Abstract
We have identified three new Haemophilus influenzae mutations causing cells to exhibit extreme hypercompetence at all stages of growth. The mutations are in murE, which encodes the meso-diaminopimelate-adding enzyme of peptidoglycan synthesis. All are point mutations causing nonconservative amino acid substitutions, two at a poorly conserved residue (G435→R and G435→W) and the third at a highly conserved leucine (L361→S). The mutant strains have very similar phenotypes and do not exhibit any defects in cell growth, permeability, or sensitivity to peptidoglycan antibiotics. Cells retain the normal specificity of DNA uptake for the H. influenzae uptake signal sequence. The mutations do not bypass genes known to be needed for competence induction but do dramatically increase expression of genes required for the normal pathway of DNA uptake. We conclude that the mutations do not act by increasing cell permeability but by causing induction of the normal competence pathway via a previously unsuspected signal.
Natural competence allows bacteria to take up DNA from their environment, but both the mechanisms of DNA translocation and its evolutionary functions are poorly understood. We have been investigating competence in the gram-negative bacterium Haemophilus influenzae, in which partial competence is induced by nutrient or oxygen limitation arising at the onset of stationary phase and full competence is induced by transfer of exponentially growing cells to a starvation medium.
At least eight genes are known to be specifically required for DNA uptake in H. influenzae, with roles in regulation, DNA binding, and DNA transport. Other genes are also required for these processes or for recombination but have additional functions not specific to competence. Most of these genes were identified either by screening randomly mutagenized cells for loss of competence or by directed insertional inactivation of candidate genes. However, the sxy gene was identified by selection for competence-inducing mutations, which cause cells to take up DNA during growth at low density in rich medium, conditions that normally preclude competence (18, 32). We have now screened additional aliquots of the same mutagenized culture and isolated four mutant strains that have normal sxy genes but are even more hypercompetent than the sxy mutants. Analysis shows that these strains all carry mutations in the murE gene, which encodes an essential step in peptidoglycan synthesis but has not previously been implicated in competence.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. All H. influenzae strains are descendants of the original Rd strain (1). Standard methods for H. influenzae are described by Barcak et al. (2). H. influenzae strains were routinely grown at 37°C in Difco brain heart infusion supplemented with NAD at 2 μg/ml and hemin at 10 μg/ml (sBHI) (16). Antibiotics for the H. influenzae experiments were used in broth and in 1.2% agar (Bacto) plates at the following concentrations: novobiocin, 2.5 μg/ml; streptomycin, 250 μg/ml; kanamycin, 7 μg/ml; and chloramphenicol, 2 μg/ml. Escherichia coli strains were grown in Luria-Bertani broth and plates with the following antibiotics where appropriate: ampicillin, 100 μg/ml; chloramphenicol, 25 μg/ml; and spectinomycin, 50 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| H. influenzae | ||
| KW20 | Wild type | H. O. Smith |
| MAP7 | str kan nov nal spc vio stv | J. Setlow; 17 |
| RR514 | str | 32 |
| RR520 | kan | 32 |
| RR563 | sxy-1 | 32 |
| RR749 | murE749; G1303→A | This study |
| RR750 | murE750; G1303→A | This study |
| RR751 | murE751; T1082→C | This study |
| RR752 | murE752; G1303→T | This study |
| RR769 | rec-2::mini-Tn10kan murE749 | This study |
| RR797 | Cmr cassette from pKRP10 inserted into HindIII site at bp 1195829 in strain RR749 | This study |
| RR804 | murE749 transformed into RR514 background | This study |
| RR806 | murE750 + CAT | This study |
| RR807 | murE751 + CAT | This study |
| RR808 | murE752 + CAT | This study |
| RR809 | murE+ + CAT | This study |
| RR829 | crp::mini-Tn10kan murE749 | This study |
| RR830 | icc::spec murE749 | This study |
| RR839 | sxy::lacZ fusion | 1a |
| RR867 | Insertion and duplication of comA; comA::lacZ Cmr | 19 |
| RR868 | rec2::lacZ fusion | 19 |
| RR876 | sxy::lacZ fusion murE749 | This work |
| RR878 | comA::lacZ fusion murE749 | This work |
| RR879 | rec2::lacZ fusion murE749 | This work |
| E. coli | ||
| DH5α | sup44 recA1 | |
| GM2163 | dam | New England Biolabs |
| Plasmids | ||
| pRRnov1 | H. influenzae n-vobiocin resistance allele of gyrA | 32 |
| pWJC3 | Kmr cassette | 9 |
| p836B | murE749KpnI-BglII fragment from RR797 | This work |
| p848M | murE+ KpnI-BglII fragment from RR805 in pSU40 | This work |
| p836B-20 | 6,845-bp HindIII fragment from p836B subcloned into pSU20 | This work |
| p848M-20 | 6,845-bp HindIII fragment from p848M subcloned into pSU20 | This work |
| pmurE::Kan | Kmr cassette from pWJC3 inserted into PstI site of p848M-20 | This work |
| pAM120 | Tn916 vector | 14 |
| pKRP10 | Cmr cassette | 33 |
EMS mutagenesis and screening.
Two aliquots of a culture of H. influenzae strain KW20, previously treated with EMS (methanesulfonic acid ethyl ester; Sigma) and stored frozen at −80°C, were screened for hypercompetent mutants by transformation during early exponential growth as previously described (32). Briefly, mutagenized cultures were thawed, diluted, and grown for two cell doublings in sBHI. Under these conditions, wild-type cells do not become competent. The cells were then incubated with pRRnov1 DNA that had been cut with KpnI and XbaI to free the insert and were plated on novobiocin plates. Rare Novr transformant colonies were tested for hypercompetence twice using the colony transformation assay described below, first selecting for a Kanr marker carried by DNA of strain RR520 and then for the closely linked Strr marker carried by DNA of strain RR514. This resulted in the isolation of nine Strr Kans hypercompetent isolates. DNA from these was used to backcross the hypercompetence mutation into the wild-type strain KW20 by screening Strr transformants with the colony competence assay.
