Abstract
Transcription of the tetA(K) tetracycline resistance determinant encoded by an IS257-flanked cointegrated copy of a pT181-like plasmid, located within the chromosomal mec region of a methicillin-resistant Staphylococcus aureus isolate, has been investigated. The results demonstrated that transcription of tetA(K) in this strain is directed by both an IS257-derived hybrid promoter, which is stronger than the native tetA(K) promoter in the autonomous form of pT181, and a complete outwardly directed promoter identified within one end of IS257. Despite lower gene dosage, the chromosomal configuration was shown to afford a higher level of resistance than that mediated by pT181 in an autonomous multicopy state. Furthermore, competition studies revealed that a strain carrying the chromosomal tetA(K) determinant exhibited a higher level of fitness in the presence of tetracycline but not in its absence. This finding suggests that tetracycline has been a selective factor in the emergence of strains carrying a cointegrated pT181-like plasmid in their chromosomes. The results highlight the potential of IS257 to influence the expression of neighboring genes, a property likely to enhance its capacity to mediate advantageous genetic rearrangements.
The staphylococcal insertion element IS257 has been found in association with determinants encoding resistance to antiseptics and disinfectants, aminoglycosides, bleomycin, cadmium, mercury, mupirocin, tetracycline, trimethoprim, and virginiamycin in both Staphylococcus aureus and coagulase-negative staphylococci (7). IS257, also known as IS431 (3), is 789 or 790 bp in length, is bounded by imperfect 27-bp terminal inverted repeats, and contains a single gene encoding a transposase (30). This element is a member of the IS6 family of bacterial insertion sequences, which contains representatives from both gram-positive and gram-negative bacteria (19). IS257-mediated cointegrate formation is thought to be responsible for the incorporation of the above-mentioned resistance determinants into staphylococcal chromosomes and plasmids through the process of nonresolved replicative transposition (33).
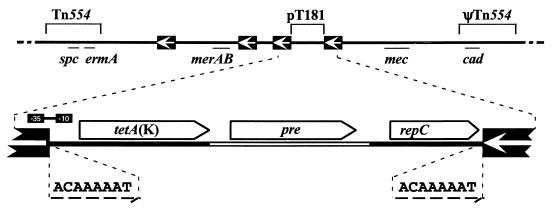
Four copies of IS257 are present in the region of the chromosome associated with methicillin resistance (the mec region) of the Australian clinical isolate SK1660 (8). Two of these copies flank genes mediating resistance to mercurial compounds, whereas the other copies flank a tetracycline resistance determinant (see Fig. 1). The latter structure corresponds to a cointegrated copy of a pT181-like plasmid that appears to have been cointegrated into the chromosome as a consequence of IS257 insertion between the replication initiation gene, repC, and tetA(K) (9), which encodes an efflux pump conferring tetracycline resistance (10). Consistent with such an insertion site, nucleotide sequencing of an equivalent segment from a similar methicillin-resistant S. aureus (MRSA) strain, ANS46, revealed the presence of 8-bp target duplications at the extremities of the integrated plasmid, corresponding to a sequence located between repC and tetA(K) (20, 34). Retrospective studies have revealed that staphylococcal strains isolated in Australian hospitals prior to 1970 commonly contained an autonomous pT181-like plasmid, whereas later isolates, such as SK1660 and ANS46, typically possessed the chromosomally cointegrated form of the plasmid (9). Identically organized plasmid cointegrates have also been detected in the chromosomal mec region of MRSA strains isolated in the United States and Greece (34).
FIG. 1.
Genetic organization of pT181 in the mec region of the chromosome of MRSA strains ANS46 and SK1660 (map adapted from Gillespie et al. [8] and Matthews et al. [20]). The IS257-plasmid junctions in SK1660 were amplified using the primer pairs IS257-719 and pT181-2334BamHI, and IS257-685BglII and pT181-3936 (Table 1) and sequenced. The plasmid sequence obtained, shown as a solid line in the enlarged view, corresponds to nt 1 to 2335 and nt 3975 to 4439 of the pT181 sequence (GenBank entry J01764). The following genes are shown: cad, cadmium resistance; ermA, resistance to macrolides, lincosamides, and streptogramin B; mec, methicillin resistance; merAB, mercury resistance; and spc, spectinomycin resistance. The locations of Tn554 and a related structure, ΨTn554, are indicated. Copies of IS257 are represented by solid boxes; the arrows indicate the direction of IS257 transposase transcription. The pT181 genes shown, with directions of transcription indicated by boxes with pointed ends, are pre, plasmid recombination; repC, replication initiation; and tetA(K), tetracycline resistance. The approximate positions of the −35 and −10 sequences for the IS257 hybrid promoter are shown. Eight-base pair target duplication sequences are denoted by dashed arrows.
In addition to facilitating the capture of resistance genes, IS257 has been shown to play a role in the expression of the trimethoprim resistance gene, dfrA. In strains exhibiting high-level trimethoprim resistance, transcription of dfrA is directed by a hybrid promoter consisting of a −35 sequence encoded within the end of IS257 and a −10 sequence located in the adjacent sequence (15). Sequence analysis has suggested that an analogous IS257-derived hybrid promoter might also be responsible for transcription of tetA(K) in the chromosomal pT181-like plasmid cointegrate of strains such as SK1660 (33). To further investigate the potential of IS257 to influence the expression of genes with which it is associated, we have analyzed the transcription of the tetA(K) gene from SK1660. A comparative analysis of tetA(K) promoter strengths, levels of tetracycline resistance, growth rates, and competitive fitness of strains carrying the chromosomally cointegrated and autonomous forms of pT181 was performed so as to gain insight into the possible reasons for the emergence of strains carrying the cointegrate structure.
MATERIALS AND METHODS
Bacterial strains, plasmids, and primers.
The S. aureus and Escherichia coli strains and plasmids and the oligonucleotides used in this study are listed in Table 1. All strains were cultured at 37°C in Luria-Bertani (LB) medium (31) containing, where appropriate, ampicillin (100 μg/ml), chloramphenicol (10 μg/ml), spectinomycin (50 μg/ml), or tetracycline (various concentrations). Recombinant plasmids were initially electroporated into E. coli DH5α (Table 1).
