Abstract
Inhibition of eukaryotic translation initiation through unscheduled RNA clamping of the DEAD-box (DDX) RNA helicases eIF4A1 and eIF4A2 has been documented for pateamine A (PatA) and rocaglates—two structurally different classes of compounds that share overlapping binding sites on eIF4A. Clamping of eIF4A to RNA causes steric blocks that interfere with ribosome binding and scanning, rationalizing the potency of these molecules since not all eIF4A molecules need to be engaged to elicit a biological effect. In addition to targeting translation, PatA and analogs have also been shown to target the eIF4A homolog, eIF4A3—a helicase necessary for exon junction complex (EJC) formation. EJCs are deposited on mRNAs upstream of exon-exon junctions and, when present downstream from premature termination codons (PTCs), participate in nonsense-mediated decay (NMD), a quality control mechanism aimed at preventing the production of dominant-negative or gain-of-function polypeptides from faulty mRNA transcripts. We find that rocaglates can also interact with eIF4A3 to induce RNA clamping. Rocaglates also inhibit EJC-dependent NMD in mammalian cells, but this does not appear to be due to induced eIF4A3-RNA clamping, but rather a secondary consequence of translation inhibition incurred by clamping eIF4A1 and eIF4A2 to mRNA.
Keywords: rocaglates, eIF4A1, eIF4A3, molecular staples, translation, NMD
INTRODUCTION
Eukaryotic RNA helicases have been implicated in all facets of RNA metabolism. They utilize the energy derived from NTP hydrolysis to remodel nucleic acids, modify nucleic acid–protein complexes, and disrupt protein–protein interactions. They are identifiable by a set of conserved motifs that are essential to their catalytic activity, and based on these have been grouped into one of two superfamilies—SF1 and SF2 (Caruthers and McKay 2002; Jankowsky 2011). Within SF2, the DEAD-box RNA helicases (DDX; named after their ATP-binding/hydrolysis Asp–Glu–Ala–Asp signature core) are the more abundant subgroup. Mammalian DDX family members include three homologous proteins: (i) eukaryotic initiation factor (eIF) 4A1 (DDX2A), (ii) eIF4A2 (DDX2B), and (iii) eIF4A3 (DDX48). In humans, eIF4A1 and eIF4A2 are implicated in translation initiation and share 90% amino acid identity, while eIF4A3 is a component of the exon junction complex (EJC) and shares 66% amino acid identity with eIF4A1.
eIF4A1 plays a critical role in cap-dependent translation (Rogers et al. 2002). It is among the more abundant initiation factors (Duncan et al. 1987; Galicia-Vazquez et al. 2012; Kulak et al. 2014) with ∼5% of total eIF4A1 present in a complex with the eIF4E cap binding protein and eIF4G, thus forming the eIF4F complex. Following binding to the cap, eIF4F prepares the mRNA template to receive a 43S preinitiation complex (40S ribosome and associated factors) by resolving local secondary structure (Pelletier and Sonenberg 2019). Secondary structure within mammalian mRNA 5′ leader regions place a higher demand on eIF4A1 helicase activity for ribosome recruitment and thus is a discriminatory feature that curbs initiation by decreasing the competitiveness of mRNAs for limiting amounts of eIF4F (Svitkin et al. 2001; Pelletier and Sonenberg 2019; Steinberger et al. 2020). Inhibiting eIF4F activity has shown itself to be a viable anti-cancer strategy and is thought to be the consequence of preferential inhibition of mRNAs encoding critical regulators of the cell cycle, cell survival, angiogenesis, and immune responses (Bhat et al. 2015).
The role of human eIF4A2 in ribosome recruitment appears similar to eIF4A1, and both proteins can recycle through the eIF4F complex (Yoder-Hill et al. 1993; Robert et al. 2020). The eIF4A2 gene has been targeted for ablation using CRISPR in NIH 3T3 and Hap1 cells with no consequence on cell proliferation (Galicia-Vazquez et al. 2015; Naineni et al. 2021). As well, mice lacking eIF4A2 are viable, whereas those lacking eIF4A1 are not, indicating the latter to be essential (Senechal et al. 2021). However, variants in EIF4A2 have been associated with neurodevelopmental disorders (Paul et al. 2023). Pateamine and rocaglates are small molecules that have been found to be quite potent at blocking translation initiation (Supplemental Fig. S1)—both of which target eIF4A1 and eIF4A2 (Chu et al. 2019; Naineni et al. 2021). Pateamines and rocaglates are natural products that, although structurally quite different, each act as molecular staples to clamp eIF4A1 and eIF4A2 onto RNA (Iwasaki et al. 2016; Iwasaki et al. 2019; Naineni et al. 2021). Unscheduled eIF4A1/2 clamping to RNA exerts several biological effects; among which are inhibition of ribosome recruitment, 40S scanning blockade, and depletion of functional eIF4F complexes (Iwasaki et al. 2016; Chu et al. 2020). Like with eIF4A1, clamping of eIF4A2 to RNA is induced by PatA and rocaglate family members, but because of its lower abundance levels (Galicia-Vazquez et al. 2012; Kulak et al. 2014), eIF4A2 contributes in a minor way to the biological activity of these compounds (Chu et al. 2019; Naineni et al. 2021).
eIF4A3 is a critical RNA binding component of the EJC. The proteins Y14 and MAGOH bind as a heterodimer to eIF4A3 and inhibit its ATPase activity, thus stabilizing eIF4A3 in the RNA-bound conformation (Schlautmann and Gehring 2020). The EJC, deposited ∼20–24 nt upstream of the spliced exon-exon junction, is removed upon the first round of translation and has been implicated in splicing regulation, nucleo-cytoplasmic mRNA export, and stimulation of translation (Schlautmann and Gehring 2020). The EJC also plays a critical role in nonsense-mediated decay (NMD), an mRNA surveillance mechanism that is triggered when premature stop codons (PTC) are present ∼50–55 nt upstream of the EJC. NMD thus serves to protect cells from producing truncated protein products having potential gain-of-function or dominant-negative potential that could prove detrimental to cell survival (Gorbalenya et al. 1989).
Beyond inhibiting translation initiation, PatA and the active analog, DMDA-PatA, have been shown to also interact with eIF4A3 and to block NMD (Dang et al. 2009). The recently elucidated structure of the PatA analog, DMPatA, in complex with eIF4A1•AMPPNP•poly r(AG)5 RNA has shown that DMPatA wedges itself between RNA and eIF4A1, interacting with RNA bases and amino acids F163, Q195, and D198 (Fig. 1A; Naineni et al. 2021). These same three residues are also present within eIF4A3 (F168, Q200, D203) (Andersen et al. 2006) and explain the ability of PatA and DMDA-PatA to interact with eIF4A3 (Fig. 1A; Dang et al. 2009). Although structurally very different, rocaglates and PatA analogs share the same binding site in eIF4A1 (Iwasaki et al. 2019; Naineni et al. 2021). Here, we address whether rocaglates can also interact with eIF4A3, and if so, whether this interaction affects the NMD response.
