Key Points
-
•
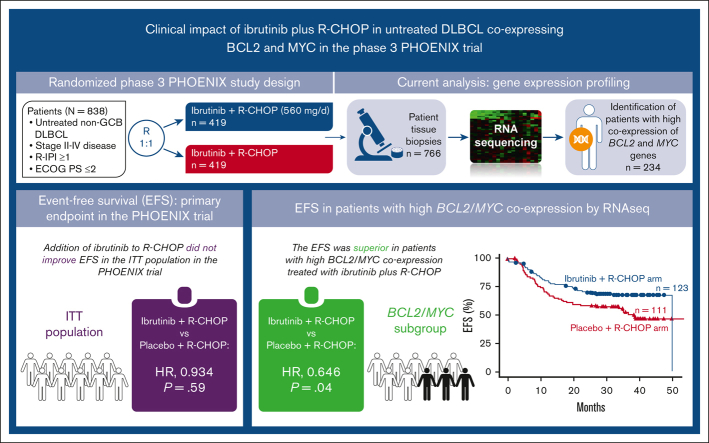
In the PHOENIX trial, the addition of ibrutinib to R-CHOP did not improve the survival of patients with previously untreated non-GCB DLBCL.
-
•
This study identified a patient subset with high BCL2/MYC coexpression using RNA sequencing, with improved EFS after R-CHOP with ibrutinib.
Visual Abstract
Abstract
Diffuse large B-cell lymphoma (DLBCL), with high coexpression of BCL2 and MYC proteins (DE lymphoma), is considered an adverse prognostic indicator associated mostly with non-germinal center B-cell–like (non-GCB) DLBCL. BCL2/MYC overexpression is associated with B-cell receptor (BCR) pathway activation; consequently, DE DLBCL may be sensitive to BCR inhibitors. We assessed whether high BCL2/MYC coexpression by RNA sequencing could identify a patient subset responsive to ibrutinib using baseline biopsies from the PHOENIX trial, which evaluated the addition of ibrutinib to rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) in untreated non-GCB DLBCL. BCL2/MYC RNA expression was correlated with lower event-free survival (EFS) and overall survival (OS) using Kaplan–Meier estimates with Cox regression and log-rank testing. In total, 234 of 766 (30.5%) patients had high BCL2/MYC coexpression: 123 of 386 (31.9%) received ibrutinib plus R-CHOP and 111 of 380 (29.2%) received R-CHOP. EFS was superior with ibrutinib plus R-CHOP compared with R-CHOP alone in patients with high BCL2/MYC coexpression, but there was no significant impact on OS. However, EFS and OS showed clinically meaningful improvement with ibrutinib plus R-CHOP over R-CHOP alone in patients aged <60 years with high BCL2/MYC coexpression. We observed a significant association between high BCL2/MYC coexpression and activated B-cell-like and MYD88L265P/CD79B-mutated subtypes of DLBCL. Consequently, high BCL2/MYC coexpression identified a subset of non-GCB DLBCL that may be preferentially responsive to ibrutinib and warrants further investigation. This trial was registered at www.clinicaltrials.gov as #NCT01855750.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common type of non-Hodgkin lymphoma (NHL), representing 25% to 30% of all NHL cases worldwide.1 It is recognized that DLBCL is a heterogenous disease characterized by distinct, pathologic subtypes and gene expression profiles impacting clinical outcomes.2,3 Studies have shown that rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) chemoimmunotherapy, the standard frontline treatment for DLBCL,4 is effective in approximately 60% of patients,5 emphasizing the need for alternative therapeutic options.
Gene expression profiling (GEP) identified 2 molecular DLBCL subtypes according to the differentiation stage of the B-cell the cancer originated from: germinal center B-cell–like (GCB) and activated B-cell–like (ABC), together comprising 80% to 85% of cases, with the remaining cases considered unclassified.6,7 Using the Hans algorithm, DLBCL can also be dichotomously classified by immunohistochemistry (IHC) into GCB and non-GCB (non-GCB includes ABC by GEP plus several unclassified cases).8 Patients with DLBCL of ABC origin (and most non-GCB by IHC) have constitutive activation of the NF-κB pathway required for survival of this type of lymphoma,9 and they are at higher risk of treatment failure following R-CHOP therapy.3,7,10
More recent molecular subclassification attempts of DLBCL tumors have proposed to further subdivide the ABC subgroup into genetic subtypes such as MCD (characterized by MYD88L265P and/or CD79B mutations), BN2 (characterized by BCL6 fusions and/or NOTCH2 mutations), N1 (characterized by NOTCH1 mutations), and A53 (aneuploid with TP53 inactivation), among others.11, 12, 13 These genetic subtypes have distinct pathogeneses and phenotypic properties and may respond differently to therapies,11,13, 14, 15 which remains to be proven prospectively.
