Abstract
Gene sequences encoding the enzymes UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) from many bacterial sources were analyzed. It was shown that whereas gram-negative bacteria have only one murA gene, gram-positive bacteria have two distinct genes encoding these enzymes which have possibly arisen from gene duplication. The two murA genes of the gram-positive organism Streptococcus pneumoniae were studied further. Each of the murA genes was individually inactivated by allelic replacement. In each case, the organism was viable despite losing one of its murA genes. However, when attempts were made to construct a double-deletion strain, no mutants were obtained. This indicates that both genes encode active enzymes that can substitute for each other, but that the presence of a MurA function is essential to the organism. The two genes were further cloned and overexpressed, and the enzymes they encode were purified. Both enzymes catalyzed the transfer of enolpyruvate from phosphoenolpyruvate to UDP-N-acetylglucosamine, confirming they are both active UDP-N-acetylglucosamine enolpyruvyl transferases. The catalytic parameters of the two enzymes were similar, and they were both inhibited by the antibiotic fosfomycin.
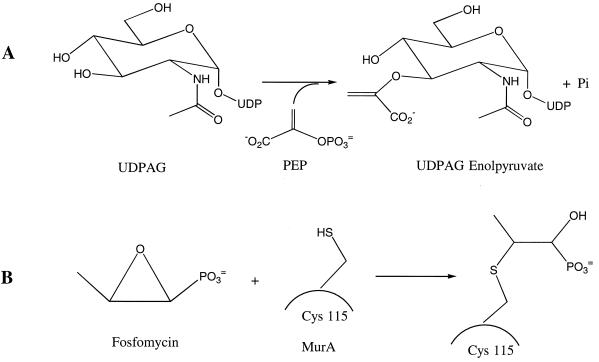
UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) catalyzes the first committed step in bacterial cell wall biosynthesis (9, 20). The enzyme transfers an enolpyruvyl group from phosphoenolpyruvate (PEP) to UDP-N-acetylglucosamine (UDPAG) to form UDP-N-acetylglucosamine enolpyruvate (Fig. 1A). This is a precursor to UDP-N-acetylmuramate, an essential building block for the bacterial cell wall. MurA is inhibited by the antibiotic fosfomycin (Fig. 1B) (10), and because of its importance in peptidoglycan biosynthesis, it is of interest as a target for the design of novel antibacterial agents.
FIG. 1.
(A) The enolpyruvyl transfer reaction catalyzed by MurA. (B) Inactivation of MurA by fosfomycin as a result of the covalent linkage between Cys-115 of MurA and fosfomycin.
MurA (sometimes called MurZ) from Escherichia coli has been studied in great depth. There is one copy of the murA gene in E. coli (16), and this has been shown to be essential by gene deletion experiments (3). The E. coli murA gene has been overexpressed, and the MurA enzyme has been purified and kinetically characterized (16). The Kms of UDPAG and PEP were reported to be 15 and 0.4 μM, respectively, while the kcat was 3.8 s−1 (11). The crystal structure of the enzyme, either ligand free or complexed with UDPAG and fosfomycin, has been solved (21, 22). This shows the enzyme to be made up of two domains, with the active site located in a deep cavity between them, and identifies catalytically and structurally important residues. One catalytically important residue (Cys-115) has been investigated in detail. It was shown that this residue is the site of alkylation by fosfomycin, a finding that has been confirmed by kinetics and mutagenesis studies (Fig. 1B) (15, 27). Mutations in the cysteine residue (C115S) inactivated the enzyme, confirming that this is an important residue in catalysis (27). Another residue (Asp-305) is indicated by the structural data as being the general base involved in the deprotonation of the substrate UDPAG (22).
The importance of MurA as an antibacterial target led us to investigate the MurA enzymes from gram-positive pathogens. While there is only one murA gene in E. coli, we have identified in gram-positive bacteria two murA genes, which have arisen by a possible gene duplication. To investigate if the two genes present in the gram-positive bacteria both encode active enzymes, we performed an in-depth study of the two genes from the gram-positive bacterium Streptococcus pneumoniae. Gene deletion experiments were used to investigate the essentiality of the murA genes. The genes were then overexpressed, the enzymes that they encode were purified, and the biochemical activities of the enzymes were investigated.
MATERIALS AND METHODS
General materials.
S. pneumoniae R6 is a nonencapsulated strain (13). pJS3 (2) was the kind gift of Sanford Lacks (Brookhaven National Laboratory). Biochemical reagents were from Sigma. Enzymes and reagents used in cloning were from Gibco BRL (Gaithersburg, Md.). DNA preparations were performed with assorted kits from Qiagen (Valencia, Calif.). Column chromatography was performed on an AKTA FPLC system (Pharmacia) with Pharmacia Hi-Load columns. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analyses were performed on Novex (San Diego, Ca) 4 to 12% bis-Tris gels. The N-terminal amino acid sequence of the purified enzyme was deduced by automated Edman degradation with a Hewlett-Packard model G1000A sequencer. The molecular weight of the enzyme was determined by matrix-assisted laser-desorption ionization–time of flight (MALDI-TOF) mass spectrometry using a Voyager RP Biospectrometer (PerSeptive Biosystems, Framingham, Mass.).
