Abstract
Background
Self-reported symptom studies rapidly increased understanding of SARS-CoV-2 during the COVID-19 pandemic and enabled monitoring of long-term effects of COVID-19 outside hospital settings. Post-COVID-19 condition presents as heterogeneous profiles, which need characterisation to enable personalised patient care. We aimed to describe post-COVID-19 condition profiles by viral variant and vaccination status.
Methods
In this prospective longitudinal cohort study, we analysed data from UK-based adults (aged 18–100 years) who regularly provided health reports via the Covid Symptom Study smartphone app between March 24, 2020, and Dec 8, 2021. We included participants who reported feeling physically normal for at least 30 days before testing positive for SARS-CoV-2 who subsequently developed long COVID (ie, symptoms lasting longer than 28 days from the date of the initial positive test). We separately defined post-COVID-19 condition as symptoms that persisted for at least 84 days after the initial positive test. We did unsupervised clustering analysis of time-series data to identify distinct symptom profiles for vaccinated and unvaccinated people with post-COVID-19 condition after infection with the wild-type, alpha (B.1.1.7), or delta (B.1.617.2 and AY.x) variants of SARS-CoV-2. Clusters were then characterised on the basis of symptom prevalence, duration, demography, and previous comorbidities. We also used an additional testing sample with additional data from the Covid Symptom Study Biobank (collected between October, 2020, and April, 2021) to investigate the effects of the identified symptom clusters of post-COVID-19 condition on the lives of affected people.
Findings
We included 9804 people from the COVID Symptom Study with long COVID, 1513 (15%) of whom developed post-COVID-19 condition. Sample sizes were sufficient only for analyses of the unvaccinated wild-type, unvaccinated alpha variant, and vaccinated delta variant groups. We identified distinct profiles of symptoms for post-COVID-19 condition within and across variants: four endotypes were identified for infections due to the wild-type variant (in unvaccinated people), seven for the alpha variant (in unvaccinated people), and five for the delta variant (in vaccinated people). Across all variants, we identified a cardiorespiratory cluster of symptoms, a central neurological cluster, and a multi-organ systemic inflammatory cluster. These three main clusers were confirmed in a testing sample. Gastrointestinal symptoms clustered in no more than two specific phenotypes per viral variant.
Interpretation
Our unsupervised analysis identified different profiles of post-COVID-19 condition, characterised by differing symptom combinations, durations, and functional outcomes. Our classification could be useful for understanding the distinct mechanisms of post-COVID-19 condition, as well as for identification of subgroups of individuals who might be at risk of prolonged debilitation.
Funding
UK Government Department of Health and Social Care, Chronic Disease Research Foundation, The Wellcome Trust, UK Engineering and Physical Sciences Research Council, UK Research and Innovation London Medical Imaging & Artificial Intelligence Centre for Value-Based Healthcare, UK National Institute for Health Research, UK Medical Research Council, British Heart Foundation, UK Alzheimer's Society, and ZOE.
Introduction
SARS-CoV-2 is a highly infectious and rapidly mutating coronavirus. Since the first human cases in December, 2019, it has infected nearly half a billion people worldwide. Early SARS-CoV-2 variants caused lower respiratory tract infections and were associated with high morbidity and mortality.1 The symptoms of COVID-19 have evolved as new variants emerged.2 Many survivors report persistent symptoms after acute illness that affect their quality of life. The prevalence of long-term symptoms varies with different SARS-CoV-2 variants.3
Research in context.
Evidence before this study
We searched PubMed with the keywords (“Long-COVID*” OR “post?covid*” OR “post?COVID*” OR “postCOVID*” OR “postCovid*”) AND (“cluster*” OR “endotype*” OR “phenotype*” OR “sub?type*” OR “subtype”) for articles published in English from database inception to June 15, 2022. Of the 161 results returned, 24 either provided descriptions of subtypes or proposed phenotypes of long COVID (ie, ongoing COVID and post-COVID-19 condition) or post-COVID-19 condition. In 16 of these studies, phenotypes were manually divided into subgroups; in six of these studies, unsupervised methods were used for patient clustering and automatic semantic phenotyping. The remaining two reports detailed uncommon presentations of long COVID or post-COVID-19 condition. Overall, two to eight symptom profiles (clusters) were identified, with three recurring clusters. A cardiopulmonary syndrome was the predominant observation, manifesting as exertional intolerance and dyspnoea in ten of the 24 studies; fatigue in eight of the 24 studies; autonomic dysfunction, tachycardia, or palpitations in five of the 24 studies; lung radiological abnormalities, including fibrosis, in two of the 24 studies; and chest pain in one of the 24 studies. Another common presentation was persistent general autoimmune activation and proinflammatory state (n=2), comprising mild multi-organ sequelae (n=2), gastrointestinal symptoms (n=2), dermatological symptoms (n=2), or fever (n=1). A third syndrome defined by neurological or neuropsychiatric symptoms was also reported, with symptoms including brain fog or dizziness (n=2), poor memory or cognition (n=2), mental health issues such as mood disorders (n=5), headache (n=2), central sensitisation (n=1), paraesthesia (n=1), autonomic dysfunction (n=1), fibromyalgia (n=2), and chronic pain or myalgias (n=6). Unsupervised clustering methods identified two-to-six different post-COVID phenotypes, mapping to the ones described previously. 14 other reports identified by our search focused on possible causes or mechanisms of disease underlying one or more manifestations of long COVID or post-COVID-19 conditions. Dysregulation of immune responses was a potential common cause. To our knowledge, no studies of the symptom profile of post-COVID-19 condition informed by the causative variant and vaccination status have been published. Also, no studies in which longitudinally collected symptoms have been modelled as time-series data with the aim of characterising post-COVID-19 condition have been published.
Added value of this study
We used a large dataset of prospectively collected self-reported symptoms to identify symptom profiles for post-COVID-19 condition in unvaccinated people infected with the wild-type or alpha (B.1.1.7) variant of SARS-CoV-2 and for vaccinated people infected with the delta (B.1.617.2 and AY.x) variants of SARS-CoV-2. We identified three main symptom profiles—a cluster defined by central neurological symptoms, a cluster dominated by cardiorespiratory symptoms, and a third more heterogeneous cluster showing systemic inflammatory symptoms—which were consistent across variants and by vaccination status. These symptom profiles differed between variants only in the proportion of individuals affected and symptom duration overall.
Implications of all the available evidence
We show the existence of different phenotypes of post-COVID-19 condition, which share commonalities across SARS-CoV-2 variant types in both symptoms and how they developed as SARS-CoV-2 evolved. These differing phenotypes reflect different underlying pathophysiological mechanisms. These insights could help with the development of personalised diagnosis and treatment, and could help policy makers to plan for the delivery of care for people living with post-COVID-19 condition.
In the UK in October, 2020, the term ongoing COVID was introduced to describe otherwise unexplained signs and symptoms 4–12 weeks after SARS-CoV-2 infection, and post-COVID-19 syndrome (known also as post-COVID condition as defined by WHO, and used hereafter in this Article) was introduced to describe signs and symptoms that persisted more than 12 weeks after infection.4 Together, ongoing COVID and post-COVID-19 condition are commonly known as long COVID.
In June, 2022, the UK Office of National Statistics estimated that 2 million people had long COVID in the UK.5 Long COVID has diverse manifestations and results in arising health-care needs.6 It adversely affects quality of life in two-thirds of people.2 Appropriate medical and social support is thus required. Vaccination campaigns helped to mitigate not only the acute effects of SARS-CoV-2 infection, but also the risk of developing long COVID and acute and long-term severe symptoms.7 In the UK, the COVID-19 vaccination campaign started in December, 2020,8 and effectively reduced the risk of developing long-lasting symptoms.3
In previous studies9, 10, 11 distinct presentations of post-COVID-19 condition were reported, and it was thus proposed that post-COVID-19 condition should not be regarded as a discrete condition. Several phenotypes were discerned either empirically or via automatic methods distinguished by the causes, phases of the disease, symptom manifestation, severity, outcome, and potential therapies.9, 10, 11 Use of semi-supervised and unsupervised methods enabled identification of up to eight clusters of symptoms,12 which could be separated into profiles of phenotypic abnormalities with enrichments in pulmonary, cardiovascular, neuropsychiatric, and constitutional symptoms such as fatigue and fever.
