Abstract
We have shown previously that Escherichia coli and Salmonella enterica serovar Typhimurium strains carrying a deletion of the uvrB-bio region are hypersensitive to the mutagenic and toxic action of 6-hydroxylaminopurine (HAP) and related base analogs. This sensitivity is not due to the uvrB excision repair defect associated with this deletion because a uvrB point mutation or a uvrA deficiency does not cause hypersensitivity. In the present work, we have investigated which gene(s) within the deleted region may be responsible for this effect. Using independent approaches, we isolated both a point mutation and a transposon insertion in the moeA gene, which is located in the region covered by the deletion, that conferred HAP sensitivity equal to that conferred by the uvrB-bio deletion. The moeAB operon provides one of a large number of genes responsible for biosynthesis of the molybdenum cofactor. Defects in other genes in the same pathway, such as moa or mod, also lead to the same HAP-hypersensitive phenotype. We propose that the molybdenum cofactor is required as a cofactor for an as yet unidentified enzyme (or enzymes) that acts to inactivate HAP and other related compounds.
The biological effects of many mutagenic agents are due to DNA base modifications, both in the DNA and the DNA precursor pool. A group of mutagens containing a preformed modified base, often referred to as base analogs (14), have received increasing attention recently. For example, 8-oxoguanine, in the form of 8-oxo-dGTP or 8-oxo-GTP, is a spontaneously arising guanine oxidation product that contributes substantially to the infidelity of DNA replication (13, 35, 37) or transcription (58). Specialized systems protecting the cell against 8-oxoguanine have been found in organisms from bacteria to humans (for reviews, see references 13 and 37), including the MutT enzyme, which is capable of hydrolyzing 8-oxo-dGTP, an activity referred to as pool sanitizing (2, 35). Other examples of mutagenic precursor pool contaminants are 5-hydroxy-dCTP (12) and 2-hydroxy-dATP (15), both oxidative stress products. The human MutT homolog hMTH1 has strong activity towards 2-hydroxy-dATP, suggesting that it, in addition to 8-oxo-GTP, could be an important threat if not actively removed (15). In addition, base analogs can be useful tools for probing the mechanisms of mutation avoidance during DNA replication, including base-base discrimination by DNA polymerases (51, 54).
An important group of base analogs are the N-hydroxy derivatives of adenine and cytidine (see reference 27 for a review). For example, 6-hydroxylaminopurine (N-6-hydroxyadenine) (HAP) and 2-amino-6-hydroxylaminopurine (AHAP) are very powerful mutagens in phage, bacteria, yeast, and eukaryotic cells (42, 43), and they have been termed universal mutagens (42). These adenine derivatives are active when provided as bases or, in some organisms, nucleosides, as they are apparently converted efficiently into the corresponding deoxynucleoside triphosphates (dNTPs), which are then incorporated into DNA by DNA polymerase. Because of the ambiguous base pairing properties of these dNTPs, their incorporation is highly mutagenic.
We have previously performed studies on the genetic requirements of HAP mutagenesis in the bacterium Escherichia coli for the purpose of understanding at which levels cells may try to prevent mutagenesis by this and related agents (43). We found little or no protection by the exonucleolytic proofreading (dnaQ gene) or the postreplicative mismatch repair system (encoded by the mutHLS genes), two systems that play important roles in preventing mutations resulting from the mispairings of normal bases (50). This lack of discrimination is likely one of the reasons for the strong mutagenic potential of HAP.
However, a strain carrying a deletion of the chromosomal uvrB-bio region had hypersensitivity to HAP for both mutagenesis and toxicity, implying the existence of a protective system. The deletion also conferred sensitivity to AHAP (43) and other analogs (31). Sensitivity of a uvrB-bio deletion strain for AHAP and related compounds has also been found in Salmonella enterica serovar Typhimurium (23–25). However, hypersensitivity is not conferred by the uvrB5 point mutation (43) or two different uvrA deficiencies (23, 24, 43). This argues against the uvrABC excision repair system being responsible for protection against HAP and related compounds. We concluded that certain genes in the uvrB-bio region, other than uvrB, were responsible for the observed sensitivity. In the present report, we investigate the nature of the gene or genes within this region of the E. coli chromosome that are responsible for this enhanced base analog sensitivity. The results point to an important role of the molybdenum cofactor, presumably through the action of a molybdenum cofactor-containing enzyme activity.
MATERIALS AND METHODS
Bacterial strains, phage stocks, and plasmids.
Table 1 lists many of the E. coli strains used in this study along with their genotypes. Δ(uvrB-bio) of strain NR10107 is bioΔ261 of strain C261 (5). Strains NR10835 and NR10836 were created by introducing the F′pro-lac from strains CC105 and CC106 (9) into KA796 by conjugation. For convenience, these F′s were designated F′CC105 and F′CC106, respectively. NR11958 was obtained by localized mutagenesis of the bio region as described below. NR12071 is identical to NR11958 but contains, in addition, transposon insertion zbi-29::Tn10. It was constructed by inserting zbi-3058::Tn10 from CAG12034 by P1 transduction into NR11958, followed by retransduction of the moeA120 zbi-3058::Tn10 duet into KA796 (∼45% linkage). Retention of the moeA (mut-1) allele was ascertained by checking individual transductants for HAP sensitivity using the spot test described below. NR12006 is a spontaneous streptomycin-resistant derivative of KA796. The derivation of NR12383 carrying a mini-Tn10cam insertion in moeA is described in detail below. Strains LCB382 and R876, JRG94, and JRG97 carrying established defects in the moaA, modC, and moeA genes, respectively, were obtained from the E. coli Genetic Stock Center (Yale University). The mol alleles were transferred by P1 transduction into KA796 (yielding NR13031, NR13035, NR13939, and NR13043; Table 1) by first linking them with up with a nearby Tn10 insertion and then transducing the mol-Tn10 combination into KA796. modC4 was transferred with nadA57::Tn10, moaA1 and moaA18 were transferred with zbi-29::Tn10, and moeA5 was transferred with zbh-3058::Tn10. In all cases, the presence of the mol allele was monitored based on the chlorate resistance it confers (see below). mol+ isolates from the same transduction were also saved (NR13032, NR12036, NR13040, and NR13044; Table 1). All strains were grown at 37°C. Kohara phages (30) were obtained from Y. Kohara (National Institute of Genetics, Mishima, Japan). Plasmid pMoeA1 is plasmid pBluescript KS(−) containing the E. coli moeA gene. It was constructed by isolating the 1.3-kb AvaI-StuI fragment containing the moeA gene (see Fig. 1B) from Kohara phage 207 (30) and inserting it, after 5′-end filling to generate blunt ends, into the HincII site of pBluescript KS(−).