Phylogenetic analysis.
Homologous protein sequences were retrieved from public and proprietary genomic sequence databases using the software BLASTP and TBLASTN (1). The partial genome of S. pneumoniae type 4 from The Institute for Genome Research was used as the source of MurA1 and MurA2 described here. The proteins were aligned using the program CLUSTALW version 1.7 (23) with the BLOSUM62 (8) similarity matrix and gap opening and extension penalties of 10.0 and 0.05, respectively. The multiple sequence alignments were refined manually using the program SEQLAB of the Wisconsin Package, version 9.0 (Genetics Computer Group, Madison, Wis.).
Phylogenetic trees were constructed by maximum likelihood (ML), neighbor-joining (N-J), and maximum parsimony (MP) methods for each set of alignments. The software PUZZLE version 4.0 was used for ML tree construction (23). N-J trees were based on pairwise distances between amino acid sequences using the programs NEIGHBOR and PROTDIST of the PHYLIP 3.57c package (J. Felsenstein, PHYLIP [Phylogeny Inference Package] version 3.57c [http://evolution.genetics.washington.edu/phylip.html]). The “Dayhoff” program option was invoked in the latter program, which estimates based on the Dayhoff 120 matrix (6). The programs SEQBOOT and CONSENSE were used to estimate the confidence limits of branching points from 1,000 bootstrap replications. MP analysis was done using the software package PAUP* (24). Given the large size of the data set, it was not possible to exhaustively search for the total number of minimal length trees. Instead, the numbers and lengths of minimal trees were estimated from 100 replicate random heuristic searches, while confidence limits of branch points were estimated by 1,000 bootstrap replications.
Generation of S. pneumoniae allelic replacement mutants.
S. pneumoniae 0100993 chromosomal DNA was prepared as follows. Four milliliters of S. pneumoniae culture with an optical density at 650 nm of 0.6 to 0.7 was spun down, resuspended in 100 μl of 1 M Tris-HCl (pH 8)–0.25 M EDTA–5% sodium deoxycholate–40% glucose, and incubated at 37°C for 20 min; 10 μl of 10% (wt/vol) SDS was added, and the suspension was incubated at 37°C for a further 10 min. The mixture was phenol-chloroform extracted three times, followed by a single chloroform extraction. The DNA was precipitated with isopropanol, washed twice in 70% ethanol, and resuspended in water. Chromosomal DNA fragments flanking murA1 and murA2 were PCR amplified from chromosomal DNA and purified as previously described (19). These were then used to make antibiotic-resistant gene-targeting constructs as described in Results, which were used to transform S. pneumoniae R6 competent cells prepared according to published protocols. A total of 106 cells were incubated with 500 ng of allelic replacement cassette at 30°C for 30 min and then transferred to 37°C for 90 min to allow expression of antibiotic resistance (7). Cells were plated in AGCH medium (12) containing erythromycin (1 μg/ml) or chloramphenicol (2.5 μg/ml) as appropriate and incubated at 37°C for 36 h in 5% CO2. Transformation efficiencies were 103 transformants per μg of control DNA.
If antibiotic-resistant S. pneumoniae R6 colonies were obtained, they were picked and grown overnight in Todd-Hewitt broth supplemented with 0.5% yeast extract. Chromosomal DNA was prepared and examined by Southern blot analysis and diagnostic PCR to verify that the appropriate chromosomal DNA rearrangement had occurred. In the former, flanking DNA fragments labeled using the ECL (enhanced chemiluminescence) system (Amersham Life Science Limited) were used as probes to chromosomal DNA restricted with appropriate enzymes and blotted using standard methodologies. In the latter, DNA primers designed to hybridize within ermAM or cat as appropriate were paired with primers hybridizing to distal chromosomal sequences to generate DNA amplification products of characteristic size.
Whole-cell antimicrobial activity.
Whole-cell antimicrobial activity was determined by broth microdilution. Fosfomycin was dissolved in dimethyl sulfoxide and diluted 1:10 in water to produce a 256-μg/ml stock solution. Using a 96-well microtiter plate, a Microlab AT Plus 2 (Hamilton Co., Reno, Nev.) serially diluted 50 μl of the stock solution into cation-adjusted Mueller-Hinton broth (Becton Dickinson, Cockeysville, Md.). After the fosfomycin was diluted, a 50-μl aliquot of the test isolate (∼106 CFU/ml) was added to each well of the microtiter plate. The final test concentrations ranged from 0.06 to 64 μg/ml. Inoculated plates were incubated at 35°C in ambient air for 18 to 24 h. The MIC was determined as the lowest concentration of fosfomycin that inhibited visible growth.
Cloning and overexpression.