We hypothesised that, among the heterogeneous presentation of post-COVID-19 condition, distinct phenotypes and symptom profiles could be identified that are potentially due to differing pathophysiology and vary by SARS-CoV-2 variant and vaccination status at the time of infection. We also hypothesised that identification of discrete illness profiles could help to guide prognostication and treatment approaches (including personalised management). To these ends, we did an unsupervised clustering analysis of self-reported symptoms and their impact on daily living in community-based people with post-COVID-19 condition.
Methods
Study overview and data sources
This prospective longitudinal study was based on data prospectively collected in the UK in the COVID Symptom Study (CSS), which were self-reported via an app developed by ZOE (London, UK) in collaboration with scientists from King's College London (London UK), Massachusetts General Hospital (Boston, MA, USA), Uppsala University (Uppsala, Sweden), and Lund University (Lund, Sweden).13 The CSS is described in the appendix (p 3).
Briefly, participants self-reported symptoms (both free text and in response to targeted questions; appendix pp 3–5), SARS-CoV-2 testing and results, vaccination status, hospital attendance, and demographic and comorbidity data. People could also use the app to proxy-report for others, but we have limited our analysis to self-reported information. The initial list of 20 explicit questions about symptoms was expanded after Nov 4, 2020, to include 34 symptoms in response to evolving knowledge of COVID-19 infection.
In addition to data logged through the CSS app, further data were available from a subset of participants who had been recruited to the CSS Biobank. Participants in the CSS Biobank responded to an online questionnaire enquiring about the effect of ongoing COVID-19 symptoms on their daily lives (considered retrospectively), as part of investigations into the effect of the pandemic on mental and physical health. The questionnaire contained 32 questions, split into five domains (medical history, symptoms during the pandemic, lifestyle, individual experiences of long COVID, and life before and after the pandemic). We included responses to a subset of six questions related to the long-term effects of COVID-19 and symptom severity (appendix p 6) in this study to assess the association between symptom profiles and the lives of people with long COVID. We also invited CSS Biobank participants experiencing post-COVID-19 condition and their families to join a volunteer advisory panel to discuss our research findings and to provide clinical and social context for them.
The app and CSS were approved in the UK by Kings College London's ethics committee. All app users provided informed consent for use of their data for COVID-19 research. The CSS Biobank was approved by the UK National Health Service Health Research Authority and licensed under the Human Tissue Authority. CSS Biobank participants were invited to join from the app user base and provided informed consent to participate in the additional questionnaire and sample collection studies, and for linkage to app-collected data. All research was done in full compliance with the Declaration of Helsinki and its updates.
Procedures
In this study, we used CSS data reported between March 24, 2020, and Dec 8, 2021, and CSS Biobank data gathered between October, 2020, and April, 2021. We defined people with post-COVID-19 condition as those reporting symptoms for longer than 12 weeks after their first positive PCR or lateral flow test within the study period, as per the UK National Institute for Health and Care Excellence definition.14 We defined long COVID as reporting symptoms for longer than 4 weeks from the initial positive test. Because we focused on post-COVID-19 condition, we did not formally model the profile of people with long COVID; we merely did descriptive analyses of the demographic profile of those individuals.
To study the effect of vaccination on symptom profiling in post-COVID-19 condition, we divided participants into two groups: vaccinated at time of SARS-CoV-2 infection and unvaccinated at time of SARS-CoV-2 infection. The different fractions of vaccinated participants for the various strains are explained by the rollout of vaccination in the UK, which began on Dec 8, 2020. Post-vaccination infection was defined as a positive test at least 7 days after the first dose of a SARS-CoV-2 vaccine. Time since vaccination (beyond 7 days) and number of doses were not considered in the analyses.
We analysed both CSS and CSS Biobank data separately for three periods of the pandemic in the UK, which were defined according to the predominant circulating SARS-CoV-2 strain (ie, if the same strain accounted for ≥80% of tested samples) as documented by COG-UK (appendix p 7).15 In the UK, wild-type SARS-CoV-2 was predominant from March 24 to Nov 29, 2020, the alpha (B.1.1.7) variant from Jan 10 to April 25, 2021, and the delta (B.1.617.2 and AY.x) variants from May 26 to Dec 8, 2021.
We included data for people who had a positive SARS-CoV-2 test (either RT-PCR or lateral flow antigen test) within one of the windows with a predominant circulating strain, had logged that they felt “physically normal” for at least 2 weeks before the test (except for the immediate 7 days before the test), reported symptoms for at least 28 days after diagnosis (symptoms experienced up to 7 days before a positive test were considered symptoms at the onset of the illness), and logged their symptoms at least weekly until they reported feeling healthy for more than 4 weeks (as in a previous study16 of long COVID in a similar sample; appendix p 8). Further analyses to characterise post-COVID-19 condition included only people whose illness lasted 84 days or longer. Exclusion criteria were inconsistent non-illness-related information (eg, BMI <10 kg/m2 or >95 kg/m2), proxy-reporting, being age younger than 18 years or older than 100 years, and reporting a positive test within 7 days of any dose of vaccination, including boosters (because the protective effects of vaccination were unlikely to have begun within this timeframe).14
The CSS data were split into training and testing sets for symptom cluster optimisation and assessment of the clinical relevance of any observed clusters and cluster reproducibility in a new population. The testing set included 50% of the CSS Biobank population and a matched population (1:1) from the CSS who were not recruited to the CSB Biobank. The training set comprised the remaining 50% of the CSS Biobank population and all remaining eligible participants from the CSS population (appendix p 9).
Data analysis and clustering estimation
We did a Mann-Whitney test (Bonferroni correction, p=2·4 × 107) to assess whether differences in symptom prevalence existed between vaccinated and unvaccinated populations for the alpha and delta variants independently (vaccination was not available in the general population when the wild-type variant was predominant). Before the clustering analysis, three clinicians (MFÖ, ELD, and CJS) independently categorised reported symptoms into domains (upper respiratory, abdominal, cardiorespiratory, immune-related or cutaneous, central neurological, and systemic or inflammatory) for descriptive purposes. Discrepancies in the allocation of domains were discussed and resolved jointly. This grouping was used to visualise and describe symptom cluster profiles predicted by the model, but was not used otherwise for model optimisation. For people infected with the wild-type SARS-CoV-2 variant, we considered only 20 symptoms (related to the 20 direct questions in the CSS app during the period when the wild-type variant was predominant). For people infected with either the alpha or delta variants, we considered all 34 symptoms for which data were available (appendix p 3).
Using an unsupervised time-series clustering approach, we clustered the individuals with post-COVID-19 condition according to their symptom patterns for vaccinated and unvaccinated populations and for each viral variant considered. Sample sizes were sufficient only for analyses of the unvaccinated wild-type, unvaccinated alpha variant, and vaccinated delta variant groups. We estimated the symptom time series per individual by using symptom aggregation per week—ie, we computed the frequency of each symptom per week as the sum of reports during 7 days. We treated missing reports within a week as missing data. Thus, missing symptom reports were linearly interpolated independently per symptom within each week. We used a model previously used in people with COVID-1916 (which relies on multivariate time-series clustering based on a principle-component analysis model, Mc2PCA)17 to estimate symptom clusters. This method allowed for the clustering of time series with distinct duration by establishing comparisons between the samples with covariance matrices, thus enabling the comparison of people who reported heterogeneous symptoms.
We used a K-means approach to optimise the number of clusters through an iterative process. The cluster estimation was based on the projection computed using the singular value decomposition of the average of the covariance matrices, where the first six dimensions were used. The process was repeated until the convergence criterion was reached: the error after projection is below 10–4. The optimal number of clusters, hereafter defined as optimised clusters, per variant and per population (vaccinated vs unvaccinated) was identified via the Bayesian information criterion, considering the global minimum or the first local minimum alternatively that were obtained across ten random initialisations. We did a bootstrapping analysis for the wild-type variant (50 randomly selected samples for training and testing, respectively) to assess the robustness of the number of optimised clusters. For the alpha and delta variants, only ten bootstraps were done in view of the small sample size of their testing sets (due to a low number of CSS Biobank samples).
