Abstract
Multidrug-resistant strains of Mycobacterium tuberculosis are a serious and continuing human health problem. Such strains may contain as many as four or five different mutations, and M. tuberculosis strains that are resistant to both streptomycin and rifampin contain mutations in the rpsL and rpoB genes, respectively. Coexisting mutations of this kind in Escherichia coli have been shown to interact negatively (S. L. Chakrabarti and L. Gorini, Proc. Natl. Acad. Sci. USA 72:2084–2087, 1975; S. L. Chakrabarti and L. Gorini, Proc. Natl. Acad. Sci. USA 74:1157–1161, 1977). We investigated this possibility in Mycobacterium smegmatis by analyzing the frequency and nature of spontaneous mutants that are resistant to either streptomycin or rifampin or to both antibiotics. Mutants resistant to streptomycin were isolated from characterized rifampin-resistant mutants of M. smegmatis under selection either for one or for both antibiotics. Similarly, mutants resistant to rifampin were isolated from streptomycin-resistant strains. The second antibiotic resistance mutation occurred at a lower frequency in both cases. Surprisingly, in both cases a very high rate of reversion of the initial antibiotic resistance allele was detected when single antibiotic selection was used; the majority of strains resistant to only one antibiotic were isolated by this process. Determinations of rates of mutation to antibiotic resistance in M. smegmatis showed that the frequencies were enhanced up to 104-fold during stationary phase. If such behavior is also typical of slow-growing pathogenic mycobacteria, these studies suggest that the generation of multiply drug-resistant strains by successive mutations may be a more complex genetic phenomenon than suspected.
The rapid emergence of multidrug-resistant Mycobacterium tuberculosis (MDRTB) strains has renewed interest in studies of the development of antibiotic resistance in mycobacteria. Streptomycin was the first antibiotic shown to be active against M. tuberculosis and was responsible for the successful treatment of millions of patients (3). The drug acts on ribosomes and causes aberrant proofreading leading to misreading of the genetic code and inhibition of initiation of translation (23). Mutations associated with streptomycin resistance (Strr) have been identified in two targets, the 16S rRNA gene (rrs) and the gene (rpsL) encoding ribosomal protein S12; both types of mutants have been characterized in M. tuberculosis (11). Rifampin has also been effectively employed in the treatment of tuberculosis; rifampin inhibits transcription by binding to the β-subunit of RNA polymerase (14), and rifampin-resistant (Rifr) mutants of M. tuberculosis have been found to harbor mutations in the rpoB gene, encoding this subunit (25). The two types of mutation have been shown to be present simultaneously in many strains of MDRTB identified in recent years (16). Early studies with Escherichia coli by Chakrabarti and Gorini (5, 6) had shown that there is antagonism between rpsL and rpoB mutations. We have analyzed mutation to Strr and Rifr and the appearance of double mutants to examine whether a similar type of antagonism is manifested in Mycobacterium smegmatis. Such antagonism was confirmed, but in addition we noted that mutation rates increased significantly during the postexponential growth phase of M. smegmatis.
(This work was presented as a poster at the 99th General Meeting of the American Society for Microbiology held in Chicago, Ill., from 30 May to 3 June 1999.)
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
All Strr and/or Rifr mutants were isolated from the antibiotic-sensitive M. smegmatis mc26. Revertants of histidine-requiring auxotrophs were isolated from M. smegmatis his5 (13). Cells were grown at 30°C in tryptic soy broth (TSB) medium (Difco) containing 0.5% glycerol. For solid medium, agar was added at 15 g per liter and glycerol was omitted. Revertants of M. smegmatis his5 were isolated by plating on 7H10 agar medium (BBL) containing 0.5% glycerol (without supplement). Viable counts were determined by plating appropriate dilutions of liquid cultures onto solid medium without antibiotic.
Isolation of spontaneous Strr and/or Rifr mutants.
M. smegmatis (108 CFU) was spread on plates containing 100 μg of streptomycin/ml and/or 500 μg of rifampin/ml. The plates were incubated at 30°C until colonies started to appear (about 3 to 5 days). The colonies were purified and their resistance characteristics were confirmed by restreaking onto agar plates containing appropriate antibiotics.
Detection of mutations in the rpsL and rpoB genes.