BCL2 and MYC genes are key regulators of cell survival16 and proliferation,17 respectively, and deregulation of these genes is a well-known adverse prognostic factor in patients with DLBCL treated with standard-of-care therapy. Overexpression of BCL2 and MYC genes can occur because of chromosomal translocations, copy number aberrations, and transcriptional/translational changes.18, 19, 20, 21 Based on the underlying molecular aberrations, DLBCL featuring BCL2 and MYC overexpression can be placed into 2 categories. Several studies using IHC have identified that a significant proportion of DLBCL cases are characterized by high coexpression of BCL2 and MYC proteins, which is associated with poor prognosis following R-CHOP treatment.20, 21, 22 DLBCL with high coexpression of BCL2 and MYC proteins, or double-expressor (DE) lymphoma,2 is found primarily in the non-GCB (or ABC) subtype of DLBCL.22 These DLBCLs are genetically distinct from those with MYC and BCL2 chromosomal rearrangements (double-hit), which are almost all of GCB origin and are now classified by the World Health Organization as high-grade B-cell lymphoma.2
Ibrutinib, a first-in-class, once-daily, oral, covalent Bruton’s tyrosine kinase (BTK) inhibitor, is approved in the United States for the treatment of adult patients with various B-cell malignancies, including chronic lymphocytic leukemia/small lymphocytic lymphoma, Waldenström’s macroglobulinemia, previously treated mantle cell lymphoma, and previously treated marginal zone lymphoma.23 The preferential antitumor activity of ibrutinib against the non-GCB subtype of DLBCL was demonstrated in an early phase clinical trial,24 likely owing to the reduction in NF-κB activity through the inhibition of BTK.25 Improved responses with ibrutinib plus R-CHOP were also reported in a small phase 1b trial in previously untreated CD20-positive NHL.26,27
In the phase 3 PHOENIX trial (NCT01855750) in patients with previously untreated non-GCB DLBCL, the addition of ibrutinib to R-CHOP did not improve event-free survival (EFS; primary end point) or overall survival (OS) in the intent-to-treat (ITT) and ABC populations.28 A preplanned analysis showed a significant interaction between treatment effect and age, with patients aged <60 years deriving significant clinical benefit from ibrutinib plus R-CHOP compared with placebo plus R-CHOP (EFS: hazard ratio [HR], 0.579; 95% confidence interval [CI], 0.380-0.881; P = .0099; OS: HR, 0.330; 95% CI, 0.162-0.673; P = .0013).28 No survival benefit was seen in patients ≥60 years, likely because of increased toxicity in the ibrutinib plus R-CHOP arm, leading to reduced R-CHOP administration and, consequently, therapeutic benefit.28
As MYC and BCL2 overexpression in ABC DLBCL can be correlated with the activation of B-cell receptor (BCR) and NF-κB signaling,25,29,30 and the BCR signaling pathway may be more active in BCL2/MYC DE DLBCL,31 targeting these pathways with BCR inhibitors might improve outcomes in patients with high BCL2/MYC coexpression. Moreover, a recent study that characterized the genetic subtypes of DLBCL in the PHOENIX trial identified MCD and N1 subtypes (2 of 4 genetic subtypes of ABC DLBCL) to be associated with clinical benefit with ibrutinib plus R-CHOP.12 We hypothesized that high BCL2/MYC coexpression may identify a subset of patients with non-GCB DLBCL DE lymphoma in the PHOENIX trial who were more responsive to ibrutinib treatment. We performed an exploratory analysis using baseline tumor biopsies from the PHOENIX trial to evaluate the expression levels of MYC and BCL2 using RNA sequencing (RNA-seq) in both treatment arms. We used RNA-seq rather than IHC because of the small amount of tissue available for the analysis. Our goal was to identify patients with high BCL2/MYC coexpression and to determine whether there was a correlation between the expression data and EFS and OS in the overall population and in patients aged <60 and ≥60 years, respectively.
Methods
Patients and study design
A detailed description of the PHOENIX study has been published previously; the study was approved by the institutional review board or independent ethics committee at each participating institution and conducted in accordance with ethical principles defined by the Declaration of Helsinki and the International Conference on Harmonisation Guidelines for Good Clinical Practice. All patients provided written informed consent.28 Briefly, this randomized, placebo-controlled, phase 3 study enrolled patients with previously untreated non-GCB DLBCL (N = 838) to receive either ibrutinib (oral 560 mg/day) plus R-CHOP (intravenous rituximab 375 mg/m2, cyclophosphamide 750 mg/m2, doxorubicin 50 mg/m2, vincristine 1.4 mg/m2 [maximum total, 2 mg], and oral prednisone [or equivalent] 100 mg) for 6 or 8 21-day cycles, or placebo plus R-CHOP.28 The primary end point was EFS in the ITT population (non-GCB per IHC assessment) and the ABC DLBCL subgroup (ABC per GEP assessment). The secondary end points were progression-free survival (PFS), OS, and safety. An exploratory analysis evaluated the efficacy and safety by age (<60 and ≥60 years). Response was assessed by investigators using computed tomography scans, per the Revised Response Criteria for Malignant Lymphoma.32
Gene expression analysis by RNA-seq
Pretreatment formalin-fixed paraffin-embedded biopsy samples were collected at baseline. RNA was extracted, prepared with TruSeq RNA Access, and profiled on an Illumina NextSeq 500 instrument (Q2 Solutions; IQVIA, Morrisville, NC) with 100bp paired-end reads and 30 to 60 million read pairs per library. Raw sequence reads were aligned to the hs37d5 genome using STAR v2.5. 1b and gene-level quantification was performed using RSEM v1.2.23.
The median transcript per million (TPM) mapped reads for BCL2 and MYC gene expression across all patients with RNA-seq data (n = 766) was used as a cutoff between high and low expression for each gene. The expression level determined by RNA-seq was compared with matching BCL2 IHC data from 184 patients for whom IHC data were available. Based on high or low BCL2 expression by IHC (low expression, <50% positive lymphoma cells; high expression, ≥50% positive lymphoma cells), an RNA-seq cutoff that minimized the sum of the percentage misclassified in each group was determined. This cutoff value was the same as the median RNA-seq expression value for BCL2 (13.3 TPM; supplemental Figure 1), validating the use of a median TPM value for the larger data set. The median TPM value of MYC was 9.31. The relationship between expression by RNA-seq and survival (EFS and OS) in the 2 study arms was analyzed using Cox regression to determine HRs and log-rank testing to assess significance. The association between HTG EdgeSeq (for ABC/non-ABC subtyping; HTG Molecular Diagnostics, Tucson, AZ) cell of origin calls28,33 and DE lymphoma status was determined using Fisher’s exact test. The calls for the MCD genetic subtype were derived from the published literature,12 and the association with DE lymphoma was tested using the statistical method described above.
Mutation detection and analysis
For biopsies from the non-China cohort of the PHOENIX study, targeted sequencing was performed at an average depth of 707× on 99 genes that were recurrently mutated in patients with lymphoma (summarized in Wilson et al12) using the NuGEN Ovation Custom Target Enrichment System (NuGen, San Carlos, CA). Whole exome sequencing in the PHOENIX China cohort was performed using a SureSelect Human All Exon V6 capture kit and SureSelectXT Reagent Kit (Agilent, Santa Clara, CA) for library preparation. Sequencing was performed using the NovaSeq 6000 System (Illumina, San Diego, CA) with an average depth of 167×. Variants were called, annotated, and filtered as previously published,34 but using Mutect2 and rejecting any variants that were multiallelic and/or did not pass standard Mutect2 filters (either “PASS” or “clustered_events” alone was required in the “filter” field). In addition, for results from the NuGEN Ovation Target Enrichment System, UMI-tools 0.3.6 was used for deduplication, and more stringent filtering criteria were applied: a minimum sequencing depth of 70 and a minimum alternate allele frequency of 0.09 were required for variants called from those samples. Gene-/variant-level mutation frequencies in patients with high BCL2/MYC coexpression by RNA-seq were compared with no BCL2/MYC coexpression, using Fisher exact test to identify genes associated with high BCL2/MYC coexpression.
