Abstract
The gene encoding sulfide-quinone reductase (SQR; E.C.1.8.5.′), the enzyme catalyzing the first step of anoxygenic photosynthesis in the filamentous cyanobacterium Oscillatoria limnetica, was cloned by use of amino acid sequences of tryptic peptides as well as sequences conserved in the Rhodobacter capsulatus SQR and in an open reading frame found in the genome of Aquifex aeolicus. SQR activity was also detected in the unicellular cyanobacterium Aphanothece halophytica following sulfide induction, with a Vmax of 180 μmol of plastoquinone-1 (PQ-1) reduced/mg of chlorophyll/h and apparent Km values of 20 and 40 μM for sulfide and quinone, respectively. Based on the conserved sequences, the gene encoding A. halophytica SQR was also cloned. The SQR polypeptides deduced from the two cyanobacterial genes consist of 436 amino acids for O. limnetica SQR and 437 amino acids for A. halophytica SQR and show 58% identity and 74% similarity. The calculated molecular mass is about 48 kDa for both proteins; the theoretical isoelectric points are 7.7 and 5.6 and the net charges at a neutral pH are 0 and −14 for O. limnetica SQR and A. halophytica SQR, respectively. A search of databases showed SQR homologs in the genomes of the cyanobacterium Anabaena PCC7120 as well as the chemolithotrophic bacteria Shewanella putrefaciens and Thiobacillus ferrooxidans. All SQR enzymes contain characteristic flavin adenine dinucleotide binding fingerprints. The cyanobacterial proteins were expressed in Escherichia coli under the control of the T7 promoter. Membranes isolated from E. coli cells expressing A. halophytica SQR performed sulfide-dependent PQ-1 reduction that was sensitive to the quinone analog inhibitor 2n-nonyl-4-hydroxyquinoline-N-oxide. The wide distribution of SQR genes emphasizes the important role of SQR in the sulfur cycle in nature.
Of the many organisms performing plant-type oxygenic photosynthesis, only cyanobacteria can facultatively shift to anoxygenic, bacterium-type photosynthesis with sulfide (H2S) as the electron donor in a photosystem I-dependent reaction (3, 15, 26). This shift, first discovered in the cyanobacterium Oscillatoria limnetica (11), occurs after 2 h of incubation in the presence of sulfide and light and requires protein synthesis (23). The induced cells perform sulfide-dependent CO2 fixation (10, 11, 15, 23), H2 evolution (5), or N2 fixation (4), depending on the growth and physiological conditions.
The discovery of anoxygenic photosynthesis in O. limnetica formed the basis for the understanding of an important and unique trait of cyanobacteria. In marked contrast to other plant-type phototrophs, which are sulfide sensitive, these organisms grow and form mats in anaerobic, sulfide-rich niches characteristic of many natural habitats and polluted water (25, 26, 38). The unique capacity to shift between the two types of photosynthesis has been suggested to represent a primitive relic in the evolution of photosynthesis (25).
Photooxidation of sulfide coupled to CO2 reduction is not unique to cyanobacteria. Sulfide is the most widely used electron donor among photolithotrophic bacteria (7, 8, 17). The transfer of electrons from sulfide directly into the quinone pool was proposed and supported by the inhibition exerted by quinone analogs as well as energetic considerations (6, 8, 42).
Tracking of the inducible factor that enables photosynthetic sulfide oxidation in O. limnetica led to the discovery of sulfide-quinone reductase (SQR; E.C.1.8.5.′), a novel enzyme that transfers electrons from sulfide into the quinone pool (36). The SQR was solubilized from membranes of sulfide-induced O. limnetica cells and purified in an active form (2). The isolated active SQR is a hydrophobic membrane enzyme composed of a single polypeptide with an apparent molecular mass of 57 kDa (as determined by sodium dodecyl sulfate [SDS]-polyacrylamide gel electrophoresis [PAGE]). It was shown to have high affinities for sulfide (Km = 8 μM) and quinone (Km = 31 μM for plastoquinone-1 [PQ-1]), to contain a flavin cofactor, and to be sensitive to quinone analogs and KCN (2, 34). The N-terminal sequence was found to contain the characteristic features of an NAD or flavin adenine dinucleotide (FAD) binding domain (2, 44).
SQR recently has been proven to be widely spread among anoxygenic phototrophs. SQR activity has been detected in purple “nonsulfur” bacteria (Rhodobacter capsulatus) (35), purple sulfur bacteria (Allochromatium vinosum) (34), green sulfur bacteria (Chlorobium) (37), and green gliding (“nonsulfur”) bacteria (Chloroflexus aurantiacus) (34); in the nonphotosynthetic chemoautotrophs Paracoccus denitrificans (31), Wolinella succinogenes, and Aquifex aeolicus; and in the mitochondria of the sulfide-tolerating marine worm Arenicola marina (34).
The SQR of the purple bacterium R. capsulatus was isolated and purified, its gene was cloned, sequenced, and functionally expressed in Escherichia coli (33) and, recently, it was shown that the enzyme is essential for the sulfide-dependent growth of R. capsulatus (32). The R. capsulatus SQR polypeptide consists of 427 amino acids and has a molecular mass of 47 kDa and a net charge of +1. The amino acid sequence of its N terminus shows high similarity (48% identity and 72% similarity) to the amino acid sequence of the O. limnetica N terminus (2), including the FAD binding fingerprint. The complete protein sequence contains two additional FAD binding motifs (33). The Rhodobacter and the cyanobacterial enzymes are rather divergent. For example, they are expected to differ in their quinone binding sites. The natural quinone acceptor for SQR is most probably ubiquinone in R. capsulatus but plastoquinone in the cyanobacterial system (34).
Recently, a gene encoding a mitochondrial polypeptide that exhibits SQR activity was cloned from the fission yeast Schizosaccharomyces pombe. This enzyme, HMT2 (heavy metal tolerance), was proposed to function in the detoxification of endogenous sulfide (43). HMT2 shares high similarity with sequences of unknown functions from the genomes of nematodes, fruit flies, mice, rats, and humans and only low similarity (∼20%) with R. capsulatus SQR (43). In addition to demonstrating further the distribution of SQR in eukaryotes, this recent publication suggests two possible roles for SQR—utilization and detoxification of sulfide—raising the question of whether one type of enzyme or more types are involved.