The rpsL gene (GenBank accession no. L34681) was amplified by PCR with primers L1 (5′-CGG TAG ATG CCA ACC ATC CAG CA-3′) and L2 (5′-CCT TGC GTG GCA TCA GCC CTT CT-3′), generating a fragment of 393 bp containing the complete gene. The rpoB gene (GenBank accession no. U24494) was amplified by PCR with primers B1 (5′-GGA CGT GGA GGC GAT CAC ACC-3′) and B2 (5′-CGT AGC GAC CGA CAC CAT CTG-3′), generating a fragment of 553 bp containing the region from codons 482 to 666. This segment includes the so-called rifampin resistance-determining region (17). The fragments were amplified from single M. smegmatis colonies added to the PCR mixture with a toothpick. The reaction mixture contained 1.5 mM MgCl2, 150 μM deoxynucleoside triphosphates, 5 U of Taq polymerase enzyme, and 25 pmol of each oligonucleotide primer in a total volume of 50 μl. The PCR cycling conditions were as follows: 1 cycle of 95°C for 5 min and 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min. This was followed by strand elongation for 10 min at 72°C. The PCR product was purified with the QIAquick PCR purification kit (Qiagen), and the purified DNA fragment was sequenced with dye-labeled terminators and primer (L1, L2, B1, or B2), using the AmpliTaq Prism kit (Applied Biosystems).
Nucleotide sequence analysis of the hisD gene.
The hisD gene of M. smegmatis his5 and its revertants were PCR amplified with primers F1 (5′-GTT GAC GGT GGC CGA CGG AT-3′) and R1 (5′-CTC GTT GGT GTT CAG GCG CA-3′), generating a fragment of 1,499 bp containing the complete gene (Fig. 1). PCR conditions were as described above. The PCR product was sequenced with additional primers F2 (5′-ATC CGT CGA GCG TCG TGA TG-3′) and F3 (5′-CCA CCA CCA AGC ACG TCG AG-3′).
FIG. 1.
PCR strategy used to analyze the nucleotide sequence of hisD. The 1,338-nucleotide hisD coding sequence is represented by the rectangle, and the primers used to amplify and/or sequence the gene are indicated by arrows. The codon numbering is based on M. smegmatis hisD sequence data published by Hinshelwood and Stoker (13).
EASPCR.
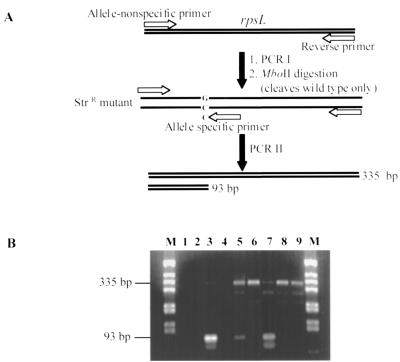
Enriched allele-specific PCR (EASPCR) was used to identify the appearance of Strr mutants in liquid culture before plating on selective media (Fig. 2). EASPCR involved two steps. Step one was to generate the rpsL gene fragments of both the wild type and mutants (PCR I), followed by MboII restriction enzyme digestion to eliminate wild-type templates. Step two involved amplification of allele-specific product (PCR II). This was done with three primers: two non-allele-specific primers (L11 [5′-GAC AAG ATC GCC AAG GTG AAG AC-3′] and L22 [5′-TCT TCT CCT TCT TCG CGC CAT AG-3′]) and one primer specific to the mutant allele (L25 [5′-CCG GAG CGC CGA GTT CGG CTA CC-3′]). PCR conditions were the same as described above except that for PCR II, the annealing temperature was 63°C and the number of cycles was 25. Two products (335 and 93 bp) were generated from the template containing mutant allele and none were generated from the wild-type template.
FIG. 2.
EASPCR to identify the appearance of rpsL mutations in liquid culture before plating on selective media. (A) Outline of the procedure. (B) EASPCR of samples taken at different times. M, PCR marker; lane 1, negative control; lane 2, wild type; lane 3, Strr mutant; lane 4, Rifr mutant (3-day culture); lane 5, Rifr mutant (8-day culture); lanes 6 to 9, same as lanes 2 to 5 but without MboII digestion before PCR II.
RESULTS
Isolation of antibiotic-resistant mutants.