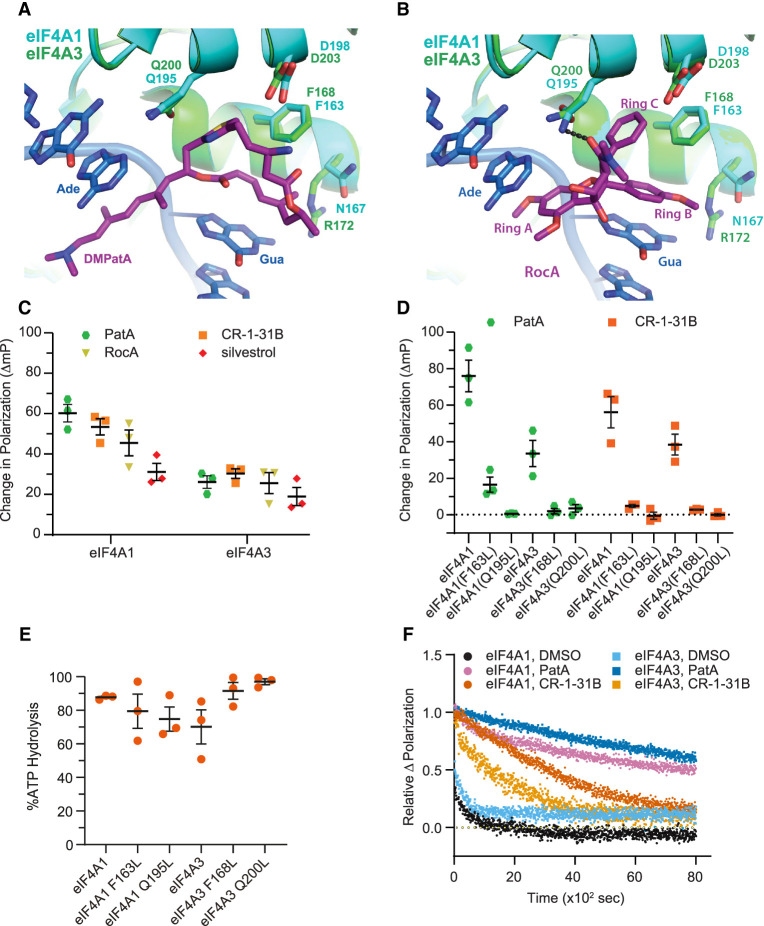
FIGURE 1.
Rocaglates induce clamping of eIF4A3 to polypurine RNA. (A) Overlay of the eIF4A1•AMPPNP•DMPatA•poly r(AG)5 RNA complex (PDB:6XKI) (Naineni et al. 2021) with the crystal structure of human eIF4A3 bound to RNA (PDB:2HYI) (Andersen et al. 2006). The relative location of amino acids from eIF4A1 and eIF4A3 interacting with, or in proximity to, DMPatA are shown. (Ade) Adenine, (Gua) guanine. (B) Overlay of the eIF4A1•AMPPNP•RocA•poly r(AG)5 RNA complex (PDB:5ZC9) (Iwasaki et al. 2019) with the crystal structure of human eIF4A3 bound to RNA (PDB:2HYI) (Andersen et al. 2006). The relative location of amino acids from eIF4A1 and eIF4A3 interacting with, or in proximity to, RocA are shown. (Ade) Adenine, (Gua) guanine. (C) Rocaglates stabilize binding of eIF4A3 to poly r(AG)8. The ΔmP obtained with eIF4A1 or eIF4A3 with poly r(AG)8 RNA and the indicated compounds (10 µM) was measured. The ΔmP obtained is relative to DMSO control. n = 3 ± SD. (D) eIF4A3F168L and eIF4A3Q200L mutants are not substrates for rocaglate-induced clamping. The ΔmP obtained with eIF4A1 or eIF4A3 mutants with poly r(AG)8 RNA and the indicated compounds (10 µM) was assessed. n = 3 ± SD. (E) ATPase activity of eIF4A1 and eIF4A3 protein preparations used in this study. n = 3 ± SD. (F) Relative dissociation of complexes preformed in the presence of eIF4A1 or eIF4A3, ATP (1 mM), and compound (10 µM) measured as a function of time in the presence of 1000-fold molar excess of unlabeled poly r(AG)8 RNA.
RESULTS
The rocaglate, RocA, π-stacks with two RNA purine bases via its aryl rings A and B and is bound to eIF4A1 via interactions with F163 and Q195: aryl ring C slots between F163 and Q195, and the 2-N,N-dimethyl-carboxamide carbonyl group hydrogen bonds with the Q195 side chain (Supplemental Fig. S1a,b; Iwasaki et al. 2019). Superimposition of the eIF4A3 structure (Andersen et al. 2006) with the eIF4A1•AMPPNP•RocA•poly r(AG)5 structure (Iwasaki et al. 2019) reveals that eIF4A3 has retained the two critical amino acids required for RocA interaction (eIF4A3 numbering is F168 and Q200) (Fig. 1B), raising the possibility that rocaglates may also be targeting eIF4A3 in a manner similar to PatA compounds.
Rocaglates induce RNA clamping of eIF4A3
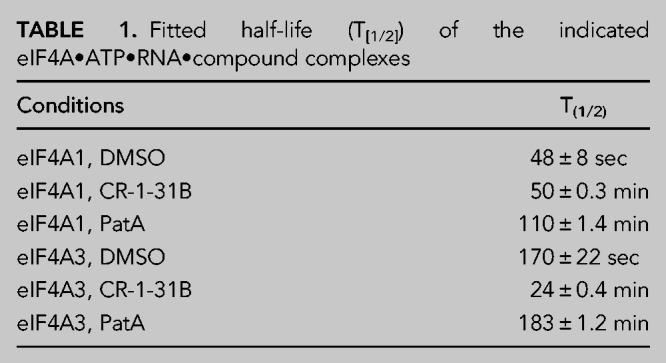
To determine if rocaglates can induce clamping of eIF4A3 to RNA, we utilized a fluorescence polarization (FP) assay using FAM-labeled poly r(AG)8 RNA (Fig. 1C). In the presence of PatA, clamping was observed for both eIF4A1 and eIF4A3, as previously reported (Dang et al. 2009). Three different rocaglates (RocA, CR-1-31B, and silvestrol) were tested for their ability to stabilize eIF4A1:poly r(AG)8 and eIF4A3:poly r(AG)8 RNA complexes. Although the change in polarization with eIF4A3 was not as pronounced as what was obtained for eIF4A1, all three rocaglates were able to induce clamping of eIF4A3 to RNA (Fig. 1C). Since structural analysis predicted that eIF4A1F163/eIF4A3F168 and eIF4A1Q195/ eIF4A3Q200 residues might play critical roles in stabilizing protein•RNA interactions in the presence of RocA (Fig. 1B), we generated missense mutations at these positions in eIF4A1 and eIF4A3. Significant reductions in pateamine- and rocaglate-induced clamping was observed with eIF4A1F163L, eIF4A3F168L, eIF4A1Q195L, and eIF4A3Q200L mutant proteins (Fig. 1D). All mutant proteins retained ATPase activity, indicating that loss of rocaglate-induced clamping was not a consequence of global protein misfolding due to the introduced missense mutations (Fig. 1E). To document the stabilizing effects of rocaglates on eIF4A3:RNA interactions, we monitored the dissociation of preformed eIF4A1•CR-1-31B•ATP•RNA or eIF4A3•CR-1-31B•ATP•RNA complexes (Fig. 1F). In the absence of compound, eIF4A1•ATP•RNA and eIF4A3•ATP•RNA complexes dissociated rapidly with half-lives of 48 and 170 sec, respectively. Dissociation of protein•RNA complexes clamped by PatA were long-lived, displaying half-lives of 110 min and 183 min for eIF4A1 and eIF4A3, respectively. Although CR-1-31B induced clamping of eIF4A1 and eIF4A3 to RNA, the eIF4A1•RNA complexes (t1/2 ∼ 50 min) were more stable than eIF4A3•RNA complexes (t1/2 ∼ 24 min) (Table 1; Fig. 1F).