The murA1 and murA2 genes were amplified by PCR from the genomic DNA of S. pneumoniae 100993. The murA1 gene (1,281 bp) was amplified in two fragments: fragment A has NcoI and EcoRI restriction ends and was amplified using the oligonucleotides 5′-GGAAACGACCATGGATAAAATTGTGGTTCAAGGTGG-3′ and 5′-CGCCTTTTGCAACTGTCATCAAGGCTG-3′. Fragment B has EcoRI and HindIII restriction ends and was amplified using the oligonucleotides 5′-GGGTGTTGAAGTAATTGAAGAAGACG-3′ and 5′-GACCGATTCCTAGAGCCAGAGTACC-3′. Following the restriction digestion, the fragments were ligated into pET28a(+) (Novagen, Madison, Wis.), and the new construct was used to transform E. coli BL21(DE3) competent cells (Stratagene, La Jolla, Calif.). The expression of murA1 was induced with 1 mM isopropylthio-β-d-galactoside (IPTG) at 18°C for 24 h.
The murA2 gene was amplified with NdeI and HindIII restriction ends using the oligonucleotides 5′-CATATGAGAAAAATTGTTATCAATG-3′ and 5′-AAGCTTAATCCTCAACAAGTCTAATATC-3′. The PCR product of the murA2 gene (1,260 bp) was first cloned into the cloning vector pCR-Blunt (Invitrogen, Carlsbad, Calif.), and the desired insert was cut from the plasmid with NdeI and HindIII. The insert was separated by 1% agarose gel, purified, and subsequently cloned into pET26b(+) (Novagen). The new construct was used to transform E. coli BL21(DE3). The expression of murA2 was induced with 0.2 mM IPTG at 30°C for 3.5 h.
Enzyme purification.
E. coli BL21(DE3) cells carrying the plasmid containing the murA1 or murA2 gene were grown, respectively, in 1 or 4 liters of LB medium containing kanamycin (0.1 mg/ml) and 2% (wt/vol) glucose. Gene expression was induced using the induction conditions described above. The harvested cells were resuspended in 100 mM Tris (pH 8.0)–5 mM dithiothreitol (DTT) and disrupted by sonication. For the MurA1 enzyme, the cell lysate was fractionated by ammonium sulfate precipitation (20, 40, and 70%), and the 70% ammonium sulfate supernatant was further purified on a Hi-Load (16/10) Q Sepharose column, equilibrated in 100 mM Tris (pH 8.0)–5 mM DTT. Protein was eluted with a gradient of 0 to 1 M KCl over 200 ml. For the MurA2 enzyme, the cell lysate was brought to 1.5 M ammonium sulfate, and the supernatant of this solution was purified on a Hi-Load (16/10) phenyl-Sepharose column, equilibrated in 100 mM Tris (pH 8)–5 mM DTT–1.5 M ammonium sulfate. Protein was eluted with a gradient of 1.5 to 0 M ammonium sulfate over 200 ml. MurA2 was further purified on a Hi-Load (16/10) Q Sepharose column under the same conditions as used for MurA1. MurA1 and MurA2 were identified by SDS-PAGE analyses and activity assays throughout the purification.
Enzyme assays.
The activity of MurA1 and MurA2 was assayed by measuring the release of Pi from the UDPAG and PEP reaction, using a malachite green assay (14) in 50 mM HEPES (pH 7.5) at room temperature. A typical assay of 200 μl contained substrates and enzyme (MurA1 at 200 nM or MurA2 at 50 nM). For kinetic measurements, the assay was performed with a five-by-five array of various substrates on a half-area 96-well microtiter plate (Costar 3696; Corning Inc., Corning, N.Y.) with a SpectraMax Plus plate reader (Molecular Devices Corp., Sunnyvale, Calif.). Actual concentrations of the substrates are listed in the footnotes to Table 1. Enzyme activity during purification was assayed with both UDPAG and PEP fixed at 1 mM.
TABLE 1.
Kinetic constants of S. pneumoniae MurA1 and MurA2
| Protein | Km(UDPAG) (μM) | Km(PEP) (μM) | kcat (s−1) | kcat/ Km(UDPAG) μM−1 s−1 | kcat/ Km(PEP) (μM−1 s−1) |
|---|---|---|---|---|---|
| MurA1a | 244 ± 20 | 37.0 ± 4.0 | 0.41 ± 0.01 | 0.0017 | 0.011 |
| MurA2b | 119 ± 16 | 11.1 ± 2.3 | 0.78 ± 0.03 | 0.0066 | 0.07 |
Kinetic constants of MurA1 were obtained from a five-by-five array of various substrates ([UDPAG] = 100, 200, 500, 1,000, and 2,000 μM; [PEP] = 50, 65, 125, 250, and 500 μM). MurA1 was at 200 nM. The data were fitted into a ping-pong model in GraFit (version 4.09; Erithacus Software Limited).