Clustering assessment and phenotype profiling
The best-performing clusters across bootstrapping (ie, the samples with the lowest Bayesian information criterion and average distance to the centroid of all samples within the same cluster, here defined as error) were used to assess the profile of the different clusters (appendix p 17). We computed the profile of the optimised clusters per variant as the average interpolated frequency of the symptoms during illness. Additionally, we assessed the proportion of people presenting with each symptom per week. We did a significance analysis to identify possible differences in symptom profile per cluster. We computed the Z score between the mean symptom profile across all included people, independent of symptom clusters, and the average profile of individuals belonging to a given cluster. We identified as most relevant the symptoms above the third quartile of the median Z scores during illness. We characterised each cluster using the set of symptoms identified by the significance analysis as a proxy for the distinctive illness phenotype within each cluster.
To assess whether the average profile per cluster was affected by intracluster variability, we compared the average profiles to an example person with the minimum distance to the centroid of the cluster among a sub-population whose age and BMI were within one SD of the means of the cluster population. Lastly, we used the Mann-Whitney test to assess differences in demographic profiles (age, gender, BMI, comorbidities; appendix p 3) across clusters for the same variant and population.
To assess the clusters' robustness to different populations, we computed the cluster prediction for an independent sample of individuals—the testing set. To assess whether such clusters were robust, we retrained the cluster model on the testing set and compared the predicted clusters from both models (ie, a model trained on the training set and a second model trained only on the testing set; appendix p 9). Agreement metrics between the two predicted sets of clusters were used for quantitative assessments (ie, calculation of Jaccard score and balanced accuracy).
To assess the clinical relevance of optimised clusters and any effects on daily living, we included information on the effects of symptoms on daily life, health-care use (both during primary infection and >12 weeks from infection), and reinfection before June, 2021. We applied a multivariate generalised linear regression model (ie, a logistic regression) to study the effect of each cluster on participants' outcomes, assessed through the CSS Biobank questionnaire. We considered age, gender, BMI, and cluster classification as covariates in the model. Odds ratios (ORs) were calculated between the predictor, which consisted of questionnaire answers binarised as reported impact (1) and no impact (0; appendix p 6), and the clusters one-hot-encoded (ie, the cluster class value was converted into a new column and assigned a 1 or 0 notation for true or false). Different models were optimised for the different questions while keeping the same covariates across models. We used the Wald test to calculate p values for the significance of the covariates, after false rate discovery correction. We used the false discovery rate instead of any other multiple correction method as it is a less conservative approach; we defined the threshold as 0·05.
All analyses were done in Python (version 3.9.7). The Python package statsmodel (version 0.13.1) was used for statistical analyses, such as the Mann-Whitney test and general linear regression. Data were extracted and curation with Exetera software version 0.6.0.18
Role of the funding source
ZOE, one of the study funders, developed the app used for data collection. ZOE employees contributed to data collection. The funders of the study had no role in study design, data analysis, data interpretation, or writing of the report.
Results
Data for 9804 (3%) participants of the 336 652 participants in the CSS who reported a positive SARS-CoV-2 test were included in our dataset (table ; appendix p 8). 1513 (15%) of these participants reported symptoms for longer than 84 days, and thus were classed as having post-COVID-19 condition. 584 (<1%) participants in the overall CSS also participated in the CSS Biobank, and 140 (24%) of these people had post-COVID-19 condition. The different proportions of vaccinated participants for the various strains are related to the rollout of vaccination in the UK, which began on Dec 8, 2020.8 We noted no significant differences in the prevalence and duration of symptoms between people who developed post-COVID-19 condition from a SARS-CoV-2 infection (alpha or delta variant) before vaccination and those who developed the condition from an infection after vaccination, suggesting that symptom prevalence and duration among individuals developing post-COVID-19 condition did not depend on the timing of infection relative to vaccination status (appendix pp 12, 17).
Table.
Demographics of the study population
|
Unvaccinated people |
Vaccinated people |
|||||
|---|---|---|---|---|---|---|
| Wild-type variant | Alpha variant | Delta variant | Alpha variant | Delta variant | ||
| Persistent symptoms at any time | ||||||
| n | 2597 | 1076 | 132 | 203 | 5396 | |
| Age, years | 56 (47 to 64) | 57 (50 to 64) | 46 (26 to 59) | 59 (52 to 66) | 57 (49 to 65) | |
| BMI, kg/m2 | 26 (23 to 31) | 26 (23 to 30) | 25 (21 to 28) | 26 (23 to 33) | 26 (23 to 30) | |
| Gender | ||||||
| Woman | 1916 (74%) | 750 (70%) | 83 (63%) | 152 | 3634 (67%) | |
| Man | 681 (26%) | 326 (30%) | 49 (37%) | 51 (25%) | 1762 (33%) | |
| At least one comorbidity | 456 (18%) | 184 (17%) | 19 (14%) | 45 (22%) | 916 (17%) | |
| Illness duration, days | 79 (40 to 348) | 71 (41 to 307) | 61 (39 to 115) | 74 (42 to 359) | 61 (39 to 133) | |
| Time since vaccination, days | NA | NA | NA | 18 (11 to 40) | 210 (168 to 243) | |
| Hospitalisation* (%) | 16 (1%) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| Ethnicity | ||||||
| White | 2300 (89%) | 921 (86%) | 125 (95%) | 191 (94%) | 5171 (96%) | |
| Black | 16 (1%) | 11 (1%) | 4 (3%) | 3 (1%) | 26 (<1%) | |
| Asian | 49 (2%) | 23 (2%) | 0 (0) | 3 (1%) | 51 (1%) | |
| Other† | 232 (9%) | 121 (11%) | 3 (2%) | 6 (3%) | 148 (3%) | |
| Persistent symptoms 28–83 days after infection | ||||||
| n | 2192 | 917 | 109 | 172 | 4501 | |
| Age, years | 54 (45 to 62) | 56 (49 to 63) | 49 (31 to 58) | 58 (49 to 64) | 55 (48 to 64) | |
| BMI, kg/m2 | 27 (23 to 31) | 26 (24 to 30) | 24 (21 to 28) | 26 (23 to 32) | 26 (23 to 30) | |
| Gender | ||||||
| Woman | 1606 (73%) | 637 (69%) | 69 (63%) | 131 (76%) | 3026 (67%) | |
| Man | 586 (27%) | 280 (31%) | 40 (37%) | 41 (24%) | 1475 (33%) | |
| At least one comorbidity | 456 (21%) | 184 (20%) | 19 (17%) | 45 (26%) | 916 (20%) | |
| Illness duration, days | 41 (33 to 55) | 43 (33 to 55) | 41 (33 to 55) | 43 (34 to 57) | 42 (34 to 55) | |
| Time since vaccination, days | NA | NA | NA | 18 (11 to 29) | 209 (167 to 242) | |
| Hospitalisation* (%) | 13 (1%) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| Ethnicity | ||||||
| White | 1960 (89%) | 783 (85%) | 104 (95%) | 164 (95%) | 4341 (96%) | |
| Black | 11 (1%) | 10 (1%) | 3 (3%) | 2 (1%) | 19 (<1%) | |
| Asian | 41 (2%) | 18 (2%) | 0 (0) | 2 (1%) | 46 (1%) | |
| Other† | 180 (8%) | 106 (12%) | 2 (2%) | 4 (2%) | 95 (2%) | |
| Persistent symptoms 84 days or more after infection | ||||||
| n | 405 | 159 | 23 | 31 | 895 | |
| Age, years | 58 (50 to 65) | 58 (52 to 65) | 41 (25 to 60) | 60 (55 to 71) | 59 (52 to 66) | |
| BMI, kg/m2 | 26 (24 to 32) | 27 (23 to 30) | 26 (20 to 28) | 26 (23 to 35) | 26 (23 to 30) | |
| Gender | ||||||
| Woman | 298 (74%) | 113 (71%) | 14 (61%) | 21 (68%) | 595 (66%) | |
| Man | 107 (26%) | 46 (29%) | 9 (39%) | 10 (32%) | 300 (34%) | |
| At least one comorbidity | 106 (26%) | 39 (25%) | 5 (22%) | 9 (29%) | 217 (24%) | |
| Illness duration, days | 355 (175 to 517) | 343 (133 to 431) | 157 (103 to 206) | 359 (143 to 419) | 152 (119 to 196) | |
| Time since vaccination, days | NA | NA | NA | 17 (10 to 47) | 211 (168 to 244) | |
| Hospitalisation* (%) | 3 (1%) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| Ethnicity | ||||||
| White | 340 (84%) | 138 (87%) | 21 (91%) | 27 (87%) | 831 (93%) | |
| Black | 5 (1%) | 1 (1%) | 1 (4%) | 1 (3%) | 7 (1%) | |
| Asian | 8 (2%) | 5 (3%) | 0 (0) | 1 (3%) | 5 (1%) | |
| Other† | 52 (13%) | 15 (9%) | 1 (4%) | 2 (6%) | 52 (6%) | |
Data are median (IQR) or n (%). NA=not applicable.