TABLE 1.
Fitted half-life (T[1/2]) of the indicated eIF4A•ATP•RNA•compound complexes
eIF4A1 and eIF4A3 are similarly targeted by different rocaglate family members
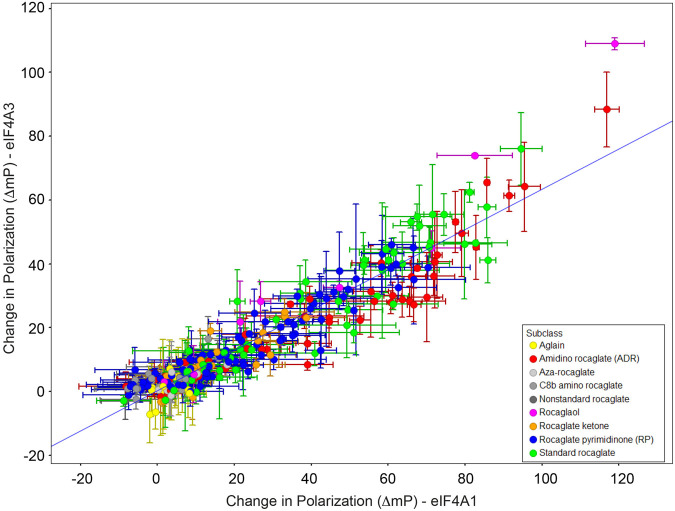
To query if rocaglate structure–activity relationships that enabled eIF4A1 clamping were similarly required for eIF4A3•RNA clamping, we compared the activity of 282 synthetic rocaglates (cyclopenta[b]benzofurans) and biosynthetically related aglains (cyclopenta[b,c]benzopyrans) in the FP assay with recombinant eIF4A1 and eIF4A3 proteins (Lajkiewicz et al. 2012; Stone et al. 2015; Wang et al. 2017). The rocaglate set can be further subclassified into various structural frameworks including rocaglate ketones, amidino rocaglates (ADRs), C8b amino rocaglates, aza-rocaglates, and rocaglate pyrimidinones (RPs) (Rodrigo et al. 2012; Wang et al. 2016; Chu et al. 2019; Zhang et al. 2019a). These scaffolds and subclasses are summarized in Supplemental Figure S2. Here, the change in polarization (ΔmP) obtained in the presence of each compound, relative to DMSO controls, was cross compared between both proteins (Fig. 2; Supplemental Table S1). The results indicate a similar rank order of activity among rocaglates toward both eIF4A1 and eIF4A3, consistent with both proteins harboring similar rocaglate binding sites.
FIGURE 2.
Scatter plot showing correlation between rocaglate-induced clamping to eIF4A1 and eIF4A3. The ΔmP obtained with eIF4A1•AMPPNP•poly r(AG)8 and eIF4A3•AMPPNP•poly r(AG)8 was measured for each compound (10 µM) in duplicate and is plotted. Pearson R2 = 0.89.
Inhibition of NMD by rocaglates
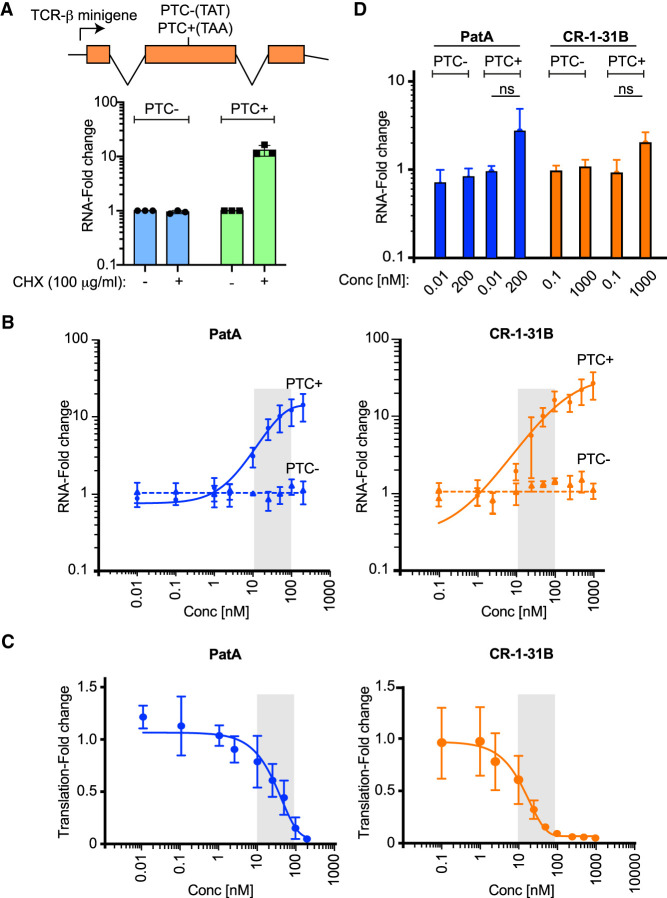
Given the ability of rocaglates to induce RNA clamping of eIF4A3, we sought to assess if rocaglates would block NMD. To this end, we used a previously described model where the T-cell receptor (TCR)-β minigene is stably integrated and constitutively expressed in HeLa cells (Carter et al. 1995; Ferraiuolo et al. 2004). The two cell lines used for NMD studies either lack (PTC−) or harbor (PTC+) a premature termination codon within the gene body—subjecting the latter to NMD regulation (Fig. 3A, top schematic). Since translation-dependence is a feature of NMD in all organisms, general inhibitors of translation can be used to block NMD (Kurosaki et al. 2019). There was no impact on TCR-β mRNA levels in the PTC− cell line, whereas TCR-β mRNA levels were significantly increased following exposure of PTC+ cells to cycloheximide (CHX) (Fig. 3A, bottom panel). Since PatA targets both eIF4A1 and eIF4A3, we also used it as a positive control in our experiments. Stabilization of TCR-β mRNA with PatA was noted upon treatment of PTC+, but not PTC− cells, as previously reported (Fig. 3B, left panel; Dang et al. 2009). Exposure of PTC− and PTC+ cells to CR-1-31B also caused a substantial stabilization of the TCR-β minigene mRNA in PTC+ cells (Fig. 3B, right panel). These results indicate that CR-1-31B inhibits NMD under these experimental conditions. Since PatA and CR-1-31B rapidly exert their effects on protein synthesis, we repeated these experiments but monitored 35S-Met incorporation into proteins 1 h following exposure of HeLa cells to compound (Fig. 3C). For both molecules, translation was blocked at doses similar to those at which NMD inhibition was observed (compare gray shaded areas in Fig. 3B,C).
FIGURE 3.