We have previously shown three functions in which SQR can be involved in cyanobacteria: (i) anoxygenic photosynthetic growth in O. limnetica (23); (ii) anaerobic respiration in O. limnetica (24); and (iii) detoxification of sulfide in Aphanothece halophytica, which survives but does not grow in the presence of sulfide (15). In contrast to O. limnetica, in which the SQR has been purified and biochemically characterized, in A. halophytica the sulfide-interacting enzyme has not yet been identified. In the present work, we describe some biochemical properties of A. halophytica SQR as well as the cloning and expression of the sqr genes of both O. limnetica and A. halophytica.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
The list of strains and plasmids used is provided in Table 1. O. limnetica was grown in aerobic growth medium (3) at 37°C as described previously (1). A. halophytica was grown in the same medium supplemented with 10 μg of vitamin B12 per liter in batches of 100 to 500 ml at 35°C without shaking. E. coli strains were grown in Luria-Bertani (LB) broth (Difco, Detroit, Mich.) or minimal medium A (12) at 37°C, unless otherwise indicated. Antibiotics were added to final concentrations of 100 μg/ml (ampicillin), 50 μg/ml (kanamycin), or 12.5 μg/ml (tetracycline).
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Characteristics or history | Source or reference |
|---|---|---|
| Strains | ||
| Oscillatoria limnetica | Filamentous | 15 |
| Aphanothece halophytica | Unicellular | 15 |
| Escherichia coli | ||
| JM109 | e14−(McrA−) recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15] | Stratagene |
| XL1 Blue MRF′ | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)]c | Stratagene |
| SOLR | e14−(McrA−) Δ(mcrCB-hsdSMR-mrr)171 sbcC recB recJ uvrC umuC::Tn5 (Kanr) lac gyrA96 relA1 thi-1 endA1 λR [F′ proAB lacIqZΔM15]c Su− (nonsuppressing) | Stratagene |
| BL21 (DE3) | E. coli B F−dcm ompT hsdS(rB− mB−) gal λ(DE3) | 39 |
| Plasmids | ||
| pBS | Multicopy cloning and sequencing vector; confers Ampr | Stratagene |
| pBS-OL | pBS with EcoRI insert of O. limnetica sqr region | This work |
| pBS-AH | pBS with EcoRI insert of A. halophytica sqr region | This work |
| pT-OL | pBS with O. limnetica sqr PCR product | This work |
| pT-AH | pBS with A. halophytica sqr PCR product | This work |
| pT7-7 | Expression vector | 40 |
| pTOLsqr | pT7-7 with O. limnetica sqr ORF | This work |
| pTAHsqr | pT7-7 with A. halophytica sqr ORF | This work |
Induction of anoxygenic photosynthesis.
O. limnetica was induced with sulfide as described previously (1). To induce anoxygenic photosynthesis in A. halophytica, cells of 5-day-old (exponential-growth-phase) cultures were harvested, washed and resuspended in anaerobic growth medium (3), and incubated in stoppered bottles flushed with N2. 3(3,4-Dichlorophenyl)-1,1-dimethylurea (10 μM) and NaHCO3 (2.5 mM) were added, and cells were induced by the addition of 1.25 mM Na2S. After 2 h of incubation at 35°C under cool-white fluorescent lamps (150 μE/m2/s), cells were washed in anaerobic growth medium and either rapidly frozen in liquid nitrogen and stored at −80°C or immediately assayed for SQR activity.
Spectroscopic assay of SQR.
SQR activity was measured as sulfide-dependent PQ-1 reduction recorded at 292 nm minus 266 nm in an SLM-Aminco DW-2000 dual-wavelength spectrophotometer as previously described (1).
Tryptic peptides of O. limnetica SQR.
SQR was purified from O. limnetica cells as previously described (2). Samples containing purified SQR were separated by SDS-PAGE and Coomassie blue stained. The gel was destained in 50% acetonitrile–200 mM ammonium bicarbonate, dried, and rehydrated with 100 mM ammonium bicarbonate containing modified trypsin (sequencing grade; Promega). After overnight incubation at 37°C, the resulting peptides were eluted from the gel, redissolved in 0.1% trifluoroacetic acid, and separated by reversed-phase high-pressure liquid chromatography on a Vydac C18 column (1 by 150 mm) with a linear gradient of 4 to 60% acetonitrile in 0.1% trifluoroacetic acid. The peptides were collected, and their amino acid sequence was determined (at the Protein Research Center, The Technion, Haifa, Israel) with a model 494A Peptide Sequencer (Applied Biosystems Division, Perkin-Elmer).
Solubilization of A. halophytica SQR.
Frozen induced cells were thawed and washed (i) with Turks Island salt solution prepared in double strength (15) and (ii) with buffer containing 20 mM sodium HEPES (pH 7.9), 0.3 mM sodium EDTA, 5 mM KCl, 5 mM MgCl2, 0.5 M glycine betaine, and 0.1 M sucrose. The washed cells were resuspended in the buffer, after the addition of bovine serum albumin (1 mg/ml) and lysozyme (3 mg/ml) to a final concentration of 25 μg of chlorophyll/ml, and incubated for 25 min at 35°C in the dark. The partially lysed cells were sonicated, and thylakoid membranes were prepared as previously described (1). Thylakoid membranes were washed and SQR was solubilized as previously described for O. limnetica SQR (2).
Gel electrophoresis and Western blotting.
SDS gel electrophoresis was carried out as described by Laemmli (19). Western blotting with polyclonal antibodies raised against denatured O. limnetica SQR (2) was carried out by use of an alkaline phosphatase detection system as previously described (30).
Isolation of cyanobacterial DNA.
Exponentially growing cyanobacterial cells in 100-ml batches were harvested and washed with 30 ml of buffer containing 50 mM Tris (pH 8.0) and 2 mM EDTA. The pellet was resuspended in 2 ml of buffer–0.8 ml of sterile glass beads (212 to 300 μm; Sigma G9143). An equal volume of phenol-chloroform mixture was added, and the cells were broken by four cycles each of 30 s of vigorous vortexing and 1 min of incubation on ice. The lysate was centrifuged at 12,000 × g for 10 min, the upper phase was collected, and the extracted DNA was washed with chloroform and precipitated with ethanol as described previously (30).
Cloning of SQR genes.
