Abstract
Extracellular vesicles (EVs) are esteemed as a promising delivery vehicle for various genetic therapeutics. They are relatively inert, non-immunogenic, biodegradable, and biocompatible. At least in rodents, they can even transit challenging bodily hurdles such as the blood-brain barrier. Constitutively shed by all cells and with the potential to interact specifically with neighboring and distant targets, EVs can be engineered to carry and deliver therapeutic molecules such as proteins and RNAs. EVs are thus emerging as an elegant in vivo gene therapy vector. Deeper understanding of basic EV biology—including cellular production, EV loading, systemic distribution, and cell delivery—is still needed for effective harnessing of these endogenous cellular nanoparticles as next-generation nanodelivery tools. However, even a perfect EV product will be challenging to produce at clinical scale. In this regard, we propose that vector transduction technologies can be used to convert cells either ex vivo or directly in vivo into EV factories for stable, safe modulation of gene expression and function. Here, we extrapolate from the current EV state of the art to a bright potential future using EVs to treat genetic diseases that are refractory to current therapeutics.
Keywords: extracellular vesicles, non-coding RNA, nanoparticle, cell targeting, ectosomes, exosomes
Graphical abstract
We describe here the use of extracellular vesicles (EVs) as RNA and protein delivery vehicles. We outline the advantages and disadvantage to using EVs as delivery vehicles and posit that EVs will emerge as a bona fide next-generation gene and cell therapy delivery approach.
EV biogenesis and therapeutic delivery potential
All eukaryotic cells release an abundance of extracellular vesicles (EVs): membrane-bound nanoparticles that are roughly spherical and range in diameter from around 50 to 500 nm.1 EVs are diverse, categorized not only by size but also by cell of origin, mode of release, molecular composition, and function. Classical EV subtypes like “ectosomes” (plasma membrane origin) and “exosomes” (endosomal origin) may be important at the cell biology level but belie incredible diversity and are difficult to distinguish after they leave the cell.2 EVs are thought to function in cell-to-cell communications by delivering nucleic acids, proteins, small molecules, and lipids between cells,3 but other modes of interaction can also be envisioned.
Notably, these molecules have been observed to retain their function in recipient cells following being transported in EVs, suggesting that EVs containing active proteins, RNAs, proteins, or DNAs can alter the biology of cells that are distant from the EV producer cells. These characteristics confer unparalleled potential to EVs in terms of safety and biocompatibility; as such, they have been the subject of extensive experimentation and captured the interest of both the public and private sectors.4 To date, several therapeutic biomolecules have been repeatedly loaded in EVs and delivered to target cells and experimentally validated in both in vitro and in vivo models.
Loading therapeutic RNAs into EVs
RNA therapeutics offer distinct advantages over zinc finger or CRISPR therapeutics, as RNAs function by endogenous cellular pathways in a transient manner and are programmable and thus relatively easy to engineer for specific diseases, and are generally not immunogenic, as is unfortunately the case with many of the emerging recombinant protein technologies. Various RNA biotypes with biological functions and therapeutic potential, such as small interfering RNAs (siRNAs), have been discovered and investigated, leading to the development of new classes of therapeutic drugs.5 RNAs can be used to impart short-term transient and longer-term epigenetic silencing, which is based on the target, e.g., targeting gene promoters can induce transcriptional gene silencing.6 Notably, mRNA-based vaccines are also now being used effectively to combat the COVID-19 pandemic.7 However, although therapeutic RNAs can be rapidly altered and produced, they must reach their intended target to be effective. For example, lipid nanoparticles (LNPs) are used in the Pfizer-BioNTech COVID-19 vaccine and for treatment of polyneuropathy targeted to the liver, but these approaches may be cytotoxic, unstable in circulation, and unsuited for delivery to other tissues.8 Moreover, cellular and subcellular delivery of RNA-based drugs is also a formidable challenge, with less than 1% of payloads reaching the cytosol of the cell.9 Potentially, packaging these RNAs into EVs, which naturally carry RNA, could be a safer and more physiologically targeted approach. As such, several attempts have been made to integrate RNA species in EVs and optimize packaging and release efficiency.