Competence assays.
The general procedures for the following competence assays have been previously described (32, 38).
(i) Spontaneous competence.
MAP7 DNA (18) at 1 μg/ml was added to cells growing in sBHI broth on a roller wheel at 37°C. After 15 min, DNase I was added at 10 μg/ml, and after five more minutes the cells were diluted and plated.
(ii) MIV induction competence.
Cells in exponential growth were collected by sterile filtration (37) and transferred to the starvation medium MIV (22), where they were shaken at 37°C for 100 min before the addition of MAP7 DNA at 1 μg/ml. The cells were incubated, treated with DNase, and plated as described above.
(iii) Colony competence assay.
Cells in a single fresh colony were resuspended in 5 ml of sBHI containing 0.1 μg of MAP7 DNA/ml, incubated without agitation for 15 min, and plated. DNase treatment was omitted.
Analysis of murE sequences.
DNA sequencing was done by the University of British Columbia Nucleic Acid-Protein Service Unit, using ABI AmpliTaq DyeDeoxy Terminator cycle-sequencing chemistry. Plasmids carrying both wild-type and mutant murE genes were sequenced, using the oligonucleotide primers A (CCACGTTGTTATCGTTTGG) and B (GCTTGAAGAATCTGTGCAAG). The wild-type murE sequence was identical to that reported by the Institute for Genomic Research (http://www.tigr.org). Preliminary sequence data for murE homologs from incomplete bacterial genomes were obtained from the National Center for Biotechnology Information (NCBI) Microbial Genomes BLAST server (http://www.ncbi.nlm.nih.gov/BLAST/unfinishedgenome.html).
PCR plus restriction assay for murE749.
Because the murE from RR749 (murE749) point mutation creates a MnlI restriction site, its presence in chromosomal DNA was easily confirmed by amplifying the murE gene with primers A and B and digesting the 587-bp product with MnlI: the wild-type allele gives fragments of 219 and 368 bp, and the mutant gives fragments of 229, 219, and 139 bp. These were visualized in a 4% agarose gel.
DNA uptake and uptake signal sequence specificity.
DNA uptake assays were done by the method of Deich and Smith (10). Aliquots (1.0 ml) of KW20 and RR804 cells at optical densities at 600 nm of 0.1 and 1.0 were incubated with 0.1 μg of 33P-labeled MAP7 DNA (labeled by nick translation to a specific activity of 6.9 × 106 cpm/μg) for 10 min at 37°C in 1.5-ml centrifuge tubes. The tubes were placed on ice, and 0.05 ml of pancreatic DNase I (1 mg/ml; Pharmacia) was added to each tube. After 5 min on ice, 0.1 ml of 5 M NaCl was added, and the cells were pelleted by centrifugation for 1 min at 4°C at maximum speed (15,000 × g) in a microcentrifuge. The pellet was resuspended in 1 ml of cold sBHI containing 0.5 M NaCl, and the cells were pelleted again for 1 min at 4°C. The pellet was resuspended in 0.2 ml of sBHI and transferred to scintillation vials containing 1 ml of scintillation fluid. The transformation frequency was measured by transforming a 1.0-ml aliquot of the same cells with 0.1 μg of unlabeled MAP7 DNA.
To examine specificity for the H. influenzae uptake signal sequence, 1.0-ml aliquots of MIV-competent cells of KW20, RR563, and RR804 were incubated for 20 min at 37°C with 0.2 μg of MAP7 DNA/ml in the presence of competing chromosomal DNAs from KW20 and E. coli strain DH5α at concentrations of 0, 0.2, 0.8, and 4.0 μg/ml. The cells were then treated with DNase I at 10 μg/ml for 5 min, and the frequency of transformation to Novr was assessed by plating.
Osmotic-shock and antibiotic sensitivity tests.
To examine the ability of cells to withstand an osmotic shock, exponentially growing and stationary-phase cells were transferred to sBHI containing 8% glucose, incubated for 30 min, diluted 100-fold in BHI and in water, and plated after 10 min. Antibiotic sensitivity tests were done by scoring colony formation on threshold concentrations of antibiotics and using zone of inhibition assays with standard disks (aztreonam [30 μg], imipenem [10 μg], and mecillinam [25 μg]).
PCR amplification, cloning, and disruption of HI1128.
To create a selectable marker tightly linked to murE, primers C (CCATCCAGCTTGTGACTGCG) and D (GCTGAGGGGAAGACACACCAAG) were used to amplify a 3.0-kb fragment containing the hypothetical gene HI1128, which was cloned into the pGEM-T vector system (Promega), giving the plasmid pGT2. A HindIII-cut Cmr cassette from pKRP10 was inserted into pGT2 at the HindIII site at bp 1195829 (HI1128), giving plasmid pGT4, which was used to construct strain RR797 by transforming its insert into RR749 and selecting for resistance to chloramphenicol. The structure of the inactivated chromosomal gene was confirmed by PCR using primers C and D.
Construction of murE749 double mutant and murE-lacZ fusion strains.
Mini-Tn10kan mutations in cya, crp, sxy, rec-2, dprA, comE, topA, icc, thdF, and rpoBC (7, 8, 13, 23, 27, 28, 36, 38) were introduced into strain RR749 by transformation with limiting amounts of the transposon-disrupted chromosomal DNA of the corresponding mutant strain, followed by selection for kanamycin-resistant cells (Table 1). Fusions of the E. coli lacZ gene to sxy, comA, and rec2 were also introduced into RR749 by transformation. The retention of the murE749 mutation in the double-mutant strains was confirmed by MnlI digests of PCR-amplified murE fragments.