Based on tryptic peptides and conserved regions, fully degenerate primers were prepared for PCR amplification with genomic DNA as a template. For O. limnetica, the forward primer, 5′-CCIACIGCITA(CT)GA(AG)CT-3′ (P1), was designed on the basis of the previously identified (2) N-terminal polypeptide stretch P15TAYEL20 (Fig. 1), with inosine in the fully degenerate positions. The reverse primer, 5′-TG(AGCT)CC(AGCT)AC(AG)TA(AGCT)GG(CT)TC-3′ (P2), was designed on the basis of the peptide H196GVYPE191, which is fully conserved among R. capsulatus SQR (33), and an open reading frame (ORF) from A. aeolicus (13) (Fig. 1). This reverse primer was also used for A. halophytica, together with the forward primer, 5′-TT(CT)GG(AGCT)CC(AGCT)GC(AGCT)TA(CT)GAATT(CT)-3′ (P3), which was designed on the basis of the conserved sequence F160GPAYEF166 (Fig. 1). The PCR was performed with Q-biotaq enzyme (Quantum) by use of an Eppendorf Mastercycler 5330 apparatus, with various annealing temperatures (touchdown PCR [29]), starting at 48°C and reducing the temperature by 2°C every 10 cycles down to 38°C. Specific PCR products were purified, cloned into T-vector that was freshly prepared as described previously (21), and sequenced.
FIG. 1.
Sequence alignment of the known SQR enzymes and putative homologs. Multiple amino acid sequence alignment of the known SQR enzymes of A. halophytica, O. limnetica, and R. capsulatus (33) as well as the putative close homologs from A. aeolicus (13), S. putrefaciens (sequence number >4287 from the genome of S. putrefaciens in the TIGR bank), T. ferrooxidans (sequence number >62 from the genome of T. ferrooxidans in the TIGR bank), Anabaena PCC7120 (sequence number C279 from the genome of Anabaena in CyanoBase), and the distant homologs from S. pombe as well as the flavoprotein subunit of A. vinosum flavocytochrome c (FCC). Residues that are identical among at least four of the sequences are indicated by black shading. The conserved FAD binding domains are indicated by overlining. The two fully conserved cysteines are marked by asterisks.
Genomic DNA libraries were constructed using a Lambda ZAPII/predigested vector/Gigapack cloning kit (Stratagene). 32P-labeled probes were used to screen the libraries. Plasmids (pBS-OL from the O. limnetica library and pBS-AH from the A. halophytica library) were excised from positive plaques. Sequencing of DNA was conducted by use of an automated DNA sequencer (ABI PRISM 377; Perkin-Elmer).
Sequence analysis and polypeptide alignments.
Sequence analysis was done using Wisconsin Package version 9.0 (Genetics Computer Group, Madison, Wis.). Preliminary sequence data were obtained from The Institute for Genomic Research website at http://www.tigr.org and the CyanoBase website at http://www.kazusa.or.jp/cyano/search.html. Polypeptide alignments were done using ClustalW (http://www2.ebi.ac.uk/clustalw) and refined by visual inspection.
Expression of SQR in E. coli.
For expression of the cyanobacterial SQRs in E. coli, the genes were amplified from the respective plasmids by PCR using the following primers: 5′-CGGAATTCATATGGCACACGTTGCAGTTAT-3′ and 5′-TCCCCCGGGGGACATCTCTATTCCCTAGTCCC-3′ for O. limnetica SQR and 5′-CCGGAATTCCATATGGCACATATCGTAATTG-3′ and 5′-CGCGGATCCGCGCAATAACTTAAACCGACTGG-3′ for A. halophytica SQR. The amplified fragments were purified and ligated to freshly prepared T-vector, resulting in pT-OL and pT-AH, respectively. An NdeI-SmaI fragment from pT-OL or an NdeI-BamHI fragment from pT-AH was inserted into the NdeI-SmaI or NdeI-BamHI restriction sites of pT7-7, resulting in pTOLsqr or pTAHsqr, respectively. Plasmid amplifications were done with E. coli JM109.
For expression of SQR, E. coli BL21(DE3) was transformed with pTOLsqr or pTAHsqr and grown in LB broth at 37°C. Expression was induced at an optical density at 600 nm of 0.6 by the addition of 0.5 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG) and further incubation for 3 h. Expression levels were determined by SDS-PAGE analysis.
Membranes were prepared from E. coli cells as described previously (33) and either tested for SQR activity or frozen in liquid nitrogen. For solubilization of SQR, membranes (containing 100 to 300 μg of protein) were resuspended in 1.15 ml of buffer containing 60 mM choline chloride, 4.5 mM Tris-Cl (pH 8.0), 110 mM sucrose, 20% glycerol, 100 mM morpholinepropanesulfonic acid (MOPS) (pH 7.0), and 1.25% n-dodecyl-β-d-maltoside (DM). After 30 min of incubation on ice, the suspension was centrifuged for 25 min at 435,000 × g. The supernatant solution was collected (DM sup). The pellet was resuspended in 1 ml of lysis buffer (50 mM Tris-Cl [pH 8.0], 100 mM NaCl, 10 mM EDTA) containing 0.5% Triton X-100. Incubation and centrifugation were repeated as above. The supernatant solution was collected (Triton sup). The pellet was resuspended in 1 ml of lysis buffer containing 8 M urea and incubated and centrifuged as above. The supernatant solution was collected (Urea sup). The pellet was resuspended in 500 μl of lysis buffer (Urea pellet). The presence of SQR was determined by both Coomassie blue staining and Western blotting after separation of the proteins by SDS-PAGE.
RESULTS AND DISCUSSION
Cloning of Oscillatoria sqr.
The cloning of O. limnetica sqr was based on the following amino acid sequence information: (i) the N-terminal sequence (2); (ii) four sequences of tryptic peptides that were obtained from purified O. limnetica SQR; and (iii) information obtained from the complete genome sequence of the hyperthermophilic chemolithotrophic bacterium A. aeolicus, which belongs to the first branching-off lineage within the phylogenetic tree of bacteria (13). In the genome of this bacterium, an ORF was identified that encodes a protein with 38% identity and 57% similarity to the R. capsulatus SQR, a remarkable homology in view of the evolutionary distance between the two species (45) (see Fig. 6). Alignment of the two sequences revealed four fully conserved regions that were used for the cloning of O. limnetica sqr. Based on all of the amino acid sequences, fully degenerate primers were designed and used in different combinations in touchdown PCR as described in Materials and Methods to obtain a specific probe for cyanobacterial sqr. A 546-bp fragment that was obtained by PCR with primer P2 (based on a Rhodobacter-Aquifex conserved domain) and primer P1 (based on part of the N-terminal sequence) was found to contain a region coding for one of the tryptic peptides of O. limnetica SQR (D160RVPIT166; Fig. 1). This fragment was then used as a probe for screening an O. limnetica genomic library. The DNA of several positive clones was sequenced and found to contain an ORF that encodes a protein that includes all the amino acid sequences of O. limnetica SQR tryptic peptides. The GenBank accession number for the O. limnetica SQR gene is AF242368.