Kinetic constants of MurA2 were obtained from a five-by-five array of various substrates ([UDPAG] = 100, 140, 250, 500, and 1,000 μM; [PEP] = 20, 33.3, 50, 100, and 200 μM). MurA2 was at 50 nM. Data fitting was the same as for MurA1.
RESULTS
Sequence and structural comparisons of UDP-N-acetylglucosamine enolpyruvyl transferases.
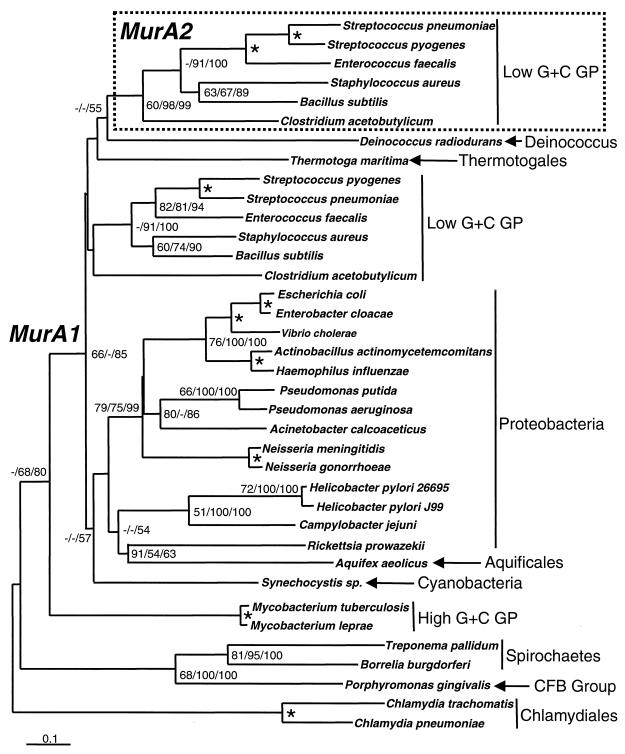
Database searches revealed that the gene encoding UDP-N-acetylglucosamine enolpyruvyl transferase exists widely in nearly all bacteria. However, in low-G+C gram-positive bacteria (Bacillus subtilis, S. pneumoniae, S. pyogenes, Staphylococcus aureus, and Clostridium acetobutylicum), two complete genes encoding UDP-N-acetylglucosamine enolpyruvyl transferase were found (Fig. 2). One of these genes (murA1) was more closely related to the murA genes found in gram-negative bacteria than the second gene copy (murA2). The extent of identical amino acids among MurA1 proteins ranged from 32 to 96% (calculated over ungapped alignments), while that for MurA2 homologs ranged from 53 to 82%. The genus Mycoplasma lacked any open reading frames (ORFs) with significant homology to UDP-N-acetylglucosamine enolpyruvyl transferase, as would be expected for an organism that contains no peptidoglycan. Both genomes were thoroughly searched at the protein (BLASTP) and nucleotide (TBLASTN) levels using B. subtilis MurA1 and MurA2 (1).
FIG. 2.
Phylogenetic tree showing divergence of UDP-N-acetylglucosamine enolpyruvyl transferase isoforms (MurA1 and MurA2). Tree was constructed by the NJ method as implemented by the program NEIGHBOR of the PHYLIP 3.57c package (see Materials and Methods). The scale bar represents 0.1 expected amino acid replacements per site as estimated by the program PROTDIST using the Dayhoff PAM substitution matrix. Numbers at the branching points represent the percent occurrence in 1,000 ML puzzling steps and 1,000 random bootstrap replications of MP and NJ methods, respectively. ML and MP analyses were done with the programs PUZZLE version 4.0 (23) and PAUP* (24), respectively. Nodes where values by all three methods were 90% or greater are labeled with an asterisk. Values less than 50% are not shown or are indicated by a dash. Major taxonomic groups of bacteria (18) are indicated with abbreviations given for gram-positive bacteria (GP) and Cytophaga-Flexibacter-Bacteroides (CFB) group.
