Abstract
Background and Objectives
After clinical evaluation, the molecular diagnosis of type 1 facioscapulohumeral dystrophy (FSHD1) relies in most laboratories on the detection of a shortened D4Z4 array at the 4q35 locus by Southern blotting. In many instances, this molecular diagnosis remains inconclusive and requires additional experiments to determine the number of D4Z4 units or identify somatic mosaicism, 4q-10q translocations, and proximal p13E-11 deletions. These limitations highlight the need for alternative methodologies, illustrated by the recent emergence of novel technologies such as molecular combing (MC), single molecule optical mapping (SMOM), or Oxford Nanopore-based long-read sequencing providing a more comprehensive analysis of 4q and 10q loci. Over the last decade, MC revealed a further increasing complexity in the organization of the 4q and 10q distal regions in patients with FSHD with cis-duplication of D4Z4 arrays in approximately 1%–2% of cases.
Methods
By using MC, we investigated in our center 2,363 cases for molecular diagnosis of FSHD. We also evaluated whether previously reported cis-duplications might be identified by SMOM using the Bionano EnFocus FSHD 1.0 algorithm.
Results
In our cohort of 2,363 samples, we identified 147 individuals carrying an atypical organization of the 4q35 or 10q26 loci. Mosaicism is the most frequent category followed by cis-duplications of the D4Z4 array. We report here chromosomal abnormalities of the 4q35 or 10q26 loci in 54 patients clinically described as FSHD, which are not present in the healthy population. In one-third of the 54 patients, these rearrangements are the only genetic defect suggesting that they might be causative of the disease. By analyzing DNA samples from 3 patients carrying a complex rearrangement of the 4q35 region, we further showed that the SMOM direct assembly of the 4q and 10q alleles failed to reveal these abnormalities and lead to negative results for FSHD molecular diagnosis.
Discussion
This work further highlights the complexity of the 4q and 10q subtelomeric regions and the need of in-depth analyses in a significant number of cases. This work also highlights the complexity of the 4q35 region and interpretation issues with consequences on the molecular diagnosis of patients or genetic counseling.
Facioscapulohumeral dystrophy (FSHD), an autosomal dominant disease, is a common muscular dystrophy, with a prevalence of 1:8,000–1:12,000.1,2 At the clinical level, the disease is characterized by a progressive and asymmetric weakening of specific muscles of the face, scapular girdle, upper arms, or pelvic girdle3 with onset during the second decade of life and variable degrees of severity.
FSHD is genetically described with at least 2 subtypes. In most of the patients (95%, FSHD1; MIM#158900), the disease is linked to the subtelomeric 4q35 locus4,5 and involves a reduction in the number of D4Z4 GC-rich macrosatellites.6 In the healthy population, the number of units comprises between 11 and up to 1067 and contains >65% of methylated CGs.8 Patients with FSHD1 carry a pathogenic contraction of the array (<10 D4Z4 repeats), associated with hypomethylation and the presence in cis of a specific type A haplotype.9 This qA region provides a polyadenylation signal required for transcription of the retrogene DUX4 encoded by the last D4Z4 unit.10,11 Type 2 FSHD (FSHD2; MIM#158901, 5% of patients) is not linked to the reduction in the number of D4Z4 units but, in 80% of cases, to a variant in the SMCHD1 gene,12 leading as in FSHD1 to D4Z4 hypomethylation.
Owing to duplication events during evolution, the 4q and 10q subtelomeric regions are 98% homologous and somatic interchromosomal rearrangements between these 2 regions result in nonstandard 4q/10q D4Z4 arrays in at least 10% of the European or Asian populations.13-15 More recently, additional rearrangements of the 4q region such as cis-duplications of the D4Z4 array have been reported in patients affected with FSHD and relatives.16 FSHD is also characterized by a high frequency of de novo cases, with 40% linked to somatic mosaicism in one of the parents.15,17
In most laboratories, FSHD diagnosis is performed by Southern blotting (SB) after digestion of DNA using EcoRI to determine the size of 4q and 10q arrays followed by electrophoresis and hybridization with the p13E-11 probe (D4S139 marker) that lies upstream of the first D4Z4 unit.18 Specific restriction enzyme of either 4q-derived (XapI/ApoI)19 or 10q-derived (BlnI)20 D4Z4 repeats are used in combination with EcoRI to discriminate between 4qTer and 10qTer-derived arrays. Determination of A-type and B-type haplotype requires an additional step with the HindIII restriction enzyme and hybridization of DNA blots with specific probes. In a significant number of situations, accurate molecular diagnosis is often challenged by technical difficulties but also 4q-10q translocations, somatic mosaicism, or more complex rearrangements, only partially resolved by SB or requiring additional manipulations. A decade ago, a molecular combing (MC)–based approach has been developed to counteract these limitations and provide in a single step, a comprehensive analysis of the 4q and 10q alleles, the sizing of the D4Z4 arrays, and associated A-type or B-type haplotypes.21
In patients clinically diagnosed with FSHD, we then identified others by MC cis-duplicated 4q35 alleles that consist in a long D4Z4 array followed by a distal small FSHD-sized repeat array.16,22 A number of these patients also carry a short D4Z4 allele or an SMCHD1 variant, likely causative of the disease. In a number of individuals (3/17 in reference 16 10/23 in reference 22), the cis-duplication is the only abnormality reported. Then, by exploring 1,029 samples by MC, we also described a broad variability in 4q and 10q subtelomeric regions in the general population with patients having atypical 10q patterns infrequently found in the healthy population.7
More recently, a novel diagnosis approach based on the use of single genomic optical mapping (SMOM) has been developed by Bionano Genomics. For this approach, long fluorescently tagged DNA molecules are stretched, imaged using fluorescence microscopy, and assembled in silico. SMOM has been validated as a reliable method for evaluation of the number of D4Z4 on chromosomes 4 or 10 and determination of associated A-type or B-type haplotypes.23,24 However, this methodology has only been partially evaluated for complex rearrangements.
After 11 years of molecular diagnosis of FSHD using MC and the exploration of 2,363 cases, we now report a total of 147 individuals, clinically affected with FSHD, carrying complex rearrangements of the 4q and 10q regions. In addition, we also compared the resolution of MC and SMOM for 3 patients carrying a 4q cis-duplication or a complex 4q or 10q genotype and provided here further details for 54 of them. Using the Bionano EnFocus FSHD 1.0 algorithm for SMOM analysis complex rearrangements including cis-duplications were not identified highlighting the need for a more in-depth analyses of the 2 alleles in the case of complex genotypes.
Methods
Details on all methods are provided in the eMethods (links.lww.com/NXG/A609).
Patient Consents
Blood samples were received at the Department of Medical Genetics, La Timone Children's Hospital in Marseille for molecular diagnosis and molecular diagnosis of facioscapulo humeral dystrophy, familial segregation analysis, or exclusion diagnosis. Informed consent was obtained from all patients or guardians for the genetic analyses, including for research purposes. Samples were provided by the Center for Biological Resources (Department of Medical Genetics, La Timone Children's hospital, Assistance Publique des hôpitaux de Marseille) with the AC 2011-1312 and N°IE-2013-710 accreditation numbers.
Data Availability
Not applicable because all available data are presented in the manuscript.
Results
Incidence of Complex Rearrangements of the 4q and 10q Subtelomeres
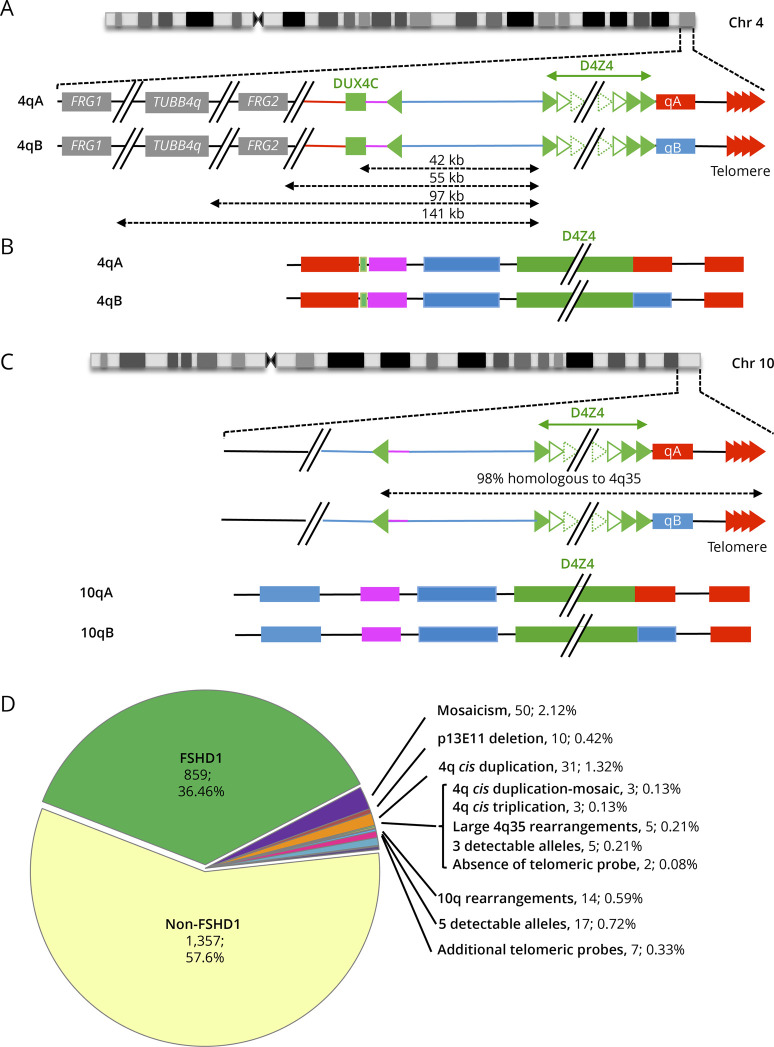
Since the first publication of the MC barcode developed for FSHD diagnosis21 (Figure 1A), we analyzed up to 2,363 individuals using this method providing a direct visualization and comprehensive analysis of 4q and 10q regions with sizing of their respective D4Z4 arrays and A-type or B-type haplotypes (Figure 1, B and C). Of these 2,363 samples, 39 prenatal tests were performed. A positive FSHD1 diagnosis was obtained for 859 patients (36.46%, FSHD1, number of D4Z4 repeated units (RU) < 10), and 1,357 cases were negative for FSHD1 (57.6%, number of RUs > 10) (Figure 1D). These 1,357 negative cases include patients with FSHD2 (i.e., patients with a clear FSHD clinical diagnosis), individuals analyzed for familial segregation studies, or individuals for which exclusion diagnosis was requested. This category also includes type 2 patients with up to date 40 patients carrying a SMCHD1 variant and showing D4Z4 hypomethylation. Of note, 10q alleles are mainly associated with a type A distal haplotype, but we identified B-type alleles on chromosome 10 in 159 samples (6.75%) (Figure 1C).
Figure 1. Overview of More Than 10 Years of Molecular Combing–Based Diagnosis of FSHD.
(A) The most distal 4q region is represented (4q35 locus) together with the proximal FRG1, TUBB4Q, and FRG2 genes. As described in reference 16, the D4Z4 array is depicted by green triangles. Sequences starting with an inverted D4Z4 repeat (green arrow) are specific to the 4q35 locus (red lines). Regions located between the D4Z4 array and the inverted D4Z4 repeat are also present on the 10q26 locus (blue bar).The 4qA (red rectangle) is characterized by the presence of the pLAM sequence distal to the last D4Z4 repeat and followed by the telomere (red arrows). The 4qB allele (blue rectangle) differs from the 4qA.16 (B) The V3 pink barcode used to distinguish the different alleles (4qA/B; 10qA/B) is based on a specific combination of 4 different colors (blue, pink, red, and green) used to label the 4q35 locus and 10q26 loci up to the telomeric sequence.21 The proximal 4q-specific region is detected by a combination of red and pink probes. The proximal 10q-specific region is identified by a blue probe. For chromosome 4, the four-color barcode comprises 1 probe detected in blue and one in pink, which hybridize the proximal region, one 6 kb red probe, which hybridizes the (TTAGGG)n telomeric ends (red). The qB-specific probe, adjacent to D4Z4, is detected in blue. (C) Schematic representation of chromosome 10q and illustration of the V3 pink barcode used to distinguish the 2 10q alleles (qA/B) based on a combination of 4 different colors and different DNA probes encompassing the distal regions up to the telomeric sequence. The barcodes for the 4q-10q homologous regions are identical. The proximal 10q-specific region is identified by a blue probe leading to a combination of blue-pink-blue probe for the 10q26 locus and red-pink-blue for the 4q35 region. The distal A-type region is identified by a red probe, the B-type allele by a blue probe.16 (D) Of the 2,363 patients analyzed since 2011 by MC, 91.3% showed a normal profile with 4 distinct alleles and absence (1,357 cases, 57.6%) or presence (859 cases, 36.46%) of D4Z4 array contraction on a 4qA allele. The authors identified 6.26% of cases with an atypical profile with 2.12% (n = 50) of samples with a mosaic D4Z4 array contraction, 0.42% (n = 10) with a deletion of the p13E-11 probe, 1.32% (n = 31) of patients with a 4qA cis-duplication,16 0.13% (n = 3) with a cis-duplication in mosaic, 0.13% (n = 3) with a cis-triplication, and 0.21% (n = 5) with a complex 4q35 rearrangement. In 0.21% (n = 5) of cases, only 3 alleles were detectable while 5 alleles were observed in 0.72% (n = 17) of cases. The authors noticed the presence of 10q26 rearrangements in 0.59% (n = 14) of cases, the absence of telomeric probes in 2 cases (0.08%), and 0.33% (n = 7) with multiple telomeric probes. FSHD = facioscapulohumeral dystrophy; MC = molecular combing.
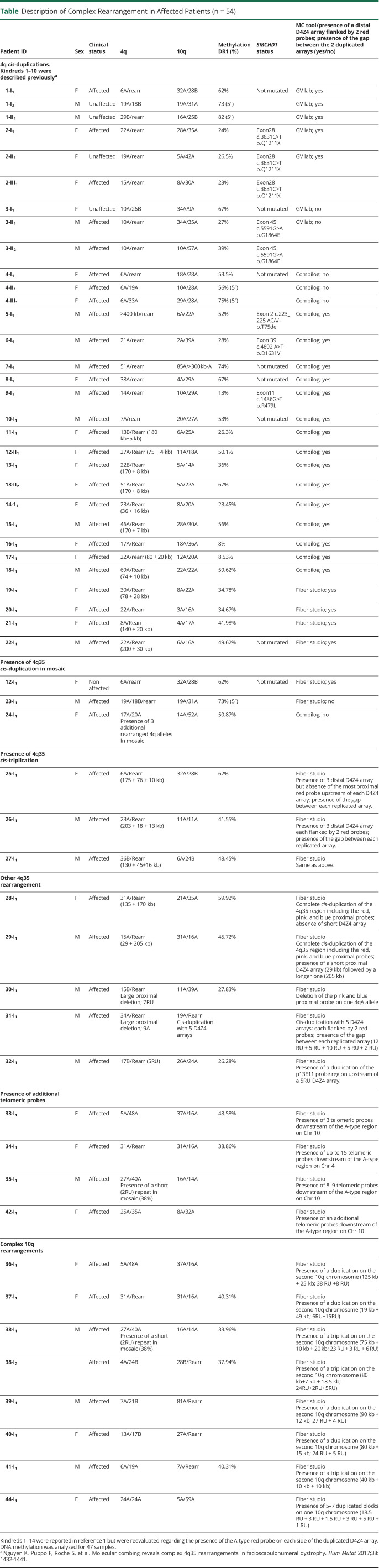
We previously reported 14 patients affected with FSHD and 4 healthy relative individuals (included in the 147 samples and presented in Table) carrying a complex 4q35 rearrangement consisting in a cis-duplication of the D4Z4 array and A-type probe.16 We further expanded this initial list of 18 individuals by reporting here 13 additional cases carrying the same type of cis-duplication with a long D4Z4 array (>10 RUs in most cases) followed by a short D4Z4 array (<10 RUs) (Table, Figure 2, A–D). Additional data are presented in eFigure 1 (links.lww.com/NXG/A608). All arrays are associated with a red A-type probe. In total, these 31 cases correspond to 1.32% of all samples processed by MC. We analyzed DNA methylation for all these 31 individuals. Of these 31 cases, 26% (n = 8) also carry short D4Z4 alleles and were classified as FSHD1; 45% (n = 14) display D4Z4 hypomethylation that segregates with a variant in SMCHD1 (classified as FSHD2) or both. For 9 individuals (29%), the rearranged allele might be directly associated with the disease.
Table.
Description of Complex Rearrangement in Affected Patients (n = 54)
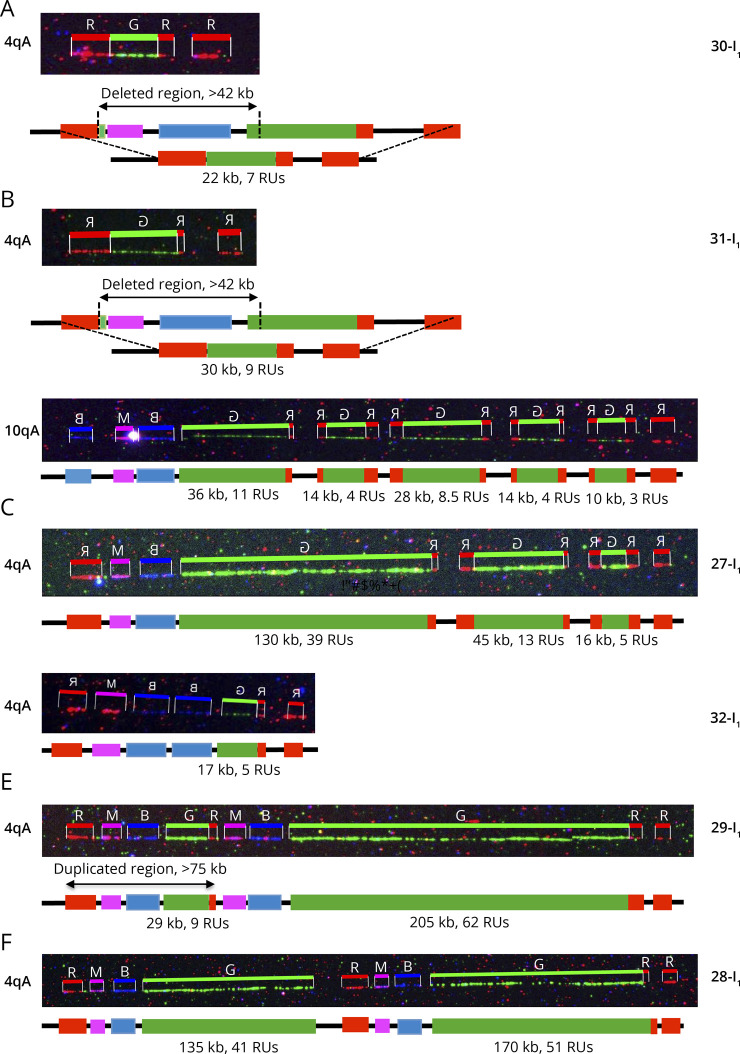
Figure 2. Presence of a Red Probe and Gap Upstream of the Duplicated D4Z4 Array Suggests Genomic Inversion.
(A) Schematic representation of the 4q35 barcode. The size of the different regions was estimated by the analysis of >1000 DNA samples previously processed by MC.7 For the 4qA allele, a probe labeled in red hybridizes the qA-specific β-satellite region of variable lengths (6.4–7.5 kb). The size of the gap between the A-type and telomeric probe is estimated around 6.6–8.8 kb.7 (B–D) Images and schematic representations of new cis-duplication identified using the Fibervision tool. For each D4Z4 array, the size in kb is indicated together with the number of D4Z4 units. (B) Absence of the red probe corresponding to the A-type region upstream of the duplicated D4Z4 array (case 19-I1). (C and D) Presence of the red probe (A-type region) upstream of the duplicated D4Z4 array (C, case 17-I1; D, case 21-I1). (E) Schematic representation of the possible mechanism leading to the 4q35 cis-duplication. The authors hypothesize a duplication and inversion of the region containing a variable number of D4Z4 repeats (<10 RUs), the A-type region, and the gap between this A-type region and the telomere followed by insertion or the duplicated region within the A-type region. Depending on the context (presence or absence of SMCHD1 variant), the long D4Z4 array might remain methylated (black dots) while the short allele might be hypomethylated (white dots). In this configuration, the DUX4 coding region is in the opposite orientation. It remains to be determined whether the recombined allele remains permissible for transcription. MC = molecular combing.
We also noticed the presence of this cis-duplication in mosaic in 2 patients (23-I1; 24-I1) and the healthy mother (12-I1) of an affected child who inherited the cis-duplicated allele (12-II1) (Table). Cis-duplication also occurs on chromosome 10q with 14 cases identified (Table). We also identified additional features that were not reported to date in a number of patients clinically diagnosed with the disease (Table). Among them, we identified 1 patient (24-I1) who carries multiple 4q chromosomes with D4Z4 arrays of different sizes. In this patient, we identified cis-duplications of large D4Z4 arrays; 1 allele consisting in a long D4Z4 array followed by a short repeated region (2 RUs) as described above but also additional alleles with triplications of D4Z4 arrays of gradual sizes with a short terminal repeated array, all followed by an A-type probe.
By looking more carefully at the cis-duplicated 4q35 region, in particular thanks to the high resolution of FiberVision, we noticed in most cases the presence of the telomeric gap (between 6.6 and 8.8 kb in size7) and A-type probe (between 6.6 and 8.8 kb in size7) upstream of the second repeat in most cases (26/31, Figure 2, A–D, eFigure 1, links.lww.com/NXG/A608). The absence of the A-type probe but presence of the distal gap was observed in 5 samples (Figure 2B). Given the presence of the gap between the 2 duplicated arrays (Figure 2, C and D), our hypothesis is that the short duplicated array is in the inverse orientation compared with the first proximal array, with insertion within the A-type region (Figure 2E).
Cis-Duplicated Alleles Are Partially Resolved by Single Molecule Optical Mapping
Given the emergence of novel methodologies for the diagnosis of the disease, in particular SMOM, we then asked whether these cis-duplications, which occur in approximately 1.32% of patients, might be detected by this methodology and resolved using the analysis pipeline designed for FSHD. We processed 4 samples previously analyzed in our laboratory by MC for identification of 4q and 10q alleles together with their respective haplotype (eTable 1, links.lww.com/NXG/A609) by SMOM.
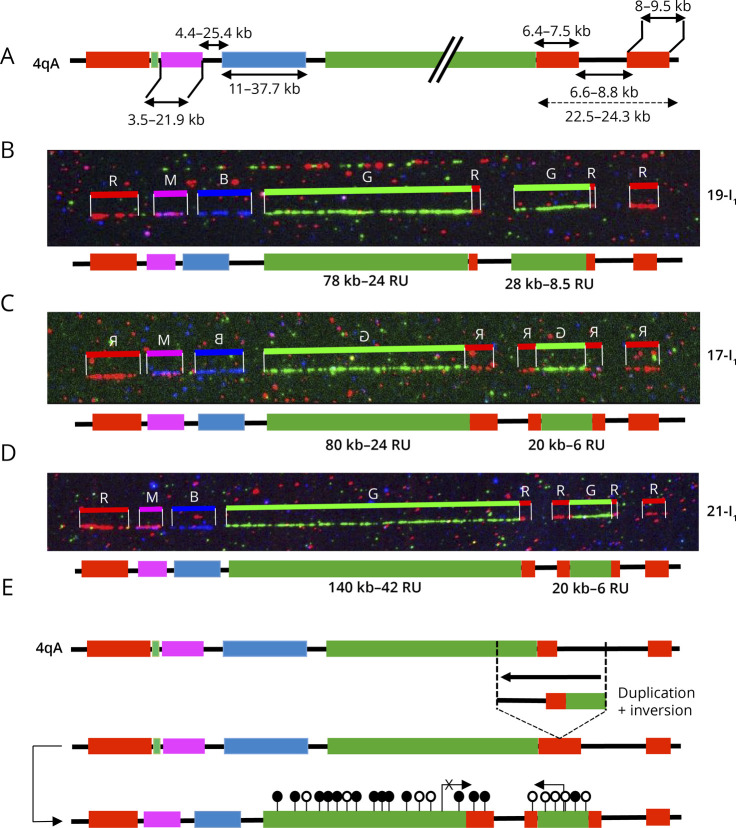
Sample 1 (2-II1, Figure 3A),16 clinically diagnosed with FSHD, was described by MC as a carrier of a 4q35 cis-duplication with a proximal type A array of 165 kb (50 RUs) followed by a distal type A array of 18 kb (5.4 RUs). The second 4qA allele carries 19 RUs.16 By SMOM and Bionano EnFocus FSHD analysis, this second allele (21 RUs, 4qA; 26x coverage) was retrieved but SMOM failed in identifying the rearranged allele and documented a 48 RUs D4Z4 array (17x coverage) and an additional 20 RUs array (3x coverage) likely corresponding to the 21 RU nonpathogenic allele. The 2 10q alleles mapped by SMOM were similar to those identified by MC (Figure 3A).
Figure 3. Comparative Analysis of 4qter and 10qter Regions by Molecular Combing and Single Molecule Optical Mapping.
(A–D) The V3 pink barcode was used to distinguish the different alleles (4qA/B, 10qA/B) by MC.21 For the samples selected, only the cis-duplicated 4q allele is presented. On scanned images, the size of the D4Z4 arrays is given in kb. A schematic representation of the 4q locus is presented with the number of D4Z4 repeats (RUs) indicated. SMOM analyses Bionano EnFocus FSHD tool is presented next to the combing images. For SMOM, the D4Z4 array is depicted in purple with only the putative cis-duplicated alleles presented. The 4qA-specific region is identified by a red square. Complete details are provided in Table. (A) Sample 1 (ID 16705) and (B) Sample 2 (ID 15906), analyzed using the GV lab software, were previously reported.16 (C and D) Samples 3 and 4 were analyzed using the FiberStudio analysis software. (A–C) Only the cis-duplicated 4q allele is presented. (A–D) The size of the D4Z4 arrays is given in kb. The number of D4Z4 repeats (RUs) is indicated underneath the schematic representation of the 4q locus. The results of SMOM are presented for the 2 4q alleles together with one of the 2 10qA allele, identified by a green square. The 4q35 allele corresponding to the duplicated locus is indicated by dashed lines. The 2 unmapped alleles, identified by MC as being 10qA are represented. (D) Representation of the shortest 4qA allele for sample 4. (E) Samples were reanalyzed using the Bionano de novo assembly tool. For identification of D4Z4 cis-duplication, distance between labels of the repeat region (purple region of SMOM Enfocus representation) was manually calculated, minus the static region of the labels around the repeat region (hg19: 19,691 for 4qA or 11,791 for 4qB). MC = molecular combing; SMOM = single molecule optical mapping.
Sample 2 (3-II2, Figure 3B)16 is clinically affected with FSHD and carries a cis-duplication that consists in a proximal 27 kb-qA array (8 RUs) followed by a 10 kb-qA array (3 RUs). The rearranged allele was not visualized by SMOM that yielded 2 healthy-type alleles (11 RUs, 46x coverage; 14 RUs, 30x coverage). The total count of repeats determined by SMOM for the cis-duplicated allele (11 RUs) are similar to the total number of RUs detected by MC but failed however to identify the interstitial A-type probe downstream of the 8 RU array.
Sample 3 (ID 21136, Figure 3C)7 is also clinically affected. MC revealed the presence of a cis-duplication of the 4q35 region consisting in a proximal array of 20 units followed by a short 9 RUs block. For this patient, the Bionano EnFocus FSHD analysis retrieved 5 different regions, 2 4q alleles, a single 10q allele, and 2 additional D4Z4 containing loci (Figure 3C). As for MC, SMOM retrieved a 25–26 RU 4q35 and 30–32 RU 10q26 alleles but failed to identify the 4q35 cis-duplication and interstitial A-type sequence and retrieved an 18 RUs qA array (Figure 3C, dashed lines). In this patient, we have also identified 2 short individual 10qA alleles of 2 and 4 RUs by MC. These alleles were not fully identified by SMOM that retrieved a 10q allele carrying 2 D4Z4 units and another allele of 35 RUS but failed to identify the distal haplotypes or 4q/10q-specific regions (Figure 3C).
For sample 4 (ID 25005, Figure 3D), the results were similar between the 2 methods, further confirming comparison with MC.21,24
Using the pipeline recommended for FSHD diagnosis, complex rearrangements, in particular cis-duplication, that are directly visualized by MC are not directly resolved by SMOM and the Bionano EnFocus FSHD tool that identifies only 1 A-type label per allele and fails to identify the more distal or additional ones. Given this limitation, all samples were reanalyzed using the SMOM de novo assembly pipeline. The interstitial A-type probes between the 2 contiguous D4Z4 arrays were retrieved for samples 1, 2, and 3 (Figure 3, A–C) revealing the duplications and allowing the sizing of D4Z4 arrays in each duplicated block (Figure 3E, eTable 1, links.lww.com/NXG/A609). The distance between labels (purple region of SMOM Enfocus representation) was manually calculated, minus the static region of the labels around the repeat region (hg19: 19,691 for 4qA or 11,791 for 4qB). This distance was then divided by 3.3 to estimate the D4Z4 units count but did not take into account the duplicated A-type label upstream of the D4Z4 array visible at least in sample 2-III1 or the gap present in all tested samples (Figure 3, A–C).
Using this methodology, the size of the more distal D4Z4 array was estimated around 10 kb (3 RUs, vs 5 RUs by MC) for sample 2-II1 (Figure 3A); 7 kb (2 RUs, vs 3 RUs by MC) for sample 3-II2 (Figure 3B), and approximately 7 RUs (vs 9 RUs by MC) for sample 3 (Figure 3C). In sample 3, the 2 additional 4q/10q-type alleles were identified, but the duplicated chromosome 10 region was not visualized. We thus concluded that in the case of complex rearrangements, such as cis-duplication, the sizing of the D4Z4 array and enumeration of D4Z4 units might result in false-negative results or erroneous molecular diagnosis, requiring additional tests and analyses.
Identification of Novel 4q35 Rearrangements in Patients With FSHD
Besides 4q35 cis-duplications, a number of additional chromosomal abnormalities were also identified by MC in individuals affected with FSHD. Our analysis of 2,363 cases led to the identification of deletions of the p13E11 region in 10 patients, a number of them reported before.7 We report here 2 additional cases in which MC revealed a large deletion of the proximal 4q35 region encompassing the pink and the red probe for an estimated size of approximately 42 kb, together with shortening of the D4Z4 array (30-I1, Figure 4A, eFigure 1, links.lww.com/NXG/A608). The second case in this category (31-I1) caught our attention as in addition to a large 4q35 proximal deletion identical to the case described above, the patient, diagnosed with FSHD at the age of 4 years also carries a complex rearrangement of the 10q distal region (31-I1, Figure 4B, eFigure 1). This rearrangement consists in 5 different D4Z4 arrays of different sizes, all flanked by red probes and separated by the gap present between the A-type tag and the telomere.
Figure 4. Complex Rearrangements at the 4q35 Locus.
(A) Presence of a large deletion encompassing the proximal pink and blue probes in a patient (30-I1, Table) affected with FSHD, with deletion of a number of D4Z4 units. The size of the deletion is estimated of more than 42 kb. (B) This patient (31-I1) carries a deletion of the proximal 4q35 region upstream of D4Z4 (>42 kb) together with a complex rearrangement of one 10q end. This rearrangement consists in the cis-duplication of 5 D4Z4 arrays of different sizes, all flanked by red probes (A-type allele) and separated by the gap present between the type A allele and the telomere. (C) This patient (27-I1) carries a triplication of D4Z4 arrays of different sizes (39 RUs, 13 RUs, 5 RUs from the centromere to the telomere). The 2 additional D4Z4 arrays are flanked by A-type probes, and all repeated arrays are separated by a gap. (D) Patient 32-I1 carries a cis-duplication of the blue probe encompassing the p13E11 probe (D4S104S1 marker), upstream of a short D4Z4 array (17 kb, 5 RUs). (E) Presence of a large duplication of the 4q35 region encompassing the proximal chromosome 4–specific region (red, pink, and blue probes) followed by a short D4Z4 array (9 RUs). This region is followed by a larger chromosome 4–specific region (red, pink, and blue probes) containing a 205 kb long D4Z4 array (62 RUs). Each D4Z4 array is followed by an A-type probe. (F) Presence of a large duplication of the 4q-specific region encompassing the proximal regions (red, pink, and blues probes), the D4Z4 array and the A-type probe. The 2 different D4Z4 arrays are of different sizes (135 kb, 41 RUs and 170 kb, 51 RUs) and above the threshold of 10 units. FSHD = facioscapulohumeral dystrophy.
We identified 3 patients affected with FSHD carrying a triplication of the D4Z4 array (Table). A representative case is presented (27-I1, Figure 4C, eFigure 1, links.lww.com/NXG/A608). In all 3 cases, patients carry 3 D4Z4 arrays of gradually decreasing size from the centromere to the telomere, with a short terminal block (<11 RUs). Additional arrays are flanked by a red probe and separated by the gap between the A-type label and the telomere as observed for cis-duplications.
Another complex genotype (32-I1) was identified. The patient carries a 4qB allele with 17 RUs and 2 identical 10qA alleles (25 RUs). By MC, we also identified a second 4q allele with 5 RUs (17 kb) but a duplication of the blue probe (encompassing the D4F104S1, p13E11 region). Because enzymes used for Southern blotting are located outside of this region, this duplication might yield a longer fragment and overestimation of the size of the array (Figure 4D, eFigure 1, links.lww.com/NXG/A608).
Patient 29-I1, clinically diagnosed with FSHD, was identified with a duplication encompassing a large part of the 4q35 locus (Figure 4E, eFigure 1, links.lww.com/NXG/A608). This case carries a large cis-duplication of the pink and blue probes, i.e., the region comprised between DUX4C and the D4F104S1 marker, estimated around 42 kb. This proximal 4q35 region is followed by a short D4Z4 array of 29 kb (9RUs) and an A-type label. In the distal part of the locus, we observed a large cis-duplication of the pink and blue probes followed by a large D4Z4 array (205 kb, 62 RUs) and an A-type probe. Similar features were observed in patient 28-I1, who carries 2 large duplicated blocks encompassing a sequence comprised between the pink and A-type probes with a proximal 41 D4Z4 units array and 51 units in the distal part (Figure 4F, eFigure 1). By Southern blotting, the duplication of the p13E11 probe would yield an additional band after EcoRI digestion.
Characterization of Rearrangements at the 10q26 Locus
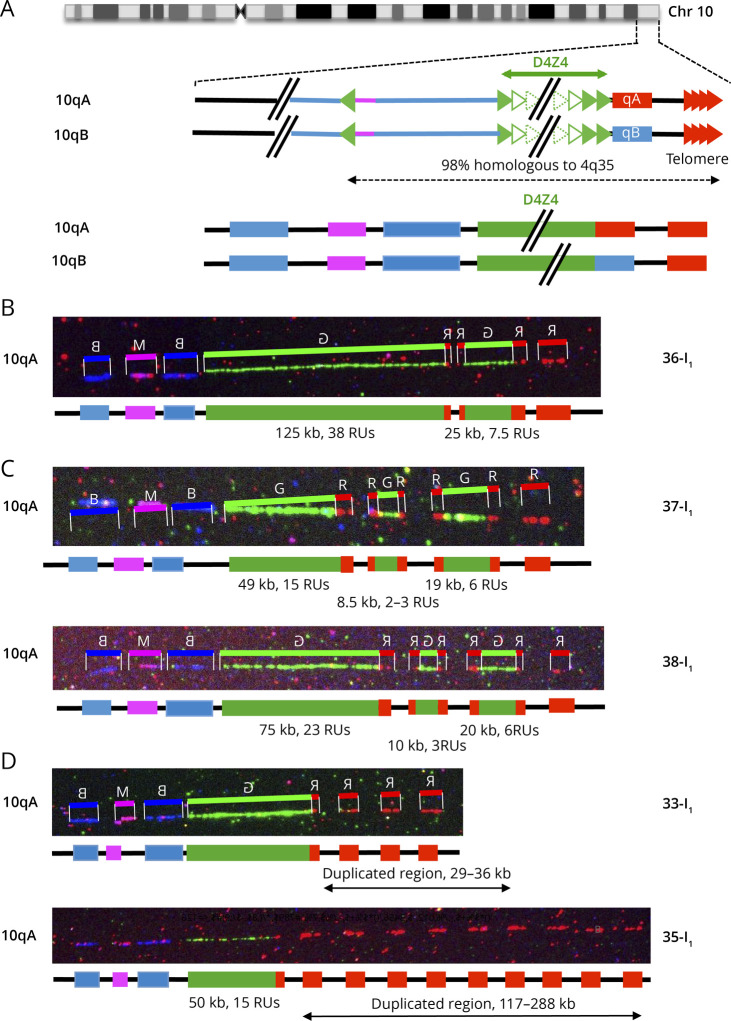
Although less frequent, we also observed duplications of the D4Z4 array on chromosome 10 (Table, Figure 5A, eFigure 1, links.lww.com/NXG/A608). We report here 3 examples of patients clinically diagnosed with FSHD carrying a 10q cis-duplication (36-I1, Figure 5B) or triplication of D4Z4 arrays (37-I1, 38-I1, Figure 5, C and D). In all cases, the duplicated region is flanked by red A-type probes and all blocks are separated by the gap present between the A-type region and the telomere suggesting that the same mechanism leads to 4q of 10q duplications. Differently of what was observed for chromosome 4, we did not systematically see a gradual decrease in the number of D4Z4 units on chromosome 10 as the smallest D4Z4 arrays in located in the middle of the rearranged region in 2 cases (37-I1, 38-I1, Figure 5, C and D). Patient 36-I1 carries a short 4qA allele (5 RUs) likely associated with the clinical signs of the disease while in the 2 other patients, the cause of the disease remains undetermined in the absence of short 4qA allele.
Figure 5. Complex Rearrangements at the 10q26 Locus.
(A) Schematic representation of chromosome 10q and illustration of the V3 pink barcode used to distinguish the 2 10q alleles (qA/B). The proximal 10q-specific region is identified by hybridization with a blue probe, with a combination of blue-pink-blue probe for the 10q26 locus and red-pink-blue for the 4q35 region as described.16 (B) Cis-duplication of the 10q26 region with 2 D4Z4 arrays of different sizes (38 RUs and 7.5 RUs) separated by a gap. The second repeated array is flanked by red probes. (C) Triplication of the D4Z4 region in 2 different cases (37-I1 and 38-I1). The 2 additional D4Z4 arrays are flanked by red probes. All 3 arrays are separated by a gap. The shortest D4Z4 array is not the most distal as observed for chromosome 4. (D) Presence of additional copies of the terminal telomeric probes in 2 cases, 33-I1 with 3 probes (estimated size, 29–36 kb) and 35-I1 with 9 probes (estimated size, 117–288 kb).
Finally, we more recently identified 3 patients displaying multiple telomeric probes downstream of D4Z4 on chromosome 4 (34-I1, Table, Figure 5D) or chromosome 10 (33-I1, 35-I1, Table, Figure 5D). The last patient (35-I1, Table, Figure 5D) carries 2 4qA alleles of 16 and 27 RUs together with a very atypical 10qA allele with a 50 kb D4Z4 array (15 RUS) followed by an A-type allele and 9 signals corresponding to the telomeric probe. The patient also presents with a mosaicism (38%) for a 2 RUs allele on chromosome 4 that might be associated with the clinical manifestation of the disease. All additional telomeric probes are all separated by a gap, likely corresponding to the region that is not covered at the distal end of each locus.
DNA Methylation Profiling in Patients Carrying Rearranged Alleles
DNA methylation is a well-known epigenetic alteration associated with chromosomal rearrangements and associated with FSHD. To define the epigenetic status of patients carrying these different rearrangements, D4Z4 methylation level was determined in 47/54 samples (Table) and for which DNA was available using sodium bisulfite sequencing for the DR1 or 5′ region in the proximal part of the D4Z4 repeat as described.8 In our experience, the level of D4Z4 methylation is above 65% in the healthy population, ranging between 40 and 60% in patients with FSHD1 and below the threshold of 40% in FSHD2. In 21 cases (45%) reported here and in the past,16 D4Z4 repeats are markedly hypomethylated (<40%) with hypomethylation associated with a presence of a SMCHD1 variant, compatible with FSHD2 (eFigure 2B, links.lww.com/NXG/A608). It is also interesting to note that among the patients for whom methylation was tested (47/54), the methylation level is decreased (<65%) compared with the healthy population in 18 cases (38%), despite a high number of D4Z4 units (>10 RUs on the shortest 4q allele) (additional data are presented in eFigure 2B and eTable 2, links.lww.com/NXG/A609). Overall, the level of methylation is correlated to the total number of repeats (eFigure 2C, eTable 2).
Discussion
The polymorphic D4Z4 repeat and 4q35 region are highly recombigenic and associated with FSHD.5,6 Somatic mosaicism for short D4Z4 alleles is found in as much as 3% of the general population15 and rearranged short D4Z4 alleles in 3% of the healthy population.25 The 10q chromosome end is nearly identical to chromosome 4qTer (98% of homology) and equally recombigenic.
In part because of the complexity of the 4q35 and 10q26 subtelomeric regions and their homology, FSHD molecular diagnosis based on Southern blotting is often challenged by technical difficulties because of the quality of biological samples but also by interpretation issues for the accurate characterization of the 4q35 pathogenic allele (sizing, discrimination of the chromosomal localization, 4q-10q translocation, mosaicism, p13E11 deletion), leading to an absence of clear molecular diagnosis in a number of patients clinically diagnosed with the disease. To bypass this limitation, we developed a MC-based strategy21 to assess the size of the D4Z4 array on both the 4q and 10q chromosomes together with the type of haplotype in a single step. After more than 10 years of molecular diagnosis of FSHD using MC and the analysis of 2,363 individual samples in our center, we report here 147 individuals carrying complex rearrangements of the 4q35 region but also of the 10q26 locus.
Among these rearrangements, we report now a list of 34 individuals (31 cases + 3 mosaic individuals) carrying a cis-duplication of D4Z4 arrays on A-type haplotypes, ranking this chromosomal anomaly as the second most frequent after somatic mosaicism in FSHD. The existence of these cis-duplications was initially identified in 14 individuals and 10 families16 and confirmed in an independent study.22 The cis-duplication was shown to segregate with SMCHD1 variants in a large proportion of cases16,22 or with a short D4Z4 allele16 but also suspected as causative of the disease,16 including for new patients reported here. All rearrangements identified in our cohort occur on A-type alleles, characterized by the presence of a stretch of β-satellite repeats9 of variable size (estimated between 6.4 and 7.5 kb)7 downstream of D4Z4 in individuals clinically diagnosed with FSHD. These rearrangements are less frequently observed for 10qA alleles, absent in the general population, and never observed for B-type alleles. Not all rearrangements segregate with a SMCHD1 variant, and 17% of these patients display a moderate to high level of DNA methylation.
Recently, SMOM has been validated for FSHD diagnosis by analysis of 36 samples previously characterized by SB. Comparison between the 2 methods revealed a high level of concordance between samples and a mean absolute difference of 10.1 kb (2–3 RUs) between the 2 methods. SMOM was also validated for identification of somatic mosaicism and haplotype testing.24 The rate of positive FSHD diagnosis is consistent between SMOM (45.7%),24,26 SB (44.1%), and MC,16,21 for the most commonly observed genotypes. However, the emergence of novel technologies revealed a further increasing complexity in the organization of the 4q and 10q distal regions and rearrangements occurring in individuals clinically affected with FSHD such as 4q or 10q cis-duplications in approximately 2% of cases.7,16,22,27 Our analysis here of 3 patients carrying 4q35 cis-duplications revealed that the Bionano EnFocus FSHD analysis tool fails to detect the interstitial A-type label together with the interstitial gap leading to an erroneous counting of D4Z4 units, retrieved as a continuous longer array. Thus, if for most of the patients, SMOM appears as a straightforward tool for FSHD1 diagnosis, it has to be kept in mind that in the case of complex rearrangements, such as cis-duplication, the sizing of the D4Z4 array and enumeration of D4Z4 units might result in false-negative results or erroneous molecular diagnosis.
The frequency and recurrence of the rearrangements associated with FSHD, compared with subtelomeric defects affecting other chromosomes, suggests a common genomic architecture and common mechanism that remained to be defined. Mitotic D4Z4 rearrangements leading to somatic mosaicism and D4Z4 array shortening likely occur via interchromatid or intrachromosomal cross-over via synthesis-dependent strand annealing.28 Because all rearranged arrays are of A-type origin, including in patients carrying a second B-type allele, rearrangements likely occur between sister chromatid exchanges during meiosis by formation of intra-allelic loops that might be facilitated by secondary structures such as hairpins or four-stranded DNA G quadruplex or tetraplex, present downstream of the last D4Z4 unit29,30 or the presence of other repetitive DNA sequences such as β satellites or microsatellites flanking the D4Z4 array. Furthermore, the 4q35 telomere replicates at the very end of S-phase.31 At this stage where transcription-replication conflict occurs, the occupancy of DNA by RNA polymerase might create obstacles to DNA replication with increased torsional stress and non-B structure formation; favoring in turn rearrangements.32,33
Our analyses strongly suggest that duplicated arrays are inverted and located tail to tail and not in the same orientation. Several mechanisms have been proposed for subtelomeric rearrangements with inverted duplications.34,35 These duplications are often caused by deletions occurring in germ cells precursors or after rearrangements involving repetitive DNA elements such as LINES or SINES36 followed by fusion of 2 uncapped sister chromatids as the result of nonhomologous end joining, formation of a dicentric chromosome, anaphase breakage, and telomere healing or telomere capture.37,38 Three mechanisms are commonly proposed to explain the origin of this type of rearrangements identified for multiple chromosome arms. One of these mechanisms suggests that the inverted duplication arises from a double strand break and a U-type fusion between the 2 sister chromatids producing a dicentric chromosome followed by recombination with the unbroken in a U-type exchange and telomere healing.34 The second mechanisms involves the presence of low copy repeats, with stretches of repetitive DNA increasing the probability of nonallelic homologous recombination and the partial folding of 1 chromosome onto itself or between homologous chromosomes, leading to a recombination event between the inverted repeats.39 This model predicts that a single copy region flanked by the inverted repeats exist between the duplicated regions on the abnormal chromosome. This is the case of the cis-duplications or triplications with the gap between each duplicated block together with the A-type red probe. In addition, replication fork stalled at replication termination sites might be associated with inverted duplications and terminal deletions. Moreover, considering the role of SMCHD1 in the epigenetic regulation of the 4q and 10q loci,22,40 but also in ATM-dependent DNA damage signaling and repair of uncapped telomeres41 or nonhomologous end joining,42 the implication of this factor remains plausible, at least in cases where the rearranged allele segregates with SMCHD1 variant or a marked hypomethylation. It is also interesting to note the presence of additional telomeric probes in 3 patients, either on chromosome 4 or 10. These results further interrogate on the recombigenic properties of these highly repetitive regions but also strengthen the importance of telomeres in the regulation of these loci as highlighted in the past.31,43,44
In all rearrangements, additional D4Z4 arrays are flanked by A-type probes likely carrying the polyadenylation site (PAS, pLAM) required for DUX4 expression.10,11 It remains to be tested whether these duplicated arrays remain permissive for DUX4 transcription in an inverted configuration.
Overall, our data further underline the complexity of FSHD molecular diagnosis, in particular when facing complex genomic cases (p13E11 deletions, somatic mosaïcism, complex rearrangements, unresolved genotypes). Even if a large number of FSHD cases are linked to the shortening of the D4Z4 array on a 4qA allele, a high number of exceptions have been described in the literature. These numerous variants cannot be ignored for a full picture of this complex pathology, the analysis of genetic data, including for the development of novel methodologies and their subsequent interpretation, genetic counseling, or development of models of the pathology, bearing in mind all current technical limitations. Thus, taking into account the recurrence of 4q rearrangements, this category of cases, all clinically diagnosed with FSHD, should be considered as a third category in patient's classification.
Acknowledgment
The authors thank Drs. Chloé Tessereau and Dana Jaber (Bionano Genomics) for help and advice in SMOM data analysis.
Glossary
- FSHD
facioscapulohumeral dystrophy
- FSHD1
type 1 FSHD
- FSHD2
type 2 FSHD
- MC
molecular combing
- SB
Southern blotting
- SMOM
single molecule optical mapping
Appendix. Authors
Study Funding
This study was funded by Association Française contre les Myopathies (AFM-Telethon, TRIM-RD and MoThARD poles). M. Delourme is the recipient of a Fellowship from the French Ministry of Higher education and Research.
Disclosure
The authors report no relevant disclosures. Go to Neurology.org/NG for full disclosures.
References
- 1.Mostacciuolo ML, Pastorello E, Vazza G, et al. Facioscapulohumeral muscular dystrophy: epidemiological and molecular study in a north-east Italian population sample. Clin Genet. 2009;75(6):550-555. doi: 10.1111/j.1399-0004.2009.01158.x [DOI] [PubMed] [Google Scholar]
- 2.Deenen JC, Arnts H, van der Maarel SM, et al. Population-based incidence and prevalence of facioscapulohumeral dystrophy. Neurology. 2014;83(12):1056-1059. doi: 10.1212/wnl.0000000000000797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Padberg GW, van Engelen BG. Facioscapulohumeral muscular dystrophy. Curr Opin Neurol. 2009;22(5):539-542. doi: 10.1097/wco.0b013e328330a572 [DOI] [PubMed] [Google Scholar]
- 4.Wijmenga C, van Deutekom JC, Hewitt JE, et al. Pulsed-field gel electrophoresis of the D4F104S1 locus reveals the size and the parental origin of the facioscapulohumeral muscular dystrophy (FSHD)-associated deletions. Genomics. 1994;19(1):21-26. doi: 10.1006/geno.1994.1006 [DOI] [PubMed] [Google Scholar]
- 5.Sarfarazi M, Wijmenga C, Upadhyaya M, et al. Regional mapping of facioscapulohumeral muscular dystrophy gene on 4q35: combined analysis of an international consortium. Am J Hum Genet. 1992;51(2):396-403. [PMC free article] [PubMed] [Google Scholar]
- 6.Deutekom JC, Wljmenga C, Tlenhoven EA, et al. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum Mol Genet. 1993;2(12):2037-2042. doi: 10.1093/hmg/2.12.2037 [DOI] [PubMed] [Google Scholar]
- 7.Nguyen K, Broucqsault N, Chaix C, et al. Deciphering the complexity of the 4q and 10q subtelomeres by molecular combing in healthy individuals and patients with facioscapulohumeral dystrophy. J Med Genet. 2019;56(9):590-601. doi: 10.1136/jmedgenet-2018-105949 [DOI] [PubMed] [Google Scholar]
- 8.Roche S, Dion C, Broucqsault N, et al. Methylation hotspots evidenced by deep sequencing in patients with facioscapulohumeral dystrophy and mosaicism. Neurol Genet. 2019;5(6):e372. doi: 10.1212/nxg.0000000000000372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemmers RJ, de Kievit P, Sandkuijl L, et al. Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat Genet. 2002;32(2):235-236. doi: 10.1038/ng999 [DOI] [PubMed] [Google Scholar]
- 10.Lemmers RJ, van der Vliet PJ, Klooster R, et al. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010;329(5999):1650-1653. doi: 10.1126/science.1189044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gabriels J, Beckers MC, Ding H, et al. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene. 1999;236(1):25-32. doi: 10.1016/s0378-1119(99)00267-x [DOI] [PubMed] [Google Scholar]
- 12.Lemmers RJ, Tawil R, Petek LM, et al. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat Genet. 2012;44(12):1370-1374. doi: 10.1038/ng.2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lemmers RJ, van der Maarel SM, van Deutekom JC, et al. Inter- and intrachromosomal sub-telomeric rearrangements on 4q35: implications for facioscapulohumeral muscular dystrophy (FSHD) aetiology and diagnosis. Hum Mol Genet. 1998;7(8):1207-1214. doi: 10.1093/hmg/7.8.1207 [DOI] [PubMed] [Google Scholar]
- 14.Matsumura T, Goto K, Yamanaka G, et al. Chromosome 4q;10q translocations; comparison with different ethnic populations and FSHD patients. BMC Neurol. 2002;2(1):7. doi: 10.1186/1471-2377-2-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Overveld PG, Lemmers RJ, Deidda G, et al. Interchromosomal repeat array interactions between chromosomes 4 and 10: a model for subtelomeric plasticity. Hum Mol Genet. 2000;9(19):2879-2884. doi: 10.1093/hmg/9.19.2879 [DOI] [PubMed] [Google Scholar]
- 16.Nguyen K, Puppo F, Roche S, et al. Molecular combing reveals complex 4q35 rearrangements in facioscapulohumeral dystrophy. Hum Mutat. 2017;38(10):1432-1441. doi: 10.1002/humu.23304 [DOI] [PubMed] [Google Scholar]
- 17.van der Maarel SM, Deidda G, Lemmers RJ, et al. De novo facioscapulohumeral muscular dystrophy: frequent somatic mosaicism, sex-dependent phenotype, and the role of mitotic transchromosomal repeat interaction between chromosomes 4 and 10. Am J Hum Genet. 2000;66(1):26-35. doi: 10.1086/302730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magdinier FUM. Facioscapulohumeral Muscular Dystrophy: Genetics. Encyclopedy of Life Science; 2018. [serial online]. [Google Scholar]
- 19.Lemmers RJL, de Kievit P, van Geel M, et al. Complete allele information in the diagnosis of facioscapulohumeral muscular dystrophy by triple DNA analysis. Ann Neurol. 2001;50(6):816-819. doi: 10.1002/ana.10057 [DOI] [PubMed] [Google Scholar]
- 20.Deidda G, Cacurri S, Piazzo N, Felicetti L. Direct detection of 4q35 rearrangements implicated in facioscapulohumeral muscular dystrophy (FSHD). J Med Genet. 1996;33(5):361-365. doi: 10.1136/jmg.33.5.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen K, Walrafen P, Bernard R, et al. Molecular combing reveals allelic combinations in facioscapulohumeral dystrophy. Ann Neurol. 2011;70(4):627-633. doi: 10.1002/ana.22513 [DOI] [PubMed] [Google Scholar]
- 22.Lemmers R, van der Vliet PJ, Vreijling JP, et al. Cis D4Z4 repeat duplications associated with facioscapulohumeral muscular dystrophy type 2. Hum Mol Genet. 2018;27(20):3488-3497. doi: 10.1093/hmg/ddy236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai Y, Li P, Wang Z, et al. Single-molecule optical mapping enables quantitative measurement of D4Z4 repeats in facioscapulohumeral muscular dystrophy (FSHD). J Med Genet. 2020;57(2):109-120. doi: 10.1136/jmedgenet-2019-106078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stence AA, Thomason JG, Pruessner JA, et al. Validation of optical genome mapping for the molecular diagnosis of facioscapulohumeral muscular dystrophy. J Mol Diagn. 2021;23(11):1506-1514. doi: 10.1016/j.jmoldx.2021.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scionti I, Greco F, Ricci G, et al. Large-scale population analysis challenges the current criteria for the molecular diagnosis of fascioscapulohumeral muscular dystrophy. Am J Hum Genet. 2012;90(4):628-635. doi: 10.1016/j.ajhg.2012.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Q, Xu X, Ding L, et al. Clinical application of single-molecule optical mapping to a multigeneration FSHD1 pedigree. Mol Genet Genomic Med. 2019;7(3):e565. doi: 10.1002/mgg3.565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitsuhashi S, Nakagawa S, Takahashi Ueda M, Imanishi T, Frith MC, Mitsuhashi H. Nanopore-based single molecule sequencing of the D4Z4 array responsible for facioscapulohumeral muscular dystrophy. Sci Rep. 2017;7(1):14789. doi: 10.1038/s41598-017-13712-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemmers RJ, Van Overveld PG, Sandkuijl LA, et al. Mechanism and timing of mitotic rearrangements in the subtelomeric D4Z4 repeat involved in facioscapulohumeral muscular dystrophy. Am J Hum Genet. 2004;75(1):44-53. doi: 10.1086/422175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciszewski L, Lu-Nguyen N, Slater A, et al. G-quadruplex ligands mediate downregulation of DUX4 expression. Nucleic Acids Res. 2020;48(8):4179-4194. doi: 10.1093/nar/gkaa146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsumagari K, Qi L, Jackson K, et al. Epigenetics of a tandem DNA repeat: chromatin DNaseI sensitivity and opposite methylation changes in cancers. Nucleic Acids Res. 2008;36(7):2196-2207. doi: 10.1093/nar/gkn055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnoult N, Schluth-Bolard C, Letessier A, et al. Replication timing of human telomeres is chromosome arm-specific, influenced by subtelomeric structures and connected to nuclear localization. PLoS Genet. 2010;6(4):e1000920. doi: 10.1371/journal.pgen.1000920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Muse T, Aguilera A. Transcription-replication conflicts: how they occur and how they are resolved. Nat Rev Mol Cell Biol. 2016;17(9):553-563. doi: 10.1038/nrm.2016.88 [DOI] [PubMed] [Google Scholar]
- 33.Achar YJ, Foiani M. Coordinating replication with transcription. Adv Exp Med Biol. 2017;1042:455-487. doi: 10.1007/978-981-10-6955-0_20 [DOI] [PubMed] [Google Scholar]
- 34.Weleber RG, Verma RS, Kimberling WJ, Fieger HG Jr., lubs HA. Duplication-deficiency of the short arm of chromosome 8 following artificial insemination. Ann Genet. 1976;19(4):241-247. [PubMed] [Google Scholar]
- 35.Weckselblatt B, Rudd MK. Human structural variation: mechanisms of chromosome rearrangements. Trends Genet. 2015;31(10):587-599. doi: 10.1016/j.tig.2015.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ballif BC, Yu W, Shaw CA, Kashork CD, Shaffer LG. Monosomy 1p36 breakpoint junctions suggest pre-meiotic breakage-fusion-bridge cycles are involved in generating terminal deletions. Hum Mol Genet. 2003;12(17):2153-2165. doi: 10.1093/hmg/ddg231 [DOI] [PubMed] [Google Scholar]
- 37.Lamb J, Harris PC, Wilkie AO, Wood WG, Dauwerse JG, Higgs DR. De novo truncation of chromosome 16p and healing with (TTAGGG)n in the alpha-thalassemia/mental retardation syndrome (ATR-16). Am J Hum Genet. 1993;52(4):668-676. [PMC free article] [PubMed] [Google Scholar]
- 38.Flint J, Wilkie AO, Buckle VJ, Winter RM, Holland AJ, McDermid HE. The detection of subtelomeric chromosomal rearrangements in idiopathic mental retardation. Nat Genet. 1995;9(2):132-140. doi: 10.1038/ng0295-132 [DOI] [PubMed] [Google Scholar]
- 39.Shaffer LG, Lupski JR. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu Rev Genet. 2000;34(1):297-329. doi: 10.1146/annurev.genet.34.1.297 [DOI] [PubMed] [Google Scholar]
- 40.Dion C, Roche S, Laberthonniere C, et al. SMCHD1 is involved in de novo methylation of the DUX4-encoding D4Z4 macrosatellite. Nucleic Acids Res. 2019;47(6):2822-2839. doi: 10.1093/nar/gkz005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vančevska A, Ahmed W, Pfeiffer V, Feretzaki M, Boulton SJ, Lingner J. SMCHD1 promotes ATM-dependent DNA damage signaling and repair of uncapped telomeres. Embo J. 2020;39(7):e102668. doi: 10.15252/embj.2019102668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang M, Li Y, Zhang X, et al. Structural maintenance of chromosomes flexible hinge domain containing 1 (SMCHD1) promotes non-homologous end joining and inhibits homologous recombination repair upon DNA damage. J Biol Chem. 2014;289(49):34024-34032. doi: 10.1074/jbc.m114.601179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robin JD, Ludlow AT, Batten K, et al. SORBS2 transcription is activated by telomere position effect-over long distance upon telomere shortening in muscle cells from patients with facioscapulohumeral dystrophy. Genome Res. 2015;25(12):1781-1790. doi: 10.1101/gr.190660.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ottaviani A, Schluth-Bolard C, Rival-Gervier S, et al. Identification of a perinuclear positioning element in human subtelomeres that requires A-type lamins and CTCF. Embo J. 2009;28(16):2428-2436. doi: 10.1038/emboj.2009.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable because all available data are presented in the manuscript.