FIG. 6.
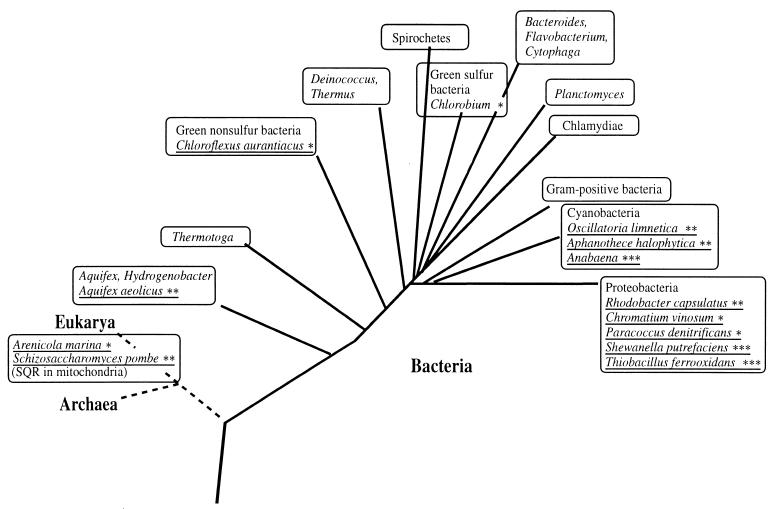
Distribution of SQRs. Schematic demonstration of the phylogenetic distribution of the SQRs known so far. Species that are already known to possess SQR are underlined. ∗, SQR detected only by membrane ∗∗, both membrane activity and cloned SQR genes; ∗∗∗, SQR-like ORFs.
SQR activity in A. halophytica.
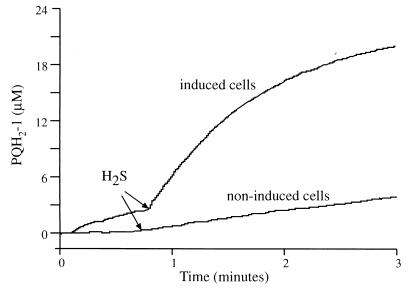
Cells of A. halophytica were previously shown to be capable of anoxygenic sulfide-dependent photosynthesis but incapable of growth on sulfide. Like that in O. limnetica, the anoxygenic reaction in A. halophytica requires sulfide induction (15). Therefore, we looked for SQR activity in A. halophytica cells that had been induced for 2 h in the presence of 1.25 mM sulfide and light. Unlike the situation in O. limnetica, in which SQR activity was observed only in thylakoids isolated from sulfide-induced cells, a high rate of PQ-1 reduction (180 μM PQ-1 reduced/mg of chlorophyll/h) was measured in intact induced cells of A. halophytica (Fig. 2). On a chlorophyll basis, this rate was about 50% higher than that reported previously for Oscillatoria thylakoids. Attempts to further increase the accessibility of the substrates (sulfide and PQ-1) by permeabilization of the cells with toluene, mild lysozyme treatment, or sonication had no effect.
FIG. 2.
SQR activity in A. halophytica. The assay mixture contained 10 mM potassium HEPES (pH 7.4), 10 mM MgCl2, 10 mM KCl, 30 μM PQ-1, and cells containing 3 μg of chlorophyll per ml. The reduction of PQ-1 was detected at A292 minus A266. Where indicated, the reaction was started by the injection of 60 μM Na2S. PQH2-1, reduced PQ-1.
To assess whether A. halophytica SQR is membrane bound, thylakoid membranes were prepared from sulfide-induced A. halophytica cultures by lysozyme treatment and mild sonication (1). Thylakoids were then washed with 5 mM sodium EDTA, and the SQR was solubilized with 10 mM sodium cholate and 25 mM DM as previously described for O. limnetica (2). Most of the SQR activity measured in intact cells or thylakoids was retained in the solubilized preparation (data not shown). The solubilized SQR had Km values of 20 and 40 μM for H2S and PQ-1, respectively, and was specifically inhibited by the quinone analog 2n-nonyl-4-hydroxyquinoline-N-oxide (NQNO) in the submicromolar range (data not shown).
Cloning of A. halophytica sqr.
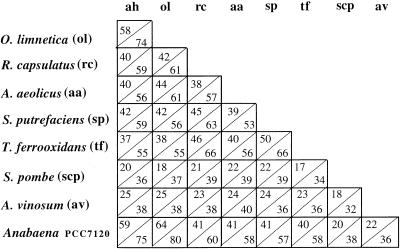
Alignment of the amino acid sequence of cloned O. limnetica SQR with those of R. capsulatus and A. aeolicus confirmed the conserved domains (Fig. 1). Based on these domains, a few more degenerate primers were constructed and used in touchdown PCR to obtain a probe specific for A. halophytica sqr. Primers P2 and P3 yielded a fragment of the expected size (111 bp). The nucleotide sequence of this fragment was 77% identical to that of O. limnetica sqr. This fragment was used as a probe for cloning the entire gene from the A. halophytica genomic library. The DNA of a few positive clones was sequenced and found to contain an ORF (GenBank accession no. AF242369) that encodes a protein that is 58% identical and 75% similar to O. limnetica SQR (Fig. 3).
FIG. 3.
Identity and similarity of SQR sequences. ah, A. halophytica. The number in the upper left corner of each box is the percent identity (from a Blast search), and the number in the lower right corner is the percent similarity (from a Blast search).
ORF downstream of sqr.
Downstream of each cyanobacterial sqr gene, a small ORF (beginning 3 bp after the termination codon of A. halophytica sqr or 195 bp downstream of the termination codon of O. limnetica sqr) encoding a 91-amino-acid peptide or a 100-amino-acid peptide was found (GenBank accession no. AF242371 and AF242370, respectively). The two ORFs share 63% identity and 69% similarity. A BLAST search for the proteins revealed highest scores (about 35% identity and 60% similarity) for several transcription factors from different sources, mostly bacteria and cyanobacteria. Since in both cyanobacterial systems SQR is an induced protein and regulatory proteins are often located in the vicinity of their targets of regulation, these small proteins may play a role in the regulation of SQR.