Results
Patients and treatment
Detailed baseline demographics and primary efficacy/safety results for the PHOENIX study have been reported previously.28 Briefly, between October 2013 and November 2015, 838 patients (ITT) with untreated non-GCB DLBCL were randomly assigned to ibrutinib plus R-CHOP (n = 419) or placebo plus R-CHOP (n = 419). Of the 747 evaluable patients, 567 (75.9%) had the ABC subtype.28 In the primary study, the median age was 62.0 years and the median time from diagnosis to treatment initiation was 27 days. More patients discontinued treatment in the ibrutinib plus R-CHOP arm than in the placebo plus R-CHOP arm (22.4% vs 13.6%), mostly because of adverse events. Median follow-up for both arms was 34.8 months.28
This exploratory analysis included 766 patients for whom RNA-seq data were available: 386 from the ibrutinib plus R-CHOP arm and 380 from the placebo plus R-CHOP arm. The baseline demographics and disease characteristics of the 766 patients included in this analysis (supplemental Table 1) were similar to those of the primary study population.28 The median age was 62.0 years, 53.7% of patients were male, median time from diagnosis was 26 days, and 68.9% of patients had ≥1 extranodal disease site. The majority of patients had advanced tumor stage (Ann Arbor III-IV, 76.2%) and had an Eastern Cooperative Oncology Group performance status (ECOG PS) score of 0 to 1 (88.0%), and 43.9% were in the revised International Prognostic Index (R-IPI) high-risk group based on information in the clinical database.
BCL2 and MYC expression by RNA-seq
Of the 766 patients for whom RNA-seq data for BCL2 or MYC expression status were available, 234 (30.5%) showed high BCL2/MYC coexpression (Table 1). The proportion of patients with high BCL2/MYC coexpression was similar between the ibrutinib plus R-CHOP (123/386 [31.9%]) and placebo plus R-CHOP (111/380 [29.2%]) treatment arms (Table 1). Similarly, the proportion of patients with high BCL2/MYC coexpression was comparable between younger (<60 years; 97/317 [30.6%]) and older (≥60 years; 137/449 [30.5%]) patients (Table 1). There were no substantial differences in baseline demographic and clinical characteristics between patients with (n = 234) and without (n = 532) high BCL2/MYC coexpression (supplemental Table 1).
Table 1.
Patients with high BCL2 and/or high MYC expression according to treatment arm and age
| n (%) | Ibrutinib + R-CHOP (n = 386) | Placebo + R-CHOP (n = 380) |
|---|---|---|
| All ages | ||
| BCL2-high | 196 (50.8) | 187 (49.2) |
| MYC-high | 199 (51.6) | 183 (48.2) |
| MYC-high + BCL2-high | 123 (31.9) | 111 (29.2) |
| <60 y | ||
| n | 149 | 168 |
| MYC-high + BCL2-high | 47 (31.5) | 50 (29.8) |
| ≥60 y | ||
| n | 237 | 212 |
| MYC-high + BCL2-high | 76 (32.1) | 61 (28.8) |
High BCL2 and MYC coexpression by RNA-seq and outcomes
Response rate
Of 123 patients with high BCL2/MYC coexpression by RNA-seq in the ibrutinib plus R-CHOP arm, 83/123 (67.5%) achieved a complete response (CR) and 28/123 (22.8%) achieved a partial response (PR) (supplemental Table 2). In the placebo plus R-CHOP arm, of 111 patients with high BCL2/MYC coexpression by RNA-seq, 72/111 (64.9%) had CR and 31/111 (27.9%) had PR (supplemental Table 2).
Survival
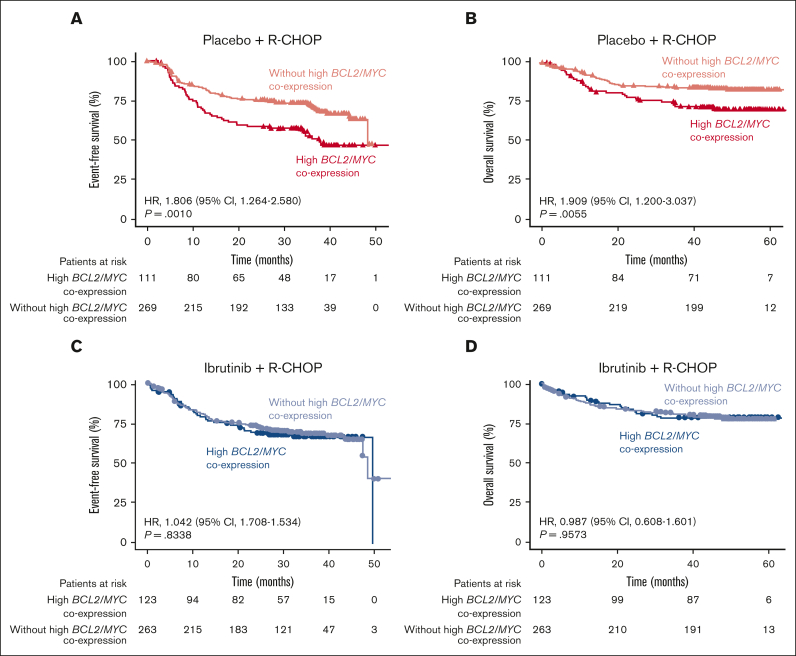
In the placebo plus R-CHOP arm, patients with high BCL2/MYC coexpression (n = 111) had poorer EFS (HR, 1.806; 95% CI, 1.264-2.580; P = .0010; Figure 1A) and OS (HR, 1.909; 95% CI, 1.200-3.037; P = .0055; Figure 1B) than those without high BCL2/MYC coexpression (n = 269). The poor prognosis of R-CHOP–treated patients with high BCL2/MYC coexpression determined by this RNA-seq analysis is consistent with the results from studies evaluating coexpression of MYC and BCL2 proteins by IHC.20, 21, 22
Figure 1.
EFS and OS in patients with or without high BCL2/MYC coexpression. Overall EFS and OS in patients with or without high BCL2/MYC coexpression determined by RNA-seq in the placebo plus R-CHOP (A,B) and ibrutinib plus R-CHOP (C,D) arms.
In contrast, in the ibrutinib plus R-CHOP arm, there was no difference in either EFS or OS between patients with high BCL2/MYC coexpression (n = 123) and those without (n = 263), suggesting a potential benefit from ibrutinib treatment (Figure 1C-D).