TABLE 1.
E. coli strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| KA796 | ara thi Δ(pro-lac) | 52 |
| NR10107 | ara thi Δ(pro-lac) Δ(uvrB-bio) | 43 |
| NR10835 | ara thi Δ(pro-lac) F′CC105 | This work |
| NR10836 | ara thi Δ(pro-lac) F′CC106 | This work |
| NR11958 | ara thi Δ(pro-lac) moeA120(Am) | This work |
| NR12006 | ara thi Δ(pro-lac) Strr | This work |
| NR12071 | ara thi Δ(pro-lac) zbi-29::Tn10 moeA120(Am) | This work |
| NR12383 | ara thi Δ(pro-lac) moeA121::mini-Tn10cam F′CC106 | This work |
| CAG12147 | nadA57::Tn10 | 57 |
| CAG18493 | zbi-29::Tn10 | 57 |
| CAG12034 | zbi-3058::Tn10 | 57 |
| CAG18531 | zbh-3108::Tn10kan | 57 |
| LCB382 | moaA1 thr-1 leuB6 fhuA2 pro-33 lacY1 purE43 glnV44(AS) gal-6 hisG1(Fs) rfbD1 galP63 Δ(gltB-gltF)500 rpsL9 malT1(λr) xylA7 mtlA2 ΔargH1 thi-1 | 46 |
| JRG94 | modC4 leuB6 bioA2 rpsL129 thi-1 gal-31 | 59 |
| JRG97 | moeA5 glnV42(AS) gal-26 | E. coli Genetic Stock Center |
| R876 | moaA18 lac-3350 bioC18 IN(rrnD-rrnE)1 | 4 |
| NR13031 | ara thi Δ(pro-lac) nadA57::Tn10 modC4 | This work |
| NR13032 | ara thi Δ(pro-lac) nadA57::Tn10 | This work |
| NR13035 | ara thi Δ(pro-lac) zbi-29::Tn10 moaA1 | This work |
| NR13036 | ara thi Δ(pro-lac) zbi-29::Tn10 | This work |
| NR13039 | ara thi Δ(pro-lac) zbi-29::Tn10 moaA18 | This work |
| NR13040 | ara thi Δ(pro-lac) zbi-29::Tn10 | This work |
| NR13043 | ara thi Δ(pro-lac) zbi-3058::Tn10 moeA5 | This work |
| NR13044 | ara thi Δ(pro-lac) zbi-3058::Tn10 | This work |
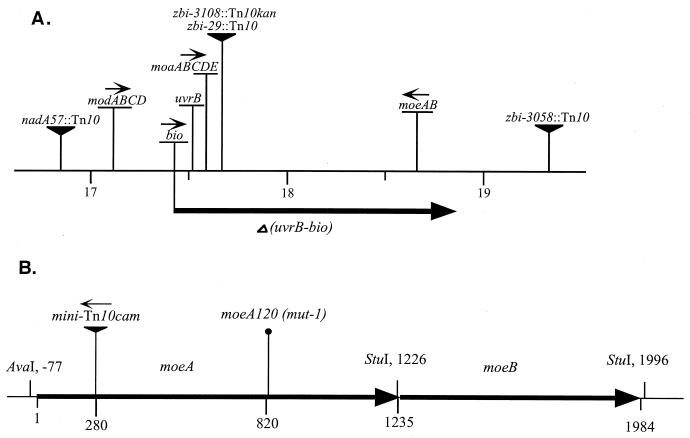
FIG. 1.
(A) The bio-uvrB region of the E. coli chromosome. The genes and transposon insertion sites relevant to this study are presented. The indicated extent of the uvrB-bio deletion is based on data from Cleary et al. (5). (B) Map of the moeAB operon. The locations of the moeA120(Am) and moeA121::mini-Tn10cam mutations are indicated. Plasmid pMoeA1 used for complementation assays contained the AvaI-StuI fragment (−77 to 1226) (see Materials and Methods).
Media.
Bacteria were cultivated on Luria broth (LB) (38) or minimal Vogel-Bonner media (60) supplemented, when necessary, with 20 μg of each amino acid or base/ml and 1 μg of the vitamins/ml. Minimal media contained 0.2% glucose as the carbon source. Solid media contained 15 g of agar/liter or, when indicated, agarose. For selection of antibiotic-resistant colonies, antibiotics were added at the following concentrations: rifampin, 100 μg/ml; ampicillin, 50 μg/ml; kanamycin, 50 μg/ml; tetracycline, 25 μg/ml; chloramphenicol, 20 μg/ml. For P1 phage titration and lysate preparation, LB was supplemented with 2 mM CaCl2. For the scoring of chlorate resistance, nutrient broth (Difco) medium containing 0.2% KClO3 was used (38). HAP was purchased from ICN Biochemicals.
Recombinational mapping using Kohara phages.
