Abstract
The biphenyl and salicylate metabolic pathways in Pseudomonas putida KF715 are chromosomally encoded. The bph gene cluster coding for the conversion of biphenyl to benzoic acid and the sal gene cluster coding for the salicylate meta-pathway were obtained from the KF715 genomic cosmid libraries. These two gene clusters were separated by 10-kb DNA and were highly prone to deletion when KF715 was grown in nutrient medium. Two types of deletions took place at the region including only the bph genes (ca. 40 kb) or at the region including both the bph and sal genes (ca. 70 kb). A 90-kb DNA region, including both the bph and sal genes (termed the bph-sal element), was transferred by conjugation from KF715 to P. putida AC30. Such transconjugants gained the ability to grow on biphenyl and salicylate as the sole sources of carbon. The bph and sal element was located on the chromosome of the recipient. The bph-sal element in strain AC30 was also highly prone to deletion; however, it could be mobilized to the chromosome of P. putida KT2440 and the two deletion mutants of KF715.
A number of biphenyl-utilizing bacteria have been isolated to date. They include gram-negative species of Pseudomonas, Achromobacter, Sphingomonas, Comamonas, Burkholderia, Ralstonia, and Alcaligenes and gram-positive Rhodococcus spp. (1, 3, 11, 37). Because these biphenyl-utilizing strains cometabolize polychlorinated biphenyls (PCB), the biochemistry of PCB degradation has been extensively studied (12). A gene cluster coding for biphenyl-PCB degradation (termed bph) was first cloned from Pseudomonas pseudoalcaligenes KF707 (14). Since then, a number of bph genes have been cloned and sequenced. Southern and sequence analyses of the bph genes revealed that some biphenyl-utilizing strains possess bph genes that are very similar, if not identical, to one another, but some share various degrees of homology (13).
The bph genes are present on bacterial chromosomes (8, 14, 16, 35), plasmids (18, 39), and transposons (22, 28, 34). The presence of similar genes in different strains implies that even chromosomal bph genes have or used to have a mechanism for mobilization to other strains. The bph genes of Pseudomonas sp. strain CB406 were mobilized following the construction in vivo of a cointegrate plasmid inserted into the broad-host-range plasmid RP4 (22). Springael and coworkers (23, 34) identified a transposon, Tn4371, carrying the bph genes encoding conversion of biphenyl to benzoic acid from Ralstonia eutrophus A5 (formerly Alcaligenes eutrophus A5) in which Tn4371 first transposed from the chromosome to indigenous IncP1 plasmid, and the plasmid carrying Tn4371 can be transferred to other strains by conjugation. A recent study shows that Tn4371 is a kind of conjugative transposon of 55 kb (24, 29). Interestingly, another smaller conjugative transposon Tn-bph coding for biphenyl catabolism resides within Tn4371. This can be transferred to the recipient strain by conjugation independently. Divergence of the bph genes among various biphenyl-utilizing strains indicates that the bph genes might be very ancient and thus have accumulated many mutations over a long historical period. In fact, some bph operons are highly rearranged and shuffled in soil bacteria (12, 26).
P. putida KF715 bphABCD genes specify conversion of biphenyl to benzoic acid and 2-hydroxypenta-2,4-dienoic acid, where bphA is composed of four subunit genes (bphA1, bphA2, bphA3, and bphA4) encoding multicomponent biphenyl dioxygenase, bphB encoding dihydrodiol dehydrogenase, bphC encoding 2,3-dihydroxybiphenyl dioxygenase (DHB dioxygenase), and bphD encoding 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid hydrolase (HOPDA hydrolase) (16, 35). The KF715 bph gene cluster is very similar to the well-characterized bphABCXD operon of P. pseudoalcaligenes KF707 in terms of gene organization as well as the restriction enzyme profiles. However, the bphX region (3.5 kb), which exists between bphC and bphD and which is involved in the conversion of 2-hydroxypenta-2,4-dienoic acid to acetyl-coenzyme A (16, 17), is missing in the KF715 bph gene cluster (Fig. 1). In the course of further study of the KF715 catabolic genes, we found that KF715 easily loses the biphenyl-utilizing ability when the cells are grown in nutrient medium. Lee et al. have cloned and sequenced the three sal genes coding for the conversion of salicylate to 2-hydroxymuconic semialdehyde in KF715 (20, 21). Furthermore, we found that the catabolic capabilities of biphenyl and salicylate are transferred from KF715 to other P. putida strains.
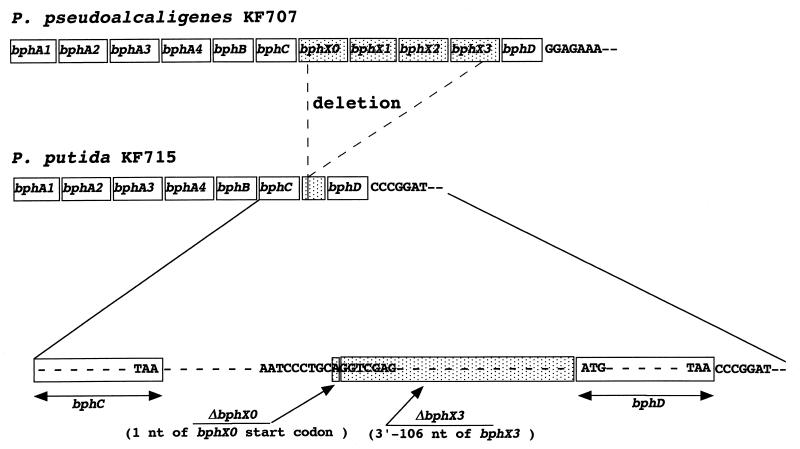
FIG. 1.
Analysis of the deletion of bphX region within the P. putida KF715 bph operon in comparison with the corresponding region of P. pseudoalcaligenes KF707. The intervening sequence between bphC and bphD is schematically depicted. The deletion point is shown by a vertical arrow, indicating that only one nucleotide A of the start codon of bphX0 and the 3′ region of 106 nucleotides of bphX3 remain. The nucleotide sequences right after the stop codon of bphD from KF707 and KF715 are presented in boxes. nt, nucleotide(s).
Here we report on the deletion and mobilization of the large chromosomal bph-sal element in P. putida KF715.
MATERIALS AND METHODS
Bacterial strains and cultivation.
P. putida KF715, used throughout this study, was originally isolated from soil in Kitakyushu, Japan, together with the well-characterized P. pseudoalcaligenes KF707 (14, 35). Some characteristics of KF715 were previously described (13, 16). P. putida KF715M1 is the KF715 mutant strain in which both the bphABCD and sal genes are deleted. P. putida KF715M2 is another KF715 derivative in which only the bph genes are deleted, but the sal genes are retained. Pseudomonas graminis KF701 (formerly Achromobacter xylosoxidans KF701), which possesses bph genes very similar to those of KF715, was described previously (13). P. putida AC30 was obtained from A. M. Chakrabarty (University of Illinois at Chicago), and a rifamcin-resistant and tryptophan-requiring mutant (AC30RifrTrp) was obtained in this study. P. putida KT2440 was donated by K. N. Timmis (GBF-National Research Center for Biotechnology, Braunschweig, Germany), and a streptomycin-resistant (Smr) and methionine-requiring mutant (KT2440SmrMet) was obtained in this study. A plasmid pCNU516 containing the salA gene was donated from Y. Kim (Chungbuk National University, Cheongju, Korea). Other strains and plasmids used in this study are listed in Table 1. Luria-Bertani (LB) broth (40) was used as a nutrient medium. Basal salt medium (BSM) (14) was also used. Rifampin or streptomycin was used at 300 or 500 μg/ml, respectively, when needed.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| P. putida KF715 | Wild type, Bph+ Sal+ Ben+ | 16 |
| P. putida KF715M1 | Bph− Sal− Ben+ (Δbph/Δsal) | This study |
| P. putida KF715M2 | Bph− Sal+ Ben+ (Δbph) | This study |
| P. putida KF715M1Bph+ | Bph+ Sal+ Ben+ | This study |
| P. putida KF715M2Bph+ | Bph+ Sal+ Ben+ | This study |
| P. putida AC30 | Ben+ | A. M. Chakrabarty |
| P. putida AC30RifrTrp | Ben+ Rifrtrp | This study |
| P. putida AC30Bph+ | Bph+ Sal+ Ben+ Rifrtrp (transconjugant acquired the bph-sal element) | This study |
| P. putida KT2440 | Ben+ | K. N. Timmis |
| P. putida KT2440SmrMet | Ben+ Smrmet | This study |
| P. putida KT2440Bph+ | Bph+ Sal+ Ben+ Smrmet (transconjugant acquired the bph-sal element) | This study |
| P. pseudoalcaligenes KF707 | Wild type, Bph+ Ben+ | 14 |
| P. graminis KF701 | Wild type, Bph+ Ben+ | 13 |
| E. coli S17-1 | pro thi recA hsdR; chromosomally integrated RP4-2-Tc::Mu-Km::Tn7 | 33 |
| E. coli DH5α | supE44 ΔlacU169(φ80 lacZ ΔM15) hsdR recA1 endA1 gyrA96 thi relA1 | 40 |
| Plasmids | ||
| pMFB2 | pKT230-bphABC(KF707); Smr; 19.7 kb | 14 |
| pNHF715 | pKT230-bphABCD(KF715); Smr; 22.3 kb | 16 |
| pCNU516 | pUC18-salA(KF715); Apr; 4.6 kb | 20, 21 |
| Super Cos1 | Apr Nmr; cosmid vector; 7.6 kb | Stratagene |
| pKTF6246 | Cosmid containing the sal genes | This study |
| pKF6337 | Cosmid containing upstream of the bph genes | This study |
| pKF6465 | Cosmid containing downstream of the bph-sal element from AC30Bph+ | This study |
| pKF6500 | Cosmid containing upstream of the bph genes | This study |
Abbreviations: Bph, biphenyl; Sal, salicylate; Ben, benzoate; Rif, rifampin; Tc, tetracycline; Km, kanamycin; Sm, streptomycin; Ap, ampicillin; Δ, deletion; r, resistance.
Conjugation.
Transfer of the KF715 Bph+ phenotype by conjugation into the recipient strain P. putida AC30RifrTrp was carried out by filter mating. Donor and recipient cells were grown overnight in LB agar medium, and both cultures (2 × 109) were suspended in 1 ml of LB broth. The cell suspensions (0.5 ml each) were mixed and put on a nitrocellulose filter and incubated on an LB agar plate for 6 h. The cells on the filter were suspended in sterilized distilled water and plated onto BSM plates containing 300 μg of rifampin and 20 μg tryptophan per ml, providing solid biphenyl in the inverted lid to select transconjugants.
Detection of loss of Bph+ and Sal+ phenotypes.
After sequential batch growth on LB medium, biphenyl-, salicylate-, and benzoate-utilizing abilities were scored for every 100 colonies.
Southern blot analysis.
Digoxigenin-11-dUTP was employed to label DNA by using a digoxigenin DNA labeling and detection kit according to the instructions of the manufacturer (Boehringer Mannheim, Mannheim, Germany). The genomic DNA was digested with appropriate restriction enzymes and subjected to electrophoresis through 0.7% agarose gels. The digested DNA fragments were transferred onto nylon membranes (Biodyne B; PALL, Port Washington, N.Y.). Hybridization was performed with the digoxigenin-labeled DNA probes.
PCR.
PCR was performed with a total volume of 50 μl which contained PCR buffer (Takara Shuzo Co., Ltd., Kyoto, Japan), template DNA (0.5 μg), 100 mM (each) deoxyribonucleotide triphosphate, 1 mM (each) oligoprimer, and 0.5 U of Taq DNA polymerase. Amplification of DNA was carried out as described elsewhere (19). The oligoprimers used for the amplification of the bphC, bphD, and salA DNA were as follows: bphC, 5′-ATGTGCATTAAAAGTTTG-3′ for the upstream sequence and 3′-AACGCCTTGTTTCGTATT-5′ for the downstream sequence; bphD, 5′-ATGACAGCGCTCACTGAA-3′ and 3′-AAAAATGCCGTCCGGATT-5′; and salA, 5′-ATGAACGCTAAGAAACCA-3′ and 3′-CGCGACGCAGTTCCCATT-5′.
Construction of cosmid libraries.
Genomic DNA of P. putida strains KF715 and AC30Bph+ were partially digested by Sau3AI. The DNA was subjected to 20% sucrose density gradient centrifugation at 85,000 × g for 16 h. After fractionation, the DNA sizes were examined by 0.3% agarose gel electrophoresis, and the 35- to 45-kb DNA was collected. The purified DNA was ligated at the BamHI site of a cosmid vector Super Cos1 and packaged in vitro into lambda phage particles by using a GIGAPACK III Gold kit (Stratagene, La Jolla, Calif.), which were infected into Escherichia coli DH5α (38). The genomic libraries were amplified by growing the cells in LB broth supplemented with ampicillin. Amplified genomic libraries were preserved in 20% glycerol at −80°C until use.
PFGE.
Agarose-embedded DNA suitable for separation by pulsed-field gel electrophoresis (PFGE) was prepared in accordance with instructions provided by the manufacturer (Bio-Rad Laboratories AG, Glattbrugg, Switzerland) with some modifications. PFGE was performed by clamped homogeneous electric-field (CHEF) electrophoresis for 24 h by the CHEF-DRII system (Bio-Rad). The gel (1%) was subjected to electrophoresis in 45 mM Tris-borate and 1 mM EDTA (pH 8.3) at 6 V/cm and 14°C.
RESULTS
Organization of the bph and sal genes in P. putida KF715.
We previously reported that a region named bphX (3.5 kb) between bphC and bphD is missing in KF715 (16), where four genes are present in P. pseudoalcaligenes KF707 (GenBank accession no. D85853) and Burkholderia cepacia LB400 (17). In KF707 they are bphX0 (encoding glutathione S-transferase; bphK in LB400), bphX1 (encoding 2-hydroxypenta-2,4-dienoate hydratase; bphH in LB400), bphX2 (acetaldehyde dehydrogenase [acylating]; bphJ in LB400), and bphX3 (encoding 4-hydroxy-2-oxovalerate aldolase; bphI in LB400). Sequence analyses of the intervening region of bphC and bphD in KF715 revealed that only one nucleotide (nucleotide A) of the start codon of bphX0 and that the 3′-terminal 106 nucleotides of bphX3 remain (Fig. 1). Thus, most of the 3.5-kb bphX region is deleted in the KF715 bph genes. The KF715-bphD and KF707-bphD genes share an identity as high as 96.0%, and both are ended with TAA in common, but their nucleotide sequence right after the stop codon is entirely different (Fig. 1).
Another catabolic gene cluster coding for the conversion of salicylate to 2-hydroxymuconic semialdehyde was cloned (termed salA-encoding salicylate hydroxylase, salB-encoding ferredoxin, and salC-encoding catechol 2,3-dioxygenase [C23O]) (20, 21). We determined the location of the sal genes. For this purpose we constructed a genomic cosmid library for KF715. First, we screened a cosmid clone expressing both DHB dioxygenase (encoded by bphC) and C23O (encoded by salC) by spraying with 2,3-dihydroxybiphenyl and catechol, respectively. From the restriction maps of the cosmids and the following Southern blot analyses with bph and sal DNA probes, it was revealed that the sal gene cluster lies 10 kb downstream of the bphABCD gene cluster.
Appearance of the Bph− and Sal− phenotypes and their characteristics.
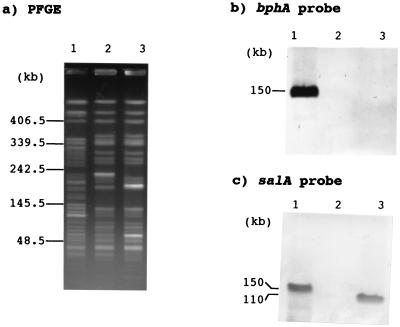
When P. putida KF715 was sequentially subcultured in LB broth, it was found that colonies unable to grow on biphenyl (Bph−) appeared at high frequency. After 80 generations, 91% of the cells tested had lost the Bph-utilizing ability. About half of such Bph− mutants derived from KF715 also had lost their ability to grow on salicylate. These results indicated that two types of deletion occurred within the KF715 genome around the bph-sal gene clusters. The genomic Southern analyses with the bphA1 DNA (see TNF-IV in Fig. 3) and salA DNA (TNF-VI), together with other parts of DNA (TNF-II and TNFVII) since the probes revealed that strain KF715M1 had lost both the bph and sal genes, including the intervening region (ca. 10 kb), and that KF715M2 had lost only the bph genes (Table 2 and Fig. 3). Shown in Fig. 2 are the PFGE profiles of SpeI digests and the following Southern blot analysis of the genomic DNA from KF715, KF715M1, and KF715M2. No hybridization was observed for the genomic DNA of both KF715M1 and KF715M2 when bphA1 was used as the probe, while KF715M2 DNA hybridized with the salA probe only. PCR experiments revealed that the bphC, bphD, and salA genes were amplified from the genomic DNA of the original KF715 and that only the salA gene was amplified from KF715M2 (data not shown). Neither bphC and bphD nor salA were amplified from KF715M1. These results are in agreement with those from the Southern blot analyses.
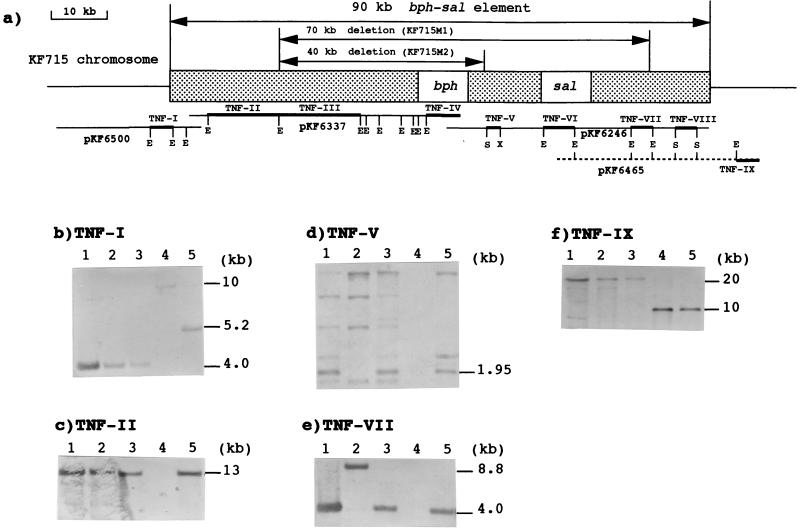
FIG. 3.
Analyses of terminal ends of the bph-sal element in P. putida KF715 and the deletion regions within the same element in KF715M1 and KF715M2. The following four cosmids were analyzed: pKT6500 containing the upstream of the bph-sal element, pKF6337 containing the bph gene cluster and its upstream region, pKF6246 containing the sal genes, and pKF6465 containing the salC gene and the downstream of the bph-sal element. The former three cosmids (pKF6500, pKF6337, and pKF6246) are derived from P. putida KF715, and pKF6465 is derived from P. putida AC30Bph+. These cosmids were digested by EcoRI, and some fragments were further digested by SalI and XhoI. The restriction sites are shown, where E, S, and X indicate EcoRI, SalI, and XhoI, respectively. The DNA fragments used as the probes are indicated by heavy bars (TNF-I to IX). The estimated sizes and the positions of the bph-sal element and the deletion regions of KF715M1 and KF715M2 are indicated by arrows. Lanes: 1, KF715; 2, KF715M1; 3, KF715M2; 4, AC30RifrTrp; 5, AC30Bph+.
TABLE 2.
Growth characteristics and genomic Southern analyses of P. putida KF715 and P. putida AC30 and their derivativesa
| Strain | Growth on:
|
Hybridization with:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bph | Sal | Ben | TNF-I | TNF-II | TNF-III | TNF-IV | TNF-V | TNF-VI | TNF-VII | TNF-VIII | TNF-IX | |
| KF715 | + | + | + | + | + | + | + | + | + | + | + | (+) |
| KF715M1 | − | − | + | + | + | − | − | (+) | − | + | + | (+) |
| KF715M2 | − | + | + | + | + | − | − | + | + | + | + | (+) |
| AC30RifrTrp | − | − | + | (+) | − | − | − | − | − | − | − | + |
| AC30Bph+ | + | + | + | + | + | + | + | + | + | + | + | + |
FIG. 2.
PFGE of the genomic DNA of P. putida KF715, KF715M1, and KF715M2 and the subsequent Southern blot analysis of bph and sal genes. The bphA1 and salA DNAs were used as the probe. The genomic DNAs were digested by SpeI. Lanes: 1, KF715; 2, KF715M1; 3, KF715M2. Molecular size markers are shown on the left.
Recovery of the Bph+ phenotype by introducing the bph genes.
The two Bph− mutant strains KF715M1 and KF715M2 were mated with E. coli S17-1 carrying pMFB2, containing the bphABC genes of P. pseudoalcaligenes KF707 (14), or pNHF715 containing the P. putida KF715 bphABCD genes (16), respectively. Both KF715M1 and KF715M2 carrying pNHF715 restored the ability to grow on biphenyl; however, the same mutant strains carrying pMFB2 failed to grow on biphenyl but produced a large amount of yellow meta-cleavage compound (HOPDA) from biphenyl (Table 2).
Deletion regions in KF715M1 and KF715M2.
From the analyses of genomic DNAs by Southern blot and PCR, it was revealed that KF715M1 had lost bph and sal genes, including the intervening region (10 kb), and that KF715M2 had lost the bph genes but retained the sal genes. Based on these findings, we analyzed KF715 cosmid DNAs carrying both the bphABCD and sal genes in comparison with the corresponding regions from KF715M1 and KF715M2. By using various DNA probes that include the upstream of bphA, the downstream of bphD, and the downstream of sal, DNA fragments which hybridized with these probes, but with different sizes, were screened (Fig. 3). In order to determine the upstream of deletion sites in KF715M1 and KF715M2, the EcoRI 13-kb DNA (TNF-II in Fig. 3) was prepared from cosmid pKF6337, which was derived from the KF715 chromosome. The TNF-II DNA probe hybridized with the same size of DNA from both KF715M1 and KF715M2 (Fig. 3c). The downstream 15-kb EcoRI DNA (TNF-III) fragment adjacent to TNF-II failed to hybridize with both KF715M1 and KF715M2 DNA (data not shown). These results indicate that the upstream deletion end of these two mutants lies just inside or just downstream of the 3′ end of the TNF-II DNA (Fig. 3). The downstream deletion site of KT715M1 was found within the 4.0-kb EcoRI fragment (TNF-VII) because this fragment was hybridized with the DNA fragment of a different size (8.8 kb) in KF715M1, while this probe hybridized with the same 4.0-kb DNA in KF715 and KF715M2 (Fig. 3e). The downstream deletion site of KF715M2 was estimated to lie just upstream of the 1.95-kb SalI-XhoI fragment (TNF-V). The TNF-V probe hybridized with DNA fragments of the same size in KF715 and KF715M2 but hybridized differently in KF715M1 (Fig. 3d). The same probe hybridized with several other DNA fragments (Fig. 3d). Preliminary sequence data showed that the 1.95-kb TNF-V DNA contains an IS-like sequence similar to IS5 (EMBL-DDBJ-GenBank database under accession no. AJ249209). We found that several copies of this IS-like sequence are present in KF715 and its derivatives (data not shown). This may be the reason why several bands appeared with the TNF-V probe. The upstream 4.6-kb DNA fragment (TNF-IV) adjacent to TNF-V DNA did not hybridize with the KF715M2 DNA (Table 2), indicating that the downstream deletion site lies just upstream of the 1.95-kb DNA. Based on these results, we estimate that ∼70-kb DNA is deleted in KF715M1, which includes the bphA upstream region (30 kb), the entire bphABCD region (10 kb), the intervening region between the bph and sal genes (10 kb), and the 20-kb region including the sal genes (Fig. 3). On the other hand, an ∼40-kb DNA region is deleted in KF715M2, which includes the bphA upstream region (30 kb) and the entire bphABCD region (10 kb). Furthermore, PFGE revealed that the SpeI digest profiles of the genomic DNA of KF715, KF715M1, and KF715M2 are different (Fig. 2a). This indicates that at least part of the genome of KF715 is highly prone to rearrangement, which may include deletion and recombination.
Self-mobilization of the bph and sal genes.
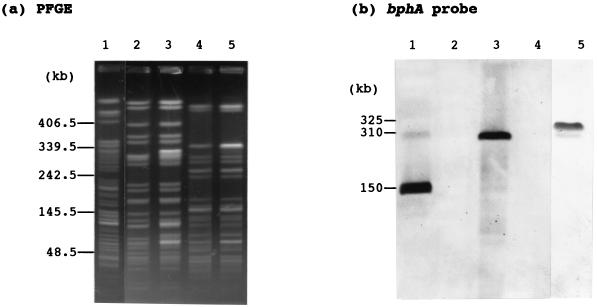
From the evidence that P. graminis KF701 possesses a bphABCD gene cluster nearly identical to that of KF715 (14), a question arises regarding whether the bph gene cluster of KF715 can be transferred to other strains. P. putida KF715 cells were filter mated with P. putida AC30RifrTrp, and Bph+ colonies were screened on a BSM agar plate containing rifampin and tryptophan in the presence of biphenyl as a sole source of carbon. As a result, Bph+ AC30 transconjugants were obtained at a frequency of as high as 10−6 per recipient cells. No colony was obtained when only donor or recipient cells were spread on the same plates as the control. All Bph+ Rifr colonies thus obtained grew on salicylate as well and were confirmed for the requirement of tryptophan. Shown in Fig. 4 are PFGE profiles of DNA fragments of one such Bph+RifrTrp strain (AC30Bph+). The SpeI digestion profile of the AC30Bph+ genome was essentially the same as that of the AC30RifrTrp strain but was different from that of KF715. Genomic Southern analyses confirmed that AC30Bph+ gained both the bph and sal genes. The two gene clusters were located on the same SpeI fragment since identical hybridization profiles were obtained when the bphA probe (Fig. 4b) and the salA probe (data not shown) were used, respectively. The SpeI DNA fragment which hybridized to the bph-sal DNAs was ca. 150 kb for KF715 and ca. 310 kb for AC30Bph+ (Fig. 4b). These results indicate that both the bph and sal gene clusters were transferred into AC30RifrTrp. We named this conjugative chromosomal DNA segment the bph-sal element.
FIG. 4.
PFGE of the genomic DNA of P. putida KF715, P. putida AC30Bph+, and P. putida KT2440Bph+ (a) and the subsequent Southern blot analysis of the bph-sal element with the bphA1 probe (b). The genomic DNA was digested by SpeI. Lanes: 1, KF715; 2, AC30RifrTrp; 3, AC30Bph+; 4, KT2440SmrMet; 5, KT2440Bph+. Molecular size markers are shown on the left.
Mobilization of the bph-sal element from AC30Bph+.
The bph-sal genes of AC30Bph+ were also highly prone to deletion. The deletion frequency of the bph-sal genes was comparable to that of KF715, but the deletion always took place in the region including both the bph and sal genes. This result is in contrast to the original KF715, in which two types of deletions, as seen in KF715M1 and KF715M2, occurred at almost the same frequency. The conjugative transfer of the Bph+ and Sal+ phenotypes from AC30Bph+ was also observed with P. putida KT2440SmrMet as the recipient, at a frequency of as high as 2 × 10−6 per recipient cell. Genomic Southern analyses of KT2440Bph+ confirmed the presence of both the bph and sal genes at the SpeI 325-kb DNA (Fig. 4b). AC30Bph+ was also mated with KF715M1 and KF715M2, respectively, resulting in the appearance of KF715M1Bph+ and KF715M2Bph+ at a frequency of 10−6 per recipients. The SpeI digests of genomic DNA from KF715M1Bph+ and KF715M2Bph+ exhibited bands hybridizing at 250- and 320-kb DNA, respectively, when the bph probe was used (data not shown).
Size of the bph-sal element.
We attempted to determine the size of the bph-sal element by using the cosmids obtained from KF715 and AC30Bph+. We constructed the physical map of this element with the restriction enzyme EcoRI (Fig. 3). The 4.0-kb EcoRI fragment (TNF-I) in a cosmid pKF6500 which is located at the 5′ region far upstream from the bph genes hybridized, with the same size, with the KF715 chromosome and with those of the two deletion mutants KF715M1 and KF715M2 as well (Fig. 3b). However, the same probe hybridized with the 5.2-kb DNA of AC30Bph+. These results indicate that the 4.0-kb DNA (TNF-I DNA) contains the upstream end of the bph-sal element and that the two deletion mutants retain the upstream end of the bph-sal element. On the other hand, cosmid pKF6465 which is from AC30Bph+ and contains the salC gene was used to determine the downstream end of the bph-sal element (Fig. 3f). The 3.7-kb EcoRI fragment (TNF-IX) which is located far downstream of the sal genes and includes the vector-borne EcoRI site was strongly hybridized with the chromosomes of AC30RifrTrp and AC30Bph+ at the same 10.0-kb DNA and hybridized with the ∼20-kb DNA of the KF715 chromosome to a lesser extent. The reason why the TNF-IV hybridized with the KF715 and the derivatives has yet to be elucidated, but the bph-sal element may be inserted within the region conserved between KF715 and AC30, such as tRNA structural genes as seen in clc element (28). The SalI 3.0-kb DNA fragment (TNF-VIII) upstream from TNF-IX DNA hybridized with both KF715 and AC30Bph+ DNA of the same size (Table 2). These results indicate that the 3′ end of the bph-sal element is located just upstream of the TNF-IX DNA fragment in pKF6465. Thus, the hybridization results allow us to assume that the size of the bph-sal element would be ca. 90 kb, which includes 40 kb upstream from the bph genes, a 10-kb bph gene cluster, a 10-kb intervening region between bph and sal, and the 30-kb region including the sal genes (Fig. 3a).
DISCUSSION
In previous studies (16, 35), we reported that the bphX region (ca. 3.5 kb conferring the conversion of 2-hydroxypenta-2,4-dienoic acid to acetyl-coenzyme A) is lacking in the KF715 bph gene cluster. In a sequence comparison with the well-characterized bph genes of P. pseudoalcaligenes KF707 (35) and B. cepacia LB400 (17), only one nucleotide (nucleotide A) of the start codon of bphX0 and the 3′-terminal 106 nucleotides of bphX3 remained, suggesting that KF715 used to have the entire bphABCXD genes as in the case of KF707 and LB400. In the present study, we have also demonstrated that two chromosomal regions, including the bph and/or sal gene clusters, are highly prone to deletion. The KF715 bph and sal gene clusters are separated by 10-kb DNA. Deletion occurs at the region that includes both the bph and sal genes (ca. 70 kb) or at the region that includes only the bph genes (ca. 40 kb). The upper deletion sites seem to be identical. No deletion was observed at the lower region containing only the sal region, and strain KF715M2, which retained only the sal genes, was stable. In contrast to KF715, other biphenyl-utilizing strains, such as P. pseudoalcaligenes KF707 and P. graminis KF701, maintain the bph genes stably. The genome of P. putida KF715 seems to be highly rearranged, not only at the bph-sal regions but at the entire genome, because the restriction profiles by SpeI digestion and following PFGE were significantly different between KF715, KF715M1, and KF715M2 (Fig. 2a). Instability of the bph-sal element may be one of such rearrangement events in strain KF715, where insertional elements could be highly involved, as shown in Yersinia pestis. A 102-kb unstable region of Y. pestis comprising a so-called high-pathogenicity island undergoes internal rearrangement, which is a consequence of the presence of numerous insertional elements in the chromosome (5).
The bph-sal element behaves like a conjugative transposon. Conjugative drug-resistant transposons were first discovered from gram-positive Enterococcus faecalis (9, 15) and Streptococcus pneumoniae (2, 32) and, later, from gram-negative Bacteroides spp., whose conjugative transposons range in size from 65 to 150 kb (4, 25). Most of them carry tetracycline resistance genes. These conjugative transposons can excise themselves from the genome in which they are integrated, transfer themselves by conjugation into a recipient cell, and integrate into the recipient's genome (4, 6, 30, 31). Recently, two conjugative catabolic transposons were also reported (24, 27). The clc gene cluster of Pseudomonas sp. strain B13 conferring 3-chlorobenzoate degradation (7, 10) is present on the B13 chromosome as a 105-kb mobile genetic element (clc element) (27, 28). This element is self-transmissible and integrates into the chromosomes of various bacterial recipients, with glycine tRNA structural gene (glyV) as the integration site (28). It has not been clear whether the insertion site of the bph-sal element is specific as seen in the clc element. However, the fact that the 4.0-kb TNF-I covering the upstream end of the bph-sal element (Fig. 3b) hybridized with ca. The 10-kb fragment of AC30 DNA (Fig. 3b) and that the 10-kb TNF-IX fragment of AC30 downstream of the bph-sal element (Fig. 3f) hybridized with ca. 20-kb fragment of KF715 DNA suggest that there may exist a hot spot of insertion, such as the conserved tRNA structural genes. The bph-sal element can be transferred from the AC30Bph+ transconjugant to another P. putida KT2440. This implies that all factors necessary for mobilization are located on the 90-kb bph-sal element. The DNA sequencing of this element is currently under way to find the genes involved in the transfer and thereby to reveal the mechanism of conjugal transfer. Occurrence of the mobile bph elements may explain why biphenyl-utilizing bacteria are widely distributed and possess bph genes that are very similar, if not identical, to one another.
ACKNOWLEDGMENTS
We thank Masataka Tsuda (Tohoku University) and Hai-Meng Tan (National University of Singapore) for their useful discussions.
This work was supported in part by CREST (Core Research for Evolutional Science and Technology) of The Japan Science and Technology Corporation.
REFERENCES
- 1.Abramowicz D A. Aerobic and anaerobic biodegradation of PCBs: a review. Crit Rev Biotechnol. 1990;10:241–251. [Google Scholar]
- 2.Ayoubi P, Kilic A, Vijayakumar M. Tn5253, the pneumococcal omega (cat tet) BM6001 element, is a composite structure of two conjugative transposons, Tn5251 and Tn5252. J Bacteriol. 1991;173:1617–1622. doi: 10.1128/jb.173.5.1617-1622.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedard D L, Unterman R, Bopp L H, Brennan M J, Haberl M L, Johnson C. Rapid assay for screening and characterizing microorganisms for the ability to degrade polychlorinated biphenyls. Appl Environ Microbiol. 1986;51:761–768. doi: 10.1128/aem.51.4.761-768.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bedzyk L A, Shoemaker N B, Young K E, Salyers A A. Insertion and excision of Bacteroides conjugative chromosomal elements. J Bacteriol. 1992;174:166–172. doi: 10.1128/jb.174.1.166-172.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchrieser C, Prentice M, Carniel E. The 102-kilobase unstable region of Yersinia pestis comprises a high-pathogenicity island linked to a pigmentation segment which undergoes internal rearrangement. J Bacteriol. 1998;180:2321–2329. doi: 10.1128/jb.180.9.2321-2329.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clewell D B, Flannagan S E. The conjugative transposons of gram-positive bacteria. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Press; 1993. pp. 369–393. [Google Scholar]
- 7.Dorn E, Hellwig M, Reineke W, Knackmuss H-J. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch Microbiol. 1974;99:61–70. doi: 10.1007/BF00696222. [DOI] [PubMed] [Google Scholar]
- 8.Erickson B D, Mondello F J. Nucleotide sequencing and transcriptional mapping of the genes encoding biphenyl dioxygenase, a multicomponent polychlorinated biphenyl-degrading enzyme in Pseudomonas strain LB400. J Bacteriol. 1992;174:2903–2912. doi: 10.1128/jb.174.9.2903-2912.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franke A E, Clewell D B. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J Bacteriol. 1981;145:494–502. doi: 10.1128/jb.145.1.494-502.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frantz B, Chakrabarty A M. Organization and nucleotide sequence determination of a gene cluster involved in 3-chlorocatechol degradation. Proc Natl Acad Sci USA. 1987;84:4460–4464. doi: 10.1073/pnas.84.13.4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furukawa K. Microbial degradation of polychlorinated biphenyls. In: Chakrabarty A M, editor. Biodegradation and detoxification of environmental pollutants. Boca Raton, Fla: CRC Press, Inc.; 1982. pp. 33–57. [Google Scholar]
- 12.Furukawa K. Molecular genetics and evolutionary relationship of PCB-degrading bacteria. Biodegradation. 1994;5:289–300. doi: 10.1007/BF00696466. [DOI] [PubMed] [Google Scholar]
- 13.Furukawa K, Hayase N, Taira K, Tomizuka N. Molecular relationship of chromosomal genes encoding biphenyl/polychlorinated biphenyl catabolism: some soil bacteria possess a highly conserved bph operon. J Bacteriol. 1989;171:5467–5472. doi: 10.1128/jb.171.10.5467-5472.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furukawa K, Miyazaki T. Cloning of gene cluster encoding biphenyl and chlorobiphenyl degradation in Pseudomonas pseudoalcaligenes. J Bacteriol. 1986;166:392–398. doi: 10.1128/jb.166.2.392-398.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gawron-Burke C, Clewell D B. A transposon in Streptococcus faecalis with fertility properties. Nature. 1982;300:281–284. doi: 10.1038/300281a0. [DOI] [PubMed] [Google Scholar]
- 16.Hayase N, Taira K, Furukawa K. Pseudomonas putida KF715 bphABCD operon encoding biphenyl and polychlorinated biphenyl degradation: cloning, analysis, and expression in soil bacteria. J Bacteriol. 1990;172:1160–1164. doi: 10.1128/jb.172.2.1160-1164.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofer B, Backhaus S, Timmis K N. The biphenyl/polychlorinated biphenyl-degradation locus (bph) of Pseudomonas sp. LB400 encodes four additional metabolic enzymes. Gene. 1994;144:9–16. doi: 10.1016/0378-1119(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 18.Hooper S W, Dockendorff T C, Sayler G S. Characteristics and restriction analysis of the 4-chlorinated biphenyl/polychlorinated biphenyl catabolic plasmid pSS50. Appl Environ Microbiol. 1989;55:1286–1288. doi: 10.1128/aem.55.5.1286-1288.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura N, Nishi A, Goto A, Furukawa K. Functional analyses of a variety of chimeric dioxygenases constructed from two biphenyl dioxygenases that are similar structurally but different functionally. J Bacteriol. 1997;179:3936–3943. doi: 10.1128/jb.179.12.3936-3943.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee J, Min K R, Kim Y-C, Kim C-K, Lim J-Y, Hoon H, Min K-H, Lee K-S, Kim Y. Cloning of salicylate hydroxylase gene and catechol 2,3-dioxygenase gene and sequencing of an intergenic sequence between the two genes of Pseudomonas putida KF715. Biochem Biophys Res Commun. 1995;211:382–388. doi: 10.1006/bbrc.1995.1825. [DOI] [PubMed] [Google Scholar]
- 21.Lee J, Oh J, Min K R, Kim Y. Nucleotide sequence of salicylate hydroxylase genes and its 5′-flanking region of Pseudomonas putida KF715. Biochem Biophys Res Commun. 1996;218:544–548. doi: 10.1006/bbrc.1996.0097. [DOI] [PubMed] [Google Scholar]
- 22.Lloyd-Jones G, De Jong C, Ogden R C, Duetz W A, Williams P A. Recombination of the bph (biphenyl) genes from pWW100 and their deletion during growth on benzoate. Appl Environ Microbiol. 1994;60:691–696. doi: 10.1128/aem.60.2.691-696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merlin C, Springael D, Mergeay M, Toussaint A. Organization of the bph gene cluster of transposon Tn4371, encoding enzymes for the degradation of biphenyl and 4-chlorobiphenyl compounds. Mol Gen Genet. 1997;253:499–506. doi: 10.1007/s004380050349. [DOI] [PubMed] [Google Scholar]
- 24.Merlin C, Springael D, Toussaint A. Tn4371: a modular structure encoding a phage-like integrase, a Pseudomonas-like catabolic pathway, and RP4-like transfer function. Plasmid. 1999;41:40–54. doi: 10.1006/plas.1998.1375. [DOI] [PubMed] [Google Scholar]
- 25.Nikolich M P, Shoemaker N B, Salyers A A. Characterization of a new type of Bacteroides conjugative transposon, TcrEmr 7853. J Bacteriol. 1994;176:6606–6612. doi: 10.1128/jb.176.21.6606-6612.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pflugmacher U, Averhoff B, Gottschalk G. Cloning, sequencing, and expression of isopropylbenzene degradation genes from Pseudomonas sp. strain JR1: identification of isopropylbenzene dioxygenase that mediates trichloroethene oxidation. Appl Environ Microbiol. 1996;62:3967–3977. doi: 10.1128/aem.62.11.3967-3977.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ravatn R, Studer S, Springael D, Zehnder A J B, van der Meer J R. Chromosomal integration, tandem amplification, and deamplification in Pseudomonas putida F1 of a 105-kilobase genetic element containing the chlorocatechol degradative genes from Pseudomonas sp. strain B13. J Bacteriol. 1998;180:4360–4369. doi: 10.1128/jb.180.17.4360-4369.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ravatn R, Studer S, Zehnder A J B, van der Meer J R. Int-B13, an unusual site-specific recombinase of the bacteriophage P4 integrase family, is responsible for chromosomal insertion of the 105-kilobase clc element of Pseudomonas sp. strain B13. J Bacteriol. 1998;180:5505–5514. doi: 10.1128/jb.180.21.5505-5514.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salyers A A, Shoemaker N B, Stevens A M, Li L-Y. Conjugative transposons: an unusual and divers set of integrated gene transfer elements. Microbiol Rev. 1995;59:579–590. doi: 10.1128/mr.59.4.579-590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott J R. Sex and the single circle: conjugative transposon. J Bacteriol. 1992;174:6005–6010. doi: 10.1128/jb.174.19.6005-6010.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shoemaker N B, Salyer A A. A cryptic 65-kilobase-pair transposonlike element isolated from Bacteroides unifornis has homology with Bacteroides conjugal tetracycline resistance elements. J Bacteriol. 1990;172:1694–1702. doi: 10.1128/jb.172.4.1694-1702.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shoemaker N B, Smith M D, Guild W R. DNase resistant transfer of chromosomal cat and tet insertions by filter mating in pneumococcus. Plasmid. 1980;3:80–87. doi: 10.1016/s0147-619x(80)90036-0. [DOI] [PubMed] [Google Scholar]
- 33.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 34.Springael D, Kreps S, Mergeay M. Identification of a catabolic transposon, Tn4371, carrying biphenyl and 4-chlorobiphenyl degradation genes in Alcaligenes eutrophus A5. J Bacteriol. 1993;175:1674–1681. doi: 10.1128/jb.175.6.1674-1681.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taira K, Hirose J, Hayashida S, Furukawa K. Analysis of bph operon from the polychlorinated biphenyl-degrading strain of Pseudomonas pseudoalcaligenes KF707. J Biol Chem. 1992;267:4844–4853. [PubMed] [Google Scholar]
- 36.Tsuda M, Tan H-M, Nishi A, Furukawa K. Mobile catabolic genes in bacteria. J Biosci Bioeng. 1999;87:401–410. doi: 10.1016/s1389-1723(99)80086-3. [DOI] [PubMed] [Google Scholar]
- 37.Unterman R. A history of PCB biodegradation. In: Crawford R L, Crawford D L, editors. Bioremediation: principles and applications. New York, N.Y: Cambridge University Press; 1996. pp. 209–253. [Google Scholar]
- 38.Wahl G M, Lewis K A, Ruiz J C, Rotherberg B, Zhao J, Evans G A. Cosmid vectors for rapid genomic walking, restriction mapping, and gene transfer. Proc Natl Acad Sci USA. 1987;84:2160–2164. doi: 10.1073/pnas.84.8.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamada A, Kishi H, Sugiyama K, Hatta T, Nakamura K, Masai E, Fukuda M. Two nearly identical aromatic compound hydrolase genes in a strong polychlorinated biphenyl degrader, Rhodococces sp. strain RHA1. Appl Environ Microbiol. 1998;64:2006–2012. doi: 10.1128/aem.64.6.2006-2012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]