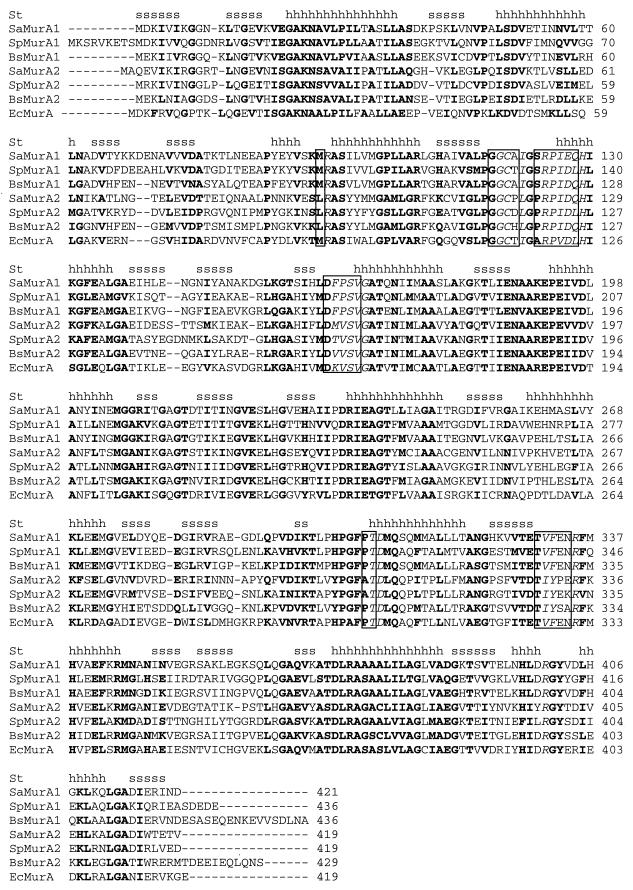
The duplicate gene copies found in low-G+C gram-positive bacteria encoded roughly similar-sized polypeptides (419 to 436 amino acids in length). Comparisons to the crystal structure of E. coli MurA showed that major structural features and residues involved in ligand interactions were highly conserved among MurA1 and MurA2 proteins (Fig. 3). These structural similarities indicate that it is unlikely that one copy is a pseudogene.
FIG. 3.
Multiple sequence alignment of UDP-N-acetylglucosamine enolpyruvyl transferases (MurA1 and MurA2) from B. subtilis (Bs), S. aureus (Sa), S. pneumoniae (Sp), and E. coli (Ec). Shown are the locations of α helices (h) and β sheets (s) in the determined structure (St) of E. coli MurA (22). Highly conserved residues (from CLUSTALW output) are boldface, while residues of E. coli MurA involved in ligand interactions are in italics and boxed.
To investigate if both genes encode active enzymes, we decided to study the two genes of S. pneumoniae more closely. The two MurA enzymes from this organism showed 45% identity to each other at the amino acid level. The genes were not close to each other in the S. pneumoniae genome and were not clustered with any other peptidoglycan synthesis enzymes. MurA1 is flanked by two small ORFs which might form a single transcriptional unit (operon). The upstream ORF is putative and unique to S. pneumoniae. The downstream ORF is 64 amino acids long and is homologous to EPUA protein, a possible membrane-bound nuclease required for DNA transformation. MurA2 is upstream of a small hypothetical protein about 165 amino acids long which shows very low homology to Bacillus licheniformis hypothetical protein P20. There is no downstream linked ORF.
Essentiality.
We first attempted to inactivate each S. pneumoniae murA gene by allelic replacement. Regions up- and downstream of each gene ranging from 406 to 555 bp were amplified by PCR and used to make constructs in which they flanked an antibiotic resistance gene. In the case of murA1, the constitutively expressed erythromycin resistance gene cassette ermAM from pAMβ1 was used (17), while the chloramphenicol resistance cat gene from pJS3 was used in the murA2 construct (2). To minimize the potential polar effects of gene replacement, the PCR primers were chosen so that flanking genes and potential promoters would remain intact in the deletion mutant. In addition, the cassettes were designed so that the resistance gene was inserted in the same orientation as the target gene, to ensure transcription of the downstream region. This strategy was designed to generate mutants which were nonpolar (since there is no structural or transcriptional disruption of adjacent genes), stable in the absence of selective pressure (because there are no insertion site sequence duplications to allow spontaneous excision), and null (given that there can be no residual target gene activity).
The two allelic replacement constructs were used separately to transform S. pneumoniae R6, and mutants of both murA1 and murA2 were obtained. The structure of each mutant was confirmed by diagnostic PCR and Southern hybridization analysis. MICs for fosfomycin for the S. pneumoniae R6 murA1 and murA2 strains were unchanged compared to wild-type S. pneumoniae R6.
In an attempt to make a murA1 murA2 double mutant, murA2 allelic replacement construct DNA was transformed into competent cells of the S. pneumoniae R6 murA1 mutant, but no transformants were obtained in multiple transformation experiments with positive allelic replacement and transformation controls. These results suggest that both murA genes encode active MurA enzymes, since elimination of only one still results in a viable organism.
Biochemical characterization of MurA1 and MurA2.
MurA1 and MurA2 from S. pneumoniae were investigated further at the protein level. The murA1 and murA2 genes were overexpressed, and the enzymes that they encode, MurA1 and MurA2, respectively, were purified and kinetically characterized. Overexpression of the murA1 and murA2 genes in E. coli accounted for ca. 40 and 30% of total cell protein, and under the selected induction conditions, more than 50 and 40% of the MurA1 and MurA2 enzymes, respectively, remained soluble.