Inhibition of NMD by rocaglates. (A, top) Schematic depiction of reporter construct used in the current study. (Bottom) RT-qPCR results of TCR-β minigene expression from PTC− and PTC+ HeLa cells. Cells were exposed to DMSO or cycloheximide (CHX) for 5 h after which time RNA was extracted and analyzed by RT-qPCR. Values were normalized to β-actin. n = 3 ± SD. (B) RT-qPCR results of TCR-β minigene expression from PTC− and PTC+ HeLa cells exposed to the indicated concentrations of PatA or CR-1-31B for 5 h. Following RNA extraction, levels of TCR-β were assessed by RT-qPCR and normalized to β-actin. n = 3 ± SD. (C) Inhibition of translation following exposure of PTC+ HeLa cells to compound for 1 h. Labeling with 35S-Met/Cys was undertaken for the last 15 min of incubation. Quantitation of 35S-Met/Cys incorporation into nascent protein was determined by TCA precipitation and normalized to total protein levels. n = 3 ± SD. (D) RT-qPCR results of TCR-β minigene expression from PTC− and PTC+ HeLa cells exposed to the indicated concentrations of PatA or CR-1-31B for 1 h. Following RNA extraction, levels of TCR-β were assessed by RT-qPCR and normalized to β-actin. n = 3 ± SD. ns, not significant P > 0.05.
To formally demonstrate that the inhibition of translation with PatA and CR-1-31B observed at 1 h preceded the detectable accumulation of PTC+ mRNA, we repeated the above-described experiment where we instead harvested the RNA 1 h after exposure of HeLa cells to PatA or CR-1-31B (Fig. 3D). Under these conditions, PatA's effect on PTC+ mRNA levels was modest, increasing RNA levels approximately threefold at the highest concentration tested (200 nM), in contrast to the 10-fold increase observed following 5 h exposure (compare Fig. 3D to Fig. 3B). CR-1-31B exerted no significant effect on TCR-β minigene mRNA levels following exposure of PTC+ cells to 1 µM compound for 1 h (Fig. 3D).
Probing translational effects of rocaglates on NMD
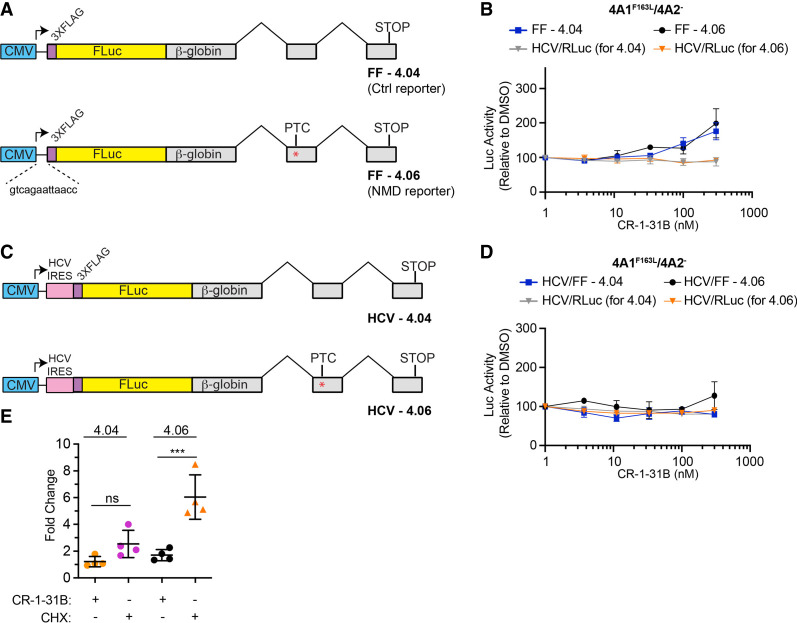
One limitation with the experiments presented so far is that the consequences on NMD observed with CR-1-31B could be attributed primarily to inhibition of translation and might be unrelated to eIF4A3-induced clamping. To tease out effects on NMD due to inhibition of eIF4A1/2 versus eIF4A3, we took advantage of a transient transfection assay involving reporter constructs containing firefly luciferase fused to the wild-type β-globin gene (Fig. 4A; Baird et al. 2018). One reporter has a PTC engineered within the second β-globin exon, subjecting the pre-mRNA to NMD (Baird et al. 2018). We noticed the transgene in these vectors had an extremely short 5′ leader region (Fig. 4A, 13 nt) with the longest stretch of polypurine bases being only four nucleotides long. We have previously shown that when present in an mRNA reporter, short regions of four polypurines mediate only a weak inhibitory response to rocaglates when compared to longer stretches of 10 contiguous purine bases (Chu et al. 2020). We therefore first directly tested the rocaglate-responsiveness of reporters containing the 13 nt comprising the 5′ leader of FF-4.04/4.06 reporters. We placed the 5′ leader nucleotides upstream of the first cistron of the FF/HCV/Ren bicistronic reporter previously used to gauge rocaglate-responsiveness of different 5′ leader sequences (Supplemental Fig. S3a; Chu et al. 2020). Messenger RNA produced in vitro was assessed for CR-1-31B and PatA response in Krebs-2 translation extracts (Supplemental Fig. S3b,c). The presence of polypurine sequences (e.g., [AG]10) within the 5′ leader region conferred CR-1-31B responsiveness (Supplemental Fig. S3b), as previously reported (Chu et al. 2020). On the other hand, the presence of polypyrimidines, or the 13 nt from the 5′ leader of FF-4.04/4.06, produced an mRNA that was significantly less responsive to CR-1-31B (Supplemental Fig. S3b). This differential response was not observed in the presence of PatA, consistent with PatA's much broader sequence targeting range; here PatA equally inhibited all reporters (Supplemental Fig. S3c; Chu et al. 2020; Naineni et al. 2021). As expected, all three reporters were found to be equally responsive to the elongation inhibitor, cycloheximide (Supplemental Fig. S3d). We therefore expect translation of the FF-4.04 and FF-4.06 NMD reporters to be minimally impacted by CR-1-31B.
FIGURE 4.
Uncoupling the effects of rocaglates on translation initiation versus NMD. (A) Reporter constructs used in these assays. In constructs FF-4.04 and FF-4.06, the FLuc cistron is fused to β-globin exons lacking (FF-4.04) or harboring (FF-4.06) a PTC (denoted by an asterisk). (B) Luciferase activity following introduction of FF-4.04 + HCV/RLuc or FF-4.06 + HCV/RLuc into 4A1F163L/4A2− Hap1 cells. Following transfections, cells were incubated for 1 d, reseeded, and exposed to compound for 6 h at which point luciferase activity was determined. n = 3 ± SD. (C) Constructs HCV-4.04 and HCV-4.06 have the HCV IRES engineered upstream of the FLuc coding region. Red asterisk indicates the location of the PTC. (D) Luciferase activity following introduction of HCV/FF-4.04 + HCV/RLuc or HCV/FF-4.06 + HCV/RLuc into 4A1F163L/4A2− Hap1 cells. Following transfections, cells were incubated for 1 d, reseeded, and exposed to compound for 6 h at which point luciferase activity was determined. n = 3 ± SD. (E) RNA levels in CR-1-31B and CHX treated FF-4.04 and FF-4.06 cells. 4A1F163L/4A2− eHAP1 cells were nucleofected with FF-4.04 or FF-4.06 and selected as a pool in G418 (1.6 mg/mL). Cells were exposed to CR-1-31B or CHX for 5 h, after which time RNA was isolated and analyzed by RT-qPCR. RNA levels were normalized to GAPDH levels and expressed relative to DMSO controls. n = 4 ± SD. (***) P = 0.0005; ns, not significant.