Expression of SQR in E. coli.
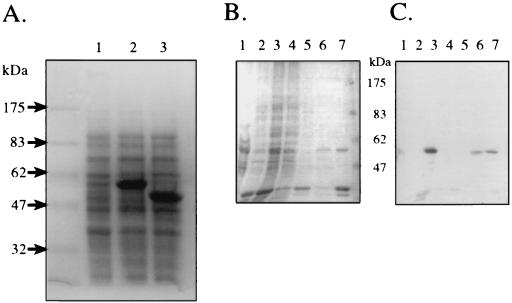
Since no molecular tools exist for either O. limnetica or A. halophytica, we expressed the cyanobacterial sqr genes in E. coli. Both genes were cloned into the pT7-7 vector under the control of the T7 promoter, yielding pTOLsqr and pTAHsqr, which contained the O. limnetica sqr and A. halophytica sqr genes, respectively (Table 1). E. coli BL21(DE3) cells containing a chromosomal T7 RNA polymerase gene inducible by IPTG were transformed with these plasmids. IPTG induction gave major bands of the expected apparent sizes (52 kDa for A. halophytica SQR and 57 kDa for O. limnetica SQR) on SDS-PAGE (Fig. 4A). In both cases, after cell disruption, these induced proteins were found in the membrane fraction (Fig. 4B). Polyclonal antibodies raised against denatured O. limnetica SQR (2) reacted with the major 57-kDa band derived from E. coli cells transformed with pTOLsqr specifically (Fig. 4B and C). The antibodies did not cross-react with the band corresponding to A. halophytica SQR (data not shown).
FIG. 4.
Overexpression of cyanobacterial SQR in E. coli (A) and solubilization (B) and antibody recognition (C) of overexpressed O. limnetica SQR. (A) Cells of E. coli BL21(DE3) transformed with either pTOLsqr or pTAHsqr were grown at 37°C in LB medium. At an optical density at 600 nm of 0.6, IPTG was added to 0.5 mM. After 2 h, 0.5-ml cultures were centrifuged, resuspended in 2% SDS loading buffer, and boiled for 5 min. Samples of 15 μl were loaded on the gels. Lane 1, noninduced BL21(DE3)/pTOLsqr; lane 2, induced BL21(DE3)/pTOLsqr; lane 3, induced BL21(DE3)/pTAHsqr. (B and C) Coomassie blue staining (B) and Western blotting (C) with polyclonal antibody raised against denatured O. limnetica SQR (2). Lane 1, O. limnetica membranes (containing 2 μg of chlorophyll); lane 2, noninduced BL21(DE3)/pTOLsqr membranes; lane 3, as in lane 2 but IPTG induced; lane 4, DM sup; lane 5, Triton sup; lane 6, Urea sup; lane 7, Urea pellet (see Materials and Methods). All samples loaded in lanes 2 to 7 were derived from membranes containing 40 μg of protein.
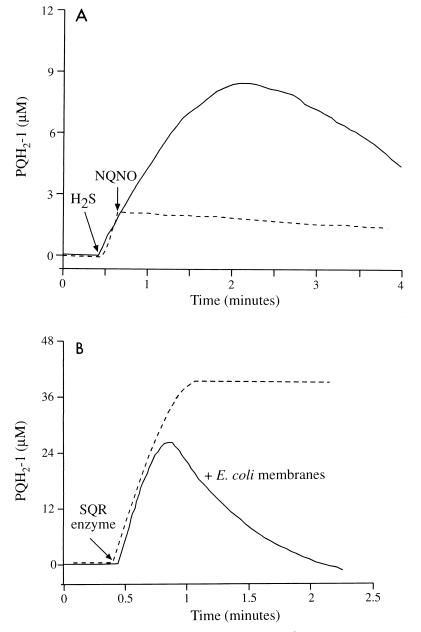
Using standard induction conditions, the overexpressed SQR accumulated in the cells in inclusion bodies in an inactive form, as indicated by the fact that the enzyme was solubilized from the E. coli membranes only in the presence of 8 M urea (30) (Fig. 4B and C, lane 6) and not by detergent treatments (DM and Triton X-100; Fig. 4B and C, lanes 4 and 5, respectively). With minimal medium A (rather than LB broth) and a lower growth and induction temperature (30°C rather than 37°C), SQR activity could be detected in membrane preparations of E. coli cells expressing A. halophytica SQR (Fig. 5A). SQR activity was not detected in membranes prepared from noninduced E. coli cells (data not shown). The estimated specific activity of the heterologously expressed A. halophytica SQR enzyme was 3.0 μmol of PQ-1 reduced/mg of SQR protein/min or 142 mol of PQ-1 reduced/mol of SQR/min. The calculation is based on the measured SQR activity in the induced E. coli membranes and on the estimated protein content of the distinct band (52 kDa) in the same membrane preparation. This activity of the heterologously expressed membrane-bound A. halophytica SQR is comparable with the activities of the purified O. limnetica SQR (1.9 μmol of PQ-1 reduced/mg of protein/min) (2) and the R. capsulatus SQR expressed in E. coli (4.0 μmol of decyl-UQ reduced/mg of protein/min) (33). SQR activity was completely inhibited by 1 μM quinone analog NQNO (Fig. 5A), as expected for cyanobacterial SQR activity (1, 2).
FIG. 5.
SQR activity in membranes of E. coli expressing A. halophytica SQR (A) and kinetics of PQ-1 reduction and reoxidation in the presence of E. coli membranes (B). (A) Membranes of induced BL21(DE3)/pTAHsqr (60 μg of protein) were incubated under anaerobic conditions in an assay mixture containing 10 mM potassium HEPES (pH 7.4), 10 mM MgCl2, 10 mM KCl, and 50 μM PQ-1. The reaction was started by the addition of 60 μM Na2S. NQNO (1 μM) was added where indicated. (B) An assay mixture containing 60 μM H2S was incubated under anaerobic conditions in either the presence (———) or the absence (–––) of noninduced E. coli membranes (60 μg of protein). Partially purified SQR obtained from O. limnetica thylakoids (40 μg of chlorophyll) was added where indicated. PQH2-1, reduced PQ-1.