RESULTS
Isolating hypercompetence mutations.
Nine new hypercompetent isolates were identified by screening additional aliquots of an EMS-mutagenized H. influenzae culture. Because the previously-identified hypercompetence mutation sxy-1 is closely linked to the streptomycin resistance locus, we determined whether these new mutations were in sxy by examining their linkages to Strr. DNA from Strr derivatives of the mutant strains was transformed into the wild-type strain KW20, and Strr transformants were screened for hypercompetence. Five of the strains showed no evidence of linkage and are the subject of this paper. The other four strains were found to carry mutations in sxy (L. Bannister and R. Redfield, unpublished data).
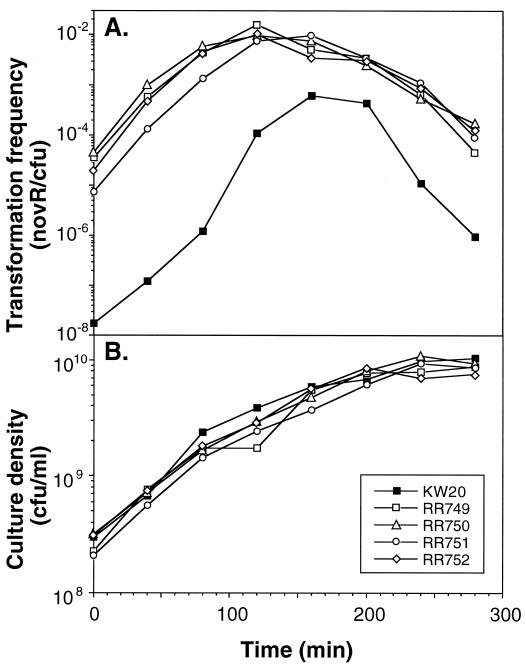
As seen in Fig. 1, the four mutant strains all grew at the same rate as their wild-type parent but exhibited greatly elevated transformation frequencies at all stages of growth. Transformation frequencies were not further increased after incubation in the competence-inducing starvation medium MIV.
FIG. 1.
Time course of growth and competence of murE mutants and the wild-type parent KW20. (A) Transformation frequencies. (B) Culture densities.
Mapping the mutation in RR749.
We initially mapped the hypercompetence mutation of strain RR749 and then demonstrated that strains 750, 751, and 752 had hypercompetence mutations in the same gene. Mapping was laborious because large numbers of potential recombinants had to be individually tested for hypercompetence.
Because the RR749 mutation was not linked to previously identified competence genes (data not shown), our first step was to isolate a selectable Tn916 transposon insertion linked to it. We introduced Tn916 into strain RR749 on the unstable plasmid pAM120 (14) and selected colonies with Tn916 insertions by plating them on tetracycline (24). DNA extracted from a pool of 10,000 independent Tetr colonies was then used to transform strain KW20 to tetracycline resistance, and the colony competence assay was used to identify colonies that had also acquired the hypercompetence mutation (to prevent transformation with multiple fragments, the DNA from the pooled colonies was used at limiting concentration). This screen generated strain RR783, in which tetracycline resistance and hypercompetence show 6% cotransformation.
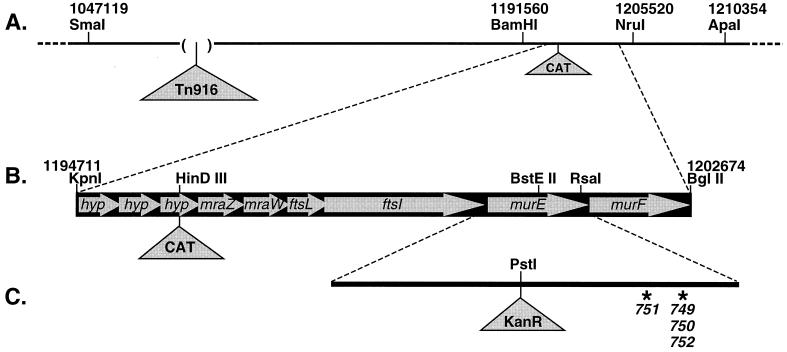
Pulsed-field (contour-clamped homogeneous electric field) gel analysis of the RR783 chromosome showed that the 18-kb Tn916 insertion was in a segment bounded by the SmaI site at bp 1047119 and the ApaI site at bp 1210354 (Fig. 2A) (25). The genome sequence was then used to predict restriction sites in this region. To find out which of these restriction sites were contained in the chromosomal segment between Tn916 and the linked hypercompetence mutation, we predigested RR783 DNA with various enzymes before using it to transform KW20 and used the colony competence assay to score hypercompetence of the Tetr transformants. Predigestion with BamHI eliminated linkage between Tn916 and the hypercompetence mutation, but predigestion with NruI did not, locating the mutation between the BamHI site at bp 1191560 and the unmethylated NruI site at bp 1205520 (Fig. 2A).
FIG. 2.
The murE region of the H. influenzae genome. (A) The 165-kb region around murE, showing the locations of the Tn916 insertion in strain RR783 and the Cmr cassette insertion in strain RR797. The parentheses around the Tn916 insertion indicate that its position is only approximate. (B) The 8.0-kb region around murE cloned in plasmids p836B and p848M. Arrows indicate the direction of transcription. (C) Locations of the RR749, RR751, and RR752 point mutations (asterisks) and of the Kanr cassette in pmurE::Kan.