To help distinguish between rocaglate-mediated effects on NMD due to eIF4A1/2 inhibition versus eIF4A3 targeting, we also took advantage of CRISPR-modified 4A1F163L/4A2− eHAP1 cells, where we had engineered a rocaglate-resistant eIF4A1 allele (F163L) and eliminated expression of eIF4A2 (Naineni et al. 2021). We introduced either FF-4.04 or FF-4.06 expression vectors, along with an HCV/Ren expression vector, into 4A1F163L/4A2− cells, 24 h later exposed cells to CR-1-31B, and measured luciferase activity 6 h later. Cells transfected with FF-4.04 or FF-4.06 showed only a slight increase (approximately twofold) in Luc expression for both reporters at 300 nM CR-1-31B, indicating this response was unrelated to any possible effects on NMD (Fig. 4B). No change in RLuc expression from the internal HCV/Ren control was observed. These results indicate that under the tested conditions, there appears to be no effect on NMD mediated through eIF4A3 engagement.
To strengthen these results, we also cloned the HCV IRES upstream of the FLuc ORF in the FF-4.04 and FF-4.06 vector backbones (Fig. 4C). The HCV IRES offers an alternative approach by which translation of FLuc can be rendered rocaglate-independent (Bordeleau et al. 2008). In 4A1F163L/4A2− eHap1 cells, neither reporter showed a change in expression over the CR-1-31B titration range tested (Fig. 4D). These results indicate that at the concentrations tested, CR-1-31B does not inhibit NMD through eIF4A3-induced clamping. We then assessed if FF-4.06 mRNA levels were altered in the presence of CR-1-31B. Exposure of 4A1F163L/4A2− eHAP1 harboring FF-4.04 or FF-4.06 to CR-1-31B or CHX for 5 h showed that levels of the NMD reporter, FF-4.06 mRNA, were increased only in the presence of CHX (Fig. 4E).
We also investigated the consequences of rocaglates on SC35 mRNA levels, an endogenous transcript whose expression is under NMD regulation (Sureau et al. 2001). Previous experiments targeting eIF4A3 with a small molecule inhibitor produced an increase in SC35 levels (Iwatani-Yoshihara et al. 2017). Treatment of 4A1F163L/4A2− eHAP1 cells with CHX or PatA for 5 h led to a significant increase in SC35 mRNA levels, relative to DMSO controls (Fig. 5). SC35 mRNA levels did not significantly change when cells were exposed to CR-1-31B. HPRT mRNA levels were not increased under any of the tested conditions (Fig. 5). These results are consistent with our previous data indicating that whereas PatA can inhibit NMD and increase levels of NMD-sensitive mRNAs, rocaglates exert their primary effect on translation and any effects on NMD stem from this initial inhibition.
FIGURE 5.
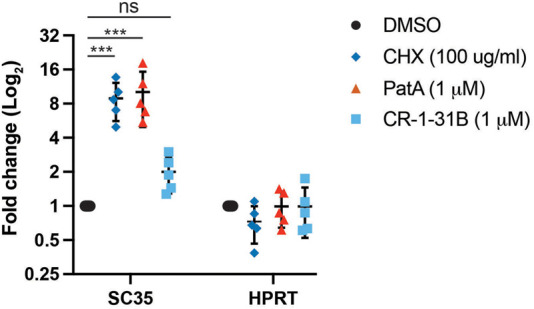
Endogenous SC35 transcript levels are not affected by CR-1-31B in 4A1F163L/4A2− eHAP1 cells. Cells were exposed to the indicated concentrations of compound for 5 h, after which RNA was isolated. SC35, HPRT and β-actin mRNA levels were determined by RT-qPCR. SC35 and HPRT mRNA levels were normalized to β-actin levels and are expressed as fold change relative to DMSO controls. n = 5 ± SD. (***) P < 0.0001; ns, not significant.
DISCUSSION
Rocaglates are potent inhibitors of eukaryotic translation initiation and exert their effects through unscheduled clamping of RNA to eIF4A1/2 (both free eIF4A and eIF4F-bound eIF4A). The eIF4A1•AMPPNP• RocA•poly r(AG)5 RNA crystal structure provided insight into the molecular basis of this clamping by identifying eIF4A1 F163 and Q195 as critical amino acids participating in rocaglate interaction (Iwasaki et al. 2019). Among DDX RNA helicase family members, only eIF4A homologs have preserved the F163/Q195 combination, rationalizing the selectivity of rocaglates for eIF4A1 and eIF4A2 (Sadlish et al. 2013; Chu et al. 2016). Herein we demonstrate that eIF4A3 is also targeted by rocaglates (Figs. 1, 2) and thus sought to determine the biological consequences. A previous study aiming to identify rocaglate targets that might exist beyond eIF4A1 and eIF4A2 used a RocA-based chemical probe designed to induce proximity (direct and indirect interacting partners of targets) labeling in rabbit reticulocyte lysates (Chen et al. 2021). This study identified DDX3X, but not eIF4A3, as a potential target of rocaglates (Chen et al. 2021). eIF4A3 may have escaped detection since reticulocytes are enucleated and would be expected to have low, if any, levels of eIF4A3.
Our results indicate that prolonged exposure of cells to rocaglates inhibits NMD (Fig. 3). Since blocking translation per se will lead to inhibition of NMD (Carter et al. 1995), we sought to determine whether the inhibition of NMD induced by rocaglates was due to translation inhibition mediated through eIF4A1/2 or perturbation of eIF4A3 activity. To this end, we took advantage of (i) reporter constructs whose translations were resistant to inhibition by rocaglates (Figs. 4A; Supplemental Fig. S3) and (ii) somatic cell genetics in which we leveraged the use of 4A1F163L/4A2− Hap1 cells engineered to harbor the rocaglate-resistant F163L eIF4A1 allele and lacking eIF4A2 (Naineni et al. 2021). In 4A1F163L/4A2− Hap1 cells, no selective increase in activity was observed from the PTC-containing construct, FF-4.06, in the presence of CR-1-31B (Fig. 4B). Additionally, grafting the HCV IRES onto the FF-4.06 reporter to allow eIF4A-independent translation failed to uncover an effect on NMD by rocaglates (Fig. 4C,D). Based on these results, we propose that rocaglates exert their effects on NMD indirectly through inhibition of translation. eIF4A3 has also been implicated in ribosome biogenesis and thus may be an exploitable vulnerability in tumors showing elevated rates of ribosome production (Kanellis et al. 2021). We have not investigated whether rocaglates impact this activity of eIF4A3.
Our results contrast with those obtained with PatA and the analog, DMDA-PatA, where NMD inhibition was observed and attributed to DMDA-PatA induced clamping of eIF4A3 (Dang et al. 2009). This was ascribed to stabilization of RNA-bound eIF4A3 which in turn stabilized the EJC and EJC•Upf1 interactions, perturbing the dynamic interplay of Upf1 with the EJC and inhibiting NMD (Dang et al. 2009). In our hands, rocaglates did not inhibit NMD through effects on eIF4A3, but rather any effect on NMD appeared to be a consequence of rocaglates inhibiting translation. The effects of PatA and DMDA-PatA on NMD may be due to higher affinity of these compounds for eIF4A3 than rocaglates and/or the broader sequence targeting range of pateamines (Naineni et al. 2021) or to the longer half-life of clamped eIF4A3•RNA•PatA complexes relative to eIF4A3•RNA•rocaglate complexes (Fig. 1F). In sum, we have shown that rocaglates can inhibit the NMD surveillance pathway but that this is a consequence of inhibiting translation rather than any effect caused by clamping of eIF4A3.
