (Avicelase) Cell from Streptomyces reticuli and analysis of protein domains. Mol Microbiol. 1992;6:3611–3621. doi: 10.1111/j.1365-2958.1992.tb01797.x. [DOI] [PubMed] [Google Scholar]
- 31.Shima S, Igarashi Y, Kodama T. Nucleotide sequence analysis of the endoglucanase-encoding gene, celCCD, of Clostridium cellulolyticum. Gene. 1991;104:33–38. doi: 10.1016/0378-1119(91)90461-j. [DOI] [PubMed] [Google Scholar]
- 32.Shima S, Igarashi Y, Kodama T. Purification and properties of two truncated endoglucanases produced in Escherichia coli harbouring Clostridium cellulolyticum endoglucanase gene celCCD. Appl Microbiol Biotechnol. 1993;38:750–754. doi: 10.1007/BF00167140. [DOI] [PubMed] [Google Scholar]
- 33.Tomme P, Kwan E, Gilkes N R, Kilburn D G, Warren R A J. Characterization of CenC, an enzyme from Cellulomonas fimi with both endo- and exoglucanase activities. J Bacteriol. 1996;178:4216–4223. doi: 10.1128/jb.178.14.4216-4223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walseth C S. Occurrence of cellulase in enzyme preparations from microorganisms. TAPPI. 1952;35:228–233. [Google Scholar]
- 35.Zverlov V V, Velikodvorskaya G V, Schwarz W H, Bronnenmeier K, Kellermann J, Staudenbauer W L. Multidomain structure and cellulosomal localisation of the Clostridium thermocellum cellobiohydrolase CbhA. J Bacteriol. 1998;180:3091–3099. doi: 10.1128/jb.180.12.3091-3099.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]