As shown in Fig. 5A, in the heterologous expression system, the quinol (PQH2-1) is reoxidized. The apparent reoxidation of the quinol results from a competitive quinol oxidant that was observed in the E. coli preparations. This unidentified oxidant was apparent also when E. coli membranes were added to the assay mixture of purified O. limnetica SQR (Fig. 5B). It was found to be insensitive to either anaerobiosis or KCN (up to 0.2 mM) but was lost upon boiling of the E. coli membranes. This oxidant is membrane bound, as it cannot be washed away but can be extracted by detergents (e.g., DM). Hence, the calculated activity of the expressed A. halophytica SQR is underestimated.
Cyanobacterial SQRs.
The two SQR polypeptides deduced from the gene sequences consist of 436 (O. limnetica SQR) and 437 (A. halophytica SQR) amino acid residues, with molecular weights of 47,744 and 48,205, respectively. Both values are significantly smaller than the apparent molecular weights obtained by SDS-PAGE (Fig. 4A). Aberrant mobility in SDS-PAGE is a property characteristic of membrane proteins (41). Indeed, cyanobacterial SQR behaves as an integral membrane protein, being solubilized in the presence of the detergent DM (2). Structure prediction analysis with PREDATOR (www.embl-heidelberg.de/cgi/predator.serv.pl) predicts 25% α-helix and 18% β-sheet structures for O. limnetica SQR and 38% α-helix and 9% β-sheet structures for A. halophytica SQR. However, hydropathy analysis with the Kyte-Doolittle hydrophobicity scale did not predict any membrane-spanning α helix. The theoretical isoelectric points are 7.7 for O. limnetica SQR and 5.6 for A. halophytica SQR, and the net charges at a neutral pH are 0 and −14, respectively. Both cyanobacterial enzymes contain, at their N termini, all 11 fingerprint residues of the βαβ ADP binding site motif characterizing the NAD or FAD binding proteins, with the exception that the 10th fingerprint residue is shifted by one position (44). Two additional segments that were suggested to be involved in FAD binding in the R. capsulatus SQR (33) and other enzymes (14) are partially conserved in the cyanobacterial enzymes (Fig. 1).
Although the two cyanobacterial species display different phenotypes in a sulfide-rich environment—O. limnetica growing anaerobically (23) and A. halophytica only surviving (15)—they share the same initial step in the sulfide oxidation pathway, which is catalyzed by the SQR enzyme. The results presented in this work show that the two enzymes are similar: the Km values of A. halophytica SQR for sulfide and PQ-1 are 20 and 40 μM, respectively, while those of purified O. limnetica SQR are 8 and 31 μM; respectively; both enzymes are inhibited by NQNO in the submicromolar range; and both enzymes are solubilized by the same protocol, previously described for O. limnetica SQR (2). Cloning of the cyanobacterial SQRs shows that the two enzymes are highly homologous. Hence, rather than being due to a difference in the SQR mechanism, the different growth phenotypes exhibited by O. limnetica and A. halophytica could be due to different SQR activity levels in cells, resulting from different expression levels. However, the data showing similar SQR activities are not in favor of this possibility. It therefore seems more likely that the different phenotypes are the outcome of other physiological parameters. For example, the two organisms may differ in unsaturated fatty acid composition and metabolism. O. limnetica possesses monounsaturated fatty acids that can be synthesized by the desaturation of long-chain fatty acids under aerobic as well as anaerobic conditions. A. halophytica possess polyunsaturated fatty acids that are synthesized only in an oxygen-dependent process (22), a possible reason for its obligate aerobic growth.
Alignment of SQRs and implications.
The cloning of O. limnetica sqr is the first molecular genetic work reported for O. limnetica. In A. halophytica, one gene, dnaK (D84421), had been cloned prior to sqr (20). In addition to these cyanobacterial sqr genes, the previously cloned R. capsulatus sqr gene (33), and the sqr-like ORF identified in the genome of A. aeolicus (13), a data bank search revealed two new sqr-like ORFs among the unfinished sequences of the TIGR bank (Fig. 3). An ORF with 42% identity and 56% similarity to O. limnetica sqr was found in the genome of Shewanella putrefaciens (sequence number >4287), and another ORF, with 38% identity and 55% similarity, was found in the genome of Thiobacillus ferrooxidans (sequence number >62). These two species are chemolithotrophic proteobacteria that can utilize sulfide as an electron donor. An additional sqr-like ORF was identified in the genome of the filamentous cyanobacterium Anabaena PCC7120 and recently published in CyanoBase (sequence number C279). The Anabaena ORF is 64% identical and 80% similar to O. limnetica sqr (Fig. 3). It has been previously suggested that sulfide is formed in Anabaena variabilis by the reduction of sulfate in vegetative cells and translocated into heterocysts, where it is utilized (16). The occurrence of SQR in Anabaena may suggest the role of sulfide as an electron donor in heterocysts, where photosystem II is known to be inactive.
Alignment of the deduced amino acid sequences from these seven prokaryotic genes allows the identification of conserved domains with possible functional or structural significance (Fig. 1). (i) All seven SQR genes encode proteins with FAD binding fingerprints at their N termini (Fig. 1) as well as two other FAD binding domains that were first recognized in R. capsulatus SQR (33). (ii) Recently, based on crystallographic data for several quinone binding proteins and sequence analysis, a structural element for the quinone binding site has been suggested (28). It is composed of a helical stretch that flanks one side of the quinone headgroup and contains a triad of close contact residues. The central residue of the triad in all cases is a conserved histidine which hydrogen bonds to one carbonyl of the quinone. The second residue, four amino acids upstream of the histidine, is an aliphatic amino acid that is in close proximity to the hydrophobic side chain of the quinone (28). Three or four residues downstream of the histidine, there is a residue that is conserved within homologous proteins but that is different between various quinone binding sites (28). In the SQRs there are two histidines (H131 and H196 in O. limnetica and H132 and H197 in A. halophytica) that are fully conserved (Fig. 1). The first conserved histidine is located in a putative helical stretch (predicted by PREDATOR) with a conserved C(X)3H(X)3A sequence and is therefore a possible candidate for the quinone binding site. (iii) In the FAD binding pocket of flavocytochrome c of A. vinosum, there are a cysteine known to covalently bind flavin and two additional cysteines that form a disulfide bridge (9). Alignment of the SQRs with the flavoprotein subunit of flavocytochrome c shows that the cysteine residue that covalently binds FAD in flavocytochrome c is not conserved in the SQRs, while the two cysteines that form the disulfide bridge in flavocytochrome c (9) are fully conserved in the SQRs (C159 and C345 of O. limnetica and C160 and C346 of A. halophytica). A third fully conserved cysteine (C127 or C128 in O. limnetica or A. halophytica, respectively) is not conserved in flavocytochrome c. (iv) Glycine has been suggested to play a structural role in many proteins (18). Sixteen glycine residues are conserved in all seven SQRs (Fig. 1).