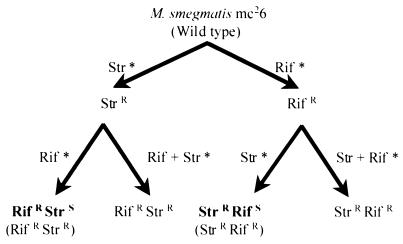
Spontaneous mutants of M. smegmatis resistant to either streptomycin or rifampin were isolated (Fig. 3), and the frequencies of appearance of mutants were determined (Table 1). About 10 independent colonies of each mutant type were selected for sequencing of the rpsL or rpoB gene in order to characterize the mutations. All rpsL mutants had mutations at codon 43, and Lys-to-Arg (AAG→AGG) transitions were more frequent than Lys-to-Thr (AAG→ACG) or Lys-to-Asn (AAG→AAT or AAC) transversions. All but one of the sequenced rpsL mutants also had a neutral base substitution (CCG→CCA) at codon 45. Mutations in rpoB were located at codon 526; His-to-Arg (CAC→CGC) transversions arose most frequently, followed by either His-to-Pro (CAC→CCC) or His-to-Tyr (CAC→TAC) transversions. In the second step, mutants resistant to streptomycin were isolated from chosen rifampin-resistant mutants and vice versa. During this step, antibiotic selection was made on solid media containing either one or both antibiotics. It was noted that the frequencies of appearance of the second antibiotic resistance were always low when simultaneous selection for streptomycin and rifampin resistance was made (Table 1).
FIG. 3.
Flow chart describing mutant isolation. Rif, rifampin; Str, streptomycin; ∗, selective medium contains the indicated drug.
TABLE 1.
Frequencies of the appearance of spontaneous mutants resistant to streptomycin and/or rifampin in M. smegmatisa
| Strain | Mutation frequency with drug | |
|---|---|---|
| Wild type | Str* | Rif* |
| mc26 | >2 × 10−4 | >2.4 × 10−5 |
| Strr mutant | Rif* | Rif + Str* |
| R43L | >4.5 × 10−5 | 9.0 × 10−7 |
| N43L | >2.5 × 10−5 | 4.2 × 10−8 |
| Rifr mutant | Str* | Str + Rif* |
| R526H | >2 × 10−3 | 2.5 × 10−7 |
| Y526H | >4 × 10−4 | 5.5 × 10−8 |
The frequencies of the appearance of resistant mutants are relative to the total CFU plated. Str, streptomycin; Rif, rifampin; ∗, selective medium contains the indicated drug.
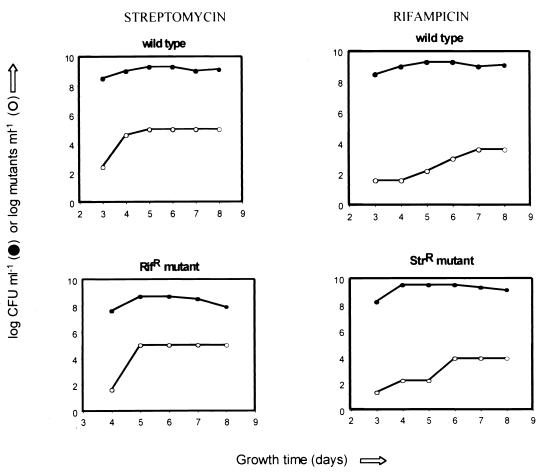
In addition, it was noted that the frequencies of appearance of mutants resistant to either streptomycin or rifampin varied as a function of the phase of growth in liquid culture when M. smegmatis cells were plated on antibiotic selection medium (Fig. 4). When the cultures were plated in late stationary phase, frequencies of appearance of mutants resistant to antibiotics as high as 10−3 mutant CFU/total CFU were observed.
FIG. 4.
Growth phase-dependent hypermutability in M. smegmatis, shown as frequencies of the appearance of resistant mutants.
Analysis of streptomycin and rifampin mutations.