To create a selectable marker more tightly linked to the hypercompetence mutation, we used PCR (with primers C and D) to amplify the segment from bp 1193842 to 1196875 and inserted a chloramphenicol resistance (Cmr) cassette from pKRP10 (33) into the HindIII site at bp 1195829 (Fig. 2A). This insertion was then transformed into RR749, producing strain RR797, in which the Cmr cassette showed 38% cotransformation with the hypercompetence mutation. Finally, we cloned an 8-kb KpnI-BglII fragment containing the mutation and the Cmr cassette into pSU40, giving plasmid p836B (Fig. 2B).
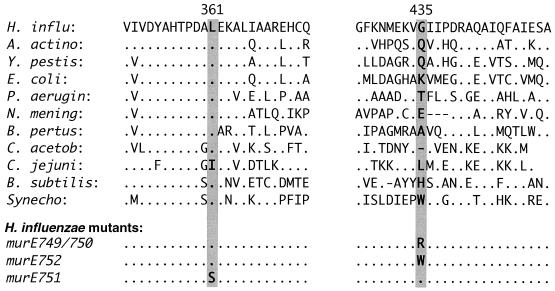
Colony competence assays of cells transformed with gel-purified subfragments of this insert mapped the hypercompetence mutation to a 300-bp BstEII-RsaI fragment within murE (HI1133). This segment was sequenced with primers A and B. Comparison to the published H. influenzae Rd sequence and to the parent strain's sequence revealed a single G→A substitution at nucleotide 1303 of the murE sequence, replacing glycine (codon 435) with arginine. This is a nonconservative substitution, and comparison with the homologous sequences of 30 other bacteria available in the NCBI Microbial Genomes BLAST database shows it to be in a nonconserved region of the protein (Fig. 3).
FIG. 3.
Comparison of murE-homologous sequences around positions 361 and 435 of the H. influenzae murE sequence. H. influ, H. influenzae; A. actino, Actinobacillus actinomycetemcomitans; Y. pestis, Yersinia pestis; P. aerug, Pseudomonas aeruginosa; N. mening, Neisseria meningitidis; B. pertus, Bordetella pertussis; C. acetob, Clostridium acetobutylicum; C. jejuni, Campylobacter jejuni; B. subtilis, Bacillus subtilis; Synecho, Synechocystis sp. strain PCC6803.
Identifying the mutations in strains RR750, RR751, and RR752.
The murE-linked Cmr cassette was transferred from strain RR783 into mutants RR750, RR751, and RR752 in two steps. It was first transformed into the wild-type strain KW20, using colony assays to confirm that the Cmr transformant retained its murE+ allele, and then DNA from this strain was used to transform RR750, RR751, and RR752 to Cmr, again using colony assays to confirm that hypercompetence was retained. Selection for the Cmr cassette was then used to clone the murE-containing 8-kb KpnI-BglII fragments from these strains into pSU40. Transformation of plasmid DNA into KW20 showed that these fragments did carry the mutations responsible for the hypercompetence of their strains, and sequencing revealed the following single mutations in the murE gene (Fig. 2C).
(i) RR750.
Strain RR750 contains a murE mutation identical to that in RR749 (nucleotide, G1303→A; amino acid, G435→R). The mutations are probably independent, because cells in the original culture had divided only two or three times between EMS mutagenesis and screening.
(ii) RR751.
Strain RR751 contains a different murE mutation (nucleotide, T1082→C; amino acid, L361→S). This is a nonconservative substitution in a conserved region of the protein. Of 30 complete or partial sequences available in the NCBI Microbial Genomes BLAST database, 22 had leucine at this position (Fig. 3).
(iii) RR752.
Strain RR752 contains a point mutation at the same position as that in murE749 but causing a different substitution (nucleotide, G1303→T; amino acid, G435→W). Like murE749, this is a nonconservative substitution.
murE749 cells take up DNA by the normal sequence-specific pathway.
The murE gene encodes the meso-diaminopimelate-adding enzyme of the peptidoglycan synthesis pathway. In principle, mutations in murE could increase competence in two ways. They could alter peptidoglycan structure in a way that directly increases the permeability of cells to DNA, bypassing the normal competence pathway of DNA uptake. Alternatively, they could cause the normal competence pathway to be induced under what are otherwise noninducing conditions.
Comparison of DNA uptake in early- and late-log-phase growth shows that murE749 cells do not constitutively take up DNA. Measurement of DNA uptake in exponential growth is limited by the assay sensitivity, but this level does not preclude the observed murE749 transformation frequency of 10−4 to 10−5. Uptake by murE749 cells increases 200-fold when the cells are maximally competent (transformation frequency, about 10−2). Stronger evidence that DNA uptake is by the normal competence pathway comes from analysis of the sequence specificity of uptake. Competent H. influenzae cells preferentially take up DNA fragments containing the abundant 9-bp Haemophilus-specific uptake signal sequence (11, 34).
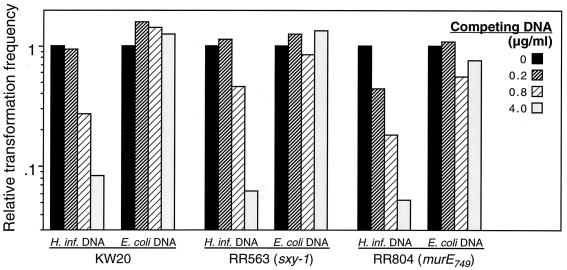
Competition experiments showed that DNA uptake by murE749 has the same uptake specificity as that of wild-type competent cells and that of the hypercompetent sxy-1 mutant RR563, which also uses the normal uptake pathway (Fig. 4). These results indicate that the murE749 mutation induces the normal competence pathway without otherwise altering the integrity of the cell envelope.
FIG. 4.
Competition for DNA uptake. Strain KW20, RR563, and RR804 cells were made competent in MIV and incubated with 200 ng of MAP7 DNA/ml and the indicated concentration of competing H. influenzae or E. coli DNA. The transformation frequencies have been normalized to that seen in the absence of competing DNA.