Most interestingly, the first eukaryotic SQR gene (hmt2) was recently cloned from the fission yeast S. pombe (43). The hmt2+ gene was cloned by complementation of a cadmium-hypersensitive S. pombe mutant. Purified HMT2 (overexpressed in E. coli) exhibited SQR activity (43). Figure 3 shows the high homology shared by all prokaryotic SQRs, as opposed to the low homology to S. pombe SQR. In addition, all SQRs, both eukaryotic and prokaryotic, share a low homology with the flavoprotein subunit of A. vinosum flavocytochrome c (Fig. 1 and 3) (9). Flavocytochrome c has previously been considered a sulfide oxidase in prokaryotes. However, its inactivation in A. vinosum was reported not to affect sulfide oxidation in this system (27). Although the homology is low, the two previously mentioned cysteines that form the disulfide bridge adjacent to the flavin in flavocytochrome c are also conserved in the eukaryotic SQR (Fig. 1). It is worth mentioning that the affinity of the eukaryotic SQR for its substrates was reported to be very low (Km of 2 mM for both sulfide and coenzyme Q2) (43), unlike the high affinity measured for the prokaryotic SQRs (Km values in the micromolar range [34]). It is possible that domains that are conserved among the prokaryotic SQRs but not in the eukaryotic SQR are involved in improving the affinity of the enzyme for its substrates.
The distribution of all the SQRs known so far, including those from cloned genes and the sqr-like ORFs and those detected only by membrane activity, is summarized in Fig. 6. The wide distribution emphasizes the important role of SQR in the sulfur cycle in nature.
ACKNOWLEDGMENTS
This work was supported by the Basic Research Foundation administered by the Israel Academy of Sciences and Humanities (award given to Y.S.) and by the Deutsche Forschungsgemeinschaft. Thanks are also due to the Moshe Shilo Minerva Center for Marine Biogeochemistry and the Massimo and Adelina Della Pergolla Chair in Life Sciences (award given to E.P.).
We thank A. Oren for critical reading of the manuscript.
REFERENCES
- 1.Arieli B, Padan E, Shahak Y. Sulfide-induced sulfide-quinone reductase activity in thylakoids of Oscillatoria limnetica. J Biol Chem. 1991;266:104–111. [PubMed] [Google Scholar]
- 2.Arieli B, Shahak Y, Taglicht D, Hauska G, Padan E. Purification and characterization of sulfide-quinone reductase, a novel enzyme driving anoxygenic photosynthesis in Oscillatoria limnetica. J Biol Chem. 1994;269:5705–5711. [PubMed] [Google Scholar]
- 3.Belkin S, Shahak Y, Padan E. Anoxygenic photosynthetic electron transport. Methods Enzymol. 1988;167:380–386. [Google Scholar]
- 4.Belkin S, Arieli B, Padan E. Sulfide-dependent electron transport in Oscillatoria limnetica. Isr J Bot. 1982;31:199–200. [Google Scholar]
- 5.Belkin S, Padan E. Sulfide-dependent hydrogen evolution in the cyanobacterium Oscillatoria limnetica. FEBS Lett. 1978;94:291–294. [Google Scholar]
- 6.Brune D C, Trüper H G. Noncyclic electron transport in chromatophores of photolithotrophically grown Rhodobacter sulfidophilus. Arch Microbiol. 1986;145:295–301. [Google Scholar]
- 7.Brune D C. Sulfur compounds as photosynthetic electron donors. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic; 1995. pp. 847–870. [Google Scholar]
- 8.Brune D C. Sulfur oxidation by phototrophic bacteria. Biochim Biophys Acta. 1989;975:189–221. doi: 10.1016/s0005-2728(89)80251-8. [DOI] [PubMed] [Google Scholar]
- 9.Chen Z W, Koh M, van Driessche G, van Beeumen J J, Bartsch R G, Meyer T E, Cusanovich M A, Mathews F S. The structure of flavocytochrome c sulfide dehydrogenase from a purple phototrophic bacterium. Science. 1994;266:430–432. doi: 10.1126/science.7939681. [DOI] [PubMed] [Google Scholar]
- 10.Cohen Y, Jørgensen B B, Padan E, Shilo M. Sulfide-dependent anoxygenic photosynthesis in the cyanobacterium Oscillatoria limnetica. Nature. 1975;257:489–492. doi: 10.1128/jb.123.3.855-861.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen Y, Padan E, Shilo M. Facultative anoxygenic photosynthesis in the cyanobacterium Oscillatoria limnetica. J Bacteriol. 1975;123:855–861. doi: 10.1128/jb.123.3.855-861.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies B D, Mingioli E S. Mutants of Escherichia coli requiring methionine or vitamin B12. J Bacteriol. 1950;60:17–28. doi: 10.1128/jb.60.1.17-28.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olsen G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 14.Eggink G, Engel H, Vriend G, Terpstra P, Witholt B. Rubredoxin reductase of Pseudomonas oleovorans. Structural relationship to other flavoprotein oxidoreductases based on one NAD and two FAD fingerprints. J Mol Biol. 1990;212:135–142. doi: 10.1016/0022-2836(90)90310-I. [DOI] [PubMed] [Google Scholar]
- 15.Garlick S, Oren A, Padan E. Occurrence of facultative anoxygenic photosynthesis among filamentous and unicellular cyanobacteria. J Bacteriol. 1977;129:623–629. doi: 10.1128/jb.129.2.623-629.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giddings T H, Jr, Wolk C P, Shomer-Ilan A. Metabolism of sulfur compounds by whole filaments and heterocysts of Anabaena variabilis. J Bacteriol. 1981;146:1067–1074. doi: 10.1128/jb.146.3.1067-1074.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen T, van Gemerden H. Sulfide utilization by purple nonsulfur bacteria. Arch Mikrobiol. 1972;86:49–56. doi: 10.1007/BF00412399. [DOI] [PubMed] [Google Scholar]