Mutants that appeared after single (rifampin or streptomycin) and double (streptomycin and rifampin) selection were tested for resistance to one or both antibiotics and then analyzed by sequencing the rpsL and rpoB genes. It was found that selection for streptomycin resistance in rifampin-resistant mutants was accompanied by a high-level reversion of rifampin resistance to sensitivity, and conversely, when streptomycin-resistant mutants were selected for resistance to rifampin, the streptomycin resistance phenotype was also preferentially lost. The reversion of Strr to Strs (rpsL) was identified by a change in codon 43 (Arg to Lys [AGG→AAG] or Asn to Lys [AAT→AAG]), and reversion of Rifr to Rifs (rpoB) was identified by a change in codon 526 (Arg to His [CGC→CAC] or Tyr to His [TAC→CAC]) (Table 2). It is of interest that the single base changes were reversions of the initial antibiotic resistance mutation. Similar antagonistic effects were observed during attempts to isolate double mutants resistant to streptomycin and ciprofloxacin in M. smegmatis (K. Lu, P. Karunakaran, and J. E. Davies, unpublished data). In companion studies, the isolation of streptomycin- and rifampin-resistant mutants of Mycobacterium phlei indicated that reversion of the nonselected phenotype also occurred in this mycobacterial species (results not shown).
TABLE 2.
Reversion of streptomycin and rifampin mutations in M. smegmatis
| Mutant | Amino acid
|
Codon alteration | Reversion | |
|---|---|---|---|---|
| Position | Exchange | |||
| Strr | ||||
| R43L | 43 | Lys→Arg | AAG→AGG | AGG→AAG |
| N43L | 43 | Lys→Asn | AAG→AAT | AAT→AAG |
| Rifr | ||||
| R526H | 526 | His→Arg | CAC→CGC | CGC→CAC |
| Y526H | 526 | His→Tyr | CAC→TAC | TAC→CAC |
At least two of each of the double mutants resistant to rifampin and streptomycin mentioned in Table 1 were characterized by sequencing the rpsL and rpoB genes. All the mutants characterized contained mutations in both the rpsL and rpoB genes. The temperature sensitivities of these mutants were tested by plating at 30 and 42°C, and all the tested mutants grew at both temperatures.
To investigate the possibility of RecA-mediated gene conversion as a cause for high-frequency reversion, a recA mutant strain (HS42) of M. smegmatis was compared to its wild-type parent (18) for reversion frequency. This experiment was performed as outlined in Fig. 3 for mc26. The results revealed no significant difference for the recA strain.
Stationary-phase hypermutability.
Increased frequencies of mutation to streptomycin or rifampin resistance in M. smegmatis were noted (Fig. 4) when cells were plated on selective medium late in stationary phase. As confirmation of this hypermutability, a His− auxotroph of M. smegmatis (his5) was tested for reversion during exponential and stationary phases of growth. The results indicate that reversion to prototrophy reached a frequency as high as 10−3 prototroph CFU/total CFU when the cultures in late stationary phase were plated on minimal medium. The mutation responsible for histidine auxotrophy in M. smegmatis his5 was located at codon 380 (Gly to Glu [GGG→GAG]) by sequencing the hisD gene (Fig. 1). Eight spontaneous his5 revertants were selected for sequencing of the hisD gene in order to map the location of the mutation. In all cases this was detected at codon 380, and all were single-base reversions of the initial mutation (Glu to Gly [GAG→GGG]).
In order to study the possibility of competition between the wild-type and resistant strains during exponential and stationary growth phases, Strr or Rifr cultures were mixed with wild-type cultures, dilutions were plated on TSB and TSB containing streptomycin or rifampin, and the ratios of wild-type and Strr or Rifr colonies were monitored. Platings at 1, 2, 4, 6, and 8 days of incubation at 30°C indicated no significant difference between the wild-type and resistant strain counts over this period.
Direct PCR analysis of mutation.
The enhanced mutation to streptomycin resistance suggested that this event might be detectable at the nucleotide level during growth in liquid culture. To accomplish this, the procedure of EASPCR was used to specifically amplify an rpsL DNA fragment of streptomycin-resistant alleles in the bacterial population. We chose to analyze the R43L mutant (AAG→AGG), as this is the most common mutation (found in 7 out of 10 isolates sequenced); the wild-type allele of this mutant can be cleaved by MboII (for the purpose of EASPCR enrichment). The results (Fig. 2) clearly indicate that the nucleotide sequence changes specific to the mutant allele started to accumulate when M. smegmatis cultures entered late stationary phase. This indicates that (as expected) the presence of selecting antibiotic was not required for the generation of resistant mutants. It was not possible to perform a similar analysis with rifampin resistance since there was no convenient restriction enzyme site specific to the wild-type rpoB allele that could be used for ASPCR enrichment.