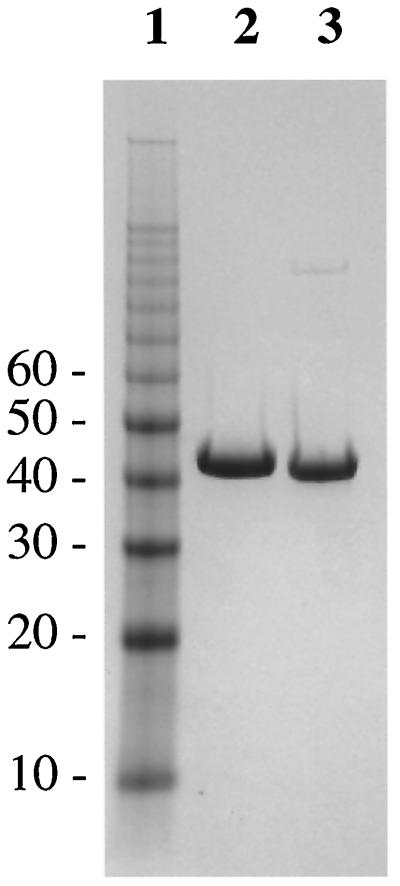
MurA1 was purified by ammonium sulfate fractionation and anion-exchange chromatography. An extra chromatography step on a hydrophobic interaction column was also included in the purification of MurA2. Approximately 33 mg of MurA1 and 21 mg of MurA2 were obtained from 1 and 4 liters of cultures, respectively, with the purity of the final preparations being 95% as judged by SDS-PAGE (Fig. 4). The identities of MurA1 and MurA2 were confirmed by N-terminal sequencing (N-terminal sequences being MDKIVVQGGD and MRKIVINGGL for MurA1 and MurA2, respectively) and MALDI-TOF mass spectrometry.
FIG. 4.
SDS-PAGE analysis of purified MurA1 and MurA2. The experiment was performed on a Novex 4 to 12% bis-Tris gel with morpholineethanesulfonic acid running buffer. Lane 1, 10-kDa molecular mass ladder (with selected molecular masses [in kilodaltons] marked); lane 2, purified MurA1 after the Hi-Load Q column; lane 3, purified MurA2 after the Hi-Load Q column.
The catalytic activities of the two enzymes were then investigated. Both MurA1 and MurA2 were able to catalyze the transfer of enolpyruvate from PEP to UDPAG, demonstrating that they are both active UDP-N-acetylglucosamine enolpyruvyl transferases as expected from the sequence analysis and gene deletion experiments. The kinetic parameters for both enzymes were determined (Table 1). The Km(UDPAG) and Km(PEP) were two- and threefold lower for MurA2 than for MurA1, while kcat was almost twice as high for MurA2 as for MurA1 (Table 1). More importantly, kcat/Km(UDPAG) and kcat/Km(PEP) values were four- and sixfold higher for MurA2 than for MurA1, indicating MurA2 is a more efficient enzyme than MurA1 under these assay conditions. Both enzymes were inhibited by the antibiotic fosfomycin, the 50% inhibitory concentrations for MurA1 and MurA2 being 2.3 and 14.9 μM, respectively.
DISCUSSION
We have shown by phylogenetic analysis that two distinct classes of UDP-N-acetylglucosamine enolpyruvyl transferase exist (Fig. 2). The first class of transferases, MurA1, occurs throughout all bacteria except gram-positive Mycoplasma spp. The second type of transferase, MurA2, exists as a duplicate gene copy in only the low-G+C gram-positive bacteria. A combination of sequence analysis, gene knockout experiments, and direct biochemical characterization was used to analyze the two copies of murA found in these gram-positive organisms. Sequence analysis indicates both genes are complete and that the enzymes they encode contain the important catalytic residues previously identified in the E. coli MurA. Structurally, and probably functionally, MurA1 and MurA2 are highly similar. Gene deletion experiments further indicate that both genes must encode active enzymes since removal of either still results in a viable organism although this activity is clearly essential for viability. This suggests that the two enzymes have the same function and that one enzyme can substitute for the other.
To demonstrate conclusively that these two genes encode active enzymes, we overexpressed the two genes and purified the two proteins from S. pneumoniae. Both MurA enzymes are active and catalyze the reaction between UDPAG and PEP to give UDP-N-acetylglucosamine enolpyruvate and Pi. Both enzymes from S. pneumoniae appear to have higher Kms for their substrates and lower kcats than the E. coli enzyme, indicating that at a kinetic level MurA1 more closely resembles MurA2 than its gram-negative counterpart. They are also both inhibited by the antibiotic fosfomycin. Fosfomycin possesses antibacterial activity against both gram-positive and gram-negative organisms. The majority of the data generated on the mode of action of this antibiotic has been focused on MurA from E. coli (for example, see references 11, 15, 16, and 26). In this study we have demonstrated that fosfomycin inhibits both MurA enzymes from S. pneumoniae, thus providing the first confirmatory evidence that MurA is also the antibacterial target of fosfomycin in this organism.
The existence of two active copies of murA gene in these gram-positive organisms raises the question of how the two copies arose. The phylogenetic tree suggests that murA2 might have evolved from a duplication of the murA1 gene in gram-positive bacteria, although the statistical support for this scenario, or any alternative, is weak. The branching of Deinococcus radiodurans and Thermotoga maritima, both not considered G+C gram-positive bacteria (18), among the low G+C gram-positive bacteria might be taken as negative evidence against the scenario of murA2 evolution by gene duplication within the gram-positive bacteria. However, there is considerable debate about the phylogenetic placement of these bacterial taxa (Cavalier-Smith [5] proposed that T. maritima might be a gram-positive bacterium) and the clustering of Deinococcus/Thermus and Thermotogales among gram-positive bacteria has been observed in other protein phylogenies such as glutamine synthetase (4).
The phylogenetic and structural analysis does support the notion that MurA1 and MurA2 proteins are closely related and are likely to have evolved somewhere in the lineage leading to the low-G+C gram-positive bacteria. MurA1 and MurA2 proteins might have evolved from an even more ancient source as suggested by Psi-BLAST homology searches (1), which reveal significant similarities between MurA1/MurA2 and 5-enolpyruvylshikimate-3-phosphate synthase, a key enzyme in the shikimate pathway.
It is also unclear why a duplication of the enolpyruvyl transfer function has arisen and whether MurA1 and MurA2 carry out identical functions at a physiological level within the cell. The two enzymes catalyze the same reaction, which is the first committed step in peptidoglycan biosynthesis. The higher kcat/Km(UDPAG) and kcat/Km(PEP) values for MurA2 from S. pneumoniae (Table 1) indicate that MurA2 is a more efficient enzyme in utilizing both substrates under the assay conditions used here, but this may not be true in vivo. Both enzymes appear to be less efficient than their E. coli counterparts (11). It would be interesting to investigate the relative levels of expression of the two murA genes within the cell, as this might shed more light on roles of their gene products in the peptidoglycan biosynthetic pathway. It is possible that although both MurA enzymes are active, only one murA gene is expressed and its expression product is utilized in S. pneumoniae.
It is interesting that the other genes encoding enzymes involved in peptidoglycan biosynthesis in gram-positive bacteria appear to be present as single copies. However, there is a further example in peptidoglycan biosynthesis of an enzyme activity that appears to have been duplicated: d-Ala-d-Ala ligase in gram-negative bacteria, which is encoded by the ddlA and ddlB genes. In this case again, the two purified enzymes show similar kinetic parameters and susceptibilities to inhibitors, and it is unclear why a duplication in activity has occurred (28).
MurA is potentially an important antibacterial target. The presence of two active copies of the enzyme in gram-positive pathogens illustrates the need for a MurA-directed antibacterial to inhibit both forms of the enzyme. This is clearly possible, as illustrated by fosfomycin, which inhibits both MurA1 and MurA2 and thus has antibacterial activity against S. pneumoniae. This observation and the fact that we have shown the enolpyruvyl transferase activity to be essential in this organism further validate the MurA enzyme as an antibacterial target.
ACKNOWLEDGMENTS
We thank Stephanie van Horn and Thomas Mathie for DNA sequencing and oligonucleotide synthesis, Gilbert Scott for N-terminal sequencing and mass spectroscopy, Wu-Schyong Liu and Wendy Crowell for preparation of Fig. 4, and Nancy Niconovich and Steve Rittenhouse for MIC determinations.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: and new generation of protein database search programs. Nucleic Acid Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballester S, Lopez P, Alonso J C, Espinosa M, Lacks S A. Selective advantage of deletions enhancing chloramphenicol transferase gene expression in Streptococcus pneumoniae plasmids. Gene. 1986;41:153–163. doi: 10.1016/0378-1119(86)90094-6. [DOI] [PubMed] [Google Scholar]
- 3.Brown E D, Vivas E I, Walsh C T, Kolter R. MurA (MurZ), the enzyme that catalyzes the first committed step in peptidoglycan biosynthesis, is essential in Escherichia coli. J Bacteriol. 1995;177:4194–4197. doi: 10.1128/jb.177.14.4194-4197.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown J R, Masuchi Y, Robb F T, Doolittle W F. Evolutionary relationships of bacterial and archaeal glutamine synthetase genes. J Mol Evol. 1994;38:566–576. doi: 10.1007/BF00175876. [DOI] [PubMed] [Google Scholar]
- 5.Cavalier-Smith T. Secondary metabolites: their function and evolution. Ciba Found Symp. 1992;171:64–87. doi: 10.1002/9780470514344.ch5. [DOI] [PubMed] [Google Scholar]
- 6.Dayhoff M O, Eck R V, Park C M. A model of evolutionary change in proteins. In: Dayhoff M O, editor. Atlas of protein sequence and structure. Vol. 5. Washington, D.C.: National Biomedical Research Foundation; 1972. pp. 89–99. [Google Scholar]
- 7.Havarstein L S, Coomaraswamy G, Morrison D A. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1995;92:11140–111404. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henikoff S, Henikoff G. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Höltje J V, Schwarz U. In: Molecular cytology of Escherichia coli. Nanninga N, editor. New York, N.Y: Academic Press; 1985. pp. 77–109. [Google Scholar]
- 10.Kahan F M, Kahan J S, Cassidy P J, Kropp H. The mechanism of action of fosfomycin (phosphonomycin) Ann N Y Acad Sci. 1974;235:364–385. doi: 10.1111/j.1749-6632.1974.tb43277.x. [DOI] [PubMed] [Google Scholar]
- 11.Kim D H, Lees W J, Kempsell K E, Lane W S, Duncan K, Walsh C T. Characterization of a Cys115 to Asp substitution in the Escherichia coli cell wall biosynthetic enzyme UDP-GlcNAc enolpyruvyl transferase (MurA) that confers resistance to inactivation by the antibiotic fosfomycin. Biochemistry. 1996;35:4923–4928. doi: 10.1021/bi952937w. [DOI] [PubMed] [Google Scholar]
- 12.Lacks S. Integration efficiency and genetic recombination in pneumococcal transformation. Genetics. 1966;53:207–235. doi: 10.1093/genetics/53.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lacks S, Hotchkiss R D. A study of the genetic material determining an enzyme activity in pneumococcus. Biochim Biophys Acta. 1960;39:508–517. doi: 10.1016/0006-3002(60)90205-5. [DOI] [PubMed] [Google Scholar]
- 14.Lanzetta P A, Alvarez L J, Reinach P S, Candia O A. An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 15.Marquardt J L, Brown E D, Lane W S, Haley T M, Ichikawa Y, Wong C H, Walsh C T. Kinetics, stoichiometry, and identification of the reactive thiolate in the inactivation of UDP-GlcNAc enolpyruvoyl transferase by the antibiotic fosfomycin. Biochemistry. 1994;33:10646–10651. doi: 10.1021/bi00201a011. [DOI] [PubMed] [Google Scholar]
- 16.Marquardt J L, Siegele D A, Kolter R, Walsh C T. Cloning and sequencing of Escherichia coli murZ and purification of its product, a UDP-N-acetylglucosamine enolpyruvyl transferase. J Bacteriol. 1992;174:5748–5752. doi: 10.1128/jb.174.17.5748-5752.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin B, Alloing G, Mejean V, Claverys J P. Constitutive expression of erythromycin resistance mediated by the ermAM determinant of plasmid pAMβ1 results from deletion of 5′ leader peptide sequences. Plasmid. 1987;18:250–253. doi: 10.1016/0147-619x(87)90068-0. [DOI] [PubMed] [Google Scholar]
- 18.Olsen G J, Woese C R, Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994;176:1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paton J C. The contribution of pneumolysin to the pathogenicity of Streptococcus pneumoniae. Trends Microbiol. 1996;4:103–106. doi: 10.1016/0966-842X(96)81526-5. [DOI] [PubMed] [Google Scholar]
- 20.Rogers H J, Perkins H R, Ward J B. Microbial cell walls and membranes. London, England: Chapman & Hall; 1980. [Google Scholar]
- 21.Schönbrunn E, Sack S, Eschenburg S, Perrakis A, Krekel F, Amrhein N, Mandelkow E. Crystal structure of UDP-N-acetylglucosamine enolpyruvyltransferase, the target of the antibiotic fosfomycin. Structure. 1996;4:1065–1075. doi: 10.1016/s0969-2126(96)00113-x. [DOI] [PubMed] [Google Scholar]
- 22.Skarzynski T, Mistry A, Wonacott A, Hutchinson S E, Kelly V A, Duncan K. Structure of UDP-N-acetylglucosamine enolpyruvyl transferase, an enzyme essential for the synthesis of bacterial peptidoglycan, complexed with substrate UDP-N-acetylglucosamine and the drug fosfomycin. Structure. 1996;4:1465–1474. doi: 10.1016/s0969-2126(96)00153-0. [DOI] [PubMed] [Google Scholar]
- 23.Strimmer K, von Haeseler A. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 24.Swofford D L. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4. Sunderland, Mass: Sinauer Associates; 1999. [Google Scholar]
- 25.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venkateswaran P S, Wu H C. Isolation and characterization of a phosphomycin-resistant mutant of Escherichia coli K-12. J Bacteriol. 1972;110:935–944. doi: 10.1128/jb.110.3.935-944.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wanke C, Amrhein N. Evidence that the reaction of the UDP-N-acetylglucosamine 1-carboxyvinyltransferase proceeds through the O-phosphothioketal of pyruvic acid bound to Cys115 of the enzyme. Eur J Biochem. 1993;218:861–870. doi: 10.1111/j.1432-1033.1993.tb18442.x. [DOI] [PubMed] [Google Scholar]
- 28.Zawadzke L E, Bugg T D H, Walsh C T. Existence of two d-alanine:d-alanine ligases in Escherichia coli: cloning and sequencing of the ddlA gene and purification and characterization of the DdlA and DdlB enzymes. Biochemistry. 1991;30:1673–1682. doi: 10.1021/bi00220a033. [DOI] [PubMed] [Google Scholar]