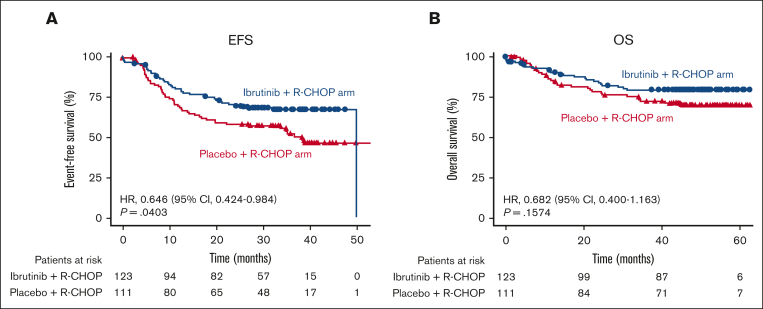
When considering all patients with high BCL2/MYC coexpression, the EFS was superior in patients who received ibrutinib plus R-CHOP (n = 123) than in placebo plus R-CHOP (n = 111) (HR, 0.646; 95% CI, 0.424-0.984; P = .0403). A trend toward better OS was observed, but the difference was not significant (HR, 0.682; 95% CI, 0.400-1.163; P = .1574; Figure 2A-B).
Figure 2.
EFS and OS in patients with high BCL2/MYC coexpression by treatment arm. Overall EFS (A) and OS (B) in patients with high BCL2/MYC coexpression by RNA-seq in the placebo plus R-CHOP and ibrutinib plus R-CHOP arms.
The primary analysis of the PHOENIX study had shown that ibrutinib plus R-CHOP had improved EFS and OS over placebo plus R-CHOP (EFS: HR, 0.579; 95% CI, 0.380-0.881; P = .0099; OS: HR, 0.330; 95% CI, 0.162-0.673; P = .0013) in patients <60 years but not in patients ≥60 years, likely because of increased treatment toxicity in older patients.28
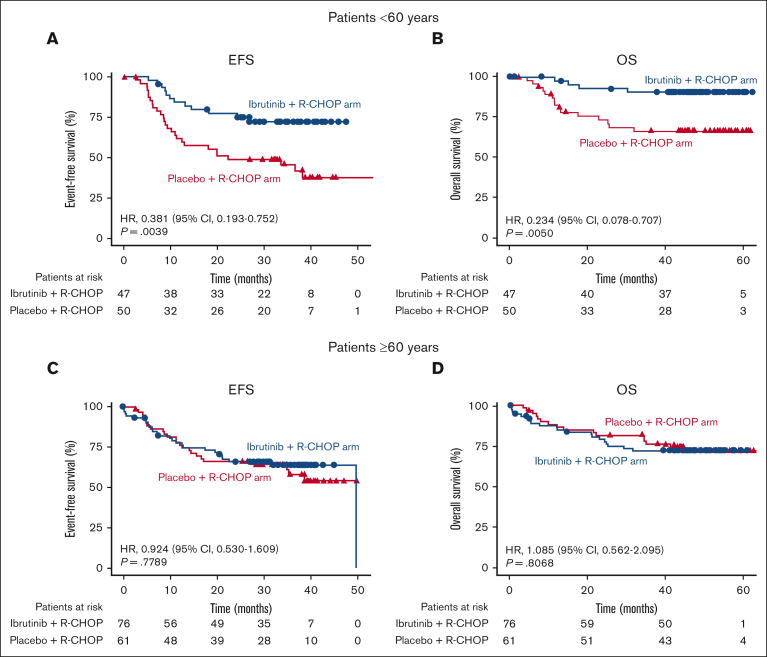
In this analysis, when the outcomes of patients with high BCL2/MYC expression were compared by age, both EFS (HR, 0.381; 95% CI, 0.193-0.752; P = .0039) and OS (HR, 0.234; 95% CI, 0.078-0.707; P = .0050) were improved in patients <60 years treated with ibrutinib plus R-CHOP (n = 47) rather than placebo plus R-CHOP (n = 50; Figure 3A-B). Interestingly, in patients ≥60 years with high BCL2/MYC coexpression (ibrutinib plus R-CHOP, n = 76; placebo plus R-CHOP, n = 61), there was no difference in EFS (HR, 0.924; 95% CI, 0.530-1.609; P = .7789) and OS (HR, 1.085; 95% CI, 0.562-2.095; P = .8068) between the treatment arms (Figure 3C-D). However, this result differed from that of the primary analysis, which reported a trend toward worse outcomes with ibrutinib plus R-CHOP than R-CHOP alone (EFS: HR, 1.228; 95% CI, 0.887-1.699; P = .2153; OS: HR, 1.440; 95% CI, 0.963-2.152; P = .0739) in patients ≥60 years.28 This was possibly because of the increased benefit of ibrutinib addition to R-CHOP in the high BCL2/MYC expression group.
Figure 3.
EFS and OS in patients with high BCL2/MYC coexpression by treatment arm and age. EFS and OS in patients with high BCL2/MYC coexpression by RNA-seq in the placebo plus R-CHOP and ibrutinib plus R-CHOP arms, aged <60 years (A,B) and ≥60 years (C,D).
Single high MYC or BCL2 expression and survival
There was no correlation with outcomes in patients with only a single high MYC or BCL2 expression. Patients with high MYC but low BCL2 expression and those with low MYC but high BCL2 expression had similar EFS and OS in the ibrutinib plus R-CHOP (n = 73) and placebo plus R-CHOP (n = 76) arms (supplemental Figure 2A-D).
DLBCL subtypes and gene mutations associated with high BCL2/MYC coexpression by RNA-seq
We found that high BCL2/MYC coexpression was significantly associated with the ABC subtype of DLBCL (P < .0001), as well as the MCD genetic subtype (P = .0016; Table 2). The proportion of patients with DE lymphoma by RNA-seq was 35.7% in ABC vs 14.5% in GCB/unclassified DLBCL, and 44% in MCD vs 28.4% in non-MCD (supplemental Table 3). The A53 genetic subtype could not be inferred because aneuploidy assessment was not possible by methods used in this analysis. We also performed Fisher’s exact test to determine which gene mutations were associated with the DE lymphoma status. Mutations in MYD88L265P, PIM1, and MYC genes were significantly more common in DE lymphoma (all P < .05, odds ratio [OR] >1; Table 3) than in non-DE lymphoma. Mutations in other genes associated with ABC DLBCL, such as CD79A, CD79B, and SYK, were also associated with positive ORs, but the association with DE status was not significant. Mutations that were less common in DE lymphoma (OR <1) than in non-DE lymphoma included BCL10, CYLD, NFKBIA, NFKB2, STAT3, PLCG2 (all P < .05), and BTK (P = .05) (Table 3).