DISCUSSION
Multidrug resistance in M. tuberculosis occurs by the accumulation of successive point mutations in various genes affecting antibiotic action (17). We have studied the development of multiple antibiotic resistance in M. smegmatis by the isolation of spontaneous mutants resistant to streptomycin and rifampin. According to previous studies by Chakrabarti and Gorini (5, 6), there exists antagonism between rpsL and rpoB mutations in E. coli. These authors showed that paired streptomycin and rifampin resistance mutations lead to a temperature-sensitive phenotype in E. coli, and they suggested a possible mechanical coupling between ribosome and RNA polymerase such that certain combinations of rpsL and rpoB mutations are unable to interact effectively. Our studies suggest that a similar antagonism may exist in M. smegmatis. This was concluded on the basis of two observations: (i) when Strr mutants were isolated from a Rifr parent, or Rifr mutants were isolated from an Strr parent, the parental resistant mutation reverted to wild type at high frequency; and (ii) when Strr M. smegmatis mutants were plated on medium containing selective concentrations of both streptomycin and rifampin, they gave rise to Strr Rifr double mutants at a significantly lower frequency than when selection was done on rifampin alone. The same was true when a Rifr parent was used and Rifr Strr double mutants were selected. The mechanism for reversion at such a high frequency is not known. Antagonism between rpsL and rpoB mutations might provide strong selection, and it is possible that other factors, such as the physiological cost of harboring resistance mutations, could play a role in this process. A recent study of the physiological cost of Rifr in M. tuberculosis indicated that the relative fitness of all but one mutant allele studied was lower than that of the antibiotic-susceptible parent (2). Gene conversion is considered unlikely to have a role, since a recA mutant of M. smegmatis showed similar reversion of the initial antibiotic resistance phenotype.
Rosenberg et al. (22) proposed a model for adaptive reversion in the lac frameshift system where starvation (stress) could stimulate the formation of double-strand breaks in a small subset of the cells. It is known that in mammalian cells, chromosomal double-strand breaks can induce gene conversion at high frequency (24). We assume that in the cases of double selection for both resistance alleles there may be compensatory mutations that permit coexistence of Strr and Rifr in M. smegmatis, but this has not been analyzed for mycobacteria. We suggest that the temperature sensitivity in E. coli with combined mutations of streptomycin and rifampin resistance, found by Chakrabarti and Gorini (6), might be due to such compensatory mutations. There is ample evidence (1) for the occurrence of compensatory mutations restoring fitness to antibiotic-resistant strains of bacteria. In many cases the compensatory mutation occurs in the same gene as the mutation to resistance or in a gene encoding a related biochemical function.
The fact that reversion of antibiotic resistance occurred at such high frequencies (Table 1), which was confirmed by the EASPCR studies (Fig. 2), suggests that M. smegmatis is capable of hypermutation under specific conditions. For example, in the experiment of Table 1, the mutation to Strr (which occurs coincidentally with reversion of Rifr to Rifs) appeared at a frequency as high as 10−3 mutant CFU/total CFU. Similar high frequencies of reversion of a histidine auxotroph to prototrophy were also found.
There are several explanations for hypermutability. First, it is possible that M. smegmatis is defective in mismatch repair (M. tuberculosis has no mutS analog) (15), which would lead to an increase in mutation rate. Since it is known that sigma factors (sigH and sigE) regulate expression of many genes in stationary phase (9), a sigma factor-associated down-regulation or the collapse of an alternative repair system could explain why hypermutation occurs only when cultures enter late stationary phase. Second, there are numerous studies demonstrating the growth-dependent alteration of mutation rates; an example is growth advantage in stationary phase, in which subpopulations of mutant cells may take over stationary-phase cultures (27). However, mixed cultures of antibiotic-sensitive and -resistant strains of M. smegmatis gave no evidence of any obvious population takeovers by the mutant strains.
Specific DNA mutases, such as DNA polymerase IV (dinB) and DNA polymerase V (umuCD′), have been shown to allow higher mutation rates under certain conditions (19). A recent survey of the existence of the DinB- and UmuC-like protein families revealed the presence of similar catalytic domains in more than 30 sequences, including that of M. tuberculosis (26). Another possibility is that hypermutation could occur as a small subpopulation of the cells undergo genomewide mutagenesis but do not survive unless a selected (adaptive) mutation is generated (12). Adaptive mutation (4) or stressful-lifestyle-associated mutation (21), which could be induced by selection, stationary phase, or stress, might also explain the hypermutation effects seen in our studies with antibiotic resistance in M. smegmatis. In addition, although we found evidence of hypermutation by EASPCR before the cells were exposed to antibiotic selection, we cannot rule out the possibility that both streptomycin and rifampin could have enhanced the selection-induced hypermutable state. In fact, streptomycin is known to be mutagenic (10, 20).