During their long COVID.
Includes data for people who did not report their ethnicity or reported ethnicities other than those specified.
Overall, we found differing numbers of clusters associated with post-COVID-19 condition per variant, each of which had a specific illness duration and symptom profile (in terms of both prevalence and duration). For the wild-type variant, we found four clusters (figure 1 ). The largest group had predominantly cardiorespiratory symptoms. People with post-COVID-19 condition who had been infected with the alpha variant while unvaccinated were clustered in seven groups, with a high prevalence of central neurological symptoms among the people in the largest cluster (appendix p 20). Similarly, among vaccinated people who developed post-COVID-19 condition after infection with the delta variant, central neurological symptoms were predominant in the largest of the five identified clusters (appendix p 24). In terms of symptom relevance (Figure 2, Figure 3, Figure 4 ), we identified three common patterns across variants: a central neurological cluster (eg, anosmia, dysosmia, brain fog, depression, delirium, and headache), often without a great association with other symptoms in infections caused by the alpha or delta variants, a cardiorespiratory cluster typically linked to severe symptoms (eg, severe dyspnoea [only mild to moderate for the wild-type variant]), and a systemic or inflammatory cluster that also often manifested with other immune-related symptoms. Additionally, abdominal symptoms were often predominantly isolated within one small cluster across the three variants (Figure 2, Figure 3, Figure 4). Immune-related and cutaneous symptoms were constricted mainly within one cluster for the alpha and delta variants (higher prevalence), with minor occurrences in a second cluster (Figure 3, Figure 4). When present, immune-related symptoms had high Z scores, contributing to cluster uniqueness, and often co-occurred with systemic and inflammatory symptoms (Figure 2, Figure 3, Figure 4). However, the overall prevalence of these symptoms was low.
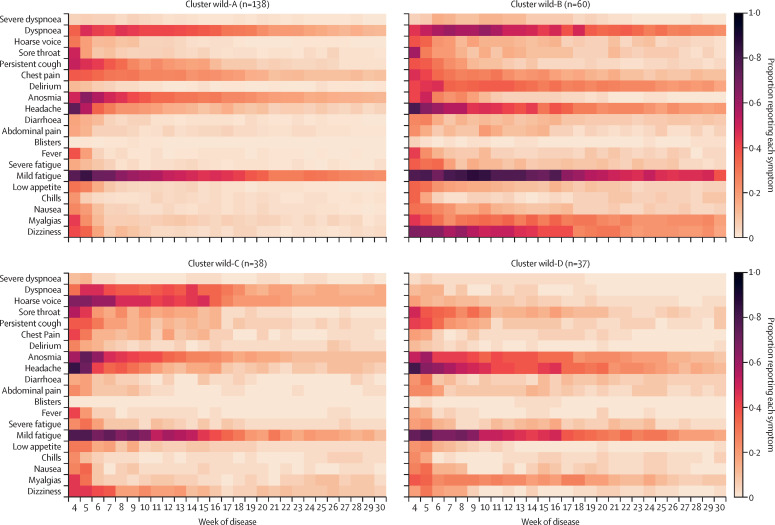
Figure 1.
Symptom prevalence profile for clusters of post-COVID-19 condition caused by wild-type SARS-CoV-2
Data are for people with symptomatic illness for at least 84 days after their initial positive SARS-CoV-2 test. The proportion of people reporting each symptom per week is encoded by the colourmap (the darker the colour, the higher the symptom prevalence).
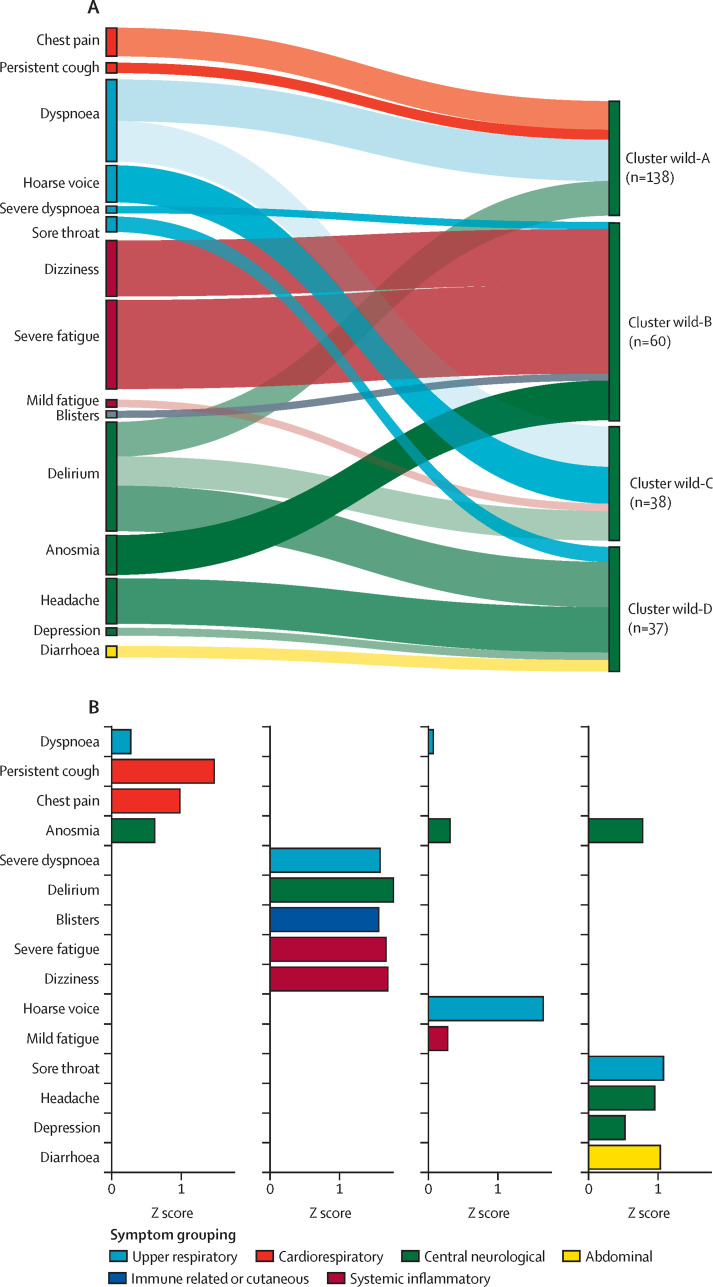
Figure 2.
Relative contribution of symptoms per cluster (A) and ranking of symptom relevance (B) in unvaccinated people with post-COVID-19 condition caused by wild-type SARS-CoV-2
Symptom selection was based on the third quartile of Z scores (only positive Z scores are shown), with the aim of highlighting the most contributing symptoms in each cluster. In A, the width of shadow connections encodes the symptom prevalence (median number of individuals reporting the symptom within the cluster), whereas colour intensity encodes the relevance of symptoms per cluster. In B, rankings are based on Z scores.