Interactions with other loci.
Insertions in a number of genes reduce or eliminate competence. If the murE mutation bypasses the normal DNA uptake pathway, transformation should be independent of both the genes that regulate competence and those directly involved in DNA uptake and recombination. To test this, we constructed double-mutant strains by transforming RR749 (murE749) with DNA from various competence mutants and selecting for the insertions, and used colony assays to examine their competence (Fig. 5).
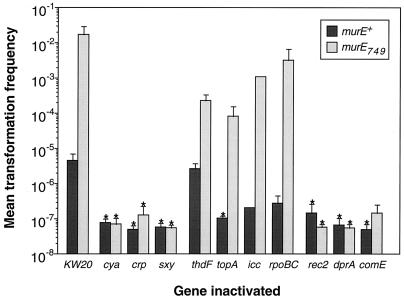
FIG. 5.
Transformation frequencies of double-mutant strains in colony assays. The error bars indicate the standard errors of the mean. The data represent one experiment (icc) or the means of two (cya, crp, rpo, rec2, dpr, and comE), three (topA and thdF), or four (KW20 and sxy) experimental points, with each point being the mean of the four colonies tested. The asterisks indicate values which are upper limits because no transformed colonies were produced by that genotype.
We tested mini-Tn10kan insertions in the regulatory genes cya (13), crp (7), sxy (38), and topA (8) and in the uptake and translocation genes comE (36), rec-2 (28), and dprA (23). We also tested the effect of a spectinomycin resistance cassette inserted in icc (27) and those of two previously characterized but unmapped mini-Tn10kan insertions in the uptake-deficient strains JG6 and JG49 (35), which have now been mapped by sequencing the DNA flanking their insertion sites. The JG6 insertion is in the homolog of the E. coli thdF gene, and the JG49 insertion is between rpoB and rpoC (R. J. Redfield, unpublished data).
Taken together, the results shown in Fig. 5 indicate that mutations that completely prevent transformation in a murE+ background (cya, crp, sxy, rec-2, comE, and dprA) also do so in a murE749 background. However, mutations that decrease but do not eliminate transformation (topA, icc, and rpoBC) reduce the transformation frequencies of murE+ and murE749 strains by roughly comparable amounts. (Although the topA mutant strain gave no transformants in this colony assay, more sensitive measurements using MIV starvation medium have shown that it reduces transformation frequencies about 105-fold but does not entirely eliminate competence (32). For the cya, sxy, topA, and rec-2 mutations, similar results were observed with murE750, murE751, and murE752 (the other combinations have not been examined). This dependence on known competence genes supports the hypothesis that the murE749 mutation increases transformation by the normal pathway.
The murE749 mutation causes induction of late-acting competence genes but not of the early-acting gene sxy.
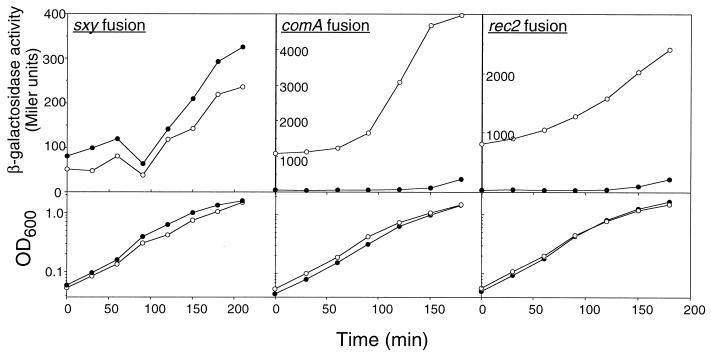
High-level expression of lacZ fusions to the DNA uptake gene comA and the DNA translocation gene rec-2 requires induction of competence by the starvation medium MIV (19). However, both fusions showed dramatically increased β-galactosidase production in a murE749 background in cells growing in rich medium (Fig. 6), confirming that the mutation increases competence by inducing the normal pathway. The comA-to-comF operon requires Sxy for its expression (39). However, expression of a fusion of lacZ to the sxy gene (Bannister and Redfield, unpublished data) was not changed by the murE749 mutation, indicating that the altered regulation of comA and rec-2 is not caused by altered expression of Sxy.
FIG. 6.
Expression of lacZ fusions to competence genes. Solid circles, murE+; open circles, murE749.
murE is an essential gene.
The murE749 mutation is likely to affect regulation of competence by decreasing the activity of the MurE protein. A gain-of-function mutation is also possible, although activities other than peptide synthetase have not been associated with any of the Mur family of proteins. A complete loss of function is highly improbable, as the gene is thought to be essential in H. influenzae, as it is in E. coli (26). To confirm that MurE is essential, we attempted to create a murE null mutant by transforming H. influenzae to kanamycin resistance with a murE::kan knockout constructed in an E. coli plasmid (the location of the Kanr insertion is shown in Fig. 2C). Although Kanr transformants could be isolated, Southern blotting analysis showed that they always contained duplications or more complex changes that preserved an intact copy of murE. This confirms that murE is essential and implies that the murE hypercompetence mutations must not cause complete loss of function of the gene product.
The murE749 mutation does not alter cell viability, permeability, or antibiotic sensitivity.