Table 2.
| DE vs ABC calls (RNA-seq vs HTG EdgeSeq), n = 705 | ||||
|---|---|---|---|---|
| Subtypes, n (%) | DE, n = 217 | OR (95% CI) | P value | Non-DE, n = 488 |
| ABC | 193 (88.9) | 3.26 (2.02-5.45) | <.0001 | 347 (71.1) |
| GCB/unclassified | 24 (11.1) | - | - | 141 (28.9) |
| DE vs MCD/N1/BN2 calls (RNA-seq vs LymphGen), n = 765 | ||||
|---|---|---|---|---|
| Subtypes, n (%) | DE, n = 234 | OR (95% CI) | P value | Non-DE, n = 531 |
| MCD | 48 (20.5) | 0.50 (0.33-0.78) | .0016 | 61 (11.5) |
| Non-MCD | 186 (79.5) | - | - | 470 (88.5) |
| N1 | 11 (4.7) | 0.67 (0.29-1.61) | .3034 | 17 (3.2) |
| Non-N1 | 223 (95.3) | - | - | 514 (96.8) |
| BN2 | 10 (4.3) | 1.63 (0.77-3.75) | .2472 | 36 (6.8) |
| Non-BN2 | 224 (95.7) | - | - | 495 (93.2) |
| ABC vs MCD/N1/BN2 calls (HTG EdgeSeq vs LymphGen), n = 709 | ||||
|---|---|---|---|---|
| Subtypes, n (%) | ABC, n = 543 | OR (95% CI) | P value | GCB/unclassified, n = 166 |
| MCD | 101 (18.6) | 0.08 (0.02-0.25) | <.0001 | 3 (1.8) |
| Non-MCD | 442 (81.4) | - | - | 163 (98.2) |
| N1 | 22 (4.1) | 0.59 (0.14-1.76) | .4785 | 4 (2.4) |
| Non-N1 | 521 (95.9) | - | - | 162 (97.6) |
| BN2 | 34 (6.3) | 1.17 (0.54-2.38) | .7186 | 12 (7.2) |
| Non-BN2 | 509 (93.7) | - | - | 154 (92.8) |
A53, aneuploid with TP53 inactivation.
Any deviations from the total number of patients in the ITT population (n = 838), the RNA-seq–surveyed subset (766 patients with DE/non-DE calls), the HTG EdgeSeq-surveyed subset (747 patients with ABC/GCB/Unclassified calls), or the subset with LymphGen calls (773 patients with MCD/N1/BN2/other calls), result from a lack of complete overlap between patients surveyed with the different assays.
A53 subtype could not be inferred as assessing aneuploidy was not allowed by methods used in the analysis.
P < .01 for the following pairwise Fisher's exact tests: DE vs MCD, DE vs ABC, and MCD vs ABC. All other Fisher's exact results had P > .20.
Table 3.
Gene mutations more frequent (OR >1) or less frequent (OR <1) in patients with DE lymphoma vs non-DE lymphoma∗
| Gene | DE, n |
Non-DE, n |
P value | OR | 95% CI | ||
|---|---|---|---|---|---|---|---|
| WT | Mut | WT | Mut | ||||
| MYD88L265P | 176 | 46 | 428 | 51 | .0006 | 2.1907 | 1.38-3.47 |
| CD79A | 212 | 10 | 468 | 11 | .1505 | 2.0047 | 0.75-5.29 |
| MYC | 198 | 24 | 450 | 29 | .0316 | 1.8791 | 1.02-3.44 |
| ERBB4 | 213 | 9 | 468 | 11 | .2238 | 1.7960 | 0.65-4.85 |
| SYK | 208 | 14 | 461 | 18 | .1718 | 1.7224 | 0.78-3.74 |
| PIM1 | 152 | 70 | 378 | 101 | .0034 | 1.7222 | 1.18-2.50 |
| MET | 203 | 19 | 453 | 26 | .1357 | 1.6295 | 0.83-3.14 |
| TRAF3 | 212 | 10 | 465 | 14 | .2745 | 1.5657 | 0.61-3.86 |
| TRAF4 | 211 | 11 | 463 | 16 | .2987 | 1.5077 | 0.62-3.53 |
| WHSC1 | 198 | 24 | 442 | 37 | .1953 | 1.4472 | 0.81-2.56 |
| CD79B | 175 | 47 | 395 | 84 | .2536 | 1.2625 | 0.83-1.91 |
| KMT2D | 142 | 80 | 330 | 149 | .1957 | 1.2473 | 0.88-1.77 |
| TNFAIP3 | 193 | 29 | 396 | 83 | .1834 | 0.7172 | 0.44-1.15 |
| EP300 | 192 | 30 | 388 | 91 | .0855 | 0.6666 | 0.41-1.06 |
| NOTCH1_N | 202 | 20 | 416 | 63 | .1317 | 0.6542 | 0.36-1.13 |
| MYD88_nonL265P | 208 | 14 | 432 | 47 | .1498 | 0.6190 | 0.31-1.18 |
| STAT3 | 210 | 12 | 430 | 49 | .0429 | 0.5019 | 0.24-0.98 |
| CARD11_nonCC | 217 | 5 | 457 | 22 | .2039 | 0.4791 | 0.14-1.32 |
| PLCG2 | 212 | 10 | 436 | 43 | .0450 | 0.4787 | 0.21-0.99 |
| TRAF2 | 218 | 4 | 461 | 18 | .2435 | 0.4704 | 0.11-1.45 |
| TXK | 216 | 6 | 452 | 27 | .1235 | 0.4655 | 0.15-1.17 |
| ITK | 219 | 3 | 463 | 16 | .2091 | 0.3968 | 0.07-1.41 |
| TRAF5 | 219 | 3 | 463 | 16 | .2091 | 0.3968 | 0.07-1.41 |
| BTK | 217 | 5 | 452 | 27 | .0515 | 0.3862 | 0.11-1.04 |
| VRK2 | 218 | 4 | 457 | 22 | .0851 | 0.3816 | 0.09-1.14 |
| CSK | 218 | 4 | 456 | 23 | .0589 | 0.3642 | 0.09-1.08 |
| NFKB2 | 217 | 5 | 450 | 29 | .0360 | 0.3580 | 0.11-0.95 |
| CYLD | 217 | 5 | 447 | 32 | .0170 | 0.3223 | 0.10-0.85 |
| MDM2 | 219 | 3 | 459 | 20 | .0662 | 0.3148 | 0.06-1.08 |
| NFKBIA | 219 | 3 | 457 | 22 | .0297 | 0.2849 | 0.05-0.96 |
| BCL10 | 218 | 4 | 448 | 31 | .0081 | 0.2655 | 0.07-0.76 |
mut, mutated; WT, wild-type.