- 18.Javadpour M M, Eilers M, Groesbeek M, Smith S O. Helix packing in polytopic membrane proteins: role of glycine in transmembrane helix association. Biophys J. 1999;77:1609–1618. doi: 10.1016/S0006-3495(99)77009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Lee B H, Hibino T, Jo J, Viale A M, Takabe T. Isolation and characterization of a dnaK genomic locus in a halotolerant cyanobacterium Aphanothece halophytica. Plant Mol Biol. 1997;35:763–775. doi: 10.1023/a:1005867420619. [DOI] [PubMed] [Google Scholar]
- 21.Marchuk D, Drumm M, Saulino A, Collins F S. Construction of T-vectors, a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res. 1991;19:1154. doi: 10.1093/nar/19.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oren A, Fattom A, Padan E, Tietz A. Unsaturated fatty acid composition and biosynthesis in Oscillatoria limnetica and other cyanobacteria. Arch Microbiol. 1985;141:138–142. [Google Scholar]
- 23.Oren A, Padan E. Induction of anaerobic, photoautotrophic growth in the cyanobacterium Oscillatoria limnetica. J Bacteriol. 1978;133:558–563. doi: 10.1128/jb.133.2.558-563.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oren A, Shilo M. Anaerobic heterotrophic dark metabolism in the cyanobacterium Oscillatoria limnetica: sulfur respiration and lactate fermentation. Arch Microbiol. 1979;122:77–84. [Google Scholar]
- 25.Padan E. Combined molecular and physiological approach to anoxygenic photosynthesis of cyanobacteria. In: Cohen Y, Rosenberg E, editors. Microbial mats: physiological ecology of benthic microbial communities. Washington, D.C.: American Society for Microbiology; 1989. pp. 277–282. [Google Scholar]
- 26.Padan E. Facultative anoxygenic photosynthesis in cyanobacteria. Annu Rev Plant Physiol. 1979;30:27–40. [Google Scholar]
- 27.Reinartz M, Tschape J, Bruser T, Trüper H G, Dahl C. Sulfide oxidation in the phototrophic sulfur bacterium Chromatium vinosum. Arch Microbiol. 1998;170:59–68. doi: 10.1007/s002030050615. [DOI] [PubMed] [Google Scholar]
- 28.Rich P, Fisher N. Quinone binding sites in membrane proteins: structure, function and applied aspects. Biochem Soc Trans. 1999;27:561–565. doi: 10.1042/bst0270561. [DOI] [PubMed] [Google Scholar]
- 29.Roux K H, Hecker K H. One-step optimization using touchdown and stepdown PCR. Methods Mol Biol. 1997;67:39–45. doi: 10.1385/0-89603-483-6:39. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Schütz M, Klughammer C, Griesbeck C, Quentmeier A, Friedrich C G, Hauska G. Sulfide-quinone reductase activity in membranes of the chemotrophic bacterium Paracoccus denitrificans GB17. Arch Microbiol. 1998;170:353–360. [Google Scholar]
- 32.Schütz M, Maldener I, Griesbeck C, Hauska G. Sulfide-quinone reductase from Rhodobacter capsulatus: requirement for growth, periplasmic localization, and extension of gene sequence analysis. J Bacteriol. 1999;181:6516–6523. doi: 10.1128/jb.181.20.6516-6523.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schütz M, Shahak Y, Padan E, Hauska G. Sulfide-quinone reductase from Rhodobacter capsulatus. Purification, cloning, and expression. J Biol Chem. 1997;272:9890–9894. doi: 10.1074/jbc.272.15.9890. [DOI] [PubMed] [Google Scholar]
- 34.Shahak Y, Schütz M, Bronstein M, Griesbeck C, Hauska G, Padan E. Sulfide-dependent anoxygenic photosynthesis in prokaryotes: sulfide-quinone reductase (SQR), the initial step. In: Peschek G A, Löffelhardt W L, Schmetterer G, editors. The phototrophic prokaryotes. New York, N.Y: Kluwer Academic/Plenum Publishers; 1998. pp. 217–228. [Google Scholar]
- 35.Shahak Y, Klughammer C, Schreiber U, Padan E, Herrmann I, Hauska G. Sulfide-quinone and sulfide-cytochrome reduction in Rhodobacter capsulatus. Photosynthesis Res. 1994;39:175–181. doi: 10.1007/BF00029384. [DOI] [PubMed] [Google Scholar]
- 36.Shahak Y, Arieli B, Binder B, Padan E. Sulfide-dependent photosynthetic electron flow coupled to proton translocation in thylakoids of the cyanobacterium Oscillatoria limnetica. Arch Biochem Biophys. 1987;259:605–615. doi: 10.1016/0003-9861(87)90527-3. [DOI] [PubMed] [Google Scholar]
- 37.Shahak Y, Arieli B, Padan E, Hauska G. Sulfide quinone reductase (SQR) activity in Chlorobium. FEBS Lett. 1992;299:127–130. doi: 10.1016/0014-5793(92)80230-e. [DOI] [PubMed] [Google Scholar]
- 38.Stal L J. Physiological ecology of cyanobacteria in microbial mats and other communities. New Phytol. 1995;131:1–32. doi: 10.1111/j.1469-8137.1995.tb03051.x. [DOI] [PubMed] [Google Scholar]
- 39.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 40.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Bio/Technology. 1992;24:280–284. [PubMed] [Google Scholar]
- 41.Taglicht D, Padan E, Schuldiner S. Overproduction and purification of a functional Na+/H+ antiporter coded by nhaA (ant) from Escherichia coli. J Biol Chem. 1991;266:11289–11294. [PubMed] [Google Scholar]
- 42.Trumpower B L. Cytochrome bc1 complexes of microorganisms. Microbiol Rev. 1990;54:101–129. doi: 10.1128/mr.54.2.101-129.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vande Weghe J G, Ow D W. A fission yeast gene for mitochondrial sulfide oxidation. J Biol Chem. 1999;274:13250–13257. doi: 10.1074/jbc.274.19.13250. [DOI] [PubMed] [Google Scholar]
- 44.Wierenga R K, Terpstra P, Hol W G J. Prediction of the occurrence of the ADP-binding β-α-β-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986;187:101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- 45.Woese C R. There must be a prokaryote somewhere: microbiology's search for itself. Microbiol Rev. 1994;58:1–9. doi: 10.1128/mr.58.1.1-9.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]