Abstract
Tubular epithelial cells (TECs) play critical roles in the development of diabetic nephropathy (DN), and can activate macrophages through the secretion of exosomes. However, the mechanism(s) of TEC-exosomes in macrophage activation under DN remains unknown. By mass spectrometry, 1,644 differentially expressed proteins, especially Dll4, were detected in the urine exosomes of DN patients compared with controls, which was confirmed by western blot assay. Elevated Epsin1 and Dll4/N1ICD expression was observed in kidney tissues in both DN patients and db/db mice and was positively associated with tubulointerstitial damage. Exosomes from high glucose (HG)-treated tubular cells (HK-2) with Epsin1 knockdown (KD) ameliorated macrophage activation, TNF-α, and IL-6 expression, and tubulointerstitial damage in C57BL/6 mice in vivo. In an in vitro study, enriched Dll4 was confirmed in HK-2 cells stimulated with HG, which was captured by THP-1 cells and promoted M1 macrophage activation. In addition, Epsin1 modulated the content of Dll4 in TEC-exosomes stimulated with HG. TEC-exosomes with Epsin1-KD significantly inhibited N1ICD activation and iNOS expression in THP-1 cells compared with incubation with HG alone. These findings suggested that Epsin1 could modulate tubular-macrophage crosstalk in DN by mediating exosomal sorting of Dll4 and Notch1 activation.
Keywords: diabetic nephropathy, Epsin1, exosome, Dll4, inflammation
Graphical abstract

Xiao and colleagues reported that Epsin1-mediated exosome-dominant Dll4 release from kidney tubular epithelial cell (TEC) activated Notch1 signaling in macrophage and hastened tubulointerstitial inflammation under hyperglycemic condition, while it could be efficiently relieved by Epsin1 knockdown.
Introduction
Diabetic nephropathy (DN) is the leading cause of end-stage renal disease worldwide.1,2 In recent years, in addition to metabolic abnormalities such as hyperglycemia, the inflammatory response in the kidney has been proven to play a crucial role in the progression of DN.3 Increasing evidence shows that tubular epithelial cells (TECs) are one of the key determinant cells in interstitial inflammation in DN.3,4,5 In addition to acting as the victim of inflammation, TECs can also transform to a proinflammatory and fibrogenic phenotype upon renal injury, thus initiating the inflammatory cascade in DN.3,5,6 Damage to TECs triggers the release of “signals” such as cytokines, resulting in the recruitment into the injured kidney and polarization of crucial macrophages.4,5,7,8 The crosstalk between TECs and macrophages modulates the progression of macrophage-mediated inflammation. However, the detailed mechanism underlying this tubular-macrophage crosstalk remains to be elucidated.
Exosomes are extracellular vesicles released by various cell types, with sizes ranging from 30 to 150 nm,9 mediating intracellular communication by delivering cell-specific cargo, including proteins, lipids, and genetic information, to recipient cells.9,10,11 Therefore, exosomes are a newly discovered communication mode between TECs and macrophages.9 Lv et al. found that BSA-treated TEC-derived exosomes could be internalized by cultured macrophages,12 and miR-20a-5p enriched in hypoxia-induced TEC-derived exosomes protected against acute tubular injury by attenuating macrophage infiltration.13 However, little is known about the role of TEC-derived exosomes in the regulation of macrophages and inflammation in the DN state.
Highly conserved Notch signaling is activated during macrophage activation and polarization in multiple inflammatory disorders.14,15,16 In diabetic kidney disease, activation of Notch in various cell types promotes DN progression.17,18,19,20,21 Notch pathway activation in podocytes induces podocyte dedifferentiation and apoptosis, resulting in glomerulosclerosis and albuminuria. In TECs, Notch signaling activation promotes fibroblast proliferation and macrophage infiltration in the renal interstitium, accelerating the development of tubulointerstitial fibrosis.22,23 Moreover, Notch pathway in macrophages interacts with nuclear factor-κB signaling and thus promotes macrophage polarization.21 The canonical cell-to-cell Notch signaling cascade is initiated by the direct interaction of Delta-like 4 (Dll4), a ligand of Notch on the cell membrane, and the Notch extracellular domain (NECD) of the adjacent cell. Ligand-receptor ligation triggers cleavage between the NECD and the Notch intracellular domain (NICD). Once separated, the sole NICD translocates into the nucleus, thus modulating the transcription of the target genes.15,24 Beyond direct cell contact, evidence has shown that Dll4 can be incorporated into exosomes. The Dll4-containing exosomes released from one cell type fuse with the receiving cells and transfer Dll4 into their cell membrane, which results in the regulation of Notch signaling.25 However, whether Dll4-containing exosome-mediated Notch signaling participates in tubular-macrophage crosstalk in DN has not yet been studied.
Epsins are a family of highly conserved, membrane-associated, ubiquitin-binding endocytic adapter proteins.26,27 Studies have shown that Epsin1 augments inflammatory signaling and aggravates endothelial cell dysfunction in atherosclerosis.26,28 Our previous study even discovered that Epsin deficiency can promote lymphangiogenesis through regulation of VEGFR3 degradation in diabetes,27 encouraging the exploration of Epsin in DN inflammation. In addition, as reported, Epsin deficiency leads to failure of membrane bending and plasma membrane invagination, and is unable to trigger and initiate endocytosis,29,30 whereas exosomes originate from early endocytosis. Therefore, investigation into the role of Epsin1 in exosomes and diabetic kidneys is warranted. More importantly, Epsin is also involved in the screening of endocytosis, and the high affinity between UIMs (one domain of Epsin) and ubiquitinated proteins enables identification and specific screening of ubiquitinated cargo, such as the Notch ligand Delta (Dl) and growth factor receptor (EGFR).31,32 Studies have already proven the essentiality of Epsin in Notch signal transduction.33 This study elucidates the requirements for ligand ubiquitination, Epsin-dependent endocytosis, and force-dependent S2 cleavage in native DSL/Notch signaling. Mechanically, Epsin targets ligands for Epsin/clathrin-mediated endocytosis, applying force across the ligand-receptor bridge to initiate "S2 cleavage" and "S3 cleavage", cutting receptors into the NICD. Then, the NICD is transferred to the nucleus and initiates transcription of Notch target genes. Therefore, whether Epsin modulates the Delta/Serrate/Lag2 ligand in TEC-exosomes remains to be determined.
Given the diverse roles of Epsin, we hypothesize that Epsin1 may be involved in TEC-to-macrophage crosstalk by regulating TEC-derived exosomal Dll4, consequently influencing the activity of the Notch1 signaling pathway, inducing macrophage phenotype shift, and accelerating tubulointerstitial inflammation under hyperglycemic conditions.
Results
Identification of enhanced Dll4 in urinary exosomes from DN patients
To explore the protein profile of renal exosomes under DN conditions, urine samples from six different persons (three DN patients vs. three normal controls) were collected, and the isolated exosomes for each group were pooled for proteome analysis. Transmission electron microscope (TEM) and nanoparticle tracking analysis (NTA) were used to identify the typical shape and size of urinary exosomes (Figures 1A and 1B). A total of 1,644 differentially expressed proteins were detected by proteomic analysis, among which 481 proteins were increased and 914 were reduced in DN patients compared with the control group (Figure 1C). In addition to vesicle budding and endocytosis pathways, inflammation-related leukocyte migration and chemokine signaling pathways were significantly changed under DN conditions (Figures 1D and 1E). Based on the functional annotation and enrichment analysis, the top 15 differentially expressed proteins according to their significant p value are listed in Figure 1F. Among them, Dll4 was significantly elevated in the urine exosomes of DN patients compared with that of the control group (Figure 1F). Western blotting further confirmed an increased abundance of the ligand Dll4 in the urine exosomes of DN patients (Figure 1G). These data suggested aberrant exosome-dependent Dll4 transfer in DN inflammation.
Figure 1.
Identification of enhanced Dll4 in urinary exosomes from DN patients
(A) Representative transmission electron microscope (TEM) image of urinary exosomes released by DN patients. Scale bar, 100 nm, 200 nm. (B) Size distribution and number of DN urinary exosomes presented with nanoparticle tracking analysis (NTA). (C) The volcano plot of proteins analyzed from urinary exosomes. The volcano plot was constructed with log2 expression of fold change and the corresponding tempered log2 p value of all proteins. Red dots represent upregulated proteins and blue dots represent downregulated proteins. (D and E) Biological process and KEGG pathway of differentially expressed proteins identified by proteome. (F) A heatmap based on top 15 differentially expressed proteins according to their significance among signal transduction mechanism. Red represents strong enrichment and green represents weak enrichment. (G–G2) Immunoblots using urinary exosome proteins (5 μg each) from normal controls(NCs) and DN patients against exosome markers (CD9, CD81, Alix) and Dll4, followed by their densitometry analysis. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Data are representative of three independent experiments and are expressed as mean ± SD.