To map the mut-1 point mutation in the bio region responsible for HAP sensitivity, recombination experiments were performed with selected members of the Kohara lambda phage set (30). A saturated culture of strain NR11958 (mut-1; 0.2 ml) containing 10 mM MgSO4 was infected at a multiplicity of infection of 0.5 with lambda phage, incubated for 20 min at 37°C, spun down, resuspended in 1 ml of LB, and incubated for one additional hour at 37°C. The cells were precipitated, resuspended in a small volume, and plated in their entirety on a minimal medium plate containing 10 μg of HAP/ml. At this HAP concentration, wild-type strains plate normally, whereas HAP-sensitive strains such as NR10107 do not grow, yielding only a background of some 20 to 30 resistant colonies. Recombination of the chromosomal mut-1 gene with the corresponding wild-type gene on the lambda phage yielded >1,000 resistant colonies per plate.
Spot test for HAP sensitivity.
Stationary bacterial cultures grown in LB (109 cells/ml) were diluted 20-fold in 10 ml of 0.9% NaCl in sterile petri dishes and transferred using a multiprong replicator device to minimal-medium plates (approximately 0.1 ml per plate) (61). After the spots on the plate had dried, a sterile filter disk was placed in the center of the plate and a HAP solution (100 μg in dimethyl sulfoxide [DMSO]) or DMSO alone was spotted onto the filter. The plates were then incubated for 12 h at 37°C and inspected for a zone of inhibition around the disk.
Localized mutagenesis.
Localized mutagenesis of the bio region of the E. coli chromosome was used in an attempt to isolate HAP-sensitive point mutants. We used the hydroxylamine-mediated localized mutagenesis method of Hong and Ames (20) with slight modifications. A P1 lysate of strain KA796 (0.5 ml; 1010 phage per ml) was added to 4.5 ml of 0.4 M hydroxylamine in 30 mM K2HPO4–70 mM KH2PO4–10 mM MgSO4–1 mM EDTA, pH 6.0. After incubation for 15 h at 30°C, the phage particles were collected on a Millipore NMW 30,000 filter by centrifugation and washed with 0.5 ml of 5 mM CaCl2. This phage preparation was used to transduce strain NR10107 [Δ(uvrB-bio)] to Bio+ by selecting on minimal-agarose plates lacking biotin. Individual Bio+ colonies were restreaked on minimal plates with 10 μg of HAP/ml or without HAP to search for HAP-sensitive clones. One HAP-sensitive mutant was obtained (NR11958; Table 1).
Isolation of a HAP-hypermutable strain by random mini-Tn10cam insertion mutagenesis.
A random library of mini-Tn10cam insertions was obtained in strain NR10835 using phage λNK1324 as described by Kleckner et al. (29). A total of 6,000 chloramphenicol-resistant colonies were inoculated individually in wells of 96-well microtiter dishes (0.2 ml per well) containing LB with 0.50 μg of HAP/ml. After overnight growth, 10 μl from each well was spotted on an LB-rifampin plate. Under these conditions, a HAP-hypermutable strain such as NR10107 produces about 30 rifampin-resistant mutants per spot, whereas a control strain produces none. Among the 6,000 isolates, one displayed high mutability in the presence of HAP but no mutability in its absence. The responsible mini-Tn10cam insertion was transduced into NR12006 and Hfr mapping was performed using the tetracycline-resistant Hfr strain set as described by Singer et al. (57). This analysis placed the insertion site in the 10- to 20-min region of the E. coli chromosome. The allele was transduced into NR10836 generating NR12383 (Table 1). P1 transductional mapping in NR12383 using known Tn10 insertions in the 10- to 20-min region (57) revealed linkage with transposons zbh-29::Tn10 (9%) and zbi-3058::Tn10 (49%).
Test for chlorate sensitivity.
Approximately 103 cells were plated on nutrient broth plates containing 0.2% KClO3 (38). The plates were incubated under anaerobic conditions using a Becton Dickinson BBL gas pack anaerobic system for 12 h, after which they were incubated aerobically for an additional 6 to 10 h. Under these conditions, chlorate-sensitive strains do not form colonies, whereas chlorate-resistant strains plate with essentially 100% efficiency.
Mutant frequency determinations.
For each strain at each concentration of HAP, 12 or 25 independent 1-ml LB cultures were started from ca. 104 cells. The cultures were grown overnight with shaking at 37°C. Mutant frequencies were determined by plating 0.1 ml on an LB-rifampin plate (to obtain the number of rifampin-resistant cells per culture) and 0.1 ml of a 10−6 dilution on LB plates (to obtain the total number of cells per culture).
PCR amplification and DNA sequencing of the moeA gene.
PCR amplification of a 1,468-bp fragment of chromosomal DNA containing the moeA gene was performed with the following oligonucleotide primers: 5′-CCATAGTATTCGTCCATTA-3′ and 5′-ATCTCCTGATCGCTGAGTT-3′. Template DNA samples were prepared by simple boiling of bacterial cells. Reaction mixtures were from a Promega PCR kit. Thirty PCR cycles were performed according to the following sequence: 40 s at 94°C, 40 s at 51°C, and 3 min at 72°C. The PCR products were purified using a QIAquick PCR purification kit (Qiagen). DNA sequencing was performed on an ABI377 Prism automatic sequencer (Perkin-Elmer) using a manufacturer-supplied protocol. Primers were chosen based on the established sequence of the moeAB operon (40). The insertion point of the moeA::mini-Tn10cam insertion was determined by DNA sequencing using the following primers homologous to the transposon ends: 5′-TGTCTATTGCTGGTTTACCG-3′ and 5′-TGGCTTCTGTTTCTATCAGC-3′.