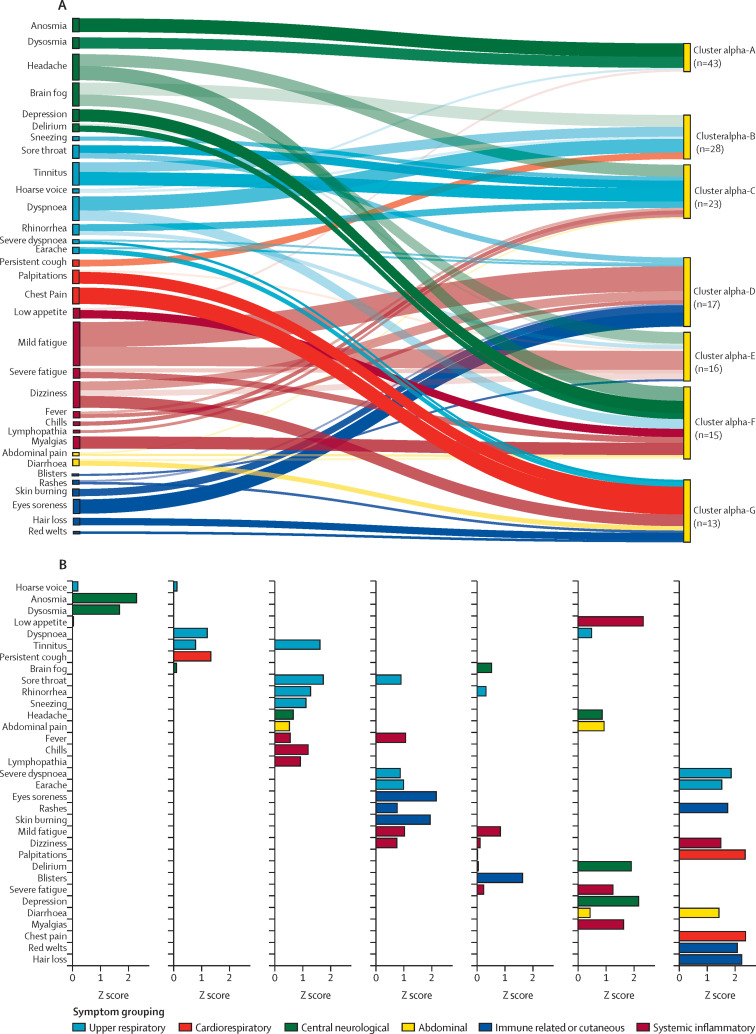
Figure 3.
Relative contribution of symptoms per cluster
(A) Ranking of symptom relevance. (B) Unvaccinated people with post-COVID-19 condition caused by the alpha SARS-CoV-2 variant. Symptom selection was based on the third quartile of Z scores (only positive Z scores are shown), with the aim of highlighting the most contributing symptoms in each cluster. In A, the width of shadow connections encodes the symptom prevalence (median number of individuals reporting the symptom within the cluster), whereas colour intensity encodes the relevance of symptoms per cluster. In B, rankings are based on Z scores.
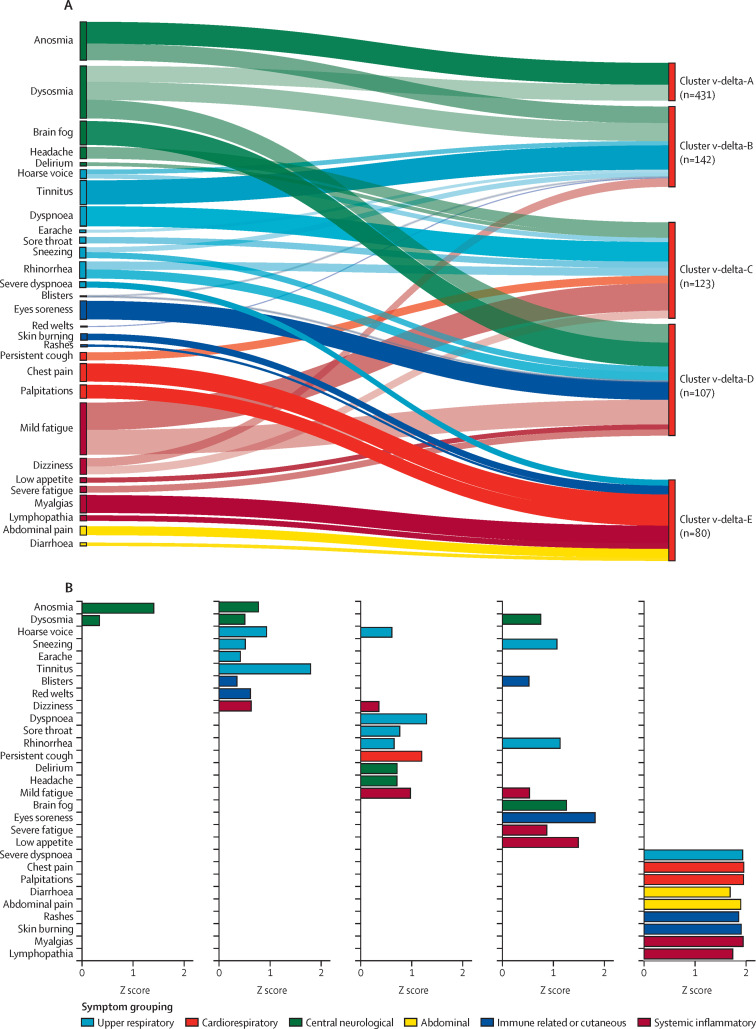
Figure 4.
Relative contribution of symptoms per cluster
(A) Ranking of symptom relevance. (B) Vaccinated people with post-COVID-19 condition caused by the delta SARS-CoV-2 variant. Symptom selection was based on the third quartile of Z scores (only positive Z scores are shown), with the aim of highlighting the most contributing symptoms in each cluster. In A, the width of shadow connections encodes the symptom prevalence (median number of individuals reporting the symptom within the cluster), whereas colour intensity encodes the relevance of symptoms per cluster. In B, rankings are based on Z scores.
The largest symptom cluster among people with post-COVID-19 condition who had been infected by the wild-type variant, cluster wild-A (n=138), was characterised by upper respiratory and central neurological symptoms (Figure 1, Figure 2; appendix p 14). The next largest cluster, wild-B (n=60), was heterogeneous and included symptoms relating to immune, central neurological, respiratory, and systemic and inflammatory systems (Figure 1, Figure 2; appendix p 14). This cluster had the most severe symptoms, including prolonged fever (appendix p 14), and 12 (20%) people had dyspnoea affecting their ability to function normally 12 weeks after infection. Cluster wild-C (n=38) was mainly associated with upper respiratory symptoms (particularly hoarse voice), anosmia, and low appetite (figure 1; appendix p 14). Lastly, cluster wild-D (n=37) had abdominal, central neurological (headache and anosmia), and upper respiratory (sore throat) symptoms that were also common to other clusters (appendix p 14). Together with cluster wild-B, this cluster's symptoms lasted longer and were of greater severity. Age, gender, and BMI did not differ significantly between wild-type clusters (appendix p 18). The appendix (p 19) contains individual symptom profiles for the optimised clusters.
The largest of the seven clusters of unvaccinated people with post-COVID-19 condition after infection with the alpha variant, cluster alpha-A (n=47, 30%), was dominated by anosmia, whereas the smallest, cluster alpha-G (n=13, 8%), was highly heterogeneous and polysymptomatic (appendix pp 14–15). Individual symptom profiles consistently qualitatively matched the average symptom profiles within each alpha cluster (appendix p 22). Abdominal symptoms were mainly confined to cluster alpha-F, but were also noted in clusters alpha-C and alpha-E (appendix pp 20–22). Median illness duration seemed to differ between clusters: it was shortest in cluster alpha-B (17 weeks [IQR 14–29]) and longest in cluster alpha-E (23 weeks [14–41]; appendix pp 14–15). Age, gender, and BMI did not differ significantly within or across clusters (appendix p 23).