MATERIALS AND METHODS
General materials
Unlabeled and FAM-labeled poly(AG)8 RNA were obtained from Integrated DNA Technologies (IDT). The generation and characterization of eIF4A1F163L/eIF4A2− Hap1 cells has been previously described (Naineni et al. 2021). Rocaglates/aglains and aza-rocaglates/aza-aglains provided from the BU-CMD collection were generated by ESIPT photocycloaddition of 3-hydroxyflavones or 3-hydroxyquinolinones, respectively, with cinnamates and were further functionalized (Roche et al. 2010; Lajkiewicz et al. 2014; Stone et al. 2015; Wang et al. 2016, 2017; Yueh et al. 2017; Zhang et al. 2019a,b). Compounds were resuspended to 10 mM in neat DMSO and stored at −80°C.
Recombinant protein purification
Recombinant eIF4A1 and eIF4A3 were purified as previously described (Lindqvist et al. 2008). Briefly, pET-15b plasmids encoding (His)6-eIF4A1 and (His)6-eIF4A3 were transformed into BL21 (DE3) competent cells. Starter cultures were grown in LB media with ampicillin (100 µg/mL) and chloramphenicol (34 µg/mL) at 37°C overnight. The cultures were then diluted 1:20 to reach a final volume of 1L and grown at 37°C until the OD600 reached 0.6–0.8. Protein expression was induced with 0.5 mM IPTG at 16°C for 16 h. Cells were harvested by centrifuging at 4000g for 30 min and resuspended in 20 mL Buffer A (20 mM Tris-HCl [pH7.5], 10% glycerol, 0.1 mM EDTA, 200 mM KCl, 0.1% Triton X-100, and 3.4 mM β-mercaptoethanol) and lysed by sonication. The insoluble material was separated by centrifuging at 24,000g for 30 min. The cleared lysate was then incubated with 1 mL Ni-NTA agarose beads (Qiagen), washed with Buffer B (Buffer A lacking β-mercaptoethanol) and incubated at 4°C for 1 h on a rotating platform. The slurry was applied to a column and washed three times with 10 mL of Buffer 1 (20 mM Tris-HCl [pH 7.5], 10% glycerol, 0.1 mM EDTA, 800 mM KCl, 20 mM imidazole) and washed three times with Buffer 2 (Buffer 1 containing 300 mM KCl). The bound proteins were eluted with 3 mL Buffer 3 (Buffer 2 containing 200 mM imidazole and 2 mM DTT). The eluted protein was dialyzed against 20 mM Tris (pH 7.5), 10% glycerol, 0.1 mM EDTA, 100 mM KCl, and 2 mM DTT.
Fluorescence polarization assays
Fluorescence polarization assays were performed as described previously (Chu et al. 2020). Briefly, 1.5 µM of indicated protein was incubated with 10 nM FAM-labeled poly(AG)8 RNA and 1 mM AMPPNP in FP buffer (14.4 mM HEPES-NaOH [pH 8], 108 mM NaCl, 1 mM MgCl2, 14.4% glycerol, 0.1% DMSO, 2 mM DTT) in the presence or absence of 10 µM compound in black, low volume 384 well plates (Corning 3820) for 30 min at RT in the dark after which the FP readings were acquired on a Pherastar FS microplate reader (BMG Labtech). For dissociation experiments, protein•ATP•FAM-poly r(AG)8 RNA•compound complexes are preformed using 1.5 µM of indicated protein, 10 nM FAM-labeled poly r(AG)8 RNA, 1 mM ATP, and either 10 µM CR-1-31B or PatA in FP buffer and incubated for 30 min at RT in the dark. For reactions containing DMSO, preformed complexes were assembled using 25 µM of indicated protein due to weak RNA binding activity of eIF4A1 and eIF4A3. After preincubation, 1000-fold molar excess (10 µM) of unlabeled poly r(AG)8 RNA was added and polarization measurements were performed with a time interval of 10 sec between each measurement. The relative dissociations were measured as a function of time. Half-lives of the complexes were calculated using the “dissociation—one phase exponential decay” nonlinear fit method with (Y0 = 1, Plateau = 0) as constraints in GraphPad 8.4.0.
ATPase assays
ATPase assays were performed using 1 µM protein, 2.5 µM poly(U) RNA, 1 µCi γ-[32P]-ATP (3000Ci/mmol) in reaction buffer (20 mM MES-KOH [pH 6.0], 2.5 mM MgCl2, 1 mM DTT, 1% glycerol, 10 mM KOAc) for 30 min. Reactions were stopped on ice and EDTA added to a final concentration of 8 mM. Reaction products were resolved on PEI cellulose TLC plates (Millipore) developed with 1 M LiCl/0.3 M NaH2PO4. ATP hydrolysis was quantified using a Storm 840 (molecular dynamics) scanner.
Nonsense-mediated decay (NMD) assays
HeLa cells harboring a (TCR)-β minigene with or without PTC were seeded at a density of 100,000/well in 12-well plates. For assays involving 4A1F163L/4A2− eHAP1, cells were seeded at a density of 250,000/well in a six well plate. Cells were treated with the indicated compound for either 1 h or 5 h. Total RNA was extracted with TRIzol (Thermo Fisher) as per the manufacturer's protocol. Complementary DNA (cDNA) was synthesized from 500 ng of total RNA using an oligo (dT)23 VN primer as per the manufacturer's recommendations. The cDNA was diluted 1:10 and later amplified using SSOFast EvaGreen mix in a CFX96 qPCR instrument (Bio-Rad). Primers used for RT-qPCR were: (i) For TCRβ, TCRβ(For); 5′GAGCTGTCACCAGACTAATAAC3′, TCRβ(Rev); 5′CTCCAGAATGAGAGAGAAATTC3′, (ii) for β-actin: β-actin(For); 5′GGCTGCTTCCAGCTCCT3′, β-actin(Rev); 5′CCACGTCACACTTCATGATG3′, (iii) for FF-4.04 and FF-4.06 mRNA: HBB(For); 5′ 5′TGGATGAAGTTGGTGGTGAG3′, HBB(REV); 5′CCTTAGGGTTGCCCATAACA3′, (iv) for GAPDH: GAPDH(For); 5′TTGGTATCGTGGAAGGACTCA3′, GAPDH(Rev); 5′TGTCATCATATTTGGCAGGTT3′, (v) for SC35: SC35(For); 5′AGTTTCTGTGGGCGTGTATT3′, SC35(Rev); 5′GGACTCTTCTTCGATGGACTATG3′, and (vi) for HPRT: HPRT(For); 5′AGGGTGTTTATTCCTCATGGAC3′, HPRT(Rev); 5′CACAGAGGGCTACAATGTGAT3′.