List of genes for which P < .3.
Discussion
This analysis examined the clinical outcomes associated with high coexpression of BCL2 and MYC using RNA-seq in the ibrutinib plus R-CHOP and placebo plus R-CHOP treatment arms from the phase 3 PHOENIX trial of non-GCB DLBCL. Our study effectively excluded patients with high BCL2/MYC expression owing to BCL2 and MYC translocations (double hits), as these are infrequent and almost always occur in GCB DLBCL.21,35 DE lymphoma is more common and is associated with the non-GCB subtype population in this trial.20, 21, 22,36
Several studies have reported high BCL2/MYC coexpression by IHC in 21% to 34% of DLBCL cases, with BCL2 and MYC high expression cutoffs of 50% to 70% and 40%, respectively.20, 21, 22,36 In this analysis, we used median expression values for BCL2 and MYC RNA expression, which corresponded to the BCL2 IHC cutoff of 50%. In the non-GCB population analyzed here, the frequency of high BCL2/MYC RNA coexpression among all patients tested was 30.5%, which is within the range reported for DE DLBCL measured by IHC in other studies.20, 21, 22,36 Notably, a high correlation between BCL2 protein expression by IHC and BCL2 RNA expression by RNA-seq was reported in experiments conducted using NHL cell lines,37 as well as in tissue samples from patients with DLBCL of the ABC and GCB subtypes.38,39
Patients with DE DLBCL without translocations usually have DE DLBCL of ABC origin and have a poor prognosis with standard R-CHOP therapy.3,22 Consistent with these reports, patients with high BCL2/MYC coexpression treated with R-CHOP in this study had worse EFS and OS than those without high BCL2/MYC coexpression (P = .0010 and P = .0055, respectively). However, when patients with high BCL2/MYC coexpression were treated with ibrutinib plus R-CHOP, there was no difference in survival compared with patients without high BCL2/MYC coexpression, suggesting that the negative prognostic impact of high BCL2/MYC coexpression may have been overcome by ibrutinib.
When considering all patients with high BCL2/MYC coexpression, there was a significantly better EFS (and a trend toward improved OS) in patients who received ibrutinib plus R-CHOP than in those who received placebo plus R-CHOP. In contrast, there was no EFS or OS benefit of ibrutinib when considering the ITT population from the PHOENIX study.28 Our results suggest that high BCL2/MYC coexpression may identify a subset of non-GCB DLBCL that particularly benefit from the addition of ibrutinib to R-CHOP. However, both the PHOENIX study and our subset analysis of patients with high BCL2/MYC coexpression showed that the benefit of ibrutinib in addition to R-CHOP was stronger in younger patients.
In this analysis, the proportion of patients with high BCL2/MYC coexpression by RNA-seq was similar between younger (<60 years, 30.6%) and older (≥60 years, 30.5%) patients. This result differs from the previously reported data showing a higher frequency of BCL2/MYC protein coexpression in older patients (68 years).20
Although DE lymphomas are not a distinct clinicopathological entity in the revised World Health Organization classification,2 patients with DLBCL featuring high MYC/BCL2 coexpression by IHC have been reported to be more likely to have advanced disease stage, high Ki67 proliferative index scores, poor ECOG PS scores, multiple extranodal disease sites, and high-risk R-IPI scores.22,40,41 In this analysis, we did not observe any major differences in the clinicopathological characteristics of patients with vs without high BCL2/MYC coexpression by RNA-seq, except for a slightly higher proportion of patients in the high BCL2/MYC group having elevated lactate dehydrogenase levels. This result may be a reflection of the non-GCB enriched population in this study, with more ABC subtypes than those included in previous studies (71% in this analysis vs 38%-48% in previous studies22,40).
The current analysis was based on the hypothesis that BCL2/MYC DE lymphomas would have, on average, greater NF-κB signaling activity, and that patients with disease driven primarily by NF-κB signaling would be more likely to respond to ibrutinib. In the traditional gene expression-based ABC/GCB subtyping scheme, the ABC subtype is generally driven more by NF-κB signaling.9,42 The same can be said for the MCD subtype in a newer genetic classification system,11 and presumably the very similar C5 cluster in an alternative genetic classification,14 enriched for MYD88L265P and CD79B mutations. Moreover, gene mutations identified in this analysis as frequently associated with DE DLBCL (MYD88L265P, PIM1, CD79B) also correspond to mutations described for the MYD88 cluster in yet another 5-molecular subtype DLBCL classification based on DNA sequencing and Bernoulli mixture-model clustering.15
Recent evidence suggests that patients with DLBCL of the MCD genetic subtype benefit from the addition of ibrutinib to R-CHOP.12 One perspective of the results reported here is that the BCL2/MYC DE population may be enriched for ABC and/or MCD subtypes of DLBCL. Supporting this statement, we found evidence of an association between high BCL2/MYC coexpression by RNA-seq and the ABC subtype (P < .0001) or MCD genetic subtype (P = .0016). We also identified gene mutations that were more frequently found in patients with high BCL2/MYC coexpression, such as MYD88L265P, PIM1, MYC (P < .05 for all) and CD79A, CD79B, and SYK (not significant), several of which have recently been reported to be associated with the MCD genetic subtype.11
The main limitation of this analysis is that because of the insufficient amount of tissue for analysis, we used an RNA-seq method rather than IHC to measure the expression of BCL2/MYC, although IHC was performed on available patient samples to partially validate the RNA-seq data. Although this method has not yet been fully validated for the characterization of DE lymphoma, it has been used for biomarker identification in relapsed or refractory non-GCB DLBCL treated with zanubrutinib43 and may prove valuable in cases where insufficient biopsy tissue precludes protein expression analysis. Another limitation was that this analysis did not evaluate BCL2/MYC rearrangements or other gene aberrations that may be associated with high BCL2/MYC RNA coexpression.