Packaging mRNAs into EVs
While EVs are emerging as a promising delivery system, it has proven challenging to effectively load therapeutic cargo into EVs. EVs can be loaded naturally during biogenesis or following EV isolation using physical or chemical methods. Electroporation has been used to load nucleic acids into EVs; however, this deteriorates the intrinsic properties of the EV membrane and causes extensive EV loss.10 As such, the most common method for mRNA loading into EVs is to transfect EV-producer cells with plasmids encoding the therapeutic mRNA. The resulting high concentration of cytoplasmic mRNA is sufficient to cause packaging of mRNA into EVs, perhaps because EVs have been found to functionally export cellular components that are in vast surplus.11 Villamizar et al. transfected mesenchymal stem cells (MSCs) with a plasmid encoding for a zinc finger transcription factor targeted to the CFTR gene promoter for the treatment of cystic fibrosis (called CFZF). The high expression of CFZF, resulting from the plasmid’s CMV promoter, was sufficient to detect both CFZF mRNA and protein in the isolated EVs.12 To increase the RNA loading output, Kojima et al. loaded catalase mRNA into EVs using a loading system called EXOtic,13 consisting of a plasmid construct encoding for CD63, a common transmembrane protein, plus the L7Ae archaeal ribosomal protein that selectively binds to the C/D box RNA structure. Next, they introduced the C/D box into the 3′ UTR of the catalase gene. When producer cells were transfected with these constructs, catalase mRNA was efficiently packed into EVs and transferred to target cell in vitro.13 A tricistronic plasmid encoding for three genes involved in exosome biogenesis (STEAP3, SDC4, L-aspartate oxidase) was also used with the EXOtic system to increase EV release. Introduced into mouse models of Parkinson’s disease, these EVs transgressed the blood-brain barrier and reduced reactive oxygen species in targeted cells in the brain. A constitutively active mutant of gap junction protein Connexin 43 (Cx43) was also included. This protein is responsible for forming gap-junction structures after fusion of two connexon hemichannels, allowing for cellular intercommunication and transfer of materials.14 It is also the most expressed Cx protein and is naturally present in EVs as hexamers organized in hemichannel structures.14,15 This protein was engineered into the EV construct and found to increase the release efficiency EV cargo into recipient cells upon contact.12,16 Indeed, CD63-fused L7Ae appears to require co-transfection of the booster tricistronic plasmid, Cx43, and a LAMP2b-fused brain targeting module to transfer nluc-C/D box mRNA.
Another method for loading RNA into EVs is to generate lipid-coated RNA particles and integrate these into purified EVs through mixing-induced partitioning.17,18 As expected, this process leads to a slight increase in EV size and a decrease in EV numbers, but it is efficient and accurate (>90%).19 However, the purification and need to pre-coat RNA with lipids introduces expense and time constraints. Therefore, while this process can be used for research purposes, it may prove challenging to scale for clinical or commercial applications.
Packaging small hairpin RNAs (shRNAs), miRNAs, and circular RNAs (circRNAs) into EVs
MicroRNAs (miRNAs) are well known to be loaded in EVs and to functionally modulate gene expression in other recipient cell types.20 This could be by direct interactions with Argonaut 2 (AGO2), which has been found to be packaged into EVs.21 Alternatively, particular proteins, such as YBX1, have been implicated in loading particular miRNAs into EVs,22 while others have suggested that there may not be a specific motif or pathway involved in miRNA recruitment into EVs.23 Yet others have found that there does not appear to be a specific miRNA packaging system for loading these RNAs into EVs.24 Due to their relatively large range of target genes, miRNAs can significantly alter the phenotype or gene expression of a cell, and therefore they can be a high-value cargo with the potential to promote, trigger, or treat diseases. Simeoli et al. were among the first to describe an endogenous pathway of EV-mediated miRNA transfer from neurons to macrophages in presence of capsaicin.20 Capsaicin incubation or nerve injury causes an increase in expression of miRNA-21 and milk fat globule-EGF factor 8 protein MFG-E8, a protein responsible for macrophage uptake. The authors demonstrated that EVs derived from capsaicin-treated neurons were taken up more readily by macrophages than the untreated control and promoted inflammatory 13phenotypes and repression of miR-21 target genes in macrophages. Activated macrophages were more likely to move toward sites of injury where the EV-releasing neurons are situated, thus demonstrating the existence and importance of EV-mediated intercellular communication mediated by miRNAs.20 EV-transferred miRNAs have also been implicated in cancer by promoting metastasis, drug resistance, proliferation, and inflammation.25
As demonstrated by the existence of EV-loaded miRNA communication pathways, miRNAs seem to be preferentially loaded in EVs relative to other RNA types, suggesting that an endogenous loading system exists within cells. AGO2 is an RNA-binding protein that binds miRNA and may be responsible for miRNA loading in EVs.26 Due to their profound regulatory potential and natural occurrence in EVs, miRNA and AGO2-binding shRNAs appear to be great candidates for EV therapeutics.