RESULTS
A previous study on the genetic factors controlling mutagenicity and sensitivity to the base analog HAP revealed that strains carrying a deletion of the uvrB-bio region were hypersensitive to this agent (43). This hypersensitivity was not due to the uvrB excision repair defect (43), and we therefore undertook a search for the identity of the responsible gene(s) within the deleted area. Due to its large size (about 2 min) and undefined right endpoint (5) (Fig. 1A), the deletion was not readily suited for mapping and identification of the gene. We therefore embarked on the isolation of additional HAP-sensitive mutant alleles that would be more suitable for mapping procedures. Below, we describe two independent approaches, localized mutagenesis of the bio region, yielding a chemically induced point mutant, and a genome-wide search for a HAP hypermutability mutant, yielding a mini-Tn10cam insertion.
HAP hypersensitivity due to a moeA point mutation.
A HAP-sensitive point mutant with a mutation in the uvrB-bio region was obtained by hydroxylamine-mediated localized mutagenesis of the bio region (see Materials and Methods). Testing of 50 Bio+ transductants of strain NR10107 [Δ(uvrB-bio)] for hypersensitivity to the toxic effects of HAP yielded one mutant, initially called the mut-1 strain, whose sensitivity was indistinguishable from that of the deletion strain (see Fig. 2). This mutant strain, NR11958 (mut-1), also proved to be hypermutable by HAP (Table 2). P1 transductional mapping using known transposon insertions in this region (57) placed the responsible gene at around 18.8 min between insertions zbh-3108::Tn10kan (or zbi-29::Tn10) and zbi-3058::Tn10, located, respectively, at 17.67 and 19.34 min of the E. coli map (39) (Fig. 1A).
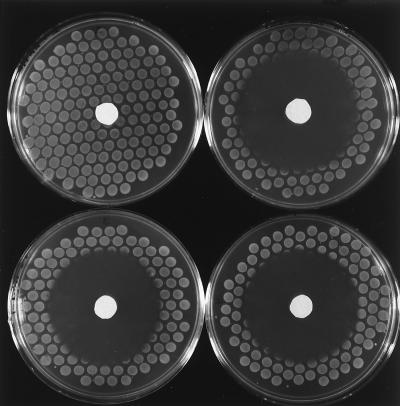
FIG. 2.
Spot tests for determination of HAP sensitivity. The test was performed as described in Materials and Methods. The strains used were KA796 (upper left), NR10107 [Δ(uvrB-bio)] (upper right), NR11958 (mut-1) (lower left), and NR12383 (moeA121::mini-Tn10cam) (lower right). See the text for the renaming of the mut-1 allele as moeA120(Am).
TABLE 2.
HAP mutability of wild-type and mutant strains
| Straina | Genotype | Avg mutant frequencyb (no. of Rifr mutants/108 cells) at a HAP concentration (μg/ml) of:
|
||
|---|---|---|---|---|
| 0 | 0.25 | 0.50 | ||
| KA796 | Wild type | 1.1 | 1.0 | 1.2 |
| NR10107 | Δ(uvrB-bio) | 0.4 | 13 | 120 |
| NR12071 | moeA120 (mut-1) | 1.8 | 16 | 710 |
| NR12383 | moeA121::mini-Tn10cam | 1.1 | 19 | 390 |
See Table 1.
Based on 12 independent cultures for each strain at each dose.
Further mapping of the mut-1 mutation was performed via recombination experiments (see Materials and Methods) with Kohara phages 206 through 211, which cover the region of the chromosome between 18.2 and 19.8 min (30, 49). This revealed that the HAP sensitivity of mut-1 could be overcome by recombination with phages 207 and 208, indicating that the putative HAP resistance gene was located on the overlap region of these two. This region has five open reading frames, the hypothetical genes ybiZ, ybiK, and yliA and the two known genes constituting the moeAB operon (18.6 min) (40, 49). The moe operon is one of five operons (moa, mob, mod, moe, and mog) responsible for molybdopterin (MPT) biosynthesis, an essential cofactor for several oxidoreductase enzymes (47). These genes, collectively called mol (53), were previously named chl (for chlorate resistance), referring to the ability of strains lacking the molybdenum cofactor to grow under anaerobic conditions in the presence of chlorate, conditions that cause the killing of wild-type E. coli due to the production of lethal chlorite (53). We found that both NR11958 (mut-1) and NR10107 [Δ(uvrB-bio)] were chlorate resistant (see Materials and Methods), whereas the parental KA796 was chlorate sensitive. This suggested that the mut-1 defect resided in moeAB.
The moeA gene was cloned from Kohara phage 207 on plasmid pMoeA1 (see Materials and Methods) and introduced into strain NR11958 (mut-1). The plasmid fully complemented the mut-1 defect because it made the strain HAP resistant and chlorate sensitive, consistent with the mut-1 mutation residing in the moeA gene. pMoeA1 did not affect the HAP sensitivity and chlorate resistance of deletion strain NR10107. As seen in Fig. 1A, the uvrB-bio deletion also comprises the moaABCD operon (as well as moeB). This observation suggests strongly that HAP resistance as observed in a wild-type strain requires the active molybdenum cofactor and argues against an individual activity of moeA towards HAP. DNA sequencing of the moeA gene of NR11958 revealed a TGG→TAG mutation creating an (amber) nonsense codon at position Trp274, resulting in a truncation of the MoeA protein (Fig. 1B). The mut-1 allele was then renamed moeA120(Am).
In addition, we tested the HAP sensitivity of established moeA mutant moeA5 (formerly called chlE5) (40). The mutation was transferred from strain JRG97 into KA796 (see Materials and Methods). This new strain was also HAP sensitive, in addition to its previously described chlorate resistance (40). Both the HAP sensitivity and chlorate resistance of this new strain were complemented by plasmid pMoeA1.
HAP hypersensitivity due to a mini-Tn10 insertion in moeA.