The largest symptom cluster among vaccinated people who developed post-COVID-19 condition after infection with the delta variant, cluster delta-A (n=431), included mainly central neurological symptoms (appendix pp 15, 24). The smallest cluster, delta-E (n=80), comprised severe symptoms (severe dysponea and cardiovascular symptoms), including systemic symptoms; it was also the only cluster showing abdominal symptoms (appendix pp 15, 24). Symptom duration was similar across all clusters in the vaccinated delta variant population (appendix p 15). In the delta-A cluster, men had significantly higher BMIs than women (p=0·0018; appendix p 27). No further demographic differences were found across clusters (appendix p 27). When we presented our findings to the CSS Biobank volunteer advisory panel, they broadly recognised the symptom clusters we defined (although no formal analysis of their responses was done).
In the testing set, we applied the clusters obtained from training on the set of wild-type infected individuals to predict the classification of individuals with post-COVID-19 condition after infection with wild-type SARS-CoV-2 (appendix p 29). To assess the robustness of clusters in different populations, we retrained a model only in the testing set, which was used to predict the clusters in the testing population. Classification obtained in the testing population showed agreement between the two models and the robustness of the clusters (Jaccard score 0·51; balanced accuracy 0·58; appendix p 30). Individuals in the wild-test-A, wild-test-B, and wild-test-D clusters showed a similar symptom profile to those in the training set. The highest-prevalence symptoms were similar between the training and testing sets, with a high prevalence of chest pain in wild-test-A, dyspnoea in wild-test-B, and diarrhoea in wild-test-D (among other common symptoms; appendix pp 29–30). The testing cluster wild-test-C seemed qualitatively different from the training cluster wild-C yet showed good agreement with the classification obtained from the training set (appendix p 30). Cluster errors between the original and retrained models were similar for the two models in clusters wild-test-A and wild-test-B, but higher in the retrained model for clusters wild-test-C and wild-test-D (median intra-cluster error 343·6 vs 341·2 for wild-test-A, 657·1 vs 650·4 for wild-test-B, 465·4 vs 768·6 for wild-test-C, and 528·6 vs 649·4 for wild-test-D).
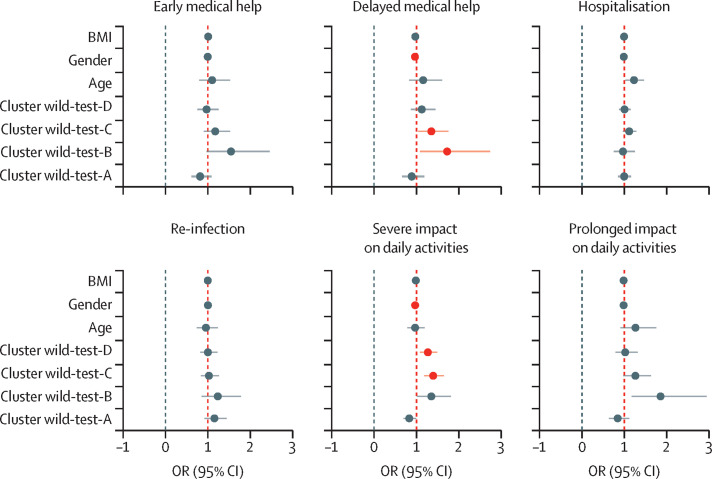
For the CSS Biobank sample, we assessed the relation between cluster classification and retrospective self-assessments of post-COVID-19 condition. Individuals in the wild-B cluster had increased odds compared with all other clusters of seeking delayed medical help for long-lasting symptoms (OR 1·75 [95% CI 1·06–2·76]; figure 5 ). People with post-COVID-19 condition in cluster wild-test-C had significantly increased odds compared with all other clusters of seeking delayed medical help for post-COVID-19 condition symptoms (1·43 [1·09–1·80]; figure 5). Individuals in clusters wild-test-C (1·42 [1·20–1·67]) and wild-test-D (1·29 [1·1–1·51]) had increased odds of severe impact on daily life (figure 5); these clusters were strongly associated with abdominal and central neurological symptoms, particularly during the first weeks of illness (Figure 1, Figure 2).
Figure 5.
Relation between predicted symptom clusters of post-COVID-19 condition caused by wild-type SARS-CoV-2 and each variable included in the multivariate model
Data points are coloured red to represent significance after false discovery rate correction for a=0·05. More detail about the questions on which these data were based is in the appendix (p 6).
Apart from the cluster classification, gender (men as reference) was the covariate with the biggest effect on the daily experience of post-COVID-19 condition among CSS Biobank participants, with women significantly more likely than men to seek delayed medical help and report severe impact on daily activities than men (after false discorvery rate correction; figure 5). Despite the significance of this covariate in the multivariate model, we found a decreased association for men (OR 0·991 [95% CI 0·985–0·998]) for prolonged illness (ie, delayed seeking of medical help and severe impact on daily activities combined), suggesting that gender is a significant covariate to explain the overall variance of the response variable.
Overall, individuals belonging to the wild-B cluster reported the greatest impact of post-COVID-19 condition in their daily experience (figure 5).
Discussion
In this study, we profiled phenotypes of post-COVID-19 condition associated with the wild-type, alpha, and delta variants of SARS-CoV-2 on the basis of a longitudinal sample of self-reported symptoms from the CSS. We identified distinct symptom profiles (or clusters), suggesting the existence of heterogeneous profiles of post-COVID-19 condition caused by these different SARS-CoV-2 strains. However, across variants, three groups of symptoms clustered consistently and were reproduced in a test dataset with additional outcome data: a primary cluster dominated by central neurological symptoms, a second cluster dominated by cardiorespiratory symptoms, and a third more heterogeneous cluster showing systemic inflammatory symptoms. This Article and our results show the value of digital health applications, such as symptom self-reporting, in the understanding and profiling of a large-scale pandemic.
Our study is among the first large-scale studies to define, in an unsupervised fashion, symptom clusters in adults with post-COVID-19 condition. The Post-Hospitalisation COVID-19 Study,19 in which long-term symptoms were assessed in patients who had been hospitalised with COVID-19, identified four symptom clusters related to physical effects and symptom severity: mild physical health impairment, moderate physical health impairment, severe mental and physical health impairment, and very severe mental and physical health impairment. Also in agreement with our findings, the National Core Study for Health and Wellbeing20 identified two symptom clusters among people in the general population at least 12 weeks after infection with SARS-CoV-2: a high symptom burden cluster and a low symptom burden cluster. The methods used in the National Core Study for Health and Wellbeing differed from those used in our study. All individuals with and without COVID-19 were included in the modelling in that study, but only 135 people reported functional limitation (ie, being unable to function as normal) after COVID-19 illness for more than 12 weeks, limiting power to identify different clusters of post-COVID-19 condition. Another study,21 which was focused on characterisation of post-COVID-19 condition in people who were hospitalised with COVID-19, identified central neurological, cardiorespiratory, and systemic and inflammatory symptoms, including gastrointestinal disorders, malaise, fatigue, musculoskeletal pain, and anaemia.
In our study, the central neurological cluster was characterised by symptoms such as anosmia and dysosmia, fatigue, brain fog, depression, delirium, and headache. This profile of symptoms in post-COVID-19 condition corresponded to the largest cluster among both unvaccinated people infected with the alpha variant and vaccinated people infected with the delta variant, and was the second largest cluster for unvaccinated people infected with the wild-type variant. Other studies have also suggested neurological involvement in post-COVID-19 condition. Notably, a study using UK Biobank data22 identified significant longitudinal effects when they compared brain-imaging data for people with post-COVID-19 condition before and after infection with SARS-CoV-2. People developing post-COVID-19 condition had reduced grey matter thickness, tissue damage in regions functionally connected to the primary olfactory cortex, and reduced global brain size.22 Furthermore, imaging of the limbic system showed the degenerative spread of SARS-CoV-2 via olfactory pathways,22 which could explain why anosmia and dysosmia are among the main symptoms of post-COVID-19 condition, as identified in our study.