Since the existence of MDRTB poses an increasing challenge in the treatment of tuberculosis, our analyses of Strr and Rifr mutants of M. smegmatis raise significant questions concerning the genetics of the development of multiple mutations to drug resistance in M. tuberculosis. Do antagonistic interactions occur between the point mutations and deletions that lead to resistance to combinations of rifampin, streptomycin, isoniazid, amikacin, and pyrazinamide in M. tuberculosis, and if so, in what manner are they compensated? Detailed comparative analyses of the genome sequences of M. tuberculosis and derived MDR strains will be revealing in this respect. We consider it unlikely that the MDR strains will consist simply of successive drug-resistant mutations. Finally, given the fact that M. tuberculosis propagates under a variety of stress conditions during its infectious process, it is reasonable to assume that these conditions may lead to hypermutation; such physiological situations will be difficult to study under normal laboratory growth and selection. Considering the number of mutations (up to six) MDRTB strains carry, as well as the fact that some of these mutations being antagonistic will lead to additional compensatory mutations, it is possible that hypermutability is inevitable to the lifestyle of M. tuberculosis. It is difficult to reconcile this with the available data which suggest that the average mutation rates for resistance to antibiotics in M. tuberculosis were on the order of 10−7 or 10−8 or even lower (about 10−10 for rifampin resistance) (7, 8), although it must be noted that the mutation frequencies reported for M. tuberculosis represent mutations per bacterium per generation, whereas our results are presented as the ratios of antibiotic-resistant to -sensitive colonies.
ACKNOWLEDGMENTS
We are grateful for generous financial support from the Canadian Bacterial Diseases Network.
We thank Neil G. Stoker for providing the His− mutant strain of M. smegmatis, Karen Lu for expert technical assistance, Fernando de la Cruz for valuable discussions, and George Spiegelman for comments on the manuscript.
REFERENCES
- 1.Andersson D I, Levin B R. The biological cost of antibiotic resistance. Curr Opin Microbiol. 1999;2:489–493. doi: 10.1016/s1369-5274(99)00005-3. [DOI] [PubMed] [Google Scholar]
- 2.Billington O J, McHugh T D, Gillespie S H. Physiological cost of rifampin resistance induced in vitro in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1999;43:1866–1869. doi: 10.1128/aac.43.8.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloom B R, Murray C J L. Tuberculosis: commentary on a reemergent killer. Science. 1992;257:1055–1064. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 4.Cairns J, Overbaugh J, Miller S. The origins of mutants. Nature. 1988;335:142–145. doi: 10.1038/335142a0. [DOI] [PubMed] [Google Scholar]
- 5.Chakrabarti S L, Gorini L. A link between streptomycin and rifampin mutation. Proc Natl Acad Sci USA. 1975;72:2084–2087. doi: 10.1073/pnas.72.6.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakrabarti S L, Gorini L. Interaction between mutations of ribosomes and RNA polymerase: a pair of strA and rif mutants individually temperature-insensitive but temperature-sensitive in combination. Proc Natl Acad Sci USA. 1977;74:1157–1161. doi: 10.1073/pnas.74.3.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.David H L. Probability distribution of drug-resistant mutants in unselected populations of Mycobacterium tuberculosis. Appl Microbiol. 1970;20:810–814. doi: 10.1128/am.20.5.810-814.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.David H L, Newman C M. Some observations on the genetics of isoniazid resistance in the tubercle bacilli. Am Rev Respir Dis. 1971;104:508–515. doi: 10.1164/arrd.1971.104.4.508. [DOI] [PubMed] [Google Scholar]