Assays in 4A1F163L/4A2− eHAP1 cells were performed essentially as described (Baird et al. 2018). Cells were plated in six-well plates and transfected when they reached 75% confluency. Plasmids pKC-4.06 (FF-4.06: FLuc-β-globin-PTCstop) or pKC-4.04 (FF-4.04: FLuc-β-globin control) were mixed with pcDNA3-HCV-hRL and transfections performed using Turbofectin (Origene). The following day, cells were reseeded into 24 well plates and allowed to recover for 24 h, after which time compounds were added for 6 h. Cells were harvested, lysed in PLB (Promega) and luciferase activity assessed on a Berthold Lumat LB 9507 luminometer (Berthold Technologies).
[35S]-Methionine labeling
Metabolic labeling was undertaken in HeLa cells harboring a (TCR)-β minigene with or without PTC. Briefly, 50,000 cells were seeded in a 24 well plate. On the following day, cells were treated with the indicated compound concentration for 1 h. De novo protein synthesis was monitored by adding [35S]-Met/Cys for the last 15 min. Cells were then washed with PBS and lysed in RIPA buffer (20 mM Tris-HCl [pH 7.6], 100 mM NaCl, 1 mM EDTA,1 mM EGTA, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM PMSF, 4 µg/mL aprotinin, 2 µg/mL leupeptin, 2 µg/mL pepstatin). Lysates were spotted onto 3 MM Whatman paper, subjected to TCA precipitation, and the amount of radioactivity determined by scintillation counting. The obtained counts were normalized to total protein levels.
In vitro translation assays
In vitro translation assays were performed using reporter mRNAs at a final concentration of 10 ng/µL in Krebs-2 cell extracts. Following incubation at 30°C for 1 h, firefly luciferase (FLuc) and Renilla luciferase (RLuc) activity were assessed using a Berthold Lumat LB 9507 luminometer (Berthold Technologies). The obtained RLUs were normalized to vehicle (DMSO) controls.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by research funding to J.P. (Canadian Institutes of Health Research [#FDN-148366]) and to J.A.P., Jr. (National Institutes of Health grants R35GM118173 and U01TR002625).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079318.122.
REFERENCES
- Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, Pedersen JS, Seraphin B, Le Hir H, Andersen GR. 2006. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science 313: 1968–1972. 10.1126/science.1131981 [DOI] [PubMed] [Google Scholar]
- Baird TD, Cheng KC, Chen YC, Buehler E, Martin SE, Inglese J, Hogg JR. 2018. ICE1 promotes the link between splicing and nonsense-mediated mRNA decay. Elife 7: e33178. 10.7554/eLife.33178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. 2015. Targeting the translation machinery in cancer. Nat Rev Drug Discov 14: 261–278. 10.1038/nrd4505 [DOI] [PubMed] [Google Scholar]
- Bordeleau ME, Robert F, Gerard B, Lindqvist L, Chen SM, Wendel HG, Brem B, Greger H, Lowe SW, Porco JA, et al. 2008. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J Clin Invest 118: 2651–2660. 10.1172/JCI34753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter MS, Doskow J, Morris P, Li S, Nhim RP, Sandstedt S, Wilkinson MF. 1995. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem 270: 28995–29003. 10.1074/jbc.270.48.28995 [DOI] [PubMed] [Google Scholar]
- Caruthers JM, McKay DB. 2002. Helicase structure and mechanism. Curr Opin Struct Biol 12: 123–133. 10.1016/S0959-440X(02)00298-1 [DOI] [PubMed] [Google Scholar]
- Chen M, Asanuma M, Takahashi M, Shichino Y, Mito M, Fujiwara K, Saito H, Floor SN, Ingolia NT, Sodeoka M, et al. 2021. Dual targeting of DDX3 and eIF4A by the translation inhibitor rocaglamide A. Cell Chem Biol 28: 475–486.e478. 10.1016/j.chembiol.2020.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J, Galicia-Vazquez G, Cencic R, Mills JR, Katigbak A, Porco JA Jr, Pelletier J. 2016. CRISPR-mediated drug-target validation reveals selective pharmacological inhibition of the RNA helicase, eIF4A. Cell Rep 15: 2340-2347. 10.1016/j.celrep.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J, Zhang W, Cencic R, Devine WG, Beglov D, Henkel T, Brown LE, Vajda S, Porco JA Jr, Pelletier J. 2019. Amidino-rocaglates: a potent class of eIF4A inhibitors. Cell Chem Biol 26: 1586–1593.e1583. 10.1016/j.chembiol.2019.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J, Zhang W, Cencic R, O'Connor PBF, Robert F, Devine WG, Selznick A, Henkel T, Merrick WC, Brown LE, et al. 2020. Rocaglates induce gain-of-function alterations to eIF4A and eIF4F. Cell Rep 30: 2481–2488.e2485. 10.1016/j.celrep.2020.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Low WK, Xu J, Gehring NH, Dietz HC, Romo D, Liu JO. 2009. Inhibition of nonsense-mediated mRNA decay by the natural product pateamine A through eukaryotic initiation factor 4AIII. J Biol Chem 284: 23613–23621. 10.1074/jbc.M109.009985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan R, Milburn SC, Hershey JW. 1987. Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J Biol Chem 262: 380–388. 10.1016/S0021-9258(19)75938-9 [DOI] [PubMed] [Google Scholar]
- Ferraiuolo MA, Lee CS, Ler LW, Hsu JL, Costa-Mattioli M, Luo MJ, Reed R, Sonenberg N. 2004. A nuclear translation-like factor eIF4AIII is recruited to the mRNA during splicing and functions in nonsense-mediated decay. Proc Natl Acad Sci 101: 4118–4123. 10.1073/pnas.0400933101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galicia-Vazquez G, Cencic R, Robert F, Agenor AQ, Pelletier J. 2012. A cellular response linking eIF4AI activity to eIF4AII transcription. RNA 18: 1373–1384. 10.1261/rna.033209.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galicia-Vazquez G, Chu J, Pelletier J. 2015. eIF4AII is dispensable for miRNA-mediated gene silencing. RNA 21: 1826–1833. 10.1261/rna.052225.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. 1989. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res 17: 4713–4730. 10.1093/nar/17.12.4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Floor SN, Ingolia NT. 2016. Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor. Nature 534: 558–561. 10.1038/nature17978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Iwasaki W, Takahashi M, Sakamoto A, Watanabe C, Shichino Y, Floor SN, Fujiwara K, Mito M, Dodo K, et al. 2019. The translation inhibitor rocaglamide targets a bimolecular cavity between eIF4A and polypurine RNA. Mol Cell 73: 738–748.e739. 10.1016/j.molcel.2018.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatani-Yoshihara M, Ito M, Ishibashi Y, Oki H, Tanaka T, Morishita D, Ito T, Kimura H, Imaeda Y, Aparicio S, et al. 2017. Discovery and characterization of a eukaryotic initiation factor 4A-3-selective inhibitor that suppresses nonsense-mediated mRNA decay. ACS Chem Biol 12: 1760–1768. 10.1021/acschembio.7b00041 [DOI] [PubMed] [Google Scholar]