The second common cluster consisted of cardiorespiratory symptoms and was the largest cluster in the wild-type period when all people were unvaccinated. This cluster could reflect lung damage, which has been identified in other studies23, 24 and is suggested in our data by the patients' reporting of dyspnoea and chest pain. A previous study23 has also identified cardiorespiratory syndrome with a high prevalence of dyspnoea, fatigue, palpitations, and chest pain in people with post-COVID-19 condition. Other studies are attempting to identify respiratory lesions in some individuals with post-COVID-19 condition, which could explain the ongoing respiratory symptoms identified in our population.24
The third common cluster was distinguished by systemic, inflammatory, and abdominal symptoms, as have been reported in previous studies of post-COVID-19 condition.23, 25 Other symptoms, reported less frequently, distinguished further clusters that differed across variants and vaccination status. Although our inability to qualitatively replicate these clusters across variants could indicate limited statistical power, it could also reflect differences in underlying processes related to the variants' properties. Some of these rarer symptoms (eg, palpitations, dizziness, and brain fog) have previously been reported in relation to long COVID phenotypes.23
Previous work26, 27 showed that vaccination reduces the risk of severe COVID-19, hospitalisation, and long COVID. Our data suggest that symptom duration and prevalence among those who develop post-COVID-19 condition do not differ significantly between unvaccinated people infected with the wild-type or alpha variant and vaccinated people infected with the delta variant. The timing of vaccination, number of doses, and vaccine type were not analysed in this study, preventing us from drawing conclusions about the relation between vaccination and post-COVID-19 effects or disentangling the normal weaning down of symptoms from the protective effects of vaccination.
The clinical relevance of our clustering results was supported by retrospective questionnaire data about the lived experience of post-COVID-19 condition. The main result from this analysis was that symptom clusters we identified differed in their relation to individuals' daily activities. We found that specific symptom profiles, such as clusters wild-B (mainly associated with systemic symptoms) and wild-test-C (mainly associated with respiratory symptoms), were related to more severe forms of post-COVID-19 conditions, which probably led people to seek medical help and to report prolonged effects on their daily lives. Cluster wild-B was also related to an overall high impact on the daily lives of people with post-COVID-19 condition. These findings also suggest that such groups of symptoms with additional neurological symptoms, such as fatigue, delirium, anosmia, and headache, could potentially be associated with prolonged disease in people with post-COVID-19 condition. Men had a decreased risk of prolonged effects of post-COVID-19 condition having a severe impact on daily activities. These results agree with those of a previous study28 of the relation between post-COVID-19 condition and gender, in which women were more likely to develop the condition than men, and had a higher prevalence of symptoms and longer symptom duration. Therefore, our clusters could be clinically useful in the future, by providing information about the potential outcomes and timescale of post-COVID-19 condition and the likelihood of needing long-term support.20 People with post-COVID-19 condition and their families (ie, the CSS Biobank volunteer advisory panel) broadly recognised the characteristics of the clusters presented here, further adding face validity.
The primary strength of this work is its uniqueness: ours is the first study to profile post-COVID-19 condition across different SARS-CoV-2 variants and in patients with varying vaccination status. The model we developed was trained on a large population and subsequently validated with a testing sample. The large sample size and the broad range of symptoms reported mean that our results could be generalised to other populations. Finally, we our results could provide meaningful insights for people with post-COVID-19 condition and enhance their understanding of their illness.
However, our study also has several limitations. The CSS population is not fully representative of the UK population (middle-aged people, women, people with higher educational status, and health-care workers are over-represented in the CSS).16, 29 We considered that this skew could have affected the likelihood of an infected person getting tested for COVID-19. However, we did not address that limitation, because our work was not focused on acute COVID-19, which might go undetected when testing is not done. We also acknowledge that different economic and cultural experiences could influence the presentation of COVID-19 and reporting of symptoms.29 Additionally, our analyses are UK-based, which limits international generalisability. Phenotypes of post-COVID-19 condition could differ genetically, environmentally, and socially, and further samples in different populations are needed. Furthermore, symptom profiles could be affected by pre-existing comorbidities.
We did not consider the ethnicity of participants as a possible covariate in the model or as a confounding effect. Future work should focus on the validation of the proposed approach in different populations with different demographic features, and on possible causes for such symptoms linked with individuals' comorbidities and demographic features. We did not do comprehensive analyses of vaccination status (including timing, number of doses, and vaccine type) and infection with the omicron variant of SARS-CoV-2, preventing additional conclusions about vaccination and subsequent variant behaviour. Future work should focus on the effect of vaccination on the development and profiling of long COVID. Our model included only the symptoms assessed by direct questioning in the CSS app, and we did not include symptoms reported as free text. Thus, we might have overlooked other symptoms that could modify the clusters. In addition, app questions were updated as the pandemic evolved, producing a difference in symptom numbers across the pandemic waves, which could affect comparison of symptom profiles across variants.
We excluded participants with extensive missing data (reports missing for more than 7 days) and conducted symptom interpolation for individuals with missing reporting for less than 7 days, which could have affected the profile of the disease. However, we consider such effects negligible; we interpolated a maximum of 7 days of missing data among 30 weeks of reported symptom data. Furthermore, the heterogeneous duration of illness across participants could have affected our analyses of symptom profiles, but was partially corrected for by the interpolation of each symptom up to the median duration within each cluster. Less strict inclusion criteria could have increased the sample size and possibly the robustness of the results. However, our strict inclusion criteria conservatively minimised possible sources of bias. Finally, future work should be focused on assessment of the effect of the symptom clusters identified on individual experiences of post-COVID-19 condition caused by vaccinated alpha and unvaccinated delta.
We used a data-driven approach to identify distinct self-reported symptom clusters in people with more than 12 weeks of persisting symptoms after SARS CoV-2 infection. Our findings could have relevance for people with post-COVID-19 condition, their health practitioners, and health-care services by providing data to verify the illness and that could help to manage expectations during the disease course. Our work adds to the emerging evidence that post-COVID-19 condition might have distinct subtypes, possibly with differing pathophysiology. An individual's post-COVID-19 condition could relate to a specific central or neurological process, to respiratory damage, or to a systemic inflammatory cause. Our study suggests that future investigations into mechanisms underlying post-COVID-19 condition should consider dividing affected individuals into different subgroups, which could increase the ability to identify distinct processes underlying these symptom clusters.
Data sharing
Data collected in the COVID Symptom Study app are shared with other health researchers through the UK National Health Service-funded Health Data Research UK and Secure Anonymised Information Linkage consortium, housed with the UK Secure Research Platform (Swansea, UK). Anonymised data can be made available to researchers, provided that they share their protocols and their research is in the public interest. Requests for access should be made via the Health Data Research Innovation Gateway (https://web.www.healthdatagateway.org/dataset/fddcb382-3051-4394-8436-b92295f14259).
Declaration of interests
AM, LP, and JCP are employees of ZOE. AM and JCP hold stocks in ZOE. CJS reports consultancy fees from ZOE. All other authors declare no competing interests.
Acknowledgments
Acknowledgments
We thank the study participants (including those who shared opinions about our work and how could we improve it to understand symptom progression and recovery) and the CSS Biobank volunteer advisory panel for their feedback on the significance of this work. This research was partly funded by the Wellcome Trust (215010/Z/18/Z), the Wellcome Engineering and Physical Sciences Research Council Centre for Medical Engineering at King's College London (WT 203148/Z/16/Z), Engineering and Physical Sciences Research Council (EP/T022205/1), and the UK Department of Health via the UK National Institute for Health and Care Research comprehensive Biomedical Research Centre award to Guy's & St Thomas's NHS Foundation Trust in partnership with King's College London and King's College Hospital NHS Foundation Trust. Further support was provided by the Chronic Disease Research Foundation (CDRF-22/2020) and the UK Department of Health via the National Core Studies, an initiative funded by UK Research and Innovation, the National Institute for Health and Care Research, and the Health and Safety Executive. The COVID-19 Longitudinal Health and Wellbeing National Core Study was funded by the UK Medical Research Council (MC_PC_20030, COV-LT-0009). The work was further supported by a grant from the UK Department of Health and Social Care to ZOE. We also acknowledge support from the UK Research and Innovation London Medical Imaging and Artificial Intelligence Centre for Value-Based Healthcare. Investigators also received support from the Wellcome Flagship Programme (WT213038/Z/18/Z and WT212904/Z/18/Z), the UK Medical Research Council, the British Heart Foundation, the UK Alzheimer's Society (AS-JF-17–011), the EU, the NIHR, and the NIHR-funded BioResource, Clinical Research Facility and Biomedical Research Centre at Guy's and St Thomas's NHS Foundation Trust (in partnership with King's College London). SO was supported by the French Government through the 3IA Côte d'Azur Investments in the Future project managed by the National Research Agency (reference number ANR-19-P3IA-0002).