The normal growth rates seen for the murE mutant strains in Fig. 1 suggest that their mutations do not significantly change the stability of the cell envelope. We have examined the abilities of the mutants to survive and recover from stationary phase and found them to be indistinguishable from that of their wild-type parent (data not shown). To look for minor permeability changes, we also examined the strains' abilities to survive an osmotic shock—transfer from medium containing 8% glucose to glucose-free medium or to water. No differences were found between strains RR749 and KW20, nor were there any differences in these strains' abilities to grow in the presence of threshold concentrations of gentamycin, an antibiotic that depends on cell envelope permeability for entry. We used disk diffusion tests to examine the sensitivities of murE+ and murE749–752 cells to antibiotics that act on the cell wall. No differences in zones of inhibition were seen with aztreonam, a specific inhibitor of the septation protein PBP3, or with imipenem or mecillinam, inhibitors of PBP2 (6, 15).
DISCUSSION
Peptidoglycan biosynthesis has not previously been implicated in competence regulation or DNA uptake in any organism, nor is anything known about the extent to which the peptidoglycan layer might limit DNA uptake.
The structure of the H. influenzae peptidoglycan has been analyzed by Burroughs et al. (5) and found to be very similar to that of E. coli. Although there have been no studies of H. influenzae peptidoglycan synthesis, the complete H. influenzae genome sequence encodes the pathways characterized in E. coli and other gram-positive and gram-negative bacteria. Peptidoglycan monomers consisting of a pentapeptide linked to a disaccharide are synthesized intracellularly by a series of four enzymatic reactions in which the products of the murC, -D, -E, and -F genes ligate successive amino acid residues to UDP-N-acetylmuramic acid (UDP-MurNAc). These disaccharide-pentapeptide subunits are translocated across the inner membrane into the periplasm, where their disaccharide backbones are joined into long chains and cross-linked by bonds between peptide side chains to form a strong flexible mesh, the murein sacculus. Growth and division of cells then involves extensive breakage of cross-links, insertion of new connections, and recycling of released monomers.
The Mur synthetases (MurC, -D, -E, and -F) are a well-defined family of proteins with closely related functions (4); they sequentially contribute l-alanine, d-glutamate, meso-2,6-diaminopimelate, and d-alanine–d-alanine to the peptide side chain. No structural information is available for MurE, but the crystal structure of MurD has been determined (3). A BLAST search (http://www.ncbi.nlm.nih.gov/blast/blast.cgi) aligns the H. influenzae MurE G435 residue (mutated in murE749, murE750, and murE752) with E. coli MurE V439, and Bertrand weakly aligns this with E. coli MurD389, which lacks defined secondary structure. However, the L361 of MurE (mutated in murE751) is highly conserved and homologous to V326 of E. coli MurD, which is in a well-structured alpha helix. Thus, the substitution of a small hydrophilic serine in murE751 for this larger hydrophobic residue might be expected to reduce catalytic function. However, the physical properties of all the murE mutant strains are indistinguishable from those of their wild-type parent, suggesting that peptidoglycan synthesis is not significantly impaired by any of the mutations.
It is not surprising that hypercompetence is not caused by nonspecifically increasing cell permeability, because the peptidoglycan network is unlikely to be the primary limit to transformation. Uptake of DNA into the cytoplasm requires that the DNA be bound on the cell surface and that it cross the outer membrane, peptidoglycan, and inner membrane. Wild-type cells in exponential growth do not become competent and do not bind DNA, but the murE mutants do, indicating that the murE mutations cause DNA-binding structures to be assembled on the cell surface. Furthermore, even major alterations of the peptidoglycan are unlikely to produce holes large enough for passive diffusion of linear DNA molecules. The extent of peptidoglycan cross-linking in H. influenzae is like that of E. coli, where regular cross-links produce a fairly homogeneous mesh with pores of about 2.06-nm diameter (5, 12). Although linear DNA (diameter, 2.0 nm) could in principle be threaded through these pores, much larger openings are likely to be required. The persistence length of DNA is approximately 50 to 80 nm (20, 21), so large double-stranded DNA molecules cannot passively move through murein. Furthermore, competent cells can efficiently take up closed circular DNAs, so a free end is not required for the initial stages of uptake.
The analysis of gene fusions indicates that the mutations cause competence by affecting gene regulation. As MurE is very unlikely to bind DNA or otherwise directly affect gene expression, the most parsimonious hypothesis is that a moderate reduction of MurE activity either generates a signal that induces competence, or eliminates a signal that normally precludes it.
A potential regulatory connection between competence and peptidoglycan synthesis exists in the peptidoglycan recycling pathway. Sugar-tripeptide intermediates (1-6-anhydro MurNAc-l-Ala-d-Glu-m-A2pm) are produced by the combined actions of the murein-specific peptidoglycan transglycosylase Slt and the endopeptidase PBP7 and brought across the inner membrane by the ampG permease. The anhydro-disaccharide-peptides are cleaved by AmpD into the free peptides and disaccharides, and the tripeptides are added to UDP-MurNAc by Mpl, bypassing the steps catalyzed by MurC, MurD, and MurE (29, 30). The intermediate is known to regulate transcription of beta-lactamase genes in some bacteria, in conjunction with the AmpR activator, and is thought to play a more general role in monitoring and regulating cell growth (31). Such regulation could include that of competence, which is induced when growth slows. H. influenzae does not have a chromosomally encoded beta-lactamases but does have the ampR homolog gcvA and the ampG, ampD, and mpl genes required for recycling. Mutations in murE might therefore increase competence by increasing the demand on the recycling pathway, reducing the concentration of the regulatory sugar tripeptide and thus relieving the postulated inhibition of competence by rapid growth. However, knockout mutations of gcvA, ampG, ampD, and mpl have no dramatic effect on competence (C. Ma and R. J. Redfield, unpublished data), so changes in peptidoglycan recycling are unlikely to be responsible for the increased competence of the murE mutants.