- 9.Fernandes N D, Wu Q-L, Kong D, Puyang X, Garg S, Husson R N. A mycobacterial extracytoplasmic sigma factor involved in survival following heat shock and oxidative stress. J Bacteriol. 1999;181:4266–4274. doi: 10.1128/jb.181.14.4266-4274.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez B, Haas F L, Wyss O. Induced host-range mutations in bacteriophage. Proc Natl Acad Sci USA. 1953;39:1052–1057. doi: 10.1073/pnas.39.10.1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finken M, Kirschner P, Meier A, Wrede A, Bottger C. Molecular basis of streptomycin resistance in Mycobacterium tuberculosis: alterations of the ribosomal protein S12 gene and point mutations within a functional 16S ribosomal RNA pseudoknot. Mol Microbiol. 1993;9:1239–1246. doi: 10.1111/j.1365-2958.1993.tb01253.x. [DOI] [PubMed] [Google Scholar]
- 12.Hall B G. Spontaneous point mutations that occur more often when advantageous than when neutral. Genetics. 1990;126:5–16. doi: 10.1093/genetics/126.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinshelwood S, Stoker N G. Cloning of mycobacterial histidine synthesis genes by complementation of a Mycobacterium smegmatis auxotroph. Mol Microbiol. 1992;6:2887–2895. doi: 10.1111/j.1365-2958.1992.tb01468.x. [DOI] [PubMed] [Google Scholar]
- 14.McClure W R, Cech C L. On the mechanism of rifampicin inhibition of RNA synthesis. J Biol Chem. 1978;253:8949–8956. [PubMed] [Google Scholar]
- 15.Mizrahi V, Andersen S J. DNA repair in Mycobacterium tuberculosis. What have we learnt from the genome sequence? Mol Microbiol. 1998;29:1331–1339. doi: 10.1046/j.1365-2958.1998.01038.x. [DOI] [PubMed] [Google Scholar]
- 16.Morris S, Bai G H, Suffys P, Portillo-Gomez L, Fairchok M, Rouse D. Molecular mechanisms of multiple drug resistance in clinical isolates of Mycobacterium tuberculosis. J Infect Dis. 1995;171:954–960. doi: 10.1093/infdis/171.4.954. [DOI] [PubMed] [Google Scholar]
- 17.Musser J M. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin Microbiol Rev. 1995;8:496–514. doi: 10.1128/cmr.8.4.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papavinasasundaram K G, Colston M J, Davis E O. Construction and complementation of a recA deletion mutant of Mycobacterium smegmatis reveals that the intein in Mycobacterium tuberculosis recA does not affect RecA function. Mol Microbiol. 1998;30:525–534. doi: 10.1046/j.1365-2958.1998.01083.x. [DOI] [PubMed] [Google Scholar]
- 19.Radman M. Enzymes of evolutionary change. Nature. 1999;401:866–869. doi: 10.1038/44738. [DOI] [PubMed] [Google Scholar]
- 20.Ren L, Rahman M S, Humayun M Z. Escherichia coli cells exposed to streptomycin display a mutator phenotype. J Bacteriol. 1999;181:1043–1044. doi: 10.1128/jb.181.3.1043-1044.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenberg S M. In pursuit of a molecular mechanism for adaptive mutation. Genome. 1994;37:893–899. doi: 10.1139/g94-127. [DOI] [PubMed] [Google Scholar]
- 22.Rosenberg S M, Harris R S, Torkelson J. Molecular handles on adaptive mutation. Mol Microbiol. 1995;18:185–189. doi: 10.1111/j.1365-2958.1995.mmi_18020185.x. [DOI] [PubMed] [Google Scholar]
- 23.Ruusala T, Kurland G C. Streptomycin preferentially perturbs ribosomal proofreading. Mol Gen Genet. 1984;198:100–104. doi: 10.1007/BF00328707. [DOI] [PubMed] [Google Scholar]
- 24.Taghian D G, Nickoloff J A. Chromosomal double-strand breaks induce gene conversion at high frequency in mammalian cells. Mol Cell Biol. 1997;17:6386–6393. doi: 10.1128/mcb.17.11.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S, Colston M J, Matter L, Schopfer K, Bodmer T. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 26.Wagner J, Gruz P, Kim S R, Yamada M, Matsui K, Fuchs R P, Nohmi T. The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol Cell. 1999;4:281–286. doi: 10.1016/s1097-2765(00)80376-7. [DOI] [PubMed] [Google Scholar]
- 27.Zambrano M M, Kolter R. GASPing for life in stationary phase. Cell. 1996;86:181–184. doi: 10.1016/s0092-8674(00)80089-6. [DOI] [PubMed] [Google Scholar]