Contributors
LSC, CHS, CJS, ELD, and MM conceived and designed the study. LSC, JD, and CHS acquired the data. LSC and MM analysed the data, which were interpreted by all authors. LSC, EM, AH, ELD, CJS, and MM drafted the Article, which was critically revised by all authors. CJS and MM supervised the study. All the authors had access to the raw data, which were verified by LSC and MM. ELD, CJS, and MM were responsible for the decision to submit for publication.
Supplementary Material
References
- 1.UK National Health Service Coronavirus (COVID-19) 2020. https://www.nhs.uk/conditions/coronavirus-covid-19/
- 2.Ayoubkhani D, Pawelek P. Office for National statistics; London: 2022. Prevalence of ongoing symptoms following coronavirus (COVID-19) infection in the UK 3 February 2022. [Google Scholar]
- 3.Antonelli M, Pujol JC, Spector TD, Ourselin S, Steves CJ. Risk of long COVID associated with delta versus omicron variants of SARS-CoV-2. Lancet. 2022;399:2263–2264. doi: 10.1016/S0140-6736(22)00941-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.UK National Institute for Health and Care Excellence COVID-19 rapid guideline: managing the long-term effects of COVID-19. 2021. https://www.nice.org.uk/guidance/NG188 [PubMed]
- 5.UK Office for National Statistics Prevalence of ongoing symptoms following coronavirus (COVID-19) infection in the UK: 22 June 2022. 2022. https://www.ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/conditionsanddiseases/bulletins/prevalenceofongoingsymptomsfollowingcoronaviruscovid19infectionintheuk/6may2022
- 6.Nurek M, Rayner C, Freyer A, et al. Recommendations for the recognition, diagnosis, and management of long COVID: a Delphi study. Br J Gen Pract. 2021;71:e815. doi: 10.3399/BJGP.2021.0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Notarte KI, Catahay JA, Velasco JV, et al. Impact of COVID-19 vaccination on the risk of developing long-COVID and on existing long-COVID symptoms: a systematic review. eClinicalMedicine. 2022;53 doi: 10.1016/j.eclinm.2022.101624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Department of Health and Social Care. Throup M, Javid S, Johnson B. UK marks one year since deploying world's first COVID-19 vaccine. 2021. https://www.gov.uk/government/news/uk-marks-one-year-since-deploying-worlds-first-covid-19-vaccine
- 9.Yong SJ, Liu S. Proposed subtypes of post-COVID-19 syndrome (or long-COVID) and their respective potential therapies. Rev Med Virol. 2021;32 doi: 10.1002/rmv.2315. [DOI] [PubMed] [Google Scholar]
- 10.Thygesen JH, Tomlinson C, Hollings S, et al. COVID-19 trajectories among 57 million adults in England: a cohort study using electronic health records. Lancet Digit Health. 2022;4:e542–e557. doi: 10.1016/S2589-7500(22)00091-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jesuthasan A, Massey F, Manji H, Zandi MS, Wiethoff S. Emerging potential mechanisms and predispositions to the neurological manifestations of COVID-19. J Neurol Sci. 2021;428 doi: 10.1016/j.jns.2021.117608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitfield E, Coffey C, Zhang H, et al. Axes of prognosis: identifying subtypes of COVID-19 outcomes. AMIA Annu Symp Proc. 2022;2021:1198–1207. [PMC free article] [PubMed] [Google Scholar]
- 13.Drew DA, Nguyen LH, Steves CJ, et al. Rapid implementation of mobile technology for real-time epidemiology of COVID-19. Science. 2020;368:1362–1367. doi: 10.1126/science.abc0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N Engl J Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.COG-UK Variants of concern (VOC) and under investigation (VUI) and any other variant by weeks and days. 2023. https://sars2.cvr.gla.ac.uk/cog-uk/
- 16.Sudre CH, Lee KA, Lochlainn MN, et al. Symptom clusters in COVID-19: a potential clinical prediction tool from the COVID Symptom Study app. Sci Adv. 2021;7 doi: 10.1126/sciadv.abd4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H. Multivariate time series clustering based on common principal component analysis. Neurocomputing. 2019;349:239–247. [Google Scholar]
- 18.Murray B, Kerfoot E, Chen L, et al. Accessible data curation and analytics for international-scale citizen science datasets. Sci Data. 2021;8:297. doi: 10.1038/s41597-021-01071-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans RA, McAuley H, Harrison EM, et al. Physical, cognitive, and mental health impacts of COVID-19 after hospitalisation (PHOSP-COVID): a UK multicentre, prospective cohort study. Lancet Respir Med. 2021;9:1275–1287. doi: 10.1016/S2213-2600(21)00383-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowyer RCE, Huggins C, Toms R, et al. Characterising patterns of COVID-19 and long COVID symptoms: evidence from nine UK longitudinal studies. Eur J Epidemiol. 2023;38:199–210. doi: 10.1007/s10654-022-00962-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Aly Z, Xie Y, Bowe B. High-dimensional characterization of post-acute sequelae of COVID-19. Nature. 2021;594:259–264. doi: 10.1038/s41586-021-03553-9. [DOI] [PubMed] [Google Scholar]
- 22.Douaud G, Lee S, Alfaro-Almagro F, et al. SARS-CoV-2 is associated with changes in brain structure in UK Biobank. Nature. 2022;604:697–707. doi: 10.1038/s41586-022-04569-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jamal SM, Landers DB, Hollenberg SM, et al. Prospective evaluation of autonomic dysfunction in post-acute sequela of COVID-19. J Am Coll Cardiol. 2022;79:2325–2330. doi: 10.1016/j.jacc.2022.03.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Couzin-Frankel J. Clues to long COVID. Science. 2022;376:1271–1275. doi: 10.1126/science.add4297. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian A, Nirantharakumar K, Hughes S, et al. Symptoms and risk factors for long COVID in non-hospitalized adults. Nature Med. 2022;28:1706–1714. doi: 10.1038/s41591-022-01909-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antonelli M, Penfold RS, Merino J, et al. Risk factors and disease profile of post-vaccination SARS-CoV-2 infection in UK users of the COVID Symptom Study app: a prospective, community-based, nested, case-control study. Lancet Infect Dis. 2022;22:43–55. doi: 10.1016/S1473-3099(21)00460-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venkatesan P. Do vaccines protect from long COVID? Lancet Respir Med. 2022;10:e30. doi: 10.1016/S2213-2600(22)00020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ortona E, Malorni W. Long COVID: to investigate immunological mechanisms and sex/gender related aspects as fundamental steps for tailored therapy. Eur Respir J. 2022;59 doi: 10.1183/13993003.02245-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Canas LS, Sudre CH, Capdevila Pujol J, et al. Early detection of COVID-19 in the UK using self-reported symptoms: a large-scale, prospective, epidemiological surveillance study. Lancet Digit Health. 2021;3:e587–e598. doi: 10.1016/S2589-7500(21)00131-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data collected in the COVID Symptom Study app are shared with other health researchers through the UK National Health Service-funded Health Data Research UK and Secure Anonymised Information Linkage consortium, housed with the UK Secure Research Platform (Swansea, UK). Anonymised data can be made available to researchers, provided that they share their protocols and their research is in the public interest. Requests for access should be made via the Health Data Research Innovation Gateway (https://web.www.healthdatagateway.org/dataset/fddcb382-3051-4394-8436-b92295f14259).