- Jankowsky E. 2011. RNA helicases at work: binding and rearranging. Trends Biochem Sci 36: 19–29. 10.1016/j.tibs.2010.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellis DC, Espinoza JA, Zisi A, Sakkas E, Bartkova J, Katsori AM, Bostrom J, Dyrskjot L, Broholm H, Altun M, et al. 2021. The exon-junction complex helicase eIF4A3 controls cell fate via coordinated regulation of ribosome biogenesis and translational output. Sci Adv 7: eabf7561. 10.1126/sciadv.abf7561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. 2014. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods 11: 319–324. 10.1038/nmeth.2834 [DOI] [PubMed] [Google Scholar]
- Kurosaki T, Popp MW, Maquat LE. 2019. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat Rev Mol Cell Biol 20: 406–420. 10.1038/s41580-019-0126-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajkiewicz NJ, Roche SP, Gerard B, Porco JA Jr. 2012. Enantioselective photocycloaddition of 3-hydroxyflavones: total syntheses and absolute configuration assignments of (+)-ponapensin and (+)-elliptifoline. J Am Chem Soc 134: 13108–13113. 10.1021/ja305342f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajkiewicz NJ, Cognetta AB III, Niphakis MJ, Cravatt BF, Porco JA Jr. 2014. Remodeling natural products: chemistry and serine hydrolase activity of a rocaglate-derived β-lactone. J Am Chem Soc 136: 2659–2664. 10.1021/ja412431g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist L, Oberer M, Reibarkh M, Cencic R, Bordeleau ME, Vogt E, Marintchev A, Tanaka J, Fagotto F, Altmann M, et al. 2008. Selective pharmacological targeting of a DEAD box RNA helicase. PLoS One 3: e1583. 10.1371/journal.pone.0001583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naineni SK, Liang J, Hull K, Cencic R, Zhu M, Northcote P, Teesdale-Spittle P, Romo D, Nagar B, Pelletier J. 2021. Functional mimicry revealed by the crystal structure of an eIF4A:RNA complex bound to the interfacial inhibitor, desmethyl pateamine A. Cell Chem Biol 28: 825–834.e826. 10.1016/j.chembiol.2020.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul MS, Duncan AR, Genetti CA, Pan H, Jackson A, Grant PE, Shi J, Pinelli M, Brunetti-Pierri N, Garza-Flores A, et al. 2023. Rare EIF4A2 variants are associated with a neurodevelopmental disorder characterized by intellectual disability, hypotonia, and epilepsy. Am J Hum Genet 110: 120–145. 10.1016/j.ajhg.2022.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Sonenberg N. 2019. The organizing principles of eukaryotic ribosome recruitment. Annu Rev Biochem 88: 307–335. 10.1146/annurev-biochem-013118-111042 [DOI] [PubMed] [Google Scholar]
- Robert F, Cencic R, Cai R, Schmeing TM, Pelletier J. 2020. RNA-tethering assay and eIF4G:eIF4A obligate dimer design uncovers multiple eIF4F functional complexes. Nucleic Acids Res 48: 8562–8575. 10.1093/nar/gkaa646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche SP, Cencic R, Pelletier J, Porco JA Jr. 2010. Biomimetic photocycloaddition of 3-hydroxyflavones: synthesis and evaluation of rocaglate derivatives as inhibitors of eukaryotic translation. Angew Chem Int Ed Engl 49: 6533–6538. 10.1002/anie.201003212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo CM, Cencic R, Roche SP, Pelletier J, Porco JA. 2012. Synthesis of rocaglamide hydroxamates and related compounds as eukaryotic translation inhibitors: synthetic and biological studies. J Med Chem 55: 558–562. 10.1021/jm201263k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GW Jr, Komar AA, Merrick WC. 2002. eIF4A: the godfather of the DEAD box helicases. Prog Nucleic Acid Res Mol Biol 72: 307–331. 10.1016/S0079-6603(02)72073-4 [DOI] [PubMed] [Google Scholar]
- Sadlish H, Galicia-Vazquez G, Paris CG, Aust T, Bhullar B, Chang L, Helliwell SB, Hoepfner D, Knapp B, Riedl R, et al. 2013. Evidence for a functionally relevant rocaglamide binding site on the eIF4A-RNA complex. ACS Chem Biol 8: 1519–1527. 10.1021/cb400158t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlautmann LP, Gehring NH. 2020. A day in the life of the exon junction complex. Biomolecules 10: 866. 10.3390/biom10060866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senechal P, Robert F, Cencic R, Yanagiya A, Chu J, Sonenberg N, Paquet M, Pelletier J. 2021. Assessing eukaryotic initiation factor 4F subunit essentiality by CRISPR-induced gene ablation in the mouse. Cell Mol Life Sci 78: 6709–6719. 10.1007/s00018-021-03940-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberger J, Shen L SJK, Naineni SK, Cencic R, Amiri M, Aboushawareb SAE, Chu J, Maiga RI, Yachnin BJ, et al. 2020. Identification and characterization of hippuristanol-resistant mutants reveals eIF4A1 dependencies within mRNA 5′ leader regions. Nucleic Acids Res 48: 9521–9537. 10.1093/nar/gkaa662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SD, Lajkiewicz NJ, Whitesell L, Hilmy A, Porco JA Jr. 2015. Biomimetic kinetic resolution: highly enantio- and diastereoselective transfer hydrogenation of aglain ketones to access flavagline natural products. J Am Chem Soc 137: 525–530. 10.1021/ja511728b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureau A, Gattoni R, Dooghe Y, Stevenin J, Soret J. 2001. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J 20: 1785–1796. 10.1093/emboj/20.7.1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, Belsham GJ, Sonenberg N. 2001. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA 7: 382–394. 10.1017/S135583820100108X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Cencic R, Whitesell L, Pelletier J, Porco JA Jr. 2016. Synthesis of Aza-rocaglates via ESIPT-mediated (3+2) photocycloaddition. Chemistry (Easton) 22: 12006–12010. 10.1002/chem.201602953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Clay A, Krishnan R, Lajkiewicz NJ, Brown LE, Sivaguru J, Porco JA Jr. 2017. Total syntheses of the isomeric aglain natural products Foveoglin A and Perviridisin B: selective excited-state intramolecular proton-transfer photocycloaddition. Angew Chem Int Ed Engl 56: 14479–14482. 10.1002/anie.201707539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder-Hill J, Pause A, Sonenberg N, Merrick WC. 1993. The p46 subunit of eukaryotic initiation factor (eIF)-4F exchanges with eIF-4A. J Biol Chem 268: 5566–5573. 10.1016/S0021-9258(18)53358-5 [DOI] [PubMed] [Google Scholar]
- Yueh H, Gao Q, Porco JA Jr, Beeler AB. 2017. A photochemical flow reactor for large scale syntheses of aglain and rocaglate natural product analogues. Bioorg Med Chem 25: 6197–6202. 10.1016/j.bmc.2017.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Chu J, Cyr AM, Yueh H, Brown LE, Wang TT, Pelletier J, Porco JA Jr. 2019a. Intercepted retro-Nazarov reaction: syntheses of amidino-rocaglate derivatives and their biological evaluation as eIF4A inhibitors. J Am Chem Soc 141: 12891–12900. 10.1021/jacs.9b06446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Liu S, Maiga RI, Pelletier J, Brown LE, Wang TT, Porco JA Jr. 2019b. Chemical synthesis enables structural reengineering of aglaroxin C leading to inhibition bias for hepatitis C viral infection. J Am Chem Soc 141: 1312–1323. 10.1021/jacs.8b11477 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.