Present models of the regulation of competence do not explain how mutations in murE could cause induction of competence genes. The rec2 and comA genes induced in the murE749 mutant belong to a group of genes preceded by a highly conserved consensus sequence called the competence regulatory element (CRE). Most genes with CRE sites are known to play roles in competence, and several have been shown to require Sxy and/or cyclic AMP receptor protein (CRP) for transcription, leading to the hypothesis that the Sxy protein is a transcription factor, acting at the CRE site and itself regulated by CRP (19, 23, 39). The strong similarity between the CRE and CRP consensus sequences (L. Macfadyen, unpublished data) suggests an alternative model in which CRP binds at the CRE sites and stimulates transcription in response to Sxy. However, neither of these models includes any obvious role for peptidoglycan synthesis.
ACKNOWLEDGMENTS
We thank Elaine Tuomanen, Terry Beveridge, Kelly MacDonald, Kevin Young, Alex Tomasz, Lawrence Macintosh, Ted Park, and Joachim-Volker Hoeltje for advice, Michelle Gwinn and Mark Chandler for strains, and Robert Hancock for advice and aztreonam. Ryan Sinotte and Lisa Oppenheim assisted with experiments. We thank the Institute for Genomic Research (http://www.tigr.org) for the H. influenzae sequence and for access to murE-homologous sequences from incomplete bacterial genomes.
REFERENCES
- 1.Alexander H, Leidy G. Determination of inherited traits of H. influenzae by deoxyribonucleic acid fractions isolated from type-specific cells. J Exp Med. 1951;93:345–359. doi: 10.1084/jem.93.4.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Bannister L A. An RNA secondary structure regulates sxy expression and competence development in Haemophilus influenzae. Ph.D. Thesis. Vancouver, Canada: University of British Columbia; 1999. [Google Scholar]
- 2.Barcak G J, Chandler M S, Redfield R J, Tomb J-F. Genetic systems in Haemophilus influenzae. Methods Enzymol. 1991;204:321–342. doi: 10.1016/0076-6879(91)04016-h. [DOI] [PubMed] [Google Scholar]
- 3.Bertrand J A, Auger G, Fanchon E, Martin L, Blanot D, van Heijenoort J. Crystal structure of UDP-N-acetylmuramoyl-L-alanine:D-glutamate ligase from Escherichia coli. EMBO J. 1997;16:3416–3425. doi: 10.1093/emboj/16.12.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouhss A, Mengin-Lecreulx D, Blanot D, van Heijenoort J, Parquet C. Invariant amino acids in the Mur peptide synthetases of bacterial peptidoglycan synthesis and their modification by site-directed mutagenesis in the UDP-MurNAc:L-alanine ligase from Escherichia coli. Biochemistry. 1997;36:11556–11563. doi: 10.1021/bi970797f. [DOI] [PubMed] [Google Scholar]
- 5.Burroughs M H, Chang Y S, Gage D A, Tuomanen E I. Composition of the peptidoglycan of Haemophilus influenzae. J Biol Chem. 1993;268:11594–11598. [PubMed] [Google Scholar]
- 6.Carlone N A, Ferrero M, Cuffini A M, Cavallo R. Imipenem: morphological changes and lethal effects on Pseudomonas aeruginosa. Drugs Exp Clin Res. 1987;13:623–629. [PubMed] [Google Scholar]
- 7.Chandler M S. The gene encoding cyclic AMP receptor protein is required for competence development in Haemophilus influenzae Rd. Proc Natl Acad Sci USA. 1992;89:1626–1630. doi: 10.1073/pnas.89.5.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandler M S, Smith R A. Characterization of the Haemophilus influenzae topA locus: DNA topoisomerase I is required for genetic competence. Gene. 1996;169:25–31. doi: 10.1016/0378-1119(95)00777-6. [DOI] [PubMed] [Google Scholar]
- 9.Chen W J, Gross L, Joho K E, McAllister W T. A modified kanamycin-resistance cassette to facilitate two-codon insertion mutagenesis. Gene. 1992;111:143–144. doi: 10.1016/0378-1119(92)90617-x. [DOI] [PubMed] [Google Scholar]
- 10.Deich R A, Smith H O. Homologous and heterologous DNA uptake in Haemophilus transformation. In: Glover S W, Butler L O, editors. Transformation, 1978. Oxford, United Kingdom: Cotswold Press; 1979. pp. 377–384. [Google Scholar]
- 11.Deich R A, Smith H O. Mechanism of homospecific DNA uptake in Haemophilus influenzae transformation. Mol Gen Genet. 1980;177:369–374. doi: 10.1007/BF00271475. [DOI] [PubMed] [Google Scholar]
- 12.Demchick P, Koch A L. The permeability of the wall fabric of Escherichia coli and Bacillus subtilis. J Bacteriol. 1996;178:768–773. doi: 10.1128/jb.178.3.768-773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorocicz I, Williams P, Redfield R J. The Haemophilus influenzae adenylate cyclase gene: cloning, sequence, and essential role in competence. J Bacteriol. 1993;175:7142–7149. doi: 10.1128/jb.175.22.7142-7149.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gawron-Burke C, Clewell D B. Regeneration of insertionally inactivated streptococcal DNA fragments after excision of transposon Tn916 in Escherichia coli: strategy for targeting and cloning of genes from gram-positive bacteria. J Bacteriol. 1984;159:214–221. doi: 10.1128/jb.159.1.214-221.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Georgopapadakou N H, Smith S A, Sykes R B. Mode of action of aztreonam. Antimicrob Agents Chemother. 1982;21:950–956. doi: 10.1128/aac.21.6.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodgal S H. Procedures for Haemophilus influenzae transformation. Methods Enzymol. 1968;12:867–876. [Google Scholar]