Our data suggested that high BCL2/MYC RNA coexpression may identify a subset of non-GCB DLBCL that is highly responsive to ibrutinib treatment. Ibrutinib plus R-CHOP improved the EFS in patients with BCL2/MYC high-expressing non-GCB DLBCL, as well as the EFS and OS in the subset aged <60 years—a finding that warrants further investigation.
Conflict-of-interest disclosure: P.W.M.J. served as a consultant or in an advisory role for Janssen, received research funding from Epizyme and Janssen, and honoraria from Bristol-Myers Squibb, Takeda, Novartis, Celgene, Janssen, Epizyme, Kite, Genmab, Kymera, Oncimmune, Immunocore, and Incyte. S.B. owns stock in Johnson & Johnson, Gilead Sciences, Celgene, Vertex, and AbbVie and is a former employee of the Janssen Pharmaceutical Companies of Johnson & Johnson, currently retired. B.H., S.M.S., S. Sun, A.J.S., S. Srinivasan, and J.V. are employees of the Janssen Pharmaceutical Companies of Johnson & Johnson. S.M.S., S. Sun, A.J.S., and J.V. own stock in Johnson & Johnson. L.H.S. served as a consultant or in an advisory role for Celgene, AbbVie, Seattle Genetics, TG Therapeutics, Janssen, Amgen, Roche/Genentech, Gilead Sciences, Lundbeck, Apobiologix, Karyopharm Therapeutics, Kite Pharma, Merck, Takeda Pharmaceuticals, TEVA Pharmaceuticals Industries, and TG Therapeutics, received research fundings from Roche/Genentech and honoraria from Amgen, Apobiologix, AbbVie, Celgene, Gilead Sciences, Incyte Janssen-Ortho, Karyopharm Therapeutics, Kite Pharma, Lundbeck, Merck, Roche/Genentech, Sandoz, Seattle Genetics, Takeda Pharmaceuticals, TEVA Pharmaceuticals Industries, and TG Therapeutics. W.H.W. has no competing financial interest.
Acknowledgments
This study was sponsored by Janssen Research & Development, LLC. The authors thank all patients who participated in this study and the study investigators. Writing assistance was provided by Izabela Bombik and Ewa Wandzioch of Parexel, and funded by Janssen Global Services, LLC.
Authorship
Contribution: P.W.M.J., S.B., B.H., L.H.S., and W.H.W. conceptualized the study; P.W.M.J., S.B., B.H., S.M.S., S. Sun, J.V., L.H.S., and W.H.W. were involved in the investigation; and all authors analyzed the data, contributed to the writing of the manuscript, and approved the final draft for submission.
Footnotes
∗S.B.’s affiliation during this study period has been listed. He is currently retired.
†A.J.S’s affiliation during this study period and the development of the manuscript has been listed.
Part of the results were presented at the American Society of Hematology 61st Annual Meeting and Exposition; 7-10 December 2019; Orlando, FL.
The data sharing policy of the Janssen Pharmaceutical Companies of Johnson & Johnson is available at www.janssen.com/clinical-trials/transparency. Information regarding the access to data from this study (EGAS00001005554) can be found at the European Genome-Phenome Archive (EGA) site at ega-archive.org. Sequencing data from the China cohort cannot be made available because of the Chinese prohibitions on genomic data sharing.
The full-text version of this article contains a data supplement.
Supplementary Material
References
- 1.Padala SA, Kallam A. StatPearls; Treasure Island, FL: 2021. Diffuse large B cell lymphoma. [PubMed] [Google Scholar]
- 2.Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375–2390. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scott DW, Mottok A, Ennishi D, et al. Prognostic significance of diffuse large B-cell lymphoma cell of origin determined by digital gene expression in formalin-fixed paraffin-embedded tissue biopsies. J Clin Oncol. 2015;33(26):2848–2856. doi: 10.1200/JCO.2014.60.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.National Comprehensive Cancer Network (NCCN®) Clinical practice guidelines in oncology: B-cell lymphomas version 5.2022. https://www.nccn.org/guidelines/guidelines-detail?category=1&id=1480 [DOI] [PubMed]
- 5.Vaidya R, Witzig TE. Prognostic factors for diffuse large B-cell lymphoma in the R(X)CHOP era. Ann Oncol. 2014;25(11):2124–2133. doi: 10.1093/annonc/mdu109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 7.Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346(25):1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 8.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103(1):275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 9.Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor kappaB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J Exp Med. 2001;194(12):1861–1874. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lenz G, Wright G, Dave SS, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359(22):2313–2323. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitz R, Wright GW, Huang DW, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med. 2018;378(15):1396–1407. doi: 10.1056/NEJMoa1801445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson WH, Wright GW, Huang DW, et al. Effect of ibrutinib with R-CHOP chemotherapy in genetic subtypes of DLBCL. Cancer Cell. 2021;39(12):1643–1653.e3. doi: 10.1016/j.ccell.2021.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wright GW, Huang DW, Phelan JD, et al. A probabilistic classification tool for genetic subtypes of diffuse large B cell lymphoma with therapeutic implications. Cancer Cell. 2020;37(4):551–568.e14. doi: 10.1016/j.ccell.2020.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chapuy B, Stewart C, Dunford AJ, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018;24(5):679–690. doi: 10.1038/s41591-018-0016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lacy SE, Barrans SL, Beer PA, et al. Targeted sequencing in DLBCL, molecular subtypes, and outcomes: a Haematological Malignancy Research Network report. Blood. 2020;135(20):1759–1771. doi: 10.1182/blood.2019003535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaux DL, Cory S, Adams JM. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature. 1988;335(6189):440–442. doi: 10.1038/335440a0. [DOI] [PubMed] [Google Scholar]