A HAP-sensitive mutant resulting from a mini-Tn10 insertion was sought as described in Materials and Methods. Six thousand random chromosomal mini-Tn10cam insertions were obtained, and the isolates were individually tested for hypermutability in the presence of HAP, scoring for high levels of rifampin-resistant mutants. One isolate (NR12383) whose spontaneous-mutant frequency was indistinguishable from that of the wild-type strain but that was hypermutable in the presence of HAP was obtained (Table 2). The mutant was also hypersensitive to killing by HAP and was chlorate resistant (Fig. 2). Mapping the insertion using Hfr crosses and P1 transductions placed the responsible insertion between the zbi-29 (zbi-3108) and zbi-3058 markers, as was the case for moeA120 (Fig. 1A). The HAP sensitivity was complemented by plasmid pMoeA1, corroborating that the insertion was in the moeA gene. The location of the insertion site in moeA was then determined by DNA sequencing (see Materials and Methods) and was found to be between nucleotides A280 and C281 (Fig. 1B). The allele was called moeA121::mini-Tn10cam. Thus, using two independent approaches, we obtained two HAP-hypersensitive strains, each carrying a mutation in the moeA gene.
Mutations in other mol genes also confer HAP sensitivity.
The moeAB operon is only one of several mol genes and operons involved in the biosynthesis and activation of the molybdenum cofactor (47). We investigated whether HAP sensitivity is also conferred by other deficiencies in the biosynthesis of active molybdenum cofactor, including those due to mutations in moa (17.6 min) and mod (17.1 min) (Fig. 1A). These were initially tested in their original backgrounds (as obtained from the E. coli Genetic Stock Center). This testing revealed significant variations in sensitivity even in the control strains for several of these strains, and we therefore transferred the alleles into our customary KA796 background (Table 1). The results in this background showed that the moaA1, moaA18, and modC4 alleles confer HAP hypersensitivity (Fig. 3) in addition to chlorate resistance (not shown). Thus, HAP sensitivity is due to the lack of an active molybdenum cofactor. Apparently, the molybdenum cofactor is required for some enzymatic activity protecting E. coli against the toxic and mutagenic effects of HAP.
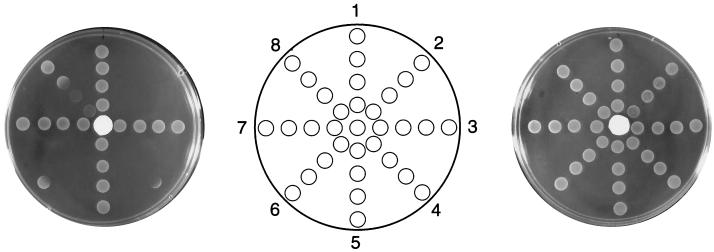
FIG. 3.
HAP sensitivities of various mol mutants defective in molybdenum cofactor synthesis. The central filter disc contained HAP (100 μg) (left plate) or no HAP (right plate). Suspensions of eight different strains were applied in a series of spots radiating out from the central disc. The strains, arranged in isogenic pairs, are NR13044 (moe+) (1), NR13043 (moeA5) (2), NR13036 (moa+) (3), NR13035 (moaA1) (4), NR13040 (moa+) (5), NR13039 (moaA18) (6), NR13032 (mod+) (7), and NR13031 (modC4) (8). See Table 1 and Materials and Methods for strain details.
DISCUSSION
In this study, we have shown that mutations in moeA, moaA, and modC lead to hypersensitivity to the base analog HAP. All these genes are involved in biosynthesis of the molybdopterin (MPT) guanine dinucleotide (MGD) cofactor, the essential molybdenum-containing cofactor for E. coli molybdoenzymes (47). Synthesis of MGD proceeds in a number of steps. The genes of the moa operon are responsible for the multistep synthesis of MPT, a dithiolene pterine derivative common to the molybdenum cofactor in all organisms. The mog gene is responsible for inserting activated molybdenum into MPT (26), and the mobAB operon is responsible for adding GMP to MPT to yield MGD (47). The mod operon encodes a high-affinity molybdenum importer (11). Among the moeAB products, MoeB is responsible for resulfurylation of MoaD, which provides the thiol groups on MPT. The function of moeA, highlighted here by two new HAP-sensitive, chlorate-resistant mutants, is not well known, but recent data have suggested a role in sulfurylation of molybdenum and generation of an activated form, possibly thiomolybdate (18). Thus, our results show that HAP hypersensitivity can result from defects in enzymes working at several stages of MGD biosynthesis. This argues that the molybdenum cofactor is required for base analog detoxification, most logically as a cofactor of a detoxifying enzyme.
At this time, the nature of the molybdenum-requiring enzyme responsible for HAP detoxification is unknown, as is the inactivating reaction. Molybdoenzymes comprise a broad, heterogeneous group, present in organisms from bacteria to humans, that assist in a variety of oxidation/reduction reactions (for a review, see reference 28). So far, nine E. coli molybdoenzymes have been described (see reference 16 for a review): DMSO reductase, TMAO (trimethylamine-N-oxide) reductase, biotin sulfoxide reductase, three nitrate reductases (NRA, NRZ, and NAP), and three formate dehydrogenases (FDH-N, FDH-H, and FDH-O). Most of these represent activities limited to anaerobic cells and controlled by the Fnr oxygen sensor (17, 56). They are therefore unlikely candidates for the HAP-inactivating activity deduced by our aerobic experiments. Nevertheless, we have considered the possibility that a low basal level of either DMSO reductase (dmsABC product) (62) or TMAO reductase (torCAD product) (36), both anaerobic activities, could be responsible for our observed effect. Both enzymes have been shown capable of reducing a wide range of S- and N-oxides, including adenosine-N1-oxide and hydroxylamine (16, 62, 64). However, defective dms or tor mutants tested by us did not show increased HAP sensitivity (data not shown). The three known aerobic E. coli molybdoenzymes are biotin sulfoxide reductase (BisC) (10, 44, 45), the minor respiratory nitrate reductase NRZ (8, 22), and the minor formate dehydrogenase FDH-O (8, 16). A defective bisC mutant (10) tested by us was not HAP sensitive (data not shown). Thus, although not all known E. coli molybdoenzymes have been tested, the activity responsible for HAP inactivation most likely represents an as yet unidentified aerobic molybdoenzyme.