- 17.Goodgal S H, Gromkova R. Separation of specific segments of transforming DNA after treatment with endodeoxyribonuclease. Proc Natl Acad Sci USA. 1973;70:503–506. doi: 10.1073/pnas.70.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodgal S H, Herriott R M. Studies on transformations of Haemophilus influenzae. I. Competence. J Gen Physiol. 1961;44:1201–1227. doi: 10.1085/jgp.44.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gwinn M L, Stellwagen A E, Craig N L, Tomb J F, Smith H O. In vitro Tn7 mutagenesis of Haemophilus influenzae Rd and characterization of the role of atpA in transformation. J Bacteriol. 1997;179:7315–7320. doi: 10.1128/jb.179.23.7315-7320.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagerman P J. Flexibility of DNA. Annu Rev Biophys Biophys Chem. 1988;17:265–286. doi: 10.1146/annurev.bb.17.060188.001405. [DOI] [PubMed] [Google Scholar]
- 21.Hansma H G, Revenko I, Kim K, Laney D E. Atomic force microscopy of long and short double-stranded, single-stranded and triple-stranded nucleic acids. Nucleic Acids Res. 1996;24:713–720. doi: 10.1093/nar/24.4.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herriott R M, Meyer E M, Vogt M. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J Bacteriol. 1970;101:517–524. doi: 10.1128/jb.101.2.517-524.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karudapuram S, Barcak G J. The Haemophilus influenzae dprABC genes constitute a competence-inducible operon that requires the product of the tfoX (sxy) gene for transcriptional activation. J Bacteriol. 1997;179:4815–4820. doi: 10.1128/jb.179.15.4815-4820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kauc L, Goodgal S H. Introduction of transposon Tn916 DNA into Haemophilus influenzae and Haemophilus parainfluenzae. J Bacteriol. 1989;171:6625–6628. doi: 10.1128/jb.171.12.6625-6628.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee J J, Smith H O, Redfield R J. Organization of the Haemophilus influenzae Rd genome. J Bacteriol. 1989;171:3016–3024. doi: 10.1128/jb.171.6.3016-3024.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lugtenberg E J, van Schijndel-van Dam A. Temperature-sensitive mutants of Escherichia coli K-12 with low activity of the diaminopimelic acid adding enzyme. J Bacteriol. 1972;110:41–46. doi: 10.1128/jb.110.1.41-46.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macfadyen L P, Ma C, Redfield R J. A 3′,5′ cyclic AMP (cAMP) phosphodiesterase modulates cAMP levels and optimizes competence in Haemophilus influenzae Rd. J Bacteriol. 1998;180:4401–4405. doi: 10.1128/jb.180.17.4401-4405.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarthy D. Cloning of the rec-2 locus of Haemophilus influenzae. Gene. 1989;75:135–143. doi: 10.1016/0378-1119(89)90390-9. [DOI] [PubMed] [Google Scholar]
- 29.Mengin-Lecreulx D, Ayala J, Bouhss A, van Heijenoort J, Parquet C, Hara H. Contribution of the Pmra promoter to expression of genes in the Escherichia coli mra cluster of cell envelope biosynthesis and cell division genes. J Bacteriol. 1998;180:4406–4412. doi: 10.1128/jb.180.17.4406-4412.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mengin-Lecreulx D, van Heijenoort J, Park J T. Identification of the mpl gene encoding UDP-N-acetylmuramate: l-alanyl-γ-d-glutamyl-meso-diaminopimelate ligase in Escherichia coli and its role in recycling of cell wall peptidoglycan. J Bacteriol. 1996;178:5347–5352. doi: 10.1128/jb.178.18.5347-5352.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park J T. Why does Escherichia coli recycle its cell wall peptides? Mol Microbiol. 1995;17:421–426. doi: 10.1111/j.1365-2958.1995.mmi_17030421.x. [DOI] [PubMed] [Google Scholar]
- 32.Redfield R J. sxy-1, a Haemophilus influenzae mutation causing greatly enhanced competence. J Bacteriol. 1991;173:5612–5618. doi: 10.1128/jb.173.18.5612-5618.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reece K S, Phillips G J. New plasmids carrying antibiotic-resistance cassettes. Gene. 1995;165:141–142. doi: 10.1016/0378-1119(95)00529-f. [DOI] [PubMed] [Google Scholar]
- 34.Smith H O, Tomb J-F, Dougherty B A, Fleischmann R D, Venter J C. Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae Rd genome. Science. 1995;269:538–540. doi: 10.1126/science.7542802. [DOI] [PubMed] [Google Scholar]
- 35.Tomb J F, Barcak G J, Chandler M S, Redfield R J, Smith H O. Transposon mutagenesis, characterization, and cloning of transformation genes of Haemophilus influenzae Rd. J Bacteriol. 1989;171:3796–3802. doi: 10.1128/jb.171.7.3796-3802.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomb J F, el Hajj H, Smith H O. Nucleotide sequence of a cluster of genes involved in the transformation of Haemophilus influenzae Rd. Gene. 1991;104:1–10. doi: 10.1016/0378-1119(91)90457-m. [DOI] [PubMed] [Google Scholar]
- 37.Williams P, Hung W L, Redfield R J. Cell transfer by filtration: evaluation of protocols for transformational competence. FEMS Microbiol Lett. 1996;137:183–187. doi: 10.1111/j.1574-6968.1996.tb08103.x. [DOI] [PubMed] [Google Scholar]
- 38.Williams P M, Bannister L A, Redfield R J. The Haemophilus influenzae sxy-1 mutation is in a newly identified gene essential for competence. J Bacteriol. 1994;176:6789–6794. doi: 10.1128/jb.176.22.6789-6794.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zulty J J, Barcak G J. Identification of a DNA transformation gene required for com101A+ expression and supertransformer phenotype in Haemophilus influenzae. Proc Natl Acad Sci USA. 1995;92:3616–3620. doi: 10.1073/pnas.92.8.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]