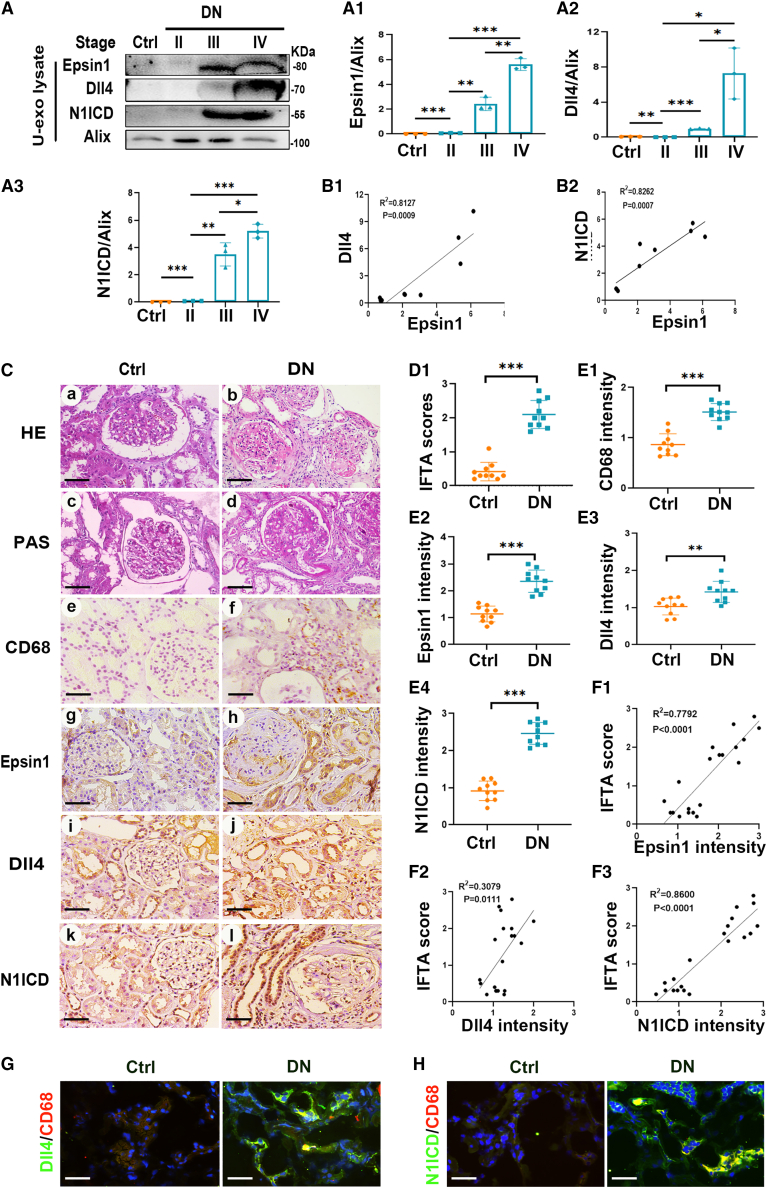
Levels of Epsin1, Dll4, and N1ICD were significantly enhanced in the urinary exosomes and kidney tissue of DN patients and were positively correlated with tubulointerstitial damage
For the purpose of probing the relationship between Epsin1 and Dll4/Notch1 signaling, urinary exosomes were collected from normal controls and patients with DN stage II to IV to assess the expression levels of Epsin1, Dll4, and N1ICD. Immunoblotting assays showed that Epsin1, Dll4, and N1ICD levels were significantly enhanced in the urinary exosomes of DN patients in parallel with DN stage progression (Figures 2A and A1–3). Correlation analysis revealed that Epsin1 expression was positively associated with Dll4 and N1ICD levels (Figures 2B1 and B2). Therefore, it is likely that Epsin1 and Dll4/Notch1 signaling in renal tubules are related to DN progression. H&E and periodic acid-Schiff (PAS) staining showed mesangial matrix proliferation, increased CD68+ macrophage infiltration, and tubular damage in the kidneys of DN patients compared with the control group (Figures 2Ca–2Cf). The protein expression of Epsin1, Dll4, and N1ICD notably increased in the kidneys of DN patients, especially in the tubulointerstitial area (Figures 2Cg–2Cl). Correlation analysis showed that there was a strong positive association between the expression of Epsin1/Dll4/N1ICD and the tubulointerstitial damage score (Figures 2D1–F3). Furthermore, immunofluorescence staining showed increased intensity of colocalization of Dll4 or N1ICD and the macrophage marker CD68 in the kidneys of DN patients (Figures 2G and 2H). These results indicate that Epsin1 and Dll4/N1ICD are crucial for macrophage activation in the kidneys of DN patients, possibly through exosome regulation pathway.
Figure 2.
Levels of Epsin1, Dll4, and N1ICD were significantly enhanced in the urinary exosomes and kidney tissue of DN patients and were positively correlated with tubulointerstitial damage
(A–B2) Immunoblot against Epsin1, Dll4, N1ICD, and the exosome marker Alix using urinary exosome preparations (U-exo), followed by their densitometric and linear correlation analysis. (C) Histologic (hematoxylin-eosin [HE] and periodic acid-Schiff [PAS] staining) and immunohistologic (CD68, Epsin1, Dll4, N1ICD immunostaining) changes. Scale bar, 50 μm. (D1) Quantitative analysis of interstitial fibrosis and tubular atrophy (IFTA) scores. (E1–E4) Relative intensity of CD68, Epsin1, Dll4, and N1ICD in the kidney tissues. (F1–F3) The linear correlations analysis of Epsin1, Dll4, and N1ICD expression levels with IFTA scores. (G and H) Immunofluorescence staining of Dll4/N1ICD (green) and the macrophage marker CD68 (red) in frozen kidney sections. Scale bar, 50 μm. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Data are representative of three independent experiments and are expressed as mean ± SD.
Expression of Epsin1 was positively associated with the level of Dll4/N1ICD and tubulointerstitial damage in the kidneys of db/db mice
As assessed by H&E and PAS staining, similar changes, including mesangial matrix proliferation, tubular damage, and interstitial macrophage infiltration, were observed in the kidneys of db/db mice compared with db/m mice (Figures 3A a–3Af, B1, and C1). Immunohistochemical staining showed that Epsin1 and Dll4/Notch1 signaling molecules were obviously increased in the renal tubules of 8-week-old db/db mice (Figures 3Ag–3An and C2–C5). In particular, the expression of Epsin1 and Dll4 was much higher than that at 12 or 16 weeks (Figure S1). To investigate the effect of Epsin1 and Dll4 on early diabetic kidney injury via the exosome regulation pathway, db/db mice at 8 weeks of age were chosen for further studies. The tubulointerstitial damage scores of db/db mice were positively correlated with the expression of Epsin1, Dll4, Notch1 and N1ICD (Figures 3D1–4). These changes were further confirmed by western blot assay and semiquantitative analysis (Figures 3E and 3E1–3E3). Immunofluorescence assays confirmed that the expression of Epsin1 and Dll4 was increased in the TECs of db/db mice (Figure 3F). In addition, the intensity of colocalization of Dll4/N1ICD with the macrophage marker F4/80 also increased in the kidneys of db/db mice (Figure 3G), indicating the activation of Dll4/N1ICD in infiltrated macrophages. These results suggest that Epsin1 contributes to tubulointerstitial injury in db/db mice, possibly via the Dll4/Notch1 signaling pathway.
Figure 3.
Expression of Epsin1 was positively associated with the level of Dll4/N1ICD and tubulointerstitial damage in the kidneys of db/db mice
(A) Histologic (HE and PAS staining) and immunohistologic (F4/80, Epsin1, Dll4, Notch1, N1ICD immunostaining) changes in mice. Scale bar, 50 μm. (B1) Quantitative analysis of IFTA scores. (C1–C5) Relative intensity of F4/80, Epsin1, Dll4, Notch1, N1ICD in the kidney tissues. (D1–D4) The linear correlations analysis of Epsin1, Dll4, Notch1, and N1ICD expression levels with IFTA scores. (E–E3) Western blot analysis of Epsin1, Dll4, N1ICD expression of kidney with animal model, followed by their densitometric analysis. (F) Immunofluorescence staining of Dll4 (red), Epsin1 (red), and the tubule marker LTL (green) in frozen kidney sections. Scale bar, 50 μm. (G) Immunostaining of Dll4 (green), N1ICD (green), and the macrophage marker F4/80 (red) in frozen kidney sections. Scale bar, 50 μm. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Data are representative of three independent experiments and are expressed as mean ± SD.
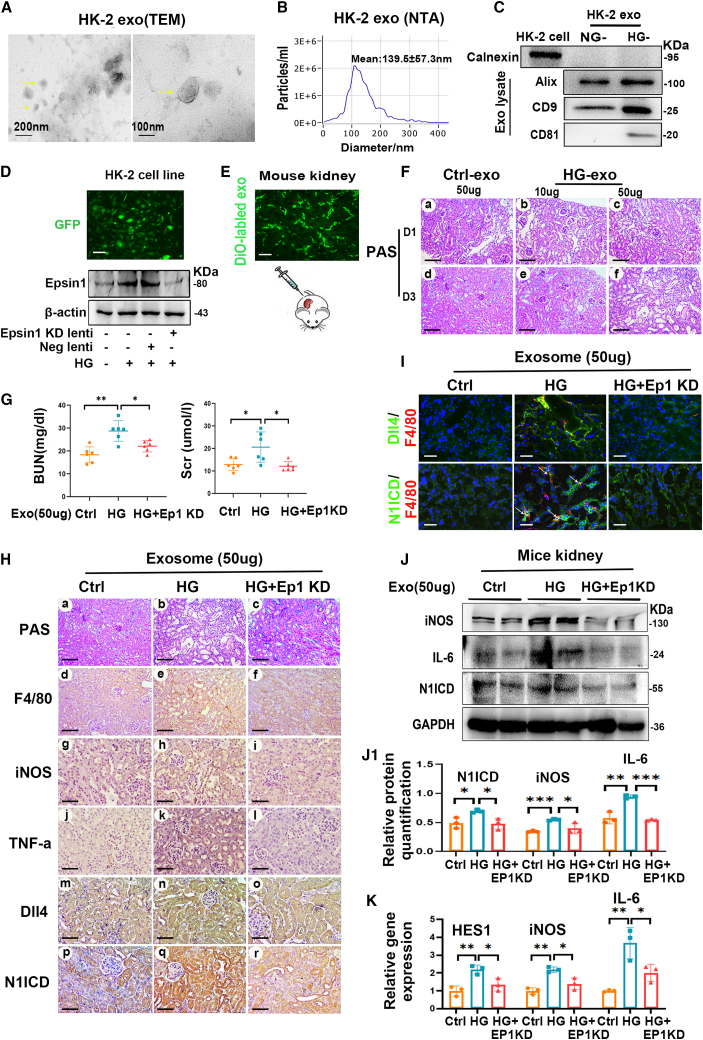
Figure 4.
Exosomes from HG-treated HK-2 cells with Epsin1 knockdown ameliorated macrophage activation and tubulointerstitial damage in vivo
(A) Typical transmission electron microscope (TEM) image of the exosomes secreted by HK-2 cells (HK-2 exo). Scale bar, 100 nm, 200 nm. (B) Size distribution and number of exosomes isolated from HK-2 cells culture medium were examined with nanoparticle tracking analysis (NTA). (C) Representative western blotting of exosome markers (including calnexin, Alix, CD9, and CD81) in HK-2-derived exosomes with normal-glucose (NG) or high-glucose (HG) treatment (also defined as NG/Ctrl-exo and HG-exo). (D) Western blotting showing that HK-2 sublines were transfected with Epsin1-knockdown (KD) lentivirus and generated Epsin1-KD exosomes upon HG treatment (called HG + Ep1 KD-exo). Scale bar, 10 μm. (E) Representative images of DiO-labeled kidney section showing the preparation of tubulointerstitial exo-injection (50 μg) mice model. Scale bar, 50 μm. (F) Representative images of PAS-stained kidney sections at day 1(D1) and day 3(D3) after Ctrl-exo or HG-exo-injection at two doses (10 μg, 50 μg), respectively. Scale bar, 50 μm. (G) Blood urea nitrogen (BUN) and serum levels of creatinine (Scr) in mice at day 3 after 50 μg exosomes administration. (H) Histological changes (PAS staining) and representative images of F4/80, iNOS, TNF-α, Dll4, and N1ICD-stained kidney sections at day 3 after 50 μg exosome administration (Ctrl-exo, HG-exo, HG + Ep1 KD-exo), respectively. Scale bar, 50 μm. (I) Immunofluorescence staining of Dll4 (green), N1ICD (green), and the macrophage marker F4/80 (red) in frozen kidney sections. Scale bar, 50 μm. (J and J1) Western blot analysis of iNOS, IL-6, N1ICD expression of kidney with exo-injected animal model, followed by their densitometric analysis. (K) Real-time quantitative PCR measuring N1ICD target gene (HES1) and M1 markers (iNOS and IL-6) in exo-injected kidney sections. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Data are representative of three independent experiments at least and are expressed as mean ± SD.
Exosomes from HG-treated HK-2 cells with Epsin1 knockdown ameliorated macrophage activation and tubulointerstitial damage in vivo
To elucidate the specific role of Epsin1 in tubulointerstitial inflammation in vivo mediated by TEC-derived exosomes, exosomes from Epsin1-knockdown (KD) HK-2 cells were collected and administered to C57BL/6 mice. Exosomes derived from HK-2 cells treated with high glucose (HG) and normal glucose (NG) were isolated and named HG-HK-2-exo and NG-exo, respectively. TEM, NTA, and western blotting were performed to characterize the exosomes (Figures 4A–4C). HK-2 cells were transfected with Epsin1-KD lentivirus and treated with HG in vitro. Obvious inhibition of the expression of Epsin1 was observed in the HK-2 cells by western blot analysis (Figure 4D). Using the DiO label, HG- HK-2-exo was apparently detectable in the tubulointerstitium 1 day after being injected into the unilateral kidney of C57BL/6 mice (Figure 4E). PAS staining revealed tubular necrosis and detachment, cellular debris accumulation and cast formation in the kidneys of 10 μg HG- HK-2-exo-treated mice 1 day after injection (Figure 4F), which were aggravated in those treated with 50 μg HG-HK-2-exo or at 3 days after injection. In addition, blood urea nitrogen (BUN), serum creatinine (Scr), and the intensity of F4/80 iNOS and Dll4/N1ICD in the kidney were dramatically upregulated in the mice treated with 50 μg HG-HK-2-exo, while these effects were significantly inhibited in those treated with exosomes of HG + Ep1-KD (Figures 4G and 4H). More interestingly, the colocalization of Dll4, N1ICD, and F4/80 was also alleviated by treatment with HG + Ep1-KD exosomes (Figure 4I). A similar phenomenon was confirmed by western blot analysis and quantitative real-time PCR (Figures 4J and 4K). Overall, Epsin1 KD prevented tubulointerstitial inflammation and damage triggered by exosomes derived from HG-treated tubular cells in vivo.
Dll4-enriched exosomes originating from HG-treated HK-2 cells promoted M1 macrophage activation
Immunoblotting showed that exosome markers (Alix, CD9 and CD81) were significantly increased in HG-HK-2-exo. Notably, enhanced expression of Dll4 was also detected in HG-HK-2-exo (Figure 5A) compared to that in NG-HK-2-exo. To investigate the effect of TEC-derived exosomes on macrophages, THP-1 and HK-2 cells were cocultured in a Transwell system. THP-1 cells were recruited as shown by DAPI staining of cells on the basolateral membrane (Figure 5B). In addition, THP-1 cells were directly treated with HG-HK-2-exo. As expected, THP-1 macrophages internalized much more HG-HK-2-exo than NG-HK-2-exo (Figures 5C and 5C1). Furthermore, notably increased expression of iNOS and IL-6 in THP-1 cells treated with HG-HK-2-exo was observed compared to that of the control. Lipopolysaccharide (LPS) treatment served as a positive control (Figures 5D, 5D1, and 5D2). Flow cytometry assays also confirmed that the expression of CD86+ in THP-1 cells increased with HG-HK-2-exo treatment, while CD206 expression remained unchanged (Figure 5E). These data suggested that Dll4 enriched in the exosomes originated from HG-treated HK-2 cells. Furthermore, exosomes derived from HG-treated HK-2 cells polarized macrophages toward a proinflammatory M1 phenotype.
Figure 5.
Dll4-enriched exosomes originating from HG-treated HK-2 cells promoted M1 macrophage activation
(A) Immunoblots against Dll4 and the exosome markers (Alix, CD9, and CD81). (B) Chemotaxis test for THP-1 macrophages when THP-1 cells were seeded in the top compartment of Transwell, separated by a porous membrane from HK-2 cells culture medium. Scale bar, 10 μm. (C) Internalization of HK-2-derived exosomes by macrophages. Exosomes were purified from the DiO-labeled HK-2 cells and were then applied to recipient THP-1 macrophages which were pretreated with 100 ng/mL PMA. Scale bar, 10 μm. (C1) Relative fluorescence intensity was calculated. (D–D2) Western blotting and quantification of iNOS, IL-6 in recipient THP-1 macrophages treated with NG-exo, HG-exo and LPS, LPS was used as a positive control. (E) Expression analysis of CD86+ and CD206+ by flow cytometry for THP-1 macrophages stimulated with NG-exo, HG-exo, and LPS. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Data are representative of three independent experiments and are expressed as mean ± SD.
Epsin1 modulated exosomal Dll4 derived from HG-stimulated HK-2 cells
To observe the effect of Epsin1 on exosomal-Dll4 transfer, we knocked down Epsin1 in HK-2 cells using Epsin1 small interfering RNA (siRNA) and analyzed the change in Dll4. In HK-2 cells, increased expression of Dll4 was observed in HG-stimulated HK-2 cells as assessed by immunoblotting, whereas no significant change in Dll4 was found after Epsin1 KD under HG conditions (Figure 6A). We then analyzed whether the expression of Dll4 changed in exosomes originating from HK-2 cells treated with HG and Epsin1 siRNA. As expected, the expression of Dll4 was dramatically increased in HG-HK-2-exosomes, while in exosomes secreted from the HG-treated HK-2 cells in which Epsin1 was knocked down, the HG-induced expression of Dll4 was reduced (Figures 6B and 6C). Furthermore, a stable HK-2 cell line overexpressing Dll4 was established (Figure 6D) and used for exosome extraction. Surprisingly, Dll4 was barely expressed in exosomes derived from Dll4-OE transfected HK-2 cells without HG treatment, whereas a remarkable increase in Dll4 expression was observed after HG stimulation (Figure 6E). In addition, another HK-2 stable cell line cotransfected with Epsin1-KD and Dll4-OE-lentivirus was successfully constructed (Figures 6F and 6F1) for exosome collection. Despite both Dll4-OE transfection and HG treatment, Epsin1 KD blocked the sorting of Dll4 into HK-2-derived exosomes (Figures 6G and 6G1), indicating that Epsin1 modulated exosomal Dll4 derived from HG-stimulated HK-2 cells.
Figure 6.
Epsin1 modulated exosomal Dll4 derived from HG-stimulated HK-2 cells
(A and A1) Western blotting for expression of Epsin1, Dll4 in HK-2 cells and their densitometric analysis. (B–C1) Immunoblotting against Alix-standardized Dll4 in exosomes secreted from Epsin1 siRNA-transfected and Epsin1-KD lentivirus-transfected HK-2 cells respectively, followed by their densitometric analysis. (D and D1) Western blotting proving HK-2 sublines were transfected with Dll4-OE lentivirus. (E and E1) Western blot analysis of exosomal Dll4 from the Dll4-OE sublines. (F and F1) Western blotting proving HK-2 sublines were transfected with both Epsin1-KD and Dll4-OE lentivirus. (G and G1) Western blot analysis of exosomal Dll4 from HK-2 subline transfected with both Epsin1-KD and Dll4-OE lentivirus. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Data are representative of three independent experiments and are expressed as mean ± SD.
Epsin1 KD in HK-2 cells alleviated macrophage N1ICD activation and M1 polarization by inhibiting HK-2-derived exosomal Dll4
To evaluate the effect of tubular Epsin1 KD on macrophage activation, HK-2 cells were cocultured with THP-1 cells using a Transwell system so that exosomes derived from TECs might pass through the basolateral membrane into the medium of THP-1 cells (Figure 7A). As illustrated in Figures 7B and 7B1–7B3, the expression of Dll4, N1ICD, and iNOS was upregulated in THP-1 macrophages cocultured with HG-treated HK-2 cells. When cocultured with HG-treated HK-2 cells transfected with Dll4 OE lentivirus, the increase in Dll4, N1ICD, and iNOS in THP-1 macrophages was further aggravated. In contrast, Epsin1 KD in HK-2 cells reversed the elevated expression of Dll4, N1ICD, and iNOS in macrophages. To better illustrate that Epsin1 in HK-2 cells could regulate M1 macrophage activation via exosomes, we treated THP-1 macrophages directly with exosomes originating from HG-treated HK-2 cells with or without Epsin1 KD (Figure 7C). The results showed that Epsin1 KD significantly inhibited the increased expression of Dll4, N1ICD and the M1 marker iNOS in THP-1 macrophages induced by HG-HK-2 exosomes (Figures 7D and 7D1–D3). These data suggested that Epsin1 KD in HK-2 cells alleviated macrophage M1 polarization by inhibiting HK-2-derived exosomal Dll4.
Figure 7.
Epsin1 knockdown in HK-2 cells alleviated macrophage N1ICD activation and M1 polarization by inhibiting HK-2-derived exosomal Dll4
(A) Transwell system where exosomes from HK-2 cells might pass through the basolateral membrane to the medium of THP-1 cells. (B–B3) Western blot analysis for expression of iNOS, Dll4, and N1ICD in THP-1 macrophages after coculturing THP-1 with HK-2 cells by Transwell system where HK-2 cells seeded upper and THP-1 cells seeded lower. (C) THP-1 cell incubation with exosomes derived from HK-2 cells. (D–D3) Immunoblot analysis for expression of iNOS, Dll4, and N1ICD in THP-1 macrophages accepting exosomes (20 μg per group) from HK-2 cell lines, respectively. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Data are representative of three independent experiments and are expressed as mean ± SD.
Discussion
In this study, we found for the first time that the expression of Epsin1 in the kidney was positively correlated with macrophage infiltration and tubulointerstitial damage in DN patients and db/db mice. Intrarenal injection of exosomes derived from TECs with Epsin1 KD dramatically ameliorated macrophage activation and tubular damage in vivo. In addition, Epsin1 modulated exosomal Dll4 release from TECs, which led to Notch1 activation, increased the M1 macrophage phenotype and hastened inflammation under hyperglycemic conditions. These data indicate that Epsin1 plays a key role in tubulointerstitial inflammation by modulating an exosome-dominant Dll4/Notch1 signaling pathway.
Exosomes, a major type of extracellular vesicles (EVs), mediate intercellular communication with neighboring or distant cells by carrying cargos including DNA, mRNA, miRNA, and proteins in both healthy and disease conditions including DN and inflammation.34,35 Liquid chromatography-tandem mass spectrometry (LC-MS/MS) proteomics analysis of human urine exosomes identified proteins from renal epithelia extending from podocytes through the proximal tubule, distal tubule, and collecting duct.36,37 Their protein or molecular cargo may reflect the pathophysiological characteristics of kidney diseases.38 Lee et al. found the exosomal microRNA signatures of urinary exosomes in DN patients by microRNA array.39 Feng et al. recently observed that CCL21 mRNA from urinary small EVs was involved in inflammation under DN.40 However, the profile of protein in the urine exosomes of DN is limited. In this study, we found that inflammation-related proteins changed critically in the urinary exosomes of DN patients, and that Dll4 expression obviously increased in the urinary exosomes of DN patients compared with controls (Figure 1). Furthermore, Dll4/N1ICD expression normalized to Alix increased with DN progression (Figure 2), suggesting a putative pathophysiological relevance. Interestingly, the exosome marker Alix presented a trend to increase first and then decrease during DN progression, which might indicate a decrease in exosome secretion in advanced DN, as reported in a previous study.41 These findings indicate that regulation of inflammation via exosomes is critical for the progression of DN, and Dll4/Notch1 signaling may be a credible candidate modulator in those exosomes.
It has been suggested that tubular epithelial cell-derived exosomes (TEC-exosomes) exert a crucial role in the progression of kidney disease, including DN.42,43,44,45 AMBP and VDAC1, which are enriched in proximal tubular cells, are obviously differentially present in urinary exosomes from DN patients.46 HNRNPA1-mediated exosomal miR-483-5p of TECs promoted renal interstitial fibrosis in STZ-induced diabetic mice.43 TEC-exosomal miR-92a-1-5p accelerated kidney damage in db/db mice,44 and TEC-exosomes have potent profibrotic activity on renal fibroblasts in diabetic conditions.45 Notably, exosomes have been demonstrated to be an important mediator of communication between TECs and macrophages in the pathogenesis of tubulointerstitial inflammation.9,12,47,48 It was found that injection of TEC-derived exosomes enriched in miR-23a or CCL2 mRNA could induce macrophage accumulation in mouse kidneys in vivo,9,12 while inhibition of miR-23a in TEC-derived exosomes decreased kidney macrophage activation under hypoxic condition.9 More importantly, Jiang et al. found that TEC-to-macrophage communication forms a feedback loop via exosome-LRG1 and TRAIL transfer under DN.49 In this study, increased Dll4 and N1ICD were observed in the renal tubulointerstitium of both DN patients and db/db mice compared with controls (Figures 2 and 3), revealing the importance of Dll4/Notch1 signaling for tubulointerstitial injury as previously reported50,51,52 and further indicating the possibility that tubule cells secreted Dll4-enriched exosomes. In addition, we found that the expression level of Epsin1 was closely related to Dll4/Notch1 signaling and tubulointerstitial injury in DN in vivo (Figures 2 and 3). In our study, injection of TEC-derived exosomes with Epn1-KD obviously decreased the F4/80 intensity of mouse kidneys, as shown by immunofluorescence and immunohistochemistry staining, as well as tubulointerstitial inflammation and activation of Dll4/Notch1 (Figure 4), which was similar to the finding that TEC-derived exosomes with inhibition of miR-19b-3p reversed renal inflammation in an adriamycin-induced chronic proteinuric kidney disease model.48 These data indicate that Epsin1 is a key modulator of tubulointerstitial inflammation in DN through regulation of exosome secretion from tubule cells, but the precise mechanism is unclear.
Epsin1, an endocytic adaptor protein, is vital for exosome cargo internalization and excretion.29,30,31,53 The ubiquitin interacting motif (UIM) domain of Epsin1 has high affinity for ubiquitinated proteins, while the ENTH domain binds to PtdIns(4,5) P2 α on the cell membrane and therefore facilitates membrane bending and endocytosis.29,31,54 Recent research has shown that Epsin can bind to Toll-like receptors (TLRs) 2 and 4 and potentiate inflammatory signaling via its UIM domain.55 Most importantly, studies have stressed that Epsin is essential for the Delta/Serrate/Lag2 (DSL) ligands-dependent Notch activation. Epsin-mediated endocytosis in signal-sending cells applies force across the ligand-receptor bridge to initiate Notch1 cleavage.33,54 Taken together, these results suggest that Epsins may participate in tubulointerstitial inflammation in DN by regulating exosomal Dll4.
It was suggested that the Notch pathway was activated in macrophages under HG conditions,56 and Dll4 activated Notch1ICD in macrophages via the canonical Notch signaling pathway.57 Beyond cell-cell contact and the canonical pathway, a novel activation of Notch signaling by Dll4 incorporation into exosomes and transfer from cell to cell at a distance was found.25,58,59 Dll4-exosomes can travel through the 3D collagen matrix and increase HEY1 and HES1 mRNA expression in distant tip cells.58 In addition, Dll4-enriched exosomes derived from tumor cells activated Notch1 in the endothelium.25 In our study, we found that Dll4 enriched in HG-TEC-exosomes, which were captured by THP-1 cells, promoted the polarization of macrophages to the M1 phenotype and increased inflammatory cytokine secretion (Figure 5). Epsin1 inhibition in TECs reduced the abundance of Dll4 in TEC-derived exosomes (Figure 6). However, whether Epsin1 modulates macrophage activation via exosomal Dll4 requires elucidation.
For further investigation, a Dll4-overexpressing TEC cell line was established to obtain Dll4-enriched exosomes. Similar to macrophages receiving HG-TEC-exosomes, macrophages receiving Dll4-overexpressing exosomes highly expressed iNOS and N1ICD. However, in the absence of Epsin1, these exosomes failed to promote macrophage activation (Figure 7). These data indicate that Epsin1 modulates tubulointerstitial inflammation via regulation of exosomal-Dll4 release from TECs, which then activates Notch1 signaling in macrophages under DN conditions. However, further investigation is needed to determine whether knocking down Epsin1 in TECs changes the expression of other functional molecules in exosomes that might modulate the inflammatory response.
In conclusion, in this study, we discovered new communication between tubular cells and macrophages via exosomal Dll4 transfer. Epsin1 is the key modulator of this noncanonical Dll4/Notch1 signal activation under DN conditions. Our study provides new insight into inflammation and potential therapeutic targets for DN.
Materials and methods
Human urinary exosome samples
This study was approved by the Ethics Committee of Second Xiangya Hospital, Central South University. Two cohorts of patients were enrolled in the study. A group of three patients with biopsy-proven DN and three age- and sex-matched healthy controls were recruited for urinary exosome proteomic analysis. In the validation cohort, six healthy controls and 12 patients with different stages of DN were enrolled. All patients provided informed consent. A whole-stream, early morning urine specimen was collected from patients and healthy volunteers. Urine samples were processed as previously reported12 by centrifugation at 2000 × g for 20 min within 2 h after collection and stored at −80°C before exosome purification. The supernatant was subsequently subjected to differential centrifugation steps at 13,500 × g for 20 min and 200,000 × g for 2 h, followed by repeated ultracentrifugation at 200,000 × g for 2 h after dilution with sterile PBS.
Human kidney biopsies
Human kidney biopsy sections were obtained from 10 type 2 DN patients who underwent renal biopsy at the Second Xiangya Hospital and signed an official agreement for this donation. An equal number of patients with glomerular minor lesions were used as controls. The Ethics Committee of Second Xiangya Hospital approved the study.
Animals
All animal studies were performed in accordance with the protocols approved by the Ethics Committee of Central South University. Diabetic male db/db mice (week-matched db/m mice as controls) and C57BL mice were purchased from the Aier Matt Experimental Animal Company (Suzhou, China). The animals were housed in a 12-h/12-h light/dark cycle at 22°C and were fed standard laboratory food and water. Diabetic db/db and db/m mice were killed, and their kidneys were harvested at 8 weeks of age. C57BL mice randomly received intrarenal injections of different doses and types of exosomes at 8 weeks of age. Blood samples and kidney tissues were collected. The details are described below.
Exosome-injected models
A total of 50 μg DiO-labeled exosomes were injected intrarenally into 8-week-old male C57BL mice to assess the tissue distribution of exosomes in vivo. Briefly, animals were anesthetized, and the inferior pole of the left kidney was exposed. Exosomes (50 μg in 60 μL of phosphate-buffered saline) were injected into two sites via a 50-G needle. At 24 h after injection, mice were euthanized. Renal tissue was excised and imaged. To specifically examine the role of TEC-exosomes in kidney injury, two doses of exosomes (10 μg and 50 μg) were injected, and renal tissues were harvested on days 1 and 3 thereafter. Then, 18 C57BL/6 mice were randomly divided into three groups: Ctrl, HG, and HG-Ep1 KD, n = 6. These mice were renally injected with 50 μg TEC-exosomes of NG treatment, HG treatment, and HG + Epsin1 KD treatment. Serum samples and kidney tissues were collected on day 3. The serum creatinine and BUN concentrations were detected with an assay kit (Jiancheng Bioengineering, China).
Transmission electron microscope
Urine or collected cell culture medium clusters were immersed in a fixative containing 2.5% glutaraldehyde and 4% paraformaldehyde in 0.1 M phosphate buffer. Sample handling and detection were performed by an electron microscopic core lab at Xiangya Hospital (H-7650, Hitachi).
Nanoparticle tracking analysis
NTA was performed using the ZetaView PMX 110 (Particle Metrix, Meerbusch, Germany). Isolated exosome samples were appropriately diluted using 1x PBS buffer to measure the particle size and concentration. Measurement data from ZetaView were analyzed using the corresponding software, ZetaView 8.04.02. Resuspension volumes and dilution factors were used to convert the yield from concentration to an absolute number of particles.
Proteomic analysis
Mass spectrometry analysis of urinary exosomes from DN patients and normal controls was performed at Jingjie PTM BioLab using 200 μg of total protein. Bioinformatic analysis of significantly differentially expressed proteins and their functional characteristics was performed.
LC-MS analysis
Briefly, exosomes were lysed to measure concentration and then digested to peptides with trypsin. After digestion, the samples were vacuum-dried, dissolved in 0.5 M TEAB and finally labeled with a TMT assay kit. The resulting tryptic peptide mixtures were subjected to LC-MS for protein identification. The peptides were dissolved in solution A (A = 0.1% formic acid, 2% acetonitrile) and injected onto a nanocolumn (EASY-nLC 1000) in a continuous acetonitrile gradient consisting of 0–20 min, 8%–23% B; 20–33 min, 23%–35% B; 33–37 min, 35%–80% B; and 37–40 min, 80% B (B = 0.1% formic acid, 90% acetonitrile). A flow rate of 300 nL/min was used to elute peptides from the nanocolumn to an emitter nanospray ionization and peptide fragmentation on a Q Exactive Plus mass spectrometer. The data-dependent scan (DDA) program was used in the way that most intense 20 parent ions were analyzed during the chromatographic run. Dynamic exclusion was set at 30 s. Secondary mass spectrometry data were retrieved using MaxQuant (v1.5.2.8). Search engines were set up to search the human_swissprot_9606 (20387 entries) database. An error of 20 ppm or 5 ppm was allowed for the first search or main search of parent ions, respectively; 0.02 Da was set for the search of secondary fragment ions. Alkylation in cysteine was selected as a fixed modification, and oxidation in methionine, acetylation, and deamidation in the N-terminus of the protein were selected as variable modifications. Quantitative comparisons are based on TMT-6plex, and the false discovery rate of protein identification and PSM identification was set to 1%.
Gene ontology biological process annotation
Protein gene ontology (GO) annotation information mainly came from the UniProt database-GOA (https://www.ebi.ac.uk/GOA/). The protein ID was first converted to the UniProtKB database ID, and then the appropriate GO annotation information was found. If there were some identified proteins that were not annotated in the database, InterProScan was used. Finally, according to GO annotation, biological process could be one of the classifications of each protein.
KEGG pathway enrichment analysis
Annotation for protein metabolic pathways was obtained from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. KEGG pathway enrichment analysis was performed using Fisher’s exact test.
Cell culture and treatment
HK-2 and THP-1 monocyte cell lines were both purchased from American Type Culture Collection (ATCC). The HK-2 cell line was cultured in Dulbecco’s modified Eagle’s medium (DMEM)/F12 (Gibco) supplemented with 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin-streptomycin (Invitrogen). The THP-1 cell line was cultured in RPMI 1640 (Gibco) supplemented with 10% FBS (Gibco) and 1% penicillin-streptomycin (Invitrogen). For all experiments, THP-1 monocytes were differentiated into macrophages using phorbol 12-myristate 13-acetate (PMA, 100 ng/mL) for 24 h. HK-2 cells were treated with HG (= 30 mM), NG (= 5.5 mM) as a control. THP-1 cells were incubated with exosomes derived from the HK-2 cell line and sublines.
Transient transfection
Epsin1 siRNA (-CCATGACGCTGATGGAGTA-) and its negative control (NC) were purchased from RiboBio (Guangzhou, China). HK-2 cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Establishment of HK-2 sublines
The lentiviruses pLV-shEPSIN1-eGFP and pLV-hDLL4-eGFP were used to stably knock down Epsin1 and overexpress Dll4, respectively. We established three sublines: an Epsin1-KD subline that stably knocked down Epsin1, a Dll4-OE subline that stably overexpressed Dll4, and a Dll4-OE and Epsin1-KD subline that both stably overexpressed Dll4 and knocked down Epsin1. All sublines were selected by puromycin (2 μg/mL) until the eGFP-labeled cells reached 100%. Every mixture cell cloning was validated before the following functional experiments. Lentiviruses were purchased from GeneChem (Shanghai, China) with their matched control lines. The concentration of puromycin at 2 μg/mL was determined by pretest study as the minimum concentration that could enable whole lentivirus-transfected cells to die within 48 h.
Purification of HK-2-exosomes
The HK-2 cell line and stable sublines established as mentioned above were cultured. After starving overnight followed by 24 h of NG or HG stimulation without serum, the resulting supernatants were subsequently subjected to differential centrifugation steps as previously described60,61 at 300 × g for 10 min, 2,000 × g for 20 min, and 10,000 × g for 30 min, concentrated using a 100-kDa molecular weight cutoff (MWCO) (Millipore) at 4,000 × g for 20 min and finally ultracentrifuged at 120,000 × g for 2 h (Type SW 40 Ti rotor, Beckman Coulter Optima XPN-100 Ultracentrifuge). The pellet was washed with PBS one time at 120,000 × g for 2 h and resuspended in sterile PBS or culture medium for the following experiments.
Exosome uptake studies
HK-2 cells were grown to 70%–80% confluence and cultured in serum free medium overnight. Then the cells were stained with DiO dye (5 μg/mL) for 30 min at 37°C and the free dye was washed away with PBS three times. HK-2 cells were then stimulated with or without HG for 24 h. Exosomes released from DiO-stained HK-2 cells were collected and purified by ultracentrifugation as described above. Exosomes released from 1 × 107 HK-2 cells were added to THP-1 macrophages for 24 h. Exosomes purified from HK-2 cells with PBS and DiO dye were used as a control to verify that no dye contamination occurred.
Transwell study
Transwells with 0.4-μm and 8-μm pores (Corning) were purchased to demonstrate the process of HK-2-exo communicating with THP-1 macrophages in different states. The HK-2 cell line and sublines were seeded in the upper chamber of the Transwell with 0.4-μm pores. After starvation overnight with FBS-free culture medium, the medium was replaced by NG or HG for 24 h, and THP-1 macrophages were washed with fresh RPMI 1640. After 24 h of coculture, THP-1 cells were collected to conduct the following experiments. For the chemotaxis test, THP-1 macrophages were cultured in the upper chamber of transwells (8 μm) and HK-2 cells were cultured in the lower chamber.
Flow cytometry analysis
THP-1 macrophages incubated with exosomes after 24 h were harvested for flow cytometry. Cells were stained with antibodies against CD11b FITC (11-0118-41; Thermo Fisher), CD86 PE (12-0869-42; Thermo Fisher), and CD206 APC (17-2069-41; Thermo Fisher), and data were analyzed with FlowJo software. For the detection of macrophage phenotypes in vitro, THP-1 macrophages were stimulated with LPS (100 ng/mL) and used as a positive control. Isotype control antibodies were used as negative controls.
Immunofluorescence staining
Formaldehyde-fixed cells or kidney sections were incubated with primary antibodies against mouse anti-CD68 (Abcam), rabbit anti-Dll4 (Abcam), activated-Notch1 (Abcam), Epsin1 (Abcam), and Rat anti-F4/80 (Abcam), followed by incubation with secondary antibodies and Lotus tetragonolobus lectin (LTL) (Invitrogen). Cell nuclei were stained with DAPI. Exosomes were labeled with DiO. Immunostained samples were visualized under a confocal microscope (FV1000, Olympus).
Morphological analysis and immunohistochemistry assay
Kidney tissues were fixed in 4% paraformaldehyde (Servicebio, Wuhan, China), sliced into 4-μm-thick paraffin-embedded sections and then stained with H&E and PAS. For immunohistochemistry staining, formalin-fixed and paraffin-embedded tissue sections were incubated with primary antibodies against Dll4, activated-Notch1, Epsin1, Clathrin, CD68, F4/80, iNOS, interleukin (IL)-6, and TNF-α (all from Abcam, Cambridge, MA) and then horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h at room temperature according to the manufacturer’s protocol. Diaminobenzidine (DAB) (Maixin) was used as an HRP-specific substrate.
Western blotting
The protein lysates from cells, exosomes, or kidney tissues were prepared following standard protocols, and the protein concentration was determined using a BCA protein assay kit(cwbiotech). Then, the protein samples (20 μg cell or tissue lysates, 5 μg exosome lysates) were separated by Bis-Tris gel electrophoresis (Invitrogen) and transferred onto polyvinylidene difluoride membranes (PVDF, Millipore) using a wet-transfer system. Membranes were blocked in 5% BSA in PBS-T (tris-buffered saline –Tween 20) for 1 h at room temperature and were incubated with primary antibodies overnight at 4°C. Then, the membranes were washed and incubated with secondary HRP-conjugated antibodies for 1 h at room temperature, and the signals were detected using an enhanced chemiluminescence advanced system (GE Healthcare). Intensity values expressed as the relative protein expression were normalized to β-actin, GAPDH, or Alix. The primary antibodies used were rabbit anti-activated Notch1 (Abcam), Dll4 (Abcam), Epsin1 (Abcam), Alix (Proteintech), calnexin (Proteintech), CD9 (Abcam), CD81 (Abcam), β-actin (Proteintech), GAPDH (Proteintech), iNOS (Proteintech), TNF-α (Abcam), and IL-6 (Proteintech). The secondary HRP-conjugated antibody used was anti-rabbit IgG (Abcam).
RNA isolation and quantitative real-time PCR
Total RNA was extracted from mouse kidney tissue using the TRIzol RNA isolation system. First-strand cDNA was synthesized using a PrimeScript RT reagent kit (Takara). Real-time PCR was performed using SYBR GreenER qPCR SuperMix (label Fisher Scientific, Waltham, MA, USA) on a Light Cycler 96 System (Roche, Roche, Switzerland). The sequences of the primer pairs were as follows: HES1 forward, GATAGCTCCCGGCATTCCAAG; HES1 rerverse, GCGCGGTATTTCCCCAACA; iNOS forward, GGAGTGACGGCAAACATGACT; iNOS reverse, TCGATGCACAACTGGGTGAAC; IL-6 forward, CTGCAAGAGACTTCCATCCAG; IL-6 reverse, AGTGGTATAGACAGGTCTGTTGG.
Statistical analysis
Data are expressed as the means ± SDs. Statistical analysis was performed using a t test or one-way ANOVA. p < 0.05 was considered statistically significant.
Data availability statement
No publicly available data or shared data are cited. All data needed to evaluate the conclusion of the current study are present in the paper and/or the supplementary materials. Additional data are available from the corresponding author on request.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (no.82170744 and 82100734), Hunan Provincial Natural Science Foundation of China (no.2021JJ40824), and the Fundamental Research Funds for the Central Universities of Central South University (no.2020zzts286).
Author contributions
J.-L.L. and L.X. designed the experiments, performed the data collection and analysis, and wrote the manuscript. L.Z. and Y.H. assisted with data analysis and manuscript writing. J.-L.L., X.-H.L., and Y.-F.L. carried out the animal experiments. J.-L.L., S.-M.Z., and Y.-E.Z. were responsible for the in vitro study. Y.L., X.-J.C., and L.-Y.H. provided the pathology analysis. Z.D., F.-Y.L., and L.S. advised on the experimental design and manuscript writing.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2023.03.027.
Supplemental information
References
- 1.Zhang L., Long J., Jiang W., Shi Y., He X., Zhou Z., Li Y., Yeung R.O., Wang J., Matsushita K., et al. Trends in chronic kidney disease in China. N. Engl. J. Med. 2016;375:905–906. doi: 10.1056/NEJMc1602469. [DOI] [PubMed] [Google Scholar]
- 2.Lin Y.C., Chang Y.H., Yang S.Y., Wu K.D., Chu T.S. Update of pathophysiology and management of diabetic kidney disease. J. Formos. Med. Assoc. 2018;117:662–675. doi: 10.1016/j.jfma.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Tang S.C.W., Yiu W.H. Innate immunity in diabetic kidney disease. Nat. Rev. Nephrol. 2020;16:206–222. doi: 10.1038/s41581-019-0234-4. [DOI] [PubMed] [Google Scholar]
- 4.Zhang S., Wang H., Liu Y., Yang W., Liu J., Han Y., Liu Y., Liu F., Sun L., Xiao L. Tacrolimus ameliorates tubulointerstitial inflammation in diabetic nephropathy via inhibiting the NFATc1/TRPC6 pathway. J. Cell. Mol. Med. 2020;24:9810–9824. doi: 10.1111/jcmm.15562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang W.X., Liu Y., Zhang S.M., Wang H.F., Liu Y.F., Liu J.L., Li X.H., Zeng M.R., Han Y.Z., Liu F.Y., et al. Epac activation ameliorates tubulointerstitial inflammation in diabetic nephropathy. Acta Pharmacol. Sin. 2022;43:659–671. doi: 10.1038/s41401-021-00689-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu B.C., Tang T.T., Lv L.L., Lan H.Y. Renal tubule injury: a driving force toward chronic kidney disease. Kidney Int. 2018;93:568–579. doi: 10.1016/j.kint.2017.09.033. [DOI] [PubMed] [Google Scholar]
- 7.Xu L., Sharkey D., Cantley L.G. Tubular GM-CSF promotes late MCP-1/CCR2-mediated fibrosis and inflammation after ischemia/reperfusion injury. J. Am. Soc. Nephrol. 2019;30:1825–1840. doi: 10.1681/ASN.2019010068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sen P., Helmke A., Liao C.M., Sörensen-Zender I., Rong S., Bräsen J.H., Melk A., Haller H., von Vietinghoff S., Schmitt R. SerpinB2 regulates immune response in kidney injury and aging. J. Am. Soc. Nephrol. 2020;31:983–995. doi: 10.1681/ASN.2019101085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z.L., Lv L.L., Tang T.T., Wang B., Feng Y., Zhou L.T., Cao J.Y., Tang R.N., Wu M., Liu H., et al. HIF-1alpha inducing exosomal microRNA-23a expression mediates the cross-talk between tubular epithelial cells and macrophages in tubulointerstitial inflammation. Kidney Int. 2019;95:388–404. doi: 10.1016/j.kint.2018.09.013. [DOI] [PubMed] [Google Scholar]
- 10.Erdbrügger U., Le T.H. Extracellular vesicles in renal diseases: more than novel biomarkers? J. Am. Soc. Nephrol. 2016;27:12–26. doi: 10.1681/ASN.2015010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karpman D., Ståhl A.L., Arvidsson I. Extracellular vesicles in renal disease. Nat. Rev. Nephrol. 2017;13:545–562. doi: 10.1038/nrneph.2017.98. [DOI] [PubMed] [Google Scholar]
- 12.Lv L.L., Feng Y., Wen Y., Wu W.J., Ni H.F., Li Z.L., Zhou L.T., Wang B., Zhang J.D., Crowley S.D., Liu B.C. Exosomal CCL2 from tubular epithelial cells is critical for albumin-induced tubulointerstitial inflammation. J. Am. Soc. Nephrol. 2018;29:919–935. doi: 10.1681/ASN.2017050523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu W., Zeng H., Chen J., Fu S., Huang Q., Xu Y., Xu A., Lan H.Y., Tang Y. miR-20a-5p is enriched in hypoxia-derived tubular exosomes and protects against acute tubular injury. Clin. Sci. 2020;134:2223–2234. doi: 10.1042/CS20200288. [DOI] [PubMed] [Google Scholar]
- 14.Shang Y., Smith S., Hu X. Role of Notch signaling in regulating innate immunity and inflammation in health and disease. Protein Cell. 2016;7:159–174. doi: 10.1007/s13238-016-0250-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keewan E., Naser S.A. The role of notch signaling in macrophages during inflammation and infection: implication in rheumatoid arthritis? Cells. 2020;9 doi: 10.3390/cells9010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei K., Korsunsky I., Marshall J.L., Gao A., Watts G.F.M., Major T., Croft A.P., Watts J., Blazar P.E., Lange J.K., Thornhill T.S., Filer A., Raza K., Donlin L.T., Accelerating Medicines Partnership Rheumatoid Arthritis & Systemic Lupus Erythematosus AMP RA/SLE Consortium. Siebel C.W., Buckley C.D., Raychaudhuri S., Brenner M.B. Notch signalling drives synovial fibroblast identity and arthritis pathology. Nature. 2020;582:259–264. doi: 10.1038/s41586-020-2222-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahn S.H., Susztak K. Getting a notch closer to understanding diabetic kidney disease. Diabetes. 2010;59:1865–1867. doi: 10.2337/db10-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin C.L., Wang F.S., Hsu Y.C., Chen C.N., Tseng M.J., Saleem M.A., Chang P.J., Wang J.Y. Modulation of notch-1 signaling alleviates vascular endothelial growth factor-mediated diabetic nephropathy. Diabetes. 2010;59:1915–1925. doi: 10.2337/db09-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonegio R., Susztak K. Notch signaling in diabetic nephropathy. Exp. Cell Res. 2012;318:986–992. doi: 10.1016/j.yexcr.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu M., Liang K., Zhen J., Zhou M., Wang X., Wang Z., Wei X., Zhang Y., Sun Y., Zhou Z., et al. Sirt6 deficiency exacerbates podocyte injury and proteinuria through targeting Notch signaling. Nat. Commun. 2017;8:413. doi: 10.1038/s41467-017-00498-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma T., Li X., Zhu Y., Yu S., Liu T., Zhang X., Chen D., Du S., Chen T., Chen S., et al. Excessive activation of notch signaling in macrophages promote kidney inflammation, fibrosis, and necroptosis. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.835879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edeling M., Ragi G., Huang S., Pavenstädt H., Susztak K. Developmental signalling pathways in renal fibrosis: the roles of Notch, Wnt and Hedgehog. Nat. Rev. Nephrol. 2016;12:426–439. doi: 10.1038/nrneph.2016.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bielesz B., Sirin Y., Si H., Niranjan T., Gruenwald A., Ahn S., Kato H., Pullman J., Gessler M., Haase V.H., Susztak K. Epithelial Notch signaling regulates interstitial fibrosis development in the kidneys of mice and humans. J. Clin. Invest. 2010;120:4040–4054. doi: 10.1172/JCI43025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai E.C. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- 25.Sheldon H., Heikamp E., Turley H., Dragovic R., Thomas P., Oon C.E., Leek R., Edelmann M., Kessler B., Sainson R.C.A., et al. New mechanism for Notch signaling to endothelium at a distance by Delta-like 4 incorporation into exosomes. Blood. 2010;116:2385–2394. doi: 10.1182/blood-2009-08-239228. [DOI] [PubMed] [Google Scholar]
- 26.Dong Y., Lee Y., Cui K., He M., Wang B., Bhattacharjee S., Zhu B., Yago T., Zhang K., Deng L., et al. Epsin-mediated degradation of IP3R1 fuels atherosclerosis. Nat. Commun. 2020;11:3984. doi: 10.1038/s41467-020-17848-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu H., Rahman H.N.A., Dong Y., Liu X., Lee Y., Wen A., To K.H., Xiao L., Birsner A.E., Bazinet L., et al. Epsin deficiency promotes lymphangiogenesis through regulation of VEGFR3 degradation in diabetes. J. Clin. Invest. 2018;128:4025–4043. doi: 10.1172/JCI96063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong Y., Wang B., Cui K., Cai X., Bhattacharjee S., Wong S., Cowan D.B., Chen H. Epsins negatively regulate aortic endothelial cell function by augmenting inflammatory signaling. Cells. 2021;10:e81918. doi: 10.3390/cells10081918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Alai M.M., Heidemann J., Skruzny M., Gieras A., Mertens H.D.T., Svergun D.I., Kaksonen M., Uetrecht C., Meijers R. Epsin and Sla2 form assemblies through phospholipid interfaces. Nat. Commun. 2018;9:328. doi: 10.1038/s41467-017-02443-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gleisner M., Kroppen B., Fricke C., Teske N., Kliesch T.T., Janshoff A., Meinecke M., Steinem C. Epsin N-terminal homology domain (ENTH) activity as a function of membrane tension. J. Biol. Chem. 2016;291:19953–19961. doi: 10.1074/jbc.M116.731612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stachowiak J.C., Schmid E.M., Ryan C.J., Ann H.S., Sasaki D.Y., Sherman M.B., Geissler P.L., Fletcher D.A., Hayden C.C. Membrane bending by protein-protein crowding. Nat. Cell Biol. 2012;14:944–949. doi: 10.1038/ncb2561. [DOI] [PubMed] [Google Scholar]
- 32.Szymanska M., Fosdahl A.M., Raiborg C., Dietrich M., Liestøl K., Stang E., Bertelsen V. Interaction with epsin 1 regulates the constitutive clathrin-dependent internalization of ErbB3. Biochim. Biophys. Acta. 2016;1863:1179–1188. doi: 10.1016/j.bbamcr.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 33.Langridge P.D., Struhl G. Epsin-dependent ligand endocytosis activates notch by force. Cell. 2017;171:1383–1396.e12. doi: 10.1016/j.cell.2017.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J., Zhang Q., Liu D., Liu Z. Exosomes: advances, development and potential therapeutic strategies in diabetic nephropathy. Metabolism. 2021;122 doi: 10.1016/j.metabol.2021.154834. [DOI] [PubMed] [Google Scholar]
- 35.Lu Y., Liu D., Feng Q., Liu Z. Diabetic nephropathy: perspective on extracellular vesicles. Front. Immunol. 2020;11:943. doi: 10.3389/fimmu.2020.00943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pisitkun T., Shen R.F., Knepper M.A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. USA. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Musante L., Tataruch D.E., Holthofer H. Use and isolation of urinary exosomes as biomarkers for diabetic nephropathy. Front. Endocrinol. 2014;5:149. doi: 10.3389/fendo.2014.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sinha N., Kumar V., Puri V., Nada R., Rastogi A., Jha V., Puri S. Urinary exosomes: potential biomarkers for diabetic nephropathy. Nephrology (Carlton) 2020;25:881–887. doi: 10.1111/nep.13720. [DOI] [PubMed] [Google Scholar]
- 39.Lee W.C., Li L.C., Ng H.Y., Lin P.T., Chiou T.T.Y., Kuo W.H., Lee C.T. Urinary exosomal MicroRNA signatures in nephrotic, biopsy-proven diabetic nephropathy. J. Clin. Med. 2020;9 doi: 10.3390/jcm9041220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng Y., Zhong X., Ni H.F., Wang C., Tang T.T., Wang L.T., Song K.Y., Tang R.N., Liu H., Liu B.C., Lv L.L. Urinary small extracellular vesicles derived CCL21 mRNA as biomarker linked with pathogenesis for diabetic nephropathy. J. Transl. Med. 2021;19:355. doi: 10.1186/s12967-021-03030-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng M., Wen J., Ma Z., Xiao L., Liu Y., Kwon S., Liu Y., Dong Z. FOXO1-Mediated downregulation of RAB27B leads to decreased exosome secretion in diabetic kidneys. Diabetes. 2021;70:1536–1548. doi: 10.2337/db20-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fakhredini F., Mansouri E., Mard S.A., Valizadeh Gorji A., Rashno M., Orazizadeh M. Effects of exosomes derived from kidney tubular cells on diabetic nephropathy in rats. Cell J. 2022;24:28–35. doi: 10.22074/cellj.2022.7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu D., Liu F., Li Z., Pan S., Xie J., Zhao Z., Liu Z., Zhang J., Liu Z. HNRNPA1-mediated exosomal sorting of miR-483-5p out of renal tubular epithelial cells promotes the progression of diabetic nephropathy-induced renal interstitial fibrosis. Cell Death Dis. 2021;12:255. doi: 10.1038/s41419-021-03460-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsai Y.C., Kuo M.C., Hung W.W., Wu P.H., Chang W.A., Wu L.Y., Lee S.C., Hsu Y.L. Proximal tubule-derived exosomes contribute to mesangial cell injury in diabetic nephropathy via miR-92a-1-5p transfer. Cell Commun. Signal. 2023;21:10. doi: 10.1186/s12964-022-00997-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wen J., Ma Z., Livingston M.J., Zhang W., Yuan Y., Guo C., Liu Y., Fu P., Dong Z. Decreased secretion and profibrotic activity of tubular exosomes in diabetic kidney disease. Am. J. Physiol. Ren. Physiol. 2020;319:F664–F673. doi: 10.1152/ajprenal.00292.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zubiri I., Posada-Ayala M., Sanz-Maroto A., Calvo E., Martin-Lorenzo M., Gonzalez-Calero L., de la Cuesta F., Lopez J.A., Fernandez-Fernandez B., Ortiz A., et al. Diabetic nephropathy induces changes in the proteome of human urinary exosomes as revealed by label-free comparative analysis. J. Proteomics. 2014;96:92–102. doi: 10.1016/j.jprot.2013.10.037. [DOI] [PubMed] [Google Scholar]
- 47.Ding C., Zheng J., Wang B., Li Y., Xiang H., Dou M., Qiao Y., Tian P., Ding X., Xue W. Exosomal MicroRNA-374b-5p from tubular epithelial cells promoted M1 macrophages activation and worsened renal ischemia/reperfusion injury. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.587693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lv L.L., Feng Y., Wu M., Wang B., Li Z.L., Zhong X., Wu W.J., Chen J., Ni H.F., Tang T.T., et al. Exosomal miRNA-19b-3p of tubular epithelial cells promotes M1 macrophage activation in kidney injury. Cell Death Differ. 2020;27:210–226. doi: 10.1038/s41418-019-0349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang W.J., Xu C.T., Du C.L., Dong J.H., Xu S.B., Hu B.F., Feng R., Zang D.D., Meng X.M., Huang C., et al. Tubular epithelial cell-to-macrophage communication forms a negative feedback loop via extracellular vesicle transfer to promote renal inflammation and apoptosis in diabetic nephropathy. Theranostics. 2022;12:324–339. doi: 10.7150/thno.63735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han F., Wang S., Chang Y., Li C., Yang J., Han Z., Chang B., Sun B., Chen L. Triptolide prevents extracellular matrix accumulation in experimental diabetic kidney disease by targeting microRNA-137/Notch1 pathway. J. Cell. Physiol. 2018;233:2225–2237. doi: 10.1002/jcp.26092. [DOI] [PubMed] [Google Scholar]
- 51.Soni H., Matthews A.T., Pallikkuth S., Gangaraju R., Adebiyi A. gamma-secretase inhibitor DAPT mitigates cisplatin-induced acute kidney injury by suppressing Notch1 signaling. J. Cell. Mol. Med. 2019;23:260–270. doi: 10.1111/jcmm.13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao H., Han Y., Jiang N., Li C., Yang M., Xiao Y., Wei L., Xiong X., Yang J., Tang C., et al. Effects of HIF-1alpha on renal fibrosis in cisplatin-induced chronic kidney disease. Clin. Sci. 2021;135:1273–1288. doi: 10.1042/CS20210061. [DOI] [PubMed] [Google Scholar]
- 53.Kajimoto T., Mohamed N.N.I., Badawy S.M.M., Matovelo S.A., Hirase M., Nakamura S., Yoshida D., Okada T., Ijuin T., Nakamura S.I. Involvement of Gbetagamma subunits of Gi protein coupled with S1P receptor on multivesicular endosomes in F-actin formation and cargo sorting into exosomes. J. Biol. Chem. 2018;293:245–253. doi: 10.1074/jbc.M117.808733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie X., Cho B., Fischer J.A. Drosophila Epsin's role in Notch ligand cells requires three Epsin protein functions: the lipid binding function of the ENTH domain, a single Ubiquitin interaction motif, and a subset of the C-terminal protein binding modules. Dev. Biol. 2012;363:399–412. doi: 10.1016/j.ydbio.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Webb A.M., Francis C.R., Judson R.J., Kincross H., Lundy K.M., Westhoff D.E., Meadows S.M., Kushner E.J. EHD2 modulates Dll4 endocytosis during blood vessel development. Microcirculation. 2021;29 doi: 10.1111/micc.12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu N., Zhang X., Zhang X., Guan Y., He R., Xue E., Zhang X., Deng W., Yu J., Wang W., Shi Q. Inhibition of Notch activity suppresses hyperglycemia-augmented polarization of macrophages to the M1 phenotype and alleviates acute pancreatitis. Clin. Sci. 2022;136:455–471. doi: 10.1042/CS20211031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fung E., Tang S.M.T., Canner J.P., Morishige K., Arboleda-Velasquez J.F., Cardoso A.A., Carlesso N., Aster J.C., Aikawa M. Delta-like 4 induces notch signaling in macrophages: implications for inflammation. Circulation. 2007;115:2948–2956. doi: 10.1161/CIRCULATIONAHA.106.675462. [DOI] [PubMed] [Google Scholar]
- 58.Sharghi-Namini S., Tan E., Ong L.L.S., Ge R., Asada H.H. Dll4-containing exosomes induce capillary sprout retraction in a 3D microenvironment. Sci. Rep. 2014;4:4031. doi: 10.1038/srep04031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marconi G.D., Diomede F., Trubiani O., Porcheri C., Mitsiadis T.A. Exosomes as carriers for notch molecules. Methods Mol. Biol. 2022;2472:197–208. doi: 10.1007/978-1-0716-2201-8_16. [DOI] [PubMed] [Google Scholar]
- 60.Liu X., Miao J., Wang C., Zhou S., Chen S., Ren Q., Hong X., Wang Y., Hou F.F., Zhou L., Liu Y. Tubule-derived exosomes play a central role in fibroblast activation and kidney fibrosis. Kidney Int. 2020;97:1181–1195. doi: 10.1016/j.kint.2019.11.026. [DOI] [PubMed] [Google Scholar]
- 61.Tang T.T., Wang B., Wu M., Li Z.L., Feng Y., Cao J.Y., Yin D., Liu H., Tang R.N., Crowley S.D., et al. Extracellular vesicle-encapsulated IL-10 as novel nanotherapeutics against ischemic AKI. Sci. Adv. 2020;6:eaaz0748. doi: 10.1126/sciadv.aaz0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No publicly available data or shared data are cited. All data needed to evaluate the conclusion of the current study are present in the paper and/or the supplementary materials. Additional data are available from the corresponding author on request.