Clement and Kunze (7) described the reduction of HAP to adenine by xanthine oxidase, a well-studied mammalian molybdoenzyme which oxidizes hypoxanthine and xanthine to uric acid (19). Apparently, HAP is a compound capable of serving as an electron acceptor for this reaction. No defined xanthine oxidase in E. coli has been described (but see reference 63). Interestingly, the sequencing of the E. coli genome revealed three open reading frames whose proteins have some homology to known xanthine oxidases (33). These proteins may be candidates for a putative HAP reductase activity. The alternative possibility of a HAP-oxidizing activity must be left open as well.
The question arises why E. coli (and Salmonella) contain an activity to inactivate HAP. The activity is strong, at least based on a comparison of HAP-induced mutant frequencies in wild-type and molybdenum cofactor-defective strains. This difference is at least 100- to 1,000-fold (24, 43) (Table 2), suggesting that the activity is capable of destroying 99 to 99.9% of all HAP. The HAP-inactivating activity may be a side reaction of the as yet undescribed oxidation/reduction activity or, alternatively, targeted primarily at destroying HAP. HAP has been shown to be produced enzymatically from adenine by hepatic microsomal N-hydroxylation (6), and similar reactions could occur in E. coli. The intracellular enzymatic usage of hydroxylamine in purine biosynthesis could also lead to HAP production (3, 34). Interestingly, HAP may also be produced by DNA oxidation resulting from oxidative stress. Experiments exposing DNA and DNA bases to peroxyl radical (ROO−), a major intracellular oxidant and oxidative stress product, showed HAP to be the major product (55).
Another suggestion that HAP may be physiologically relevant is the discovery in Saccharomyces cerevisiae of a putative dNTPase (HAM1 gene product) involved in protection against HAP (41). The defective ham1 mutant is hypersensitive to HAP, and a deoxy-HAP-triphosphatase activity for the Ham1 product is inferred based on certain homologies with known dNTPases (32). The homologous enzyme from Methanococcus jannaschii is capable of hydrolyzing nonstandard nucleotides such as xanthine triphosphate and inosine triphosphate to the corresponding monophosphates (21). Thus, protection against the mutagenic activity of HAP could take place on at least two levels by (i) oxidation/reduction of the base moiety as suggested by the present study and (ii) hydrolysis of the most dangerous form, the triphosphate.
The Δ(uvrB-bio) strain is sensitive not only to HAP but also to AHAP, hydroxylamine, and N4-hydroxycytidine (4-OH-C) in a manner that appears independent of the excision repair system (31, 42, 43). Thus, molybdoenzymes play a general role in the protection against N-hydroxylated base analogs and related compounds. It is of interest to note that a deletion of the uvrB-bio region is carried by several of the Ames tester strains of S. enterica serovar Typhimurium, such as TA1530, TA98, and TA100 (1). These strains have been shown to be hypersensitive to the mutagenic action of HAP, AHAP, and 4-OH-C (23–25). The present data suggest that the simple interpretation of hypermutability of ΔuvrB strains in terms of susceptibility of the mutagenic agent to nucleotide excision repair is not always warranted. It is noted that N-hydroxyl compounds (hydroxylamines) are the active intermediates for the mutagenic action of both aromatic nitro compounds (by reduction) and aromatic amines (by oxidation) (48).
ACKNOWLEDGMENTS
We thank David DeMarini, U.S. EPA, and Joseph Wachsman, NIEHS, for helpful comments on the manuscript and Sean Moore for providing expert technical assistance.
This research was supported by Collaborative Research Grant 971734 from the North Atlantic Treaty Organization and by Russian Fund for Fundamental Scientific Research grant no. 97-0449719.