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Genotype, description, or sequence | Reference or source |
|---|---|---|
| E. coli DH5α | supE44 ΔlacU169(φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Bethesda Research Laboratories |
| S. aureus | ||
| NCTC8325-4 | Laboratory strain | 28 |
| RN4220 | Restrictionless derivative of NCTC8325-4 | 28 |
| RN2424 | NCTC8325-4 harboring pT181 | 26 |
| SK5318 | RN4220; geh::pCL84 | This study |
| SK5319 | RN4220 harboring pT181 | This study |
| SK5323 | RN4220; geh::pSK5321 | This study |
| SK1660 | MRSA strain isolated in 1976 | 9 |
| CYL316 | RN4220 harboring pYL112Δ19 | 14 |
| Plasmids | ||
| pCL84 | S. aureus integration vector that inserts into the lipase gene (geh); includes HindIII fragment carrying tetA(K) from pT181; confers spectinomycin resistance | 14 |
| pSK5321 | pCL84 carrying 2.4-kb HindIII fragment containing IS257-tetA(K) region from SK1660, which replaces the HindIII tetA(K) fragment from pT181 | This study |
| pT181 | 4.45-kb S. aureus plasmid containing tetA(K) | 13 |
| pYL112Δ19 | Encodes the integrase that mediates chromosomal integration of pCL84 and derivatives; confers chloramphenicol resistance | 14 |
| Primersa | ||
| geh-2488 | 5′-GCAAGAGAAGAAGAATGGCG-3′ | |
| IS257-685BglII | 5′-GATCGAAGATCTACGGATTTTCGCCATGCC-3′ | |
| IS257-719 | 5′-CATGATGCTAATTTCGTGGCATGG-3′ | |
| pT181-904 | 5′-TCACCTCATATAAATTCCCCAAAT-3′ | |
| pT181-1010 | 5′-GCCAAAATAAAACGCTATAAAACAAACC-3′ | |
| pT181-1095 | 5′-CTCCTGGAATTACAAACTGGGTAAAC-3′ | |
| pT181-1578 | 5′-GACATTAAAACAATACCTACGATATC-3′ | |
| pT181-2334BamHI | 5′-GAGGGATCCTATTATTCAAACTGCTTTTCAGAACG-3′ | |
| pT181-3025 | 5′-GCTCATTAAATCTATCTTGAAACGCTG-3′ | |
| pT181-3936 | 5′-GGAGTTGAAGAAATGAAATTCAGAG-3′ |
The number of each primer refers to the 5′-most base in each instance that corresponds to a nucleotide in the previously published sequences of IS257 (GenBank entry U40381), pT181 (GenBank entry J01764), and geh (GenBank entry M12715). Restriction endonuclease recognition sites incorporated into primers, as indicated in the primer names, are underlined.
To generate SK5323 (Table 1), which carries a single copy of the tetA(K) gene behind the IS257 hybrid promoter in the RN4220 chromosome, pSK5321 (Table 1) was constructed. A 2.9-kb PCR product amplified from the SK1660 chromosome, using primers IS257-719 and pT181-3025 (Table 1), and cut with HindIII was cloned into pCL84 (Table 1) in the same orientation as the HindIII fragment carrying the tetA(K) gene, which it replaced. The pCL84 vector inserts into the lipase gene (geh) of the staphylococcal chromosome (14). Following transformation of pSK5321 into the S. aureus RN4220 derivative CYL316 (Table 1), which contains the integrase-encoding plasmid pYL112Δ19, the integrants were selected on LB agar containing 3 μg of tetracycline/ml. Loss of lipase activity in a selected transformant was confirmed on Sierra medium (1), which permits identification of lipase mutants; a precipitate surrounds colonies expressing lipase activity. SK5318 (Table 1) was constructed by integrating a copy of pCL84, which carries the tetA(K) gene and promoter region from pT181 (14), into the CYL316 chromosome. All clones constructed from the products of PCRs were sequenced to confirm that no mutations had been incorporated during amplification. Insertions into the geh gene were checked by PCR using the primers pT181-1095 and geh-2488 (Table 1), which are complementary to sequences within tetA(K) and geh, respectively. SK5323 and SK5318 were cured of pYL112Δ19 by a previously described method (18).
DNA isolation, recombinant DNA techniques, and bacterial electroporation.
Plasmid DNA was isolated from E. coli using the Quantum Prep miniprep kit (Bio-Rad) according to the manufacturer's instructions. S. aureus DNA isolations were performed as described previously (18). All restriction endonuclease digestion and ligation reactions were carried out in accordance with the manufacturers' instructions. DNA cloning was performed by standard techniques (31). PCR was undertaken using Pfu (Stratagene) or Pyrostase (Molecular Genetic Resources) enzymes, according to the manufacturers' instructions, in an MJ Research PTC-100 with Hot Bonnet. Primers were synthesized using a Beckman Oligo 1000 DNA synthesizer. Electroporation of E. coli (5) and S. aureus (32) was performed with a Bio-Rad Gene Pulser with a pulse controller.
DNA sequencing.
Automated cycle sequencing was performed by the Sydney University and Prince Alfred Macromolecular Analysis Centre or the Australian Genome Research Facility. Manual sequencing was carried out with a Sequitherm sequencing kit (Epicentre Technologies) to obtain sequence ladders for transcript mapping. Sequence data was stored and assembled with the program SEQUENCHER (Gene Codes Corporation).
Transcript mapping by primer extension.
Total cellular RNA was isolated from S. aureus strains as previously described (15). Transcript mapping was performed essentially as described by Ausubel et al. (2). Two primers were used; oligonucleotide pT181-1010 (Table 1), complementary to sequences within the tetA(K) structural gene and 145 bp from the expected hybrid promoter transcription start point (TSP), and oligonucleotide pT181-904 (Table 1), 43 bp from the expected TSP.
Northern hybridization.
Total cellular RNA was isolated from S. aureus strains using a high-speed reciprocating homogenizer (FastPREP apparatus; BIO101) and a FastRNA isolation kit (BIO101), according to the manufacturer's instructions. An approximate total RNA concentration for each sample was determined spectrophotometrically (QuantaGene; Pharmacia Biotech). Accurate estimation of the relative RNA content of individual samples was achieved by electrophoresis of aliquots through a 1.0% agarose gel in TAE buffer (40 mM Tris-HCl [pH 8.5], 5 mM sodium acetate, 1 mM EDTA), ethidium bromide staining, visualization with a Molecular Imager FX (Bio-Rad), and quantitation by volume analysis of the 16S and 23S rRNA bands in the resulting image using the software Quantity One (Bio-Rad). Aliquots containing equal amounts of total RNA (approximately 14 μg) were electrophoresed through a 2.2 M formaldehyde–1.5% agarose gel in MOPS buffer (20 mM 3-N-morpholinopropanesulfonic acid, 8 mM sodium acetate, 1 mM EDTA, pH 7.0). RNA was transferred to a Hybond N+ membrane (Amersham) via capillary action. The DNA probe consisted of a 484-bp internal tetA(K) fragment amplified from pT181 using primers pT181-1095 and pT181-1578 (Table 1). The probe was purified by using a Microcon YM-100 (Millipore) and radiolabeled with [α-32P]dCTP by the random-primed method (Ready-To-Go labeling kit; Pharmacia). Hybridization was performed by standard methods (31). The membrane was imaged with a storage phosphor screen (Kodak) and a Molecular Imager FX. Relative amounts of tetA(K) mRNA were quantitated by volume analysis.
Antimicrobial susceptibility testing.
The MIC of tetracycline for a strain was determined by the standard agar dilution method according to National Committee for Clinical Laboratory Standards guidelines for antimicrobial susceptibility testing (24).
Inhibition studies.
Overnight cultures grown in the absence or presence of 2 μg of tetracycline/ml were diluted to an optical density at 600 nm (OD600) of 0.05 and subcultured into 4 ml of LB medium in the presence of tetracycline at 0, 2, 4, 8, 16, 32, 64, and 128 μg/ml. After growth for 3.5 h, the OD600 of the culture was determined.
Growth studies.
Overnight cultures grown in LB medium were diluted to an OD600 of 0.05 and subcultured into 100 ml of LB medium, LB medium containing 1 μg of tetracycline/ml, and LB medium containing 5 μg of tetracycline/ml. The OD600 was determined at the beginning of the experiment and subsequently at 0.5-h intervals for 3.5 h.
Competition studies.
Equal proportions of overnight cultures of strains SK5319 and SK5323 were diluted to an OD600 of 0.05 and used to inoculate 10 ml of LB medium or LB medium containing 1 μg of tetracycline/ml. These mixed cultures were diluted 10,000-fold each day in fresh medium for 8 days; such dilution results in approximately 13.3 generations per day, a value confirmed by viable counts on selected days. The relative proportions of each strain were determined at the beginning of the experiment and subsequently every 24 h by spreading dilutions of the mixed culture on Sierra medium (1) and scoring for lipase activity. DNA isolations from colonies identified by lipase activity to be SK5319 confirmed the presence of pT181.
Statistical analysis.
Statistical analysis was carried out with Statview (SAS Institute Inc.). Differences between groups were evaluated by Fisher's protected least significant difference test after analysis of variance, and by repeated-measures analysis where appropriate. A significant difference was defined as a P value of <0.05.
RESULTS
Structure of the cointegrated pT181-like plasmid in the SK1660 chromosome.
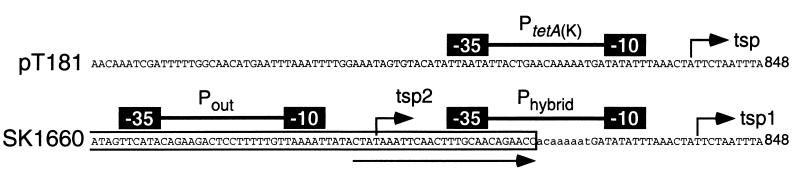
The previous identification of a putative IS257-derived hybrid promoter upstream of tetA(K) in the pT181-like chromosomal cointegrate was based on the published IS257 insertion site in the strain ANS46 (20) and assumed sequence identity between the integrated plasmid and pT181 (33). Amplification and sequencing of both IS257-plasmid junctions from SK1660 confirmed that this strain possesses the same cointegrate structure as ANS46, including the same target duplication at the extremities of the plasmid (Fig. 1). The tetA(K) gene and upstream sequence were found to be identical to those of pT181 (13). However, sequencing of the replication region of the cointegrated plasmid revealed that the repC gene contains a 10-bp duplication (corresponding to nucleotides [nt] 4370 to 4379 of pT181 [GenBank entry J01764]) which has resulted in a truncated RepC protein of only 30 amino acids, consistent with the suggestion that the repC gene of the cointegrated pT181 in ANS46 is defective (6). These findings, therefore, confirm the presence of the proposed hybrid promoter upstream of tetA(K) in SK1660, designated Phybrid, consisting of the candidate −35 and −10 sequences, TTGCAA and TATATT, respectively, separated by 17 bp (Fig. 2).
FIG. 2.
Organization of tetA(K) promoters in pT181 and SK1660. The sequence corresponds to that obtained from the tetA(K) regions of pT181 and the chromosome of SK1660. The boxed sequence represents IS257 sequence. The arrow underneath the sequence represents the terminal inverted repeat of IS257. The arrows above the sequence represent TSPs identified in this study. The solid boxes represent −10 and −35 sequences of the promoters PtetA(K), Phybrid, and Pout. The sequence in lowercase letters represents the 8-bp target duplication. The number at the end of the sequence indicates the position in the pT181 sequence (GenBank entry J01764).
Transcription of tetA(K).
To determine if the putative hybrid promoter upstream of tetA(K) in SK1660 is functional, primer extension studies were performed. For comparison, equivalent studies were undertaken with strain RN2424 (Table 1) to determine the TSP of tetA(K) in the autonomously carried plasmid pT181 (data not shown). As summarized in Fig. 2, two extension products were obtained for SK1660. The position of the more intense product corresponded to a thymine residue 117 bp upstream of the tetA(K) start codon and is consistent with a TSP expected for Phybrid, designated TSP1, thereby confirming its activity. The weaker extension product, designated TSP2, corresponded to a thymine residue 164 bp upstream from the tetA(K) start codon. Examination of the sequence upstream of TSP2 revealed the presence of an appropriately positioned candidate promoter, Pout, consisting of the −35 and −10 sequences, TTCATA and TAAAAT, respectively, separated by 18 bp. Pout represents a complete, outwardly directed promoter within one end of IS257. Thus, transcription of tetA(K) in SK1660 initiates at two sites; the majority of tetA(K) transcripts appear to initiate within the cointegrated plasmid, directed by Phybrid, whereas a smaller proportion originate within the upstream copy of IS257, directed by Pout (Fig. 2).
A single extension product was detected with RNA isolated from S. aureus RN2424, identifying a TSP at approximately the same position as that recognized for Phybrid in SK1660 (Fig. 2). It would therefore appear that the same −10 sequence is utilized by Phybrid and the native promoter for tetA(K) in the autonomous form of pT181, designated PtetA(K) (Fig. 2). A candidate −35 sequence for PtetA(K), TTAATA, is located 17 bp upstream of this −10 sequence. It should be noted that although the TSPs identified in Fig. 2 were obtained using primer pT181-904, which is complementary to sequences located upstream of tetA(K), equivalent experiments utilizing primer pT181-1010, which is complementary to sequences located within the tetA(K) coding sequence, identified the same TSPs, thereby ruling out the possibility of other promoters closer to tetA(K).
Comparative levels of tetA(K) transcription.
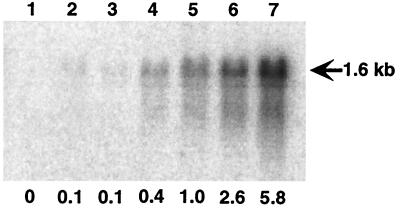
To investigate the comparative levels of tetA(K) transcription, a DNA segment from strain SK1660 encoding tetA(K) and upstream IS257-derived sequences, including Phybrid and Pout, was cloned into the integration vector, pCL84 (14), and subsequently inserted into the chromosomal lipase gene (geh) of S. aureus RN4220 to generate strain SK5323. For comparison, an equivalent fragment of pT181, encompassing tetA(K) and its native promoter, was similarly inserted into the RN4220 chromosome to generate SK5318. Northern hybridization (Fig. 3) demonstrated an approximately sixfold increase in the amount of tetA(K) mRNA in SK5323 cells carrying a single copy of the tetA(K) gene, in comparison to SK5319 cells carrying autonomous pT181 present in multiple copies; pT181 is normally maintained at approximately 20 copies per cell (27). An equivalent strong transcript from the clinical strain SK1660 was similarly detected (data not shown).
FIG. 3.
Northern blot analysis of tetA(K) transcription. The size of the transcript, shown as 1.6 kb, was determined using coelectrophoresed RNA markers. The numbers shown at the bottom indicate the amounts of transcript relative to that detected in SK5319 (lane 5). The lanes were loaded with RNA from RN4220 (lane 1), SK5318 (lanes 2 and 3), SK5319 (lanes 4 and 5), and SK5323 (lanes 6 and 7). The RNAs in lanes 3, 5, and 7 were isolated from strains grown in the presence of 2 μg of tetracycline/ml, and the RNAs in lanes 1, 2, 4, and 6 were isolated from strains grown without selection.
It therefore appears that the lower gene dosage due to the chromosomal integration of tetA(K) is compensated for by the generation of a stronger promoter. Indeed, comparison of the amount of tetA(K) transcript in SK5318 and SK5323 cells, carrying chromosomal tetA(K) behind the native and hybrid promoters, respectively, in the presence of tetracycline (Fig. 3, lanes 3 and 7), suggests an approximately 60-fold increase in promoter strength for the hybrid promoter. A twofold increase in the amount of tetA(K) transcript is seen in SK5319 and SK5323 cells grown in the presence of tetracycline (Fig. 3, lanes 4 to 7).
Comparative levels of tetracycline susceptibility.
Since tetA(K) is present as only a single copy in the chromosome of S. aureus SK1660, rather than approximately 20 copies typically carried by a cell harboring autonomous pT181 (27), and is transcribed from a different promoter configuration, we were interested in ascertaining the level of tetracycline resistance expressed by this strain. The MIC of tetracycline for SK1660 was found to be two to four times higher than that for RN2424, which contains autonomous pT181 (Table 2). However, this comparison is complicated, since the tetracycline resistance phenotype of a clinical strain such as SK1660 could reflect factors other than the genetic context of its tetA(K) gene. For example, SK1660 is thought to possess an additional determinant which mediates resistance to both tetracycline and the semisynthetic derivative, minocycline (9). A more informative comparison is provided by analysis of the tetracycline susceptibilities of tetA(K)-containing RN4220 derivatives. The MIC of tetracycline for SK5323, bearing Phybrid, was twice that for SK5319, containing pT181, and eight times higher than that for SK5318, which contains chromosomal PtetA(K) (Table 2). This pattern was observed irrespective of whether the cells were initially grown in the absence of tetracycline or preexposed to the antibiotic; preexposure resulted in reduced susceptibility values, presumably due to the protective effect of preexisting TetA(K) protein.
TABLE 2.
Tetracycline susceptibilities of strains used in this study
| Strain | tetA(K) promoter(s) [locationa] | MIC of tetracyclineb (μg/ml) |
|---|---|---|
| NCTC8325-4 | 1 | |
| RN2424 | PtetA(K) [pT181] | 32 [128] |
| SK1660 | Pout + Phybrid [chr] | 128 [256] |
| RN4220 | 1 | |
| SK5318 | PtetA(K) [chr] | 8 [32] |
| SK5319 | PtetA(K) [pT181] | 32 [128] |
| SK5323 | Pout + Phybrid [chr] | 64 [256] |
Location of tetA(K) and promoters; chr, chromosome; pT181, autonomous pT181.
Values indicate the MICs of tetracycline determined according to standard procedures for strains pregrown in the absence of tetracycline; values in brackets are MICs for strains pregrown in the presence of 2 μg of tetracycline/ml prior to the assay.
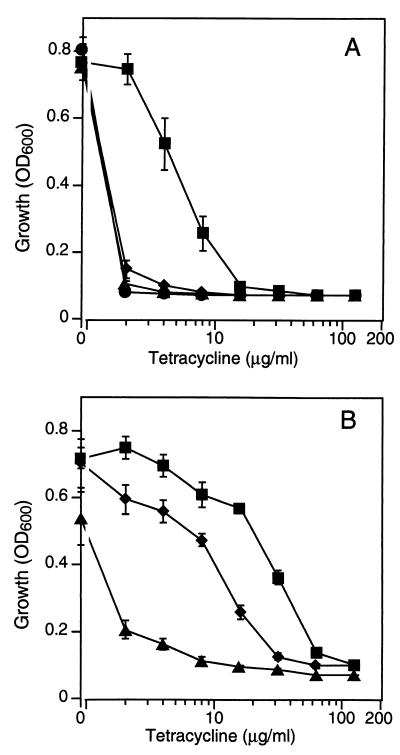
The impact of each promoter configuration on the tetracycline resistance of an entire bacterial population was investigated using growth inhibition studies (Fig. 4). Statistical analysis revealed that, in comparison to SK5319, SK5323 was significantly less inhibited in tetracycline concentrations ranging from 2 to 32 μg/ml for both preexposed and naïve cells (P < 0.05). The growth of SK5319, preexposed to tetracycline, was significantly stronger than that of SK5318 in tetracycline concentrations ranging from 2 to 128 μg/ml (P < 0.05), presumably reflecting its greater tetA(K) copy number in the multicopy pT181 plasmid. Interestingly, in the absence of preexposure to tetracycline, no significant difference in growth was detected between SK5319 and either the background strain RN4220 or SK5318 within the 3.5-h period of the experiment. This result probably reflects a lag in the expression of TetA(K), which is thought to be inducible by translation attenuation (13). The increased basal level of TetA(K) present in SK5323, resulting from enhanced transcription of tetA(K), presumably mitigates lag in this strain at this level of tetracycline.
FIG. 4.
Tetracycline resistance of S. aureus RN4220 and tetA(K)-containing derivatives. Overnight cultures grown in the absence (A) or presence (B) of tetracycline (2 μg/ml) were subcultured in the presence of tetracycline at the indicated concentrations. The OD was then determined after 3.5 h. Each data point is the mean of six experiments. Data for the following strains are shown: RN4220 (●), SK5318 (▴; PtetA(K) in the chromosome), SK5319 (⧫; PtetA(K) in pT181), and SK5323 (■; Pout and Phybrid in the chromosome); for promoter configurations, see Fig. 2. The error bars represent standard error.
Influence of promoter configuration on bacterial growth.
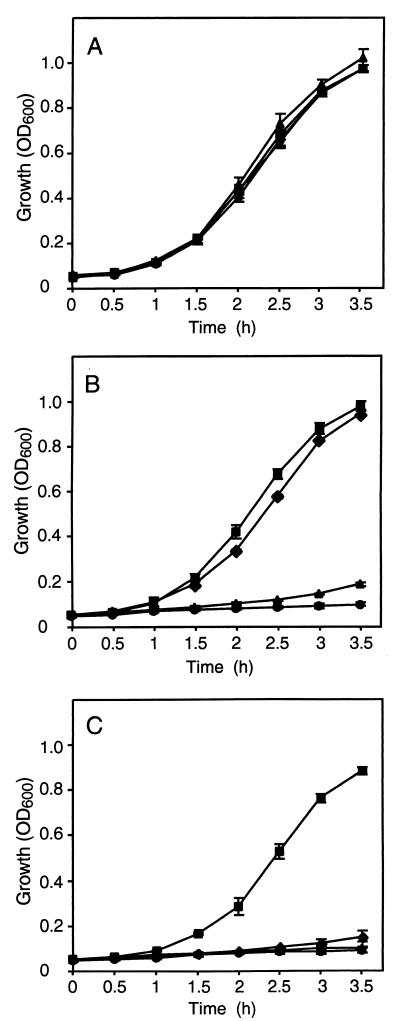
The studies described above demonstrated that a chromosomal tetA(K) gene behind Phybrid affords a higher degree of tetracycline resistance than that mediated by pT181. It is conceivable that this configuration might also be advantageous in the absence or in the presence of low levels of tetracycline, since it would be expected to relieve any burden associated with carriage of a multicopy plasmid. Growth studies revealed that in the absence of tetracycline there was no significant difference between the growth rates of the four strains in the 3.5-h period of the experiment (Fig. 5A). However, at low (Fig. 5B) and intermediate (Fig. 5C) levels of tetracycline (1 and 5 μg/ml, respectively), SK5323 cells grew significantly better than cells carrying all other promoter configurations (P < 0.0001 in all cases except SK5323 versus SK5319 at 1 μg/ml, where P was <0.05). The curves shown in Fig. 5B and C emphasize a reduced lag in the growth of SK5323, in which tetA(K) is expressed from Phybrid, in comparison to the other strains.
FIG. 5.
Relative growth of S. aureus strains carrying various tetA(K) promoter configurations. Overnight cultures were subcultured in LB medium (A), LB medium containing 1 μg of tetracycline/ml (B), and LB medium containing 5 μg of tetracycline/ml (C). Each data point is the mean of three experiments. Data for the following strains are shown: RN4220 (●), SK5318 (▴; PtetA(K) in the chromosome), SK5319 (⧫; PtetA(K) in pT181), and SK5323 (■; Pout and Phybrid in the chromosome); for promoter configurations, see Fig. 2. The error bars represent standard error.
Relative fitness of tetracycline-resistant strains.
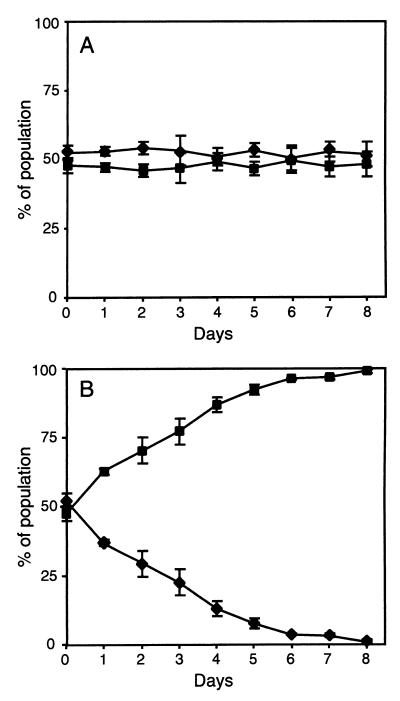
To determine if the properties associated with the hybrid promoter, viz., lower susceptibility to tetracycline and reduced lag time in its presence, result in improved relative fitness, competition assays between strains SK5323 and SK5319 were undertaken. In essence, with respect to tetA(K)-mediated tetracycline resistance, the former is equivalent to the clinical isolate SK1660, whereas the latter corresponds to its presumed progenitor carrying the autonomous pT181 plasmid. Consistent with the growth studies (Fig. 5A), in the absence of tetracycline (Fig. 6A), no significant difference between the fitnesses of SK5323 and SK5319 was detected during 8 days. Conversely, in the presence of a subinhibitory level of tetracycline (1 μg/ml [Fig. 6B]), SK5323 cells had a significantly higher level of fitness than those of strain SK5319 (P < 0.0001).
FIG. 6.
Relative fitness of S. aureus strains carrying different tetA(K) promoter configurations. Equal proportions of SK5323 (■; Pout and Phybrid in the chromosome) and SK5319 (⧫; PtetA(K) in pT181) cells were used to inoculate LB medium (A) or LB medium containing 1 μg of tetracycline/ml (B); for promoter configurations, see Fig. 2. Eight days of growth represents approximately 100 generations. Each data point is the mean of four experiments. The error bars represent standard error.
DISCUSSION
This study has confirmed that IS257-mediated cointegration of a pT181-like plasmid into the chromosome of the MRSA strain SK1660 generated a novel hybrid promoter for the tetA(K) gene such that a−35 sequence closer to optimal is utilized for transcription. This conclusion is based on remapping of the native tetA(K) promoter on pT181 to the intergenic region upstream of the gene (Fig. 2). A tetA(K) promoter was previously reported within the upstream repC gene (21), but the −35 and −10 sequences identified, TCGACT-22 bp-TGCAAA, respectively, represent a suboptimal match to the canonical promoter consensus (TTGACA-17 bp-TATAAT for −35 and −10 sequences, respectively) (11, 22). Since we were unable to detect a primer extension product corresponding to a TSP from this promoter, it is possible that the species previously identified by S1 nuclease mapping (21) was derived from a processed form of the repC transcript. Our transcript mapping also identified a TSP for a complete, outwardly directed promoter, Pout, located at one end of IS257.
Northern hybridization suggested that the chromosomal Phybrid is a considerably more powerful promoter than PtetA(K) of the multicopy plasmid pT181 (Fig. 3), so that it more than compensates for the reduced gene dosage of this genetic context. Although a proportion of the tetA(K) transcripts present in SK1660 cells initiate at Pout, image analysis of primer extension products suggests that, in comparison to Phybrid, it makes a relatively minor contribution (less than 10%). The relative strengths of these promoters probably reflect variations in their sequences. Phybrid possesses an optimal TTG trinucleotide at the start of its −35 sequence, rather than TTA in the native tetA(K) promoter of pT181 (Fig. 2 and 7). Sequence differences both upstream and in the 7 bp downstream of the −35 sequences (Fig. 2) may also contribute to the different strengths of these promoters. Pout possesses both a suboptimal −35 sequence and an 18-bp spacer region.
FIG. 7.
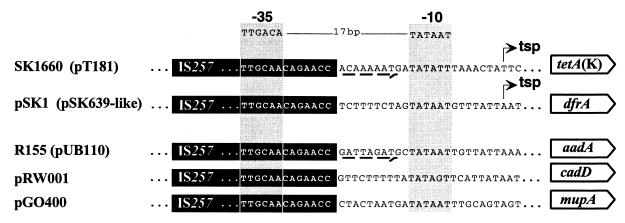
IS257 hybrid promoters. The sequences shown are from the plasmids pSK1 (29), pGO400 (23), and pRW001 (4) and the chromosomes of the MRSA strains R155 (34) and SK1660. Genes encoding resistance to the following compounds are shown: aminoglycosides (aadA), cadmium (cadD), trimethoprim (dfrA), mupirocin (mupA), and tetracycline [tetA(K)]. Where a DNA segment corresponds to a cointegrated plasmid or remnant thereof, the name of that plasmid is given in brackets. Copies of IS257 are denoted by solid boxes. The canonical promoter consensus is shown at the top. The TSPs mapped for dfrA on pSK1 (15) and tetA(K) on SK1660 (this study) are indicated. Eight-base pair target duplication sequences are denoted by dashed arrows.
The enhanced transcription afforded by the hybrid promoter was found to translate into higher levels of tetracycline resistance associated with strains carrying this configuration (Fig. 4 and Table 2). Exposure to tetracycline resulted in an approximately twofold increase in the amount of tetA(K) mRNA detected in cells carrying pT181 and the SK1660-derived chromosomal structure (Fig. 3) and a two- to fourfold increase in resistance for all tetA(K)-containing strains (Table 2). pT181 tetA(K) is thought to be regulated via translational attenuation (13), and this mechanism is expected to be operational in SK1660 also, since equivalent transcripts are produced in both cases (Fig. 3). Indeed, the equivalent levels of induction expressed by strains containing distinct promoter configurations is consistent with a posttranscriptional regulatory mechanism. It is thought that binding of the ribosomes during translation, induced by the presence of tetracycline, provides some protection from degradation of the tetA(K) message (21). This may account for the increased amount of tetA(K) mRNA detected in the Northern blot. The reduced susceptibility associated with preexposure to tetracycline might therefore be a consequence of protection afforded by existing TetA(K) protein, resulting from a combination of induced translation and message stabilization.
Competition studies suggested that the cointegration of a pT181-like plasmid into the chromosome has produced a strain with greater relative fitness than its progenitor carrying the autonomous form of the plasmid, but only in the presence of tetracycline. Since no competitive advantage was observed in media without tetracycline, it would seem likely that the cointegrate structure was selected by the presence of this antibiotic. It should be realized that this evolutionary event may not necessarily have coincided with the emergence of MRSA strains such as SK1660 around 1970. Rather, the cointegrate structure could have arisen in another host strain and subsequently been transferred into an SK1660 ancestor as part of a mec region cassette (12).
Although greater levels of resistance were found to be conferred by the chromosomally cointegrated plasmid than by its autonomous form, this property may not represent the most significant selective advantage mediated by the former configuration, since we found that a relative fitness advantage was manifested at only 1 μg of tetracycline/ml (Fig. 6B). Furthermore, as the growth curve shown in Fig. 5B illustrates, at this concentration, SK5319, harboring autonomous pT181, achieved a growth rate comparable to that of SK5323, which possesses the cointegrated form. However, SK5323 exhibited less lag than SK5319 (Fig. 5B). We therefore suggest that, in addition to the capacity to grow in the presence of higher levels of tetracycline, strains such as SK1660 are also evolutionarily advantaged by exhibiting reduced lag upon exposure to even low levels of the antibiotic. Both of these traits are likely to have contributed to the emergence of such strains through the course of evolution. Lenski and coworkers (17) have similarly noted the relationship among promoter strength, lag, and fitness in the inducible tetracycline resistance system encoded by Tn10 of E. coli.
The IS257-derived hybrid promoter driving transcription of tetA(K) in SK1660 is the second confirmed example of such a promoter; the first mediates dfrA-encoded high-level trimethoprim resistance (Fig. 7) (15). IS257 has been found to insert into several locations in pT181, with no apparent insertion site specificity (16, 25, 35). It is therefore likely that cointegrate structures equivalent to that of SK1660 found in other strains (34) are clonal in nature, rather than arising from independent IS257 transposition events. From the reported sequences, we have identified additional potential IS257-derived hybrid promoters upstream of the genes aadA (34), cadD (4), and mupA (23), which mediate resistance to aminoglycosides, cadmium, and mupirocin, respectively (Fig. 7) (33). In each case, a good match to the −10 promoter consensus sequence is present adjacent to a copy of IS257 such that it is an optimal 17 bp from the −35 sequence at the end of this element. In the chromosome of R155 and in the plasmid pRW001, the putative hybrid promoters appear to have arisen as a result of IS257-mediated cointegrative capture of a small plasmid.
In addition to the hybrid promoter, the studies described here also revealed the existence of a complete outwardly directed promoter within IS257. Despite the fact that it is considerably weaker than Phybrid, it is nonetheless possible that Pout alone could be sufficient for the transcription of neighboring genes in situations where no hybrid promoter is present. It would seem likely that IS257 plays a role in the transcription of more genes than previously recognized. The capacity of IS257 to influence adjacent gene expression enhances its potential to effect beneficial genetic rearrangements, thereby contributing to the flexibility of the staphylococcal genome and hence the ability of the organism to adapt to an environment of widespread antimicrobial use.
ACKNOWLEDGMENTS
We thank Chia Lee for providing the plasmids pCL84 and pYL112Δ19 and Melissa Brown for critical reading of the manuscript.
This work was supported in part by Project Grant 980075 from the National Health and Medical Research Council (Australia). A.E.S. was the recipient of an Australian Postgraduate Award.
REFERENCES
- 1.Atlas R M. Alphabetical listing of media-S. In: Parks L C, editor. Handbook of microbiological media. Boca Raton, Fla: CRC Press; 1993. pp. 782–862. [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J S, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1990. [Google Scholar]
- 3.Barberis-Maino L, Ryffel C, Kayser F H, Berger-Bachi B. Complete nucleotide sequence of IS431mec in Staphylococcus aureus. Nucleic Acids Res. 1990;18:5548. doi: 10.1093/nar/18.18.5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crupper S S, Worrell V, Stewart G C, Iandolo J J. Cloning and expression of cadD, a new cadmium resistance gene of Staphylococcus aureus. J Bacteriol. 1999;181:4071–4075. doi: 10.1128/jb.181.13.4071-4075.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubin D T, Matthews P R, Chikramane S G, Stewart P R. Physical mapping of the mec region of an American methicillin-resistant Staphylococcus aureus strain. Antimicrob Agents Chemother. 1991;35:1661–1665. doi: 10.1128/aac.35.8.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Firth N, Skurray R A. Mobile elements in the evolution and spread of multiple-drug resistance in staphylococci. Drug Resist Updates. 1998;1:49–58. doi: 10.1016/s1368-7646(98)80214-8. [DOI] [PubMed] [Google Scholar]
- 8.Gillespie M T, Lyon B R, Loo L S L, Matthews P R, Stewart P R, Skurray R A. Homologous direct repeat sequences associated with mercury, methicillin, tetracycline and trimethoprim resistance determinants in Staphylococcus aureus. FEMS Microbiol Lett. 1987;43:165–171. [Google Scholar]
- 9.Gillespie M T, May J W, Skurray R A. Detection of an integrated tetracycline resistance plasmid in the chromosome of methicillin-resistant Staphylococcus aureus. J Gen Microbiol. 1986;132:1723–1728. doi: 10.1099/00221287-132-6-1723. [DOI] [PubMed] [Google Scholar]
- 10.Guay G G, Khan S A, Rothstein D M. The tet(K) gene of plasmid pT181 of Staphylococcus aureus encodes an efflux protein that contains 14 transmembrane helices. Plasmid. 1993;30:163–166. doi: 10.1006/plas.1993.1045. [DOI] [PubMed] [Google Scholar]
- 11.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito T, Katayama Y, Hiramatsu K. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob Agents Chemother. 1999;43:1449–1458. doi: 10.1128/aac.43.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan S A, Novick R P. Complete nucleotide sequence of pT181, a tetracycline-resistance plasmid from Staphylococcus aureus. Plasmid. 1983;10:251–259. doi: 10.1016/0147-619x(83)90039-2. [DOI] [PubMed] [Google Scholar]
- 14.Lee C Y, Buranen S L, Ye Z. Construction of single-copy integration vectors for Staphylococcus aureus. Gene. 1991;103:101–105. doi: 10.1016/0378-1119(91)90399-v. [DOI] [PubMed] [Google Scholar]
- 15.Leelaporn A, Firth N, Byrne M E, Roper E, Skurray R A. Possible role of insertion sequence IS257 in dissemination and expression of high- and low-level trimethoprim resistance in staphylococci. Antimicrob Agents Chemother. 1994;38:2238–2244. doi: 10.1128/aac.38.10.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leelaporn A, Firth N, Paulsen I T, Skurray R A. IS257-mediated cointegration in the evolution of a family of staphylococcal trimethoprim resistance plasmids. J Bacteriol. 1996;178:6070–6073. doi: 10.1128/jb.178.20.6070-6073.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lenski R E, Souza V, Duong L P, Phan Q G, Nguyen T N, Bertrand K P. Epistatic effects of promoter and repressor functions of the Tn10 tetracycline-resistance operon on the fitness of Escherichia coli. Mol Ecol. 1994;3:127–135. doi: 10.1111/j.1365-294x.1994.tb00113.x. [DOI] [PubMed] [Google Scholar]
- 18.Lyon B R, May J W, Skurray R A. Analysis of plasmids in nosocomial strains of multiple-antibiotic-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1983;23:817–826. doi: 10.1128/aac.23.6.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matthews P R, Inglis B, Stewart P R. Clustering of resistance genes in the mec region of the chromosome of Staphylococcus aureus. In: Novick R P, editor. Molecular biology of the staphylococci. New York, N.Y: VCH; 1990. pp. 69–83. [Google Scholar]
- 21.Mojumdar M, Khan S A. Characterization of the tetracycline resistance gene of plasmid pT181 of Staphylococcus aureus. J Bacteriol. 1988;170:5522–5528. doi: 10.1128/jb.170.12.5522-5528.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moran C P, Lang N, LeGrice S F J, Lee G, Stephens M, Sonenshein A L, Pero J, Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186:339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- 23.Morton T M, Johnston J L, Patterson J, Archer G L. Characterization of a conjugative staphylococcal mupirocin resistance plasmid. Antimicrob Agents Chemother. 1995;39:1272–1280. doi: 10.1128/aac.39.6.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 4th ed. 1997. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa. [Google Scholar]
- 25.Needham C, Rahman M, Dyke K G H, Noble W C. An investigation of plasmids from Staphylococcus aureus that mediate resistance to mupirocin and tetracycline. Microbiology. 1994;140:2577–2583. doi: 10.1099/00221287-140-10-2577. [DOI] [PubMed] [Google Scholar]
- 26.Novick R, Sanchez R C, Gruss A, Edelman I. Involvement of the cell envelope in plasmid maintenance: plasmid curing during the regeneration of protoplasts. Plasmid. 1980;3:348–358. doi: 10.1016/0147-619x(80)90048-7. [DOI] [PubMed] [Google Scholar]
- 27.Novick R P. Staphylococcal plasmids and their replication. Annu Rev Microbiol. 1989;43:537–565. doi: 10.1146/annurev.mi.43.100189.002541. [DOI] [PubMed] [Google Scholar]
- 28.Novick R P. The Staphylococcus as a molecular genetic system. In: Novick R P, editor. Molecular biology of the staphylococci. New York, N.Y: VCH; 1990. pp. 1–37. [Google Scholar]
- 29.Rouch D A, Messerotti L J, Loo L S, Jackson C A, Skurray R A. Trimethoprim resistance transposon Tn4003 from Staphylococcus aureus encodes genes for a dihydrofolate reductase and thymidylate synthetase flanked by three copies of IS257. Mol Microbiol. 1989;3:161–175. doi: 10.1111/j.1365-2958.1989.tb01805.x. [DOI] [PubMed] [Google Scholar]
- 30.Rouch D A, Skurray R A. IS257 from Staphylococcus aureus: member of an insertion sequence superfamily prevalent among gram-positive and gram-negative bacteria. Gene. 1989;76:195–205. doi: 10.1016/0378-1119(89)90160-1. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 32.Schenk S, Laddaga R A. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol Lett. 1992;73:133–138. doi: 10.1016/0378-1097(92)90596-g. [DOI] [PubMed] [Google Scholar]
- 33.Skurray R A, Firth N. Molecular evolution of multiply-antibiotic-resistant staphylococci. Ciba Found Symp. 1997;207:167–183. doi: 10.1002/9780470515358.ch11. [DOI] [PubMed] [Google Scholar]
- 34.Stewart P R, Dubin D T, Chikramane S G, Inglis B, Matthews P R, Poston S M. IS257 and small plasmid insertions in the mec region of the chromosome of Staphylococcus aureus. Plasmid. 1994;31:12–20. doi: 10.1006/plas.1994.1002. [DOI] [PubMed] [Google Scholar]
- 35.Werckenthin C, Schwarz S, Roberts M C. Integration of pT181-like tetracycline resistance plasmids into large staphylococcal plasmids involves IS257. Antimicrob Agents Chemother. 1996;40:2542–2544. doi: 10.1128/aac.40.11.2542. [DOI] [PMC free article] [PubMed] [Google Scholar]