Another class of regulatory RNAs that have been observed in EVs are circRNAs.27 circRNAs are a class of single-stranded circular non-coding RNA resulting from the back splicing of exons in mRNAs.28,29 Some genes have been observed to express several times the amount of circRNA compared with the protein-coding mRNA, suggesting an important functional role, which includes transcription regulation by absorbing miRNAs, interaction with proteins, competition with pre-mRNA splicing, and, more rarely, as templates for protein translation.30 The lack of 5′ and 3′ ends protects the circRNAs from degradation by exonucleases, which ultimately confers a longer lifespan of these transcripts in the cytoplasm compared with other RNAs.31 This is also confirmed by the negative relation between cell proliferation and circRNA concentration, allegedly because circRNAs can be diluted to daughter cells after proliferation. Recently, functional circRNAs were found to be loaded into and transferred to recipient cells by EVs. The ratio between circRNA and linear RNA in EVs is higher than in the producer cells, indicating an endogenous sorting mechanism.32 Some circRNAs are highly expressed in cancer cells, and EV-packed circRNAs are demonstrated to be partially responsible for the proliferation of various cancers; as such, exosomal circRNAs (exo-circRNAs) have been considered important primarily as biomarkers for screening of cancer in early onset.32,33 However, due to their increased stability, circRNA can be packaged into EVs and transferred to target cells, where they may support protein translation for longer than a typical mRNA.34 Notably, circRNAs can be engineered with an internal ribosome entry site (IRES) to express proteins of interest.34 As circRNAs persist longer than linear RNAs, this may be one approach to generating enhanced long-term protein expression. Such an application would be especially useful in vaccine treatments to extend the exposure time of antigens to the immune system or, generally, to produce the most protein out of a therapeutic dose. While naturally occurring open reading frame (ORF)-possessing circRNAs are a minority in cells and have yet to be proven capable of translation, attempts have been made to engineer circRNAs with coding capacity.35 Wesselhoeft et al. achieved robust expression of luciferase, EGFP, erythropoietin, and CRISPR-associated endonuclease 9 (Cas9) upon transfection of a self-splicing intron-induced circRNA into HEK293 cells.34 Qu et al. created a circRNA encoding the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein and performed in vivo experiments in mice to test the immunization capacity of circRNA vaccines encapsulated in LNPs.36 Mice treated with these particles produced antibodies and T cell responses similar to those of counterparts treated with a linear mRNA.36 Overall, these results suggest that circRNAs make more proteins than linear mRNAs and therefore may improve the general efficacy of mRNA therapies and may prove useful in vaccine approaches and in treating cancer and infectious and genetic diseases. Ideally, such therapeutic circRNAs could be engineered to code for therapeutic proteins and be specifically loaded into EVs for long-lasting expression of proteins in target cells.
Packaging proteins into EVs
While the EXOtic system allows for packaging protein-coding mRNAs into EVs, others have developed a means of loading unbound therapeutic proteins into EVs. CRY2 is a plant protein that changes conformation upon exposure to blue light, and CIBN is a truncated version of CIB1, a protein with affinity for CRY2 in its excited form.37 CIB1 was attached to the cytosolic tail of EV marker CD9 and CRY2 to reporter proteins such as mCherry and GFP.38 This system, named EXPLOR, was demonstrated to cause loading of the cargo-CRY2 into EVs by reversible binding to CIB1 when producer cells are exposed to blue light. In the absence of blue light, however, the cargo-CRY2 complex was freely available in the isolated EVs. Using this approach, Choi et al. successfully loaded Cre recombinase and the nuclear factor κB (NF-κB) pathway suppressor srIκB into EVs.39
Based on this model, Osteikoetxea et al. tested whether the Cas9 protein could be loaded into EVs and compared it with three other similar loading systems based on heterodimerization upon exposure to an activating stimulus.40 These were PHIB and PIF6, which interact upon exposure to 630 nm light, and the small molecule phycocyanobilin, engineered VVD proteins with nanomagnets that interact in presence of blue light, and finally FKBP and FRB, which interact in the presence of the small molecule rapamycin. The group demonstrated that loading with CRY2-CIB1 resulted in the highest concentration of Cas9 in EV fractions, reaching more than 20 Cas9 molecules per EV.40 A noteworthy observation of this study was the data suggesting that engineering of MysPalm for protein cargo delivery appears to be more advantageous compared with engineering to tetraspanin markers such as CD9. Two possible reasons for this observation are (1) the conjugated domain may not be folded correctly in the endoplasmic reticulum (ER) when bound to a membrane protein, or (2) it could interfere with the positioning of CD9 to its natural site in the EV membrane. Supporting this notion is the observation that conjugated Cas9s were only 30%–50% as active as wild-type (WT) Cas9, indicating that these loading systems reduce the efficacy of gene editing.40 This is relevant, as the efficacy of future therapeutic EVs will depend on the amount of active therapeutic proteins delivered to target cells.
Another highly innovative method for loading proteins into EVs involved modifying cargo proteins with a WW tag that is recognized by Ndfip1, an L-domain-containing ubiquitin ligase that ubiquitinates the cargo protein, leading it into EVs during biogenesis.41 Unlike the CRY2 system, the WW tag attached to Cre was as efficient as the WT protein in performing its function. WW-Cre was functional after EV-mediated delivery to recipient cells, and WW-Cre loading into EVs occurred in a Ndfip1-dependent manner.41 This method neatly exploits the cell’s endogenous budding and protein loading system that could be expanded to load therapeutic proteins into EVs (Figure 1). This system presents some advantages compared with the CRY2-CIB1 loading method. First, the WW domain is substantially smaller than the CRY2 protein, measuring only ∼40 residues compared with the 593 residues of human CRY2. It could be argued that larger conjugated proteins pose a higher risk of interfering with the function of the cargo protein, thus limiting therapeutic effect. This occurred in a study by Osteikoetxea et al., whereby CRY2-Cas9 had only 50% editing potential compared with WT Cas9, whereas WW-Cre was as effective at recombining DNA as its WT version.40 Second, because the WW system utilizes a pathway already present in cells, it requires minimal manipulation of producer cells, which only need to be transfected with the WW-protein therapy for effective EV loading. Since producer cells would already require transfection of the therapeutic protein, the WW system upregulates protein loading without additional cell modifications, although overexpression of Ndfip1 positively correlates with WW-Cre ubiquitination. Lastly, the WW domain occurs naturally in other human proteins and therefore presumably poses a lower risk of immunological reaction compared with plant-derived CRY2-CIB1. Both systems are advantageous to other protein loading systems, as the protein is free in EVs and directly delivered to the cytoplasm of target cells.
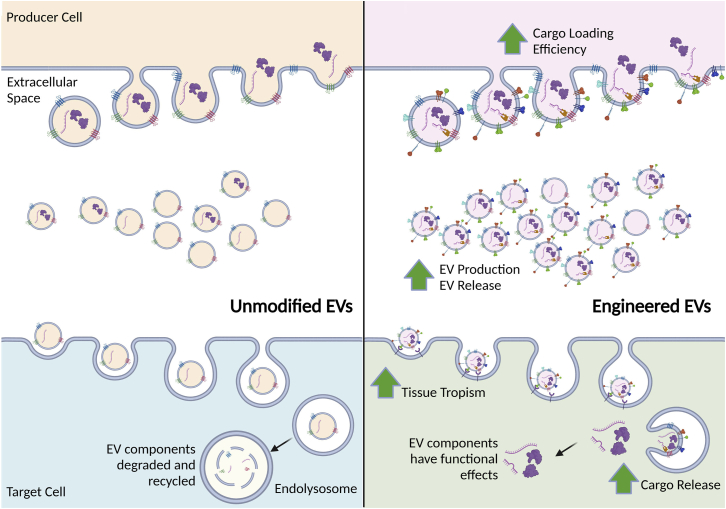
Figure 1.
Endogenous and engineered EV pathways
Unmodified EVs are naturally generated and taken up by target cells where their payloads can be taken up, recycled, or degraded. Producer cells can be modified to generate EVs packaged with RNA, protein, and potentially even DNA payloads, and these shed EVs can be taken up by target cells where their payloads are released.
Manufacturing EVs for therapeutic use
Although several ongoing clinical trials involve EVs, so far, no EV therapies have entered the market.42 Part of the reason for this may be found in the inadequacy of production and purification processes for manufacturing EVs on a large scale43 and because a majority of the current literature focuses on developing new EVs and proving their potential with in vitro and in animal studies, with little emphasis or understanding as to what is required for translating EV therapeutics.
EVs can be engineered to carry therapeutic molecules, such as proteins or RNAs, and many studies are ongoing to optimize their therapeutic potential by increasing EV production and release from producer cells, improvements in cargo loading efficiency, increased tissue tropism, and a boosting of cargo release to target cells (Figure 1). EV biogenesis initiates with the generation of early endosome by inward budding of the cell membrane followed by another inward budding of the endosomal membrane to create intraluminal vesicles (ILVs) (Figure 1). The ILVs are then either degraded by fusion with the lysosome or secreted through the endosomal sorting complex transport (ESCRT)-dependent pathway or ESCRT-independent pathway.44,45 The biogenesis of EVs is orchestrated by several ESCRT proteins and proteins involved in vesicle formation and budding; as such, silencing or promoting expression of these protein may increase the quantity of EVs released by the producer cells. This may be relevant to increase the efficiency of EV production from the producer cells and reduce costs in the manufacturing of therapeutic EVs. Colombo et al. assessed whether the silencing of several EV biogenesis-associated proteins could lead to higher EV numbers. The researchers developed shRNA targeted to CHMP4C, VPS4B, ALIX, and VTA1, four ESCRT genes involved in EV biogenesis and observed that the combination of VPS4B and CHMP4C-shRNAs, LIX and VTA1 shRNA increased the quantity of released EVs by more than 50%.46
Challenges and future prospects for EV therapeutics
The majority of experimental publications involving EVs report extracting the EVs from cells supernatant through ultracentrifugation at 100,000 × g and filtration through 0.22 μm filters. These isolated EVs are then used for various studies, including in vivo assessment. However, this approach is onerous, produces heterologous populations of EVs, and may prove challenging to translate clinically. Moreover, the relative short ½ life of less than 48 h in vivo11 indicates that multiple EV administrations would be required as a therapeutic strategy. An alternative approach would be to engineer cells, directly in vivo, to produce EVs with defined therapeutic payloads. One can envision perhaps using stealth lentiviral vector systems47 or adenoviruses to in vivo transduce cells and convert them into therapeutic EV-producing factories. Alternatively, ex vivo transduction of immune cells, converting them into EV-producing factories, could be employed, similar to what is currently done with chimeric antigen (CAR) T cell therapeutics.48 While safety will be a concern, such an approach could skirt the issue of manufacturing and batch variation issues that plague current in vitro EV systems. Added layers of safety for in vivo engineering cells as EV factories could be instilled by using tissue-specific promoters,49,50,51 selecting disease-specific therapeutic RNAs, or targeting long non-coding RNAs involved in regulating particular genes of interest that only target the disease genes and not host genes.52 Safety can also be imbued by the use of a suicide gene, such as cetuximab (Erbitux), which can recognize and impart cell death specifically on those cells expressing truncated EGFR and is routinely used clinically in CAR therapies to treat cancer.53 Lastly, perhaps next-generation therapeutics, such as using a hybrid integrating LNP approach or an LNP/adenovirus approach may prove useful in converting liver cells in vivo, for instance into therapeutic EV factories. Regardless, one fact is clear: EVs are emerging as a unique delivery vehicle that is sufficiently varied from LNPs and vector-based systems, which will no doubt change the current trajectory of gene and cell therapeutics and harken a bright future to treat various diseases. The question, however, is not if, but when.
Acknowledgments
This project was supported by National Institute of Mental Health (NIMH) R01 113407-01 and National Institute of Allergy and Infectious Diseases (NIAID) R56AI147684 to K.V.M.
Author contributions
R.C., Z.T., K.W., and K.V.M. wrote the entire article and edited collaboratively. Z.T. generated the figure.
Declaration of interests
The authors declare no conflicts of interest for this work.
Contributor Information
Ken Witwer, Email: kwitwer1@jhmi.edu.
Kevin V. Morris, Email: kevin.morris@griffith.edu.au.
References
- 1.Wang W., Li M., Chen Z., Xu L., Chang M., Wang K., Deng C., Gu Y., Zhou S., Shen Y., et al. Biogenesis and function of extracellular vesicles in pathophysiological processes of skeletal muscle atrophy. Biochem. Pharmacol. 2022;198:114954. doi: 10.1016/j.bcp.2022.114954. [DOI] [PubMed] [Google Scholar]
- 2.Buzas E.I. The roles of extracellular vesicles in the immune system. Nat. Rev. Immunol. 2022:1–15. doi: 10.1038/s41577-022-00763-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li S.P., Lin Z.X., Jiang X.Y., Yu X.Y. Exosomal cargo-loading and synthetic exosome-mimics as potential therapeutic tools. Acta Pharmacol. Sin. 2018;39:542–551. doi: 10.1038/aps.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang M., Wu S.Y. The advances and challenges in utilizing exosomes for delivering cancer therapeutics. Front. Pharmacol. 2018;9:735. doi: 10.3389/fphar.2018.00735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Damase T.R., Sukhovershin R., Boada C., Taraballi F., Pettigrew R.I., Cooke J.P. The limitless future of RNA therapeutics. Front. Bioeng. Biotechnol. 2021;9:628137. doi: 10.3389/fbioe.2021.628137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinberg M.S., Morris K.V. Transcriptional gene silencing in humans. Nucleic Acids Res. 2016;44:6505–6517. doi: 10.1093/nar/gkw139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiaie S.H., Majidi Zolbanin N., Ahmadi A., Bagherifar R., Valizadeh H., Kashanchi F., Jafari R. Recent advances in mRNA-LNP therapeutics: immunological and pharmacological aspects. J. Nanobiotechnology. 2022;20:276. doi: 10.1186/s12951-022-01478-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hou X., Zaks T., Langer R., Dong Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021;6:1078–1094. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maugeri M., Nawaz M., Papadimitriou A., Angerfors A., Camponeschi A., Na M., Hölttä M., Skantze P., Johansson S., Sundqvist M., et al. Linkage between endosomal escape of LNP-mRNA and loading into EVs for transport to other cells. Nat. Commun. 2019;10:4333. doi: 10.1038/s41467-019-12275-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnsen K.B., Gudbergsson J.M., Skov M.N., Christiansen G., Gurevich L., Moos T., Duroux M. Evaluation of electroporation-induced adverse effects on adipose-derived stem cell exosomes. Cytotechnology. 2016;68:2125–2138. doi: 10.1007/s10616-016-9952-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shrivastava S., Morris K.V. The multifunctionality of exosomes; from the garbage bin of the cell to a next generation gene and cellular therapy. Genes (Basel) 2021;12:173. doi: 10.3390/genes12020173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Villamizar O., Waters S.A., Scott T., Grepo N., Jaffe A., Morris K.V. Mesenchymal Stem Cell exosome delivered Zinc Finger Protein activation of cystic fibrosis transmembrane conductance regulator. J. Extracell. Vesicles. 2021;10:e12053. doi: 10.1002/jev2.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kojima R., Bojar D., Rizzi G., Hamri G.C.E., El-Baba M.D., Saxena P., Ausländer S., Tan K.R., Fussenegger M. Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for Parkinson's disease treatment. Nat. Commun. 2018;9:1305. doi: 10.1038/s41467-018-03733-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soares A.R., Martins-Marques T., Ribeiro-Rodrigues T., Ferreira J.V., Catarino S., Pinho M.J., Zuzarte M., Isabel Anjo S., Manadas B., P G Sluijter J., et al. Gap junctional protein Cx43 is involved in the communication between extracellular vesicles and mammalian cells. Sci. Rep. 2015;5:13243. doi: 10.1038/srep13243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gemel J., Kilkus J., Dawson G., Beyer E.C. Connecting exosomes and connexins. Cancers (Basel) 2019;11:476. doi: 10.3390/cancers11040476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shrivastava S., Ray R.M., Holguin L., Echavarria L., Grepo N., Scott T.A., Burnett J., Morris K.V. Exosome-mediated stable epigenetic repression of HIV-1. Nat. Commun. 2021;12:5541. doi: 10.1038/s41467-021-25839-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato Y.T., Umezaki K., Sawada S., Mukai S.A., Sasaki Y., Harada N., Shiku H., Akiyoshi K. Engineering hybrid exosomes by membrane fusion with liposomes. Sci. Rep. 2016;6:21933. doi: 10.1038/srep21933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y.J., Wu J.Y., Liu J., Xu W., Qiu X., Huang S., Hu X.B., Xiang D.X. Artificial exosomes for translational nanomedicine. J. Nanobiotechnology. 2021;19:242. doi: 10.1186/s12951-021-00986-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsai S.J., Atai N.A., Cacciottolo M., Nice J., Salehi A., Guo C., Sedgwick A., Kanagavelu S., Gould S.J. Exosome-mediated mRNA delivery in vivo is safe and can be used to induce SARS-CoV-2 immunity. J. Biol. Chem. 2021;297:101266. doi: 10.1016/j.jbc.2021.101266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simeoli R., Montague K., Jones H.R., Castaldi L., Chambers D., Kelleher J.H., Vacca V., Pitcher T., Grist J., Al-Ahdal H., et al. Exosomal cargo including microRNA regulates sensory neuron to macrophage communication after nerve trauma. Nat. Commun. 2017;8:1778. doi: 10.1038/s41467-017-01841-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beltrami C., Clayton A., Newbury L.J., Corish P., Jenkins R.H., Phillips A.O., Fraser D.J., Bowen T. Stabilization of urinary MicroRNAs by association with exosomes and argonaute 2 protein. Noncoding. RNA. 2015;1:151–166. doi: 10.3390/ncrna1020151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X.M., Ma L., Schekman R. Selective sorting of microRNAs into exosomes by phase-separated YBX1 condensates. Elife. 2021;10:e71982. doi: 10.7554/eLife.71982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hung M.E., Leonard J.N. A platform for actively loading cargo RNA to elucidate limiting steps in EV-mediated delivery. J. Extracell. Vesicles. 2016;5:31027. doi: 10.3402/jev.v5.31027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albanese M., Chen Y.F.A., Hüls C., Gärtner K., Tagawa T., Mejias-Perez E., Keppler O.T., Göbel C., Zeidler R., Shein M., et al. MicroRNAs are minor constituents of extracellular vesicles that are rarely delivered to target cells. PLoS Genet. 2021;17:e1009951. doi: 10.1371/journal.pgen.1009951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dilsiz N. Role of exosomes and exosomal microRNAs in cancer. Future Sci. OA. 2020;6:FSO465. doi: 10.2144/fsoa-2019-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKenzie A.J., Hoshino D., Hong N.H., Cha D.J., Franklin J.L., Coffey R.J., Patton J.G., Weaver A.M. KRAS-MEK signaling controls Ago2 sorting into exosomes. Cell Rep. 2016;15:978–987. doi: 10.1016/j.celrep.2016.03.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J., Chen D., Gu J., He X., Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 29.Ragan C., Goodall G.J., Shirokikh N.E., Preiss T. Insights into the biogenesis and potential functions of exonic circular RNA. Sci. Rep. 2019;9:2048. doi: 10.1038/s41598-018-37037-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meng S., Zhou H., Feng Z., Xu Z., Tang Y., Li P., Wu M. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol. Cancer. 2017;16:94. doi: 10.1186/s12943-017-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu L., Wang J., Khanabdali R., Kalionis B., Tai X., Xia S. Circular RNAs: isolation, characterization and their potential role in diseases. RNA Biol. 2017;14:1715–1721. doi: 10.1080/15476286.2017.1367886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lasda E., Parker R. Circular RNAs Co-precipitate with extracellular vesicles: a possible mechanism for circRNA clearance. PLoS One. 2016;11:e0148407. doi: 10.1371/journal.pone.0148407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du W.W., Li X., Ma J., Fang L., Wu N., Li F., Dhaliwal P., Yang W., Yee A.J., Yang B.B. Promotion of tumor progression by exosome transmission of circular RNA circSKA3. Mol. Ther. Nucleic Acids. 2022;27:276–292. doi: 10.1016/j.omtn.2021.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wesselhoeft R.A., Kowalski P.S., Anderson D.G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 2018;9:2629. doi: 10.1038/s41467-018-05096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miao Q., Ni B., Tang J. Coding potential of circRNAs: new discoveries and challenges. PeerJ. 2021;9:e10718. doi: 10.7717/peerj.10718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qu L., Yi Z., Shen Y., Lin L., Chen F., Xu Y., Wu Z., Tang H., Zhang X., Tian F., et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell. 2022;185:1728–1744.e16. doi: 10.1016/j.cell.2022.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kennedy M.J., Hughes R.M., Peteya L.A., Schwartz J.W., Ehlers M.D., Tucker C.L. Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yim N., Ryu S.W., Choi K., Lee K.R., Lee S., Choi H., Kim J., Shaker M.R., Sun W., Park J.H., et al. Exosome engineering for efficient intracellular delivery of soluble proteins using optically reversible protein-protein interaction module. Nat. Commun. 2016;7:12277. doi: 10.1038/ncomms12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi H., Kim Y., Mirzaaghasi A., Heo J., Kim Y.N., Shin J.H., Kim S., Kim N.H., Cho E.S., Song E., Kim P., Shin E.C., Chung K., Choi K., Choi C., In Yook J., Yoo T.H. Exosome-based delivery of super-repressor IkappaBalpha relieves sepsis-associated organ damage and mortality. Sci. Adv. 2020;6:eaaz6980. doi: 10.1126/sciadv.aaz6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osteikoetxea X., Silva A., Lázaro-Ibáñez E., Salmond N., Shatnyeva O., Stein J., Schick J., Wren S., Lindgren J., Firth M., et al. Engineered Cas9 extracellular vesicles as a novel gene editing tool. J. Extracell. Vesicles. 2022;11:e12225. doi: 10.1002/jev2.12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sterzenbach U., Putz U., Low L.H., Silke J., Tan S.S., Howitt J. Engineered exosomes as vehicles for biologically active proteins. Mol. Ther. 2017;25:1269–1278. doi: 10.1016/j.ymthe.2017.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y.S., Lin E.Y., Chiou T.W., Harn H.J. Exosomes in clinical trial and their production in compliance with good manufacturing practice. Ci Ji Yi Xue Za Zhi. 2020;32:113–120. doi: 10.4103/tcmj.tcmj_182_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li X., Corbett A.L., Taatizadeh E., Tasnim N., Little J.P., Garnis C., Daugaard M., Guns E., Hoorfar M., Li I.T.S. Challenges and opportunities in exosome research-Perspectives from biology, engineering, and cancer therapy. APL Bioeng. 2019;3:011503. doi: 10.1063/1.5087122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hessvik N.P., Llorente A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018;75:193–208. doi: 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jadli A.S., Ballasy N., Edalat P., Patel V.B. Inside(sight) of tiny communicator: exosome biogenesis, secretion, and uptake. Mol. Cell. Biochem. 2020;467:77–94. doi: 10.1007/s11010-020-03703-z. [DOI] [PubMed] [Google Scholar]
- 46.Colombo M., Moita C., van Niel G., Kowal J., Vigneron J., Benaroch P., Manel N., Moita L.F., Théry C., Raposo G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013;126:5553–5565. doi: 10.1242/jcs.128868. [DOI] [PubMed] [Google Scholar]
- 47.Milani M., Annoni A., Moalli F., Liu T., Cesana D., Calabria A., Bartolaccini S., Biffi M., Russo F., Visigalli I., et al. Phagocytosis-shielded lentiviral vectors improve liver gene therapy in nonhuman primates. Sci. Transl. Med. 2019;11:eaav7325. doi: 10.1126/scitranslmed.aav7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghassemi S., Durgin J.S., Nunez-Cruz S., Patel J., Leferovich J., Pinzone M., Shen F., Cummins K.D., Plesa G., Cantu V.A., et al. Rapid manufacturing of non-activated potent CAR T cells. Nat. Biomed. Eng. 2022;6:118–128. doi: 10.1038/s41551-021-00842-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lois C., Hong E.J., Pease S., Brown E.J., Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 50.Kuzmin D., Gogvadze E., Kholodenko R., Grzela D.P., Mityaev M., Vinogradova T., Kopantzev E., Malakhova G., Suntsova M., Sokov D., et al. Novel strong tissue specific promoter for gene expression in human germ cells. BMC Biotechnol. 2010;10:58. doi: 10.1186/1472-6750-10-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y., He Y., Wang Y., Liu M., Jiang M., Gao R., Wang G. Synthetic promoter for efficient and muscle-specific expression of exogenous genes. Plasmid. 2019;106:102441. doi: 10.1016/j.plasmid.2019.102441. [DOI] [PubMed] [Google Scholar]
- 52.Ray R.M., Hansen A.H., Slott S., Taskova M., Astakhova K., Morris K.V. Control of LDL uptake in human cells by targeting the LDLR regulatory long non-coding RNA BM450697. Mol. Ther. Nucleic Acids. 2019;17:264–276. doi: 10.1016/j.omtn.2019.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X., Chang W.C., Wong C.W., Colcher D., Sherman M., Ostberg J.R., Forman S.J., Riddell S.R., Jensen M.C. A transgene-encoded cell surface polypeptide for selection, in vivo tracking, and ablation of engineered cells. Blood. 2011;118:1255–1263. doi: 10.1182/blood-2011-02-337360. [DOI] [PMC free article] [PubMed] [Google Scholar]