REFERENCES
- 1.Ames B N, Lee F D, Durston W E. An improved bacterial test system for the detection and classification of mutagens and carcinogens. Proc Natl Acad Sci USA. 1973;70:782–786. doi: 10.1073/pnas.70.3.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bessman M J, Frick D N, O'Handley S F. The MutT proteins or “nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J Biol Chem. 1996;271:25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- 3.Budowski E I. The mechanisms of the mutagenic action of hydroxylamine. Prog Nucleic Acid Res Mol Biol. 1976;16:125–182. doi: 10.1016/s0079-6603(08)60757-6. [DOI] [PubMed] [Google Scholar]
- 4.Cleary P P, Campbell A. Deletion and complementation analysis of the biotin cluster of Escherichia coli. J Bacteriol. 1972;112:830–839. doi: 10.1128/jb.112.2.830-839.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cleary P P, Campbell A, Chang R. Location of promoter and operator sites in the biotin cluster of Escherichia coli. Proc Natl Acad Sci USA. 1972;69:2219–2223. doi: 10.1073/pnas.69.8.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clement B, Kunze T. Hepatic microsomal N-hydroxylation of adenine to 6-N-hydroxylaminopurine. Biochem Pharmacol. 1990;39:925–933. doi: 10.1016/0006-2952(90)90209-4. [DOI] [PubMed] [Google Scholar]
- 7.Clement B, Kunze T. The reduction of 6-N-hydroxylaminopurine to adenine by xanthine oxidase. Biochem Pharmacol. 1992;44:1501–1509. doi: 10.1016/0006-2952(92)90464-t. [DOI] [PubMed] [Google Scholar]
- 8.Cole J A. Nitrate reduction to ammonia by enteric bacteria: redundancy, or a strategy for survival during oxygen starvation? FEMS Microbiol Lett. 1996;136:1–11. doi: 10.1111/j.1574-6968.1996.tb08017.x. [DOI] [PubMed] [Google Scholar]
- 9.Cupples C G, Miller J H. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc Natl Acad Sci USA. 1989;86:5345–5349. doi: 10.1073/pnas.86.14.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.del Campillo-Campbell A, Campbell A. Molybdenum cofactor requirement for biotin sulfoxide reduction in Escherichia coli. J Bacteriol. 1982;149:469–478. doi: 10.1128/jb.149.2.469-478.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Earhart C. Uptake and metabolism of iron and molybdenum. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1075–1090. [Google Scholar]
- 12.Feig D I, Sowers L C, Loeb L A. Reverse chemical mutagenesis: identification of the mutagenic lesions resulting from reactive oxygen species-mediated damage to DNA. Proc Natl Acad Sci USA. 1994;91:6609–6613. doi: 10.1073/pnas.91.14.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fowler R G, Schaaper R M. The role of the mutT gene of Escherichia coli in maintaining replication fidelity. FEMS Microbiol Rev. 1997;21:43–54. doi: 10.1111/j.1574-6976.1997.tb00344.x. [DOI] [PubMed] [Google Scholar]
- 14.Freese E. The specific mutagenic effect of base analogs on phage T4. J Mol Biol. 1959;1:87–105. [Google Scholar]
- 15.Fujikawa K, Kamiya H, Yakushiji H, Fujii Y, Nakabeppu Y, Kasai H. The oxidized forms of dATP are substrates for the human Mut homologue, the hMTH1 protein. J Biol Chem. 1999;274:18201–18205. doi: 10.1074/jbc.274.26.18201. [DOI] [PubMed] [Google Scholar]
- 16.Gennis R B, Stewart V. Respiration. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 217–261. [Google Scholar]
- 17.Gunsalus R P. Control of electron flow in Escherichia coli: coordination of transcription of respiratory pathway genes. J Bacteriol. 1992;174:7069–7074. doi: 10.1128/jb.174.22.7069-7074.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasona A, Ray R M, Shanmugam K T. Physiological and genetic analyses leading to identification of a biochemical role for the moeA (molybdate metabolism) gene product in Escherichia coli. J Bacteriol. 1998;180:1466–1472. doi: 10.1128/jb.180.6.1466-1472.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hille R, Nishino T. Xanthine oxidase and xanthine dehydrogenase. FASEB J. 1995;9:995–1003. [PubMed] [Google Scholar]
- 20.Hong J-S, Ames B N. Localized mutagenesis of any specific small region of the bacterial chromosome. Proc Natl Acad Sci USA. 1971;68:3158–3162. doi: 10.1073/pnas.68.12.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang K Y, Chung J H, Kim S-H, Han Y S, Cho Y. Structure-based identification of a novel NTPase from Methanococcus jannaschii. Nat Struct Biol. 1999;6:691–696. doi: 10.1038/10745. [DOI] [PubMed] [Google Scholar]
- 22.Iobbi-Nivol C, Santini C L, Blasco F, Giordano G. Purification and further characterization of the second nitrate reductase of Escherichia coli K-12. Eur J Biochem. 1990;188:679–687. doi: 10.1111/j.1432-1033.1990.tb15450.x. [DOI] [PubMed] [Google Scholar]
- 23.Janion C. The efficiency and extent of mutagenic activity of some new mutagens of base-analogue type. Mutat Res. 1978;56:225–234. doi: 10.1016/0027-5107(78)90189-6. [DOI] [PubMed] [Google Scholar]
- 24.Janion C. On the different response of Salmonella typhimurium hisG46 and TA1530 to mutagenic action of base analogs. Acta Biochim Pol. 1979;26:171–177. [PubMed] [Google Scholar]
- 25.Janion C, Myszkowska K. Mutagenic and inhibitory properties of some new purine analogs on Salmonella typhimurium TA1530. Mutat Res. 1981;91:193–197. doi: 10.1016/0165-7992(81)90030-0. [DOI] [PubMed] [Google Scholar]
- 26.Joshi M S, Johnson J L, Rajagopalan K V. Molybdenum cofactor biosynthesis in Escherichia coli mod and mog mutants. J Bacteriol. 1996;178:4310–4312. doi: 10.1128/jb.178.14.4310-4312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khromov-Borisov N N. Naming the mutagenic nucleic acid base analogs: the Galatea syndrome. Mutat Res. 1997;379:95–103. doi: 10.1016/s0027-5107(97)00112-7. [DOI] [PubMed] [Google Scholar]