- 17.Adams JM, Harris AW, Pinkert CA, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318(6046):533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen L, Papenhausen P, Shao H. The role of c-MYC in B-cell lymphomas: diagnostic and molecular aspects. Genes (Basel) 2017;8(4) doi: 10.3390/genes8040116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu TX, Fan L, Wang L, et al. MYC or BCL2 copy number aberration is a strong predictor of outcome in patients with diffuse large B-cell lymphoma. Oncotarget. 2015;6(21):18374–18388. doi: 10.18632/oncotarget.4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green TM, Young KH, Visco C, et al. Immunohistochemical double-hit score is a strong predictor of outcome in patients with diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol. 2012;30(28):3460–3467. doi: 10.1200/JCO.2011.41.4342. [DOI] [PubMed] [Google Scholar]
- 21.Johnson NA, Slack GW, Savage KJ, et al. Concurrent expression of MYC and BCL2 in diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol. 2012;30(28):3452–3459. doi: 10.1200/JCO.2011.41.0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu S, Xu-Monette ZY, Tzankov A, et al. MYC/BCL2 protein coexpression contributes to the inferior survival of activated B-cell subtype of diffuse large B-cell lymphoma and demonstrates high-risk gene expression signatures: a report from The International DLBCL Rituximab-CHOP Consortium Program. Blood. 2013;121(20):4021–4031. doi: 10.1182/blood-2012-10-460063. quiz 4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.2019. IMBRUVICA® (ibrutinib) [prescribing information]. Pharmacyclics LLC and Janssen Biotech, Inc. [Google Scholar]
- 24.Wilson WH, Young RM, Schmitz R, et al. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat Med. 2015;21(8):922–926. doi: 10.1038/nm.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis RE, Ngo VN, Lenz G, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463(7277):88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Younes A, Thieblemont C, Morschhauser F, et al. Combination of ibrutinib with rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) for treatment-naive patients with CD20-positive B-cell non-Hodgkin lymphoma: a non-randomised, phase 1b study. Lancet Oncol. 2014;15(9):1019–1026. doi: 10.1016/S1470-2045(14)70311-0. [DOI] [PubMed] [Google Scholar]
- 27.Schaffer M, Chaturvedi S, Davis C, et al. Activity of ibrutinib plus R-CHOP in diffuse large B-cell lymphoma: response, pharmacodynamic, and biomarker analyses of a phase Ib study. Cancer Treat Res Commun. 2020;25 doi: 10.1016/j.ctarc.2020.100235. [DOI] [PubMed] [Google Scholar]
- 28.Younes A, Sehn LH, Johnson P, et al. Randomized phase III trial of ibrutinib and rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone in non-germinal center B-cell diffuse large B-cell lymphoma. J Clin Oncol. 2019;37(15):1285–1295. doi: 10.1200/JCO.18.02403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang WG, Liu ZB, Jiang XN, Lee J, Zhou XY, Li XQ. MYC protein dysregulation is driven by BCR-PI3K signalling in diffuse large B-cell lymphoma. Histopathology. 2017;71(5):778–785. doi: 10.1111/his.13287. [DOI] [PubMed] [Google Scholar]
- 30.Xia Y, Zhang X. The spectrum of MYC alterations in diffuse large B-cell lymphoma. Acta Haematol. 2020;143(6):520–528. doi: 10.1159/000505892. [DOI] [PubMed] [Google Scholar]
- 31.Bogusz AM, Kovach AE, Le LP, Feng D, Baxter RHG, Sohani AR. Diffuse large B-cell lymphoma with concurrent high MYC and BCL2 expression shows evidence of active B-cell receptor signaling by quantitative immunofluorescence. PLoS One. 2017;12(2) doi: 10.1371/journal.pone.0172364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheson BD, Pfistner B, Juweid ME, et al. Revised response criteria for malignant lymphoma. J Clin Oncol. 2007;25(5):579–586. doi: 10.1200/JCO.2006.09.2403. [DOI] [PubMed] [Google Scholar]
- 33.Balasubramanian S, Wang S, Major C, et al. Comparison of immunohistochemistry and gene expression profiling subtyping for diffuse large B-cell lymphoma in the phase III clinical trial of R-CHOP +/- ibrutinib. Br J Haematol. 2021;194(1):83–91. doi: 10.1111/bjh.17450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balasubramanian S, Hodkinson B, Schuster SJ, et al. Identification of a genetic signature enriching for response to ibrutinib in relapsed/refractory follicular lymphoma in the DAWN phase 2 trial. Cancer Med. 2022;11(1):61–73. doi: 10.1002/cam4.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le Gouill S, Talmant P, Touzeau C, et al. The clinical presentation and prognosis of diffuse large B-cell lymphoma with t(14;18) and 8q24/c-MYC rearrangement. Haematologica. 2007;92(10):1335–1342. doi: 10.3324/haematol.11305. [DOI] [PubMed] [Google Scholar]
- 36.Batlle-Lopez A, Gonzalez de Villambrosia S, Francisco M, et al. Stratifying diffuse large B-cell lymphoma patients treated with chemoimmunotherapy: GCB/non-GCB by immunohistochemistry is still a robust and feasible marker. Oncotarget. 2016;7(14):18036–18049. doi: 10.18632/oncotarget.7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Punnoose E, Peale FV, Szafer-Glusman E, et al. BCL2 expression in first-line diffuse large B-cell lymphoma identifies a patient population with poor prognosis. Clin Lymphoma Myeloma Leuk. 2021;21(4):267–278.e10. doi: 10.1016/j.clml.2020.11.004. [DOI] [PubMed] [Google Scholar]
- 38.Iqbal J, Neppalli VT, Wright G, et al. BCL2 expression is a prognostic marker for the activated B-cell-like type of diffuse large B-cell lymphoma. J Clin Oncol. 2006;24(6):961–968. doi: 10.1200/JCO.2005.03.4264. [DOI] [PubMed] [Google Scholar]
- 39.Shen Y, Iqbal J, Huang JZ, Zhou G, Chan WC. BCL2 protein expression parallels its mRNA level in normal and malignant B cells. Blood. 2004;104(9):2936–2939. doi: 10.1182/blood-2004-01-0243. [DOI] [PubMed] [Google Scholar]
- 40.Pena C, Villegas P, Cabrera ME. Double or triple-expressor lymphomas: prognostic impact of immunohistochemistry in patients with diffuse large B-cell lymphoma. Hematol Transfus Cell Ther. 2020;42(2):192–193. doi: 10.1016/j.htct.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riedell PA, Smith SM. Double hit and double expressors in lymphoma: definition and treatment. Cancer. 2018;124(24):4622–4632. doi: 10.1002/cncr.31646. [DOI] [PubMed] [Google Scholar]
- 42.Staudt LM. Oncogenic activation of NF-kappaB. Cold Spring Harb Perspect Biol. 2010;2(6):a000109. doi: 10.1101/cshperspect.a000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang H, Li Y, Oh SY, et al. Biomarker identification in relapsed/refractory non-germinal center B-cell–like diffuse large B-cell lymphoma treated with zanubrutinib. HemaSphere. 2020;4(S1):584. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.