- 28.Kisker C, Schindelin H, Rees D C. Molybdenum-cofactor-containing enzymes: structure and mechanism. Annu Rev Biochem. 1997;66:233–267. doi: 10.1146/annurev.biochem.66.1.233. [DOI] [PubMed] [Google Scholar]
- 29.Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 30.Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 31.Kozmin S G, Leroy P, Pavlov Y I. Overexpression of the yeast HAM1 gene prevents 6-N-hydroxylaminopurine mutagenesis in Escherichia coli. Acta Biochim Pol. 1998;45:645–652. [PubMed] [Google Scholar]
- 32.Kozmin S G, Schaaper R M, Shcherbakova P V, Kulikov V N, Noskov V N, Guetsova M L, Alenin V V, Rogozin I B, Makarova K S, Pavlov Y I. Multiple antimutagenesis mechanisms affect mutagenic activity and specificity of the base analog 6-N-hydroxylaminopurine in bacteria and yeast. Mutat Res. 1998;402:41–50. doi: 10.1016/s0027-5107(97)00280-7. [DOI] [PubMed] [Google Scholar]
- 33.Leimkuhler S, Kern M, Solomon P S, McEwan A G, Schwarz G, Mendel R R, Klipp W. Xanthine dehydrogenase from the phototrophic purple bacterium Rhodobacter capsulatus is more similar to its eukaryotic counterparts than to prokaryotic molybdenum enzymes. Mol Microbiol. 1998;27:853–869. doi: 10.1046/j.1365-2958.1998.00733.x. [DOI] [PubMed] [Google Scholar]
- 34.Lieberman I. Enzymatic synthesis of adenosine-5′-phosphate from inosine-5′-phosphate. J Biol Chem. 1956;223:327–339. [PubMed] [Google Scholar]
- 35.Maki H, Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 36.Méjean V, Iobbi-Nivol C, Lepelletier M, Giordano G, Chippaux M, Pascal M-C. TMAO anaerobic respiration in Escherichia coli: involvement of the tor operon. Mol Microbiol. 1994;11:1169–1179. doi: 10.1111/j.1365-2958.1994.tb00393.x. [DOI] [PubMed] [Google Scholar]
- 37.Michaels M L, Miller J H. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) J Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 39.Nichols B P, Shafiq O, Meiners V. Sequence analysis of Tn10 insertion sites in a collection of Escherichia coli strains used for genetic mapping and strain construction. J Bacteriol. 1998;180:6408–6411. doi: 10.1128/jb.180.23.6408-6411.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nohno T, Kasai Y, Saito T. Cloning and sequencing of the Escherichia coli chlEN operon involved in molybdopterin biosynthesis. J Bacteriol. 1988;170:4097–4102. doi: 10.1128/jb.170.9.4097-4102.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noskov V N, Staak K, Shcherbakova P V, Kozmin S G, Negishi K, Ono B-C, Hayatsu H, Pavlov Y I. HAM1, the gene controlling 6-N-hydroxylaminopurine sensitivity and mutagenesis in the yeast Saccharomyces cerevisiae. Yeast. 1996;12:17–29. doi: 10.1002/(SICI)1097-0061(199601)12:1%3C17::AID-YEA875%3E3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 42.Pavlov Y I, Noskov V N, Lange E K, Moiseeva E V, Pshenichnov M R, Khromov-Borisov N N. The genetic activity of N6-hydroxyadenine and 2-amino-N6-hydroxyadenine in Escherichia coli, Salmonella typhimurium and Saccharomyces cerevisiae. Mutat Res. 1991;253:33–46. doi: 10.1016/0165-1161(91)90343-7. [DOI] [PubMed] [Google Scholar]
- 43.Pavlov Y I, Suslov V V, Shcherbakova P V, Kunkel T A, Ono A, Matsuda A, Schaaper R M. Base analog N6-hydroxylaminopurine mutagenesis in Escherichia coli: genetic control and molecular specificity. Mutat Res. 1996;357:1–15. doi: 10.1016/0027-5107(96)00060-7. [DOI] [PubMed] [Google Scholar]
- 44.Pierson D E, Campbell A. Cloning and nucleotide sequence of bisC, the structural gene for biotin sulfoxide reductase in Escherichia coli. J Bacteriol. 1990;172:2194–2198. doi: 10.1128/jb.172.4.2194-2198.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pollock V V, Barber M J. Biotin sulfoxide reductase. Heterologous expression and characterisation of a functional molybdopterin guanine dinucleotide-containing enzyme. J Biol Chem. 1997;272:3355–3362. doi: 10.1074/jbc.272.6.3355. [DOI] [PubMed] [Google Scholar]
- 46.Puig J, Azoulay E, Gendre J, Richard E. Genetic studies of mutants of the chlA region in Escherichia coli K-12. C R Acad Sci. 1969;268:183–184. [PubMed] [Google Scholar]
- 47.Rajagopalan K V. Biosynthesis of the molybdenum cofactor. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 674–679. [Google Scholar]
- 48.Rosenkranz H S, Mermelstein R. Mutagenicity and genotoxicity of nitroarenes. All nitro-containing chemicals were not created equal. Mutat Res. 1983;114:217–267. doi: 10.1016/0165-1110(83)90034-9. [DOI] [PubMed] [Google Scholar]
- 49.Rudd K E. Linkage map of Escherichia coli K-12, edition 10: the physical map. Microbiol Mol Biol Rev. 1998;62:985–1019. doi: 10.1128/mmbr.62.3.985-1019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schaaper R M. Base selection, proofreading and mismatch repair during DNA replication in Escherichia coli. J Biol Chem. 1993;268:23762–23765. [PubMed] [Google Scholar]
- 51.Schaaper R M, Dunn R L. Effect of Escherichia coli dnaE antimutator mutants on mutagenesis by the base analog N4-aminocytidine. Mutat Res. 1998;402:23–28. doi: 10.1016/s0027-5107(97)00278-9. [DOI] [PubMed] [Google Scholar]
- 52.Schaaper R M, Danforth B N, Glickman B W. Rapid repeated cloning of mutant lac repressor genes. Gene. 1985;39:181–189. doi: 10.1016/0378-1119(85)90312-9. [DOI] [PubMed] [Google Scholar]
- 53.Shanmugam K T, Stewart V, Gunsalus R P, Boxer D H, Cole J A, Chippaux M, DeMoss J A, Giordano G, Lin E C C, Rajagopalan K V. Proposed nomenclature for the genes involved in molybdenum metabolism in Escherichia coli and Salmonella typhimurium. Mol Microbiol. 1992;6:3452–3454. doi: 10.1111/j.1365-2958.1992.tb02215.x. [DOI] [PubMed] [Google Scholar]
- 54.Shcherbakova P V, Pavlov Y I. 3′→5′ exonucleases of DNA polymerases epsilon and delta correct base analog induced DNA replication errors on opposite DNA strands in Saccharomyces cerevisiae. Genetics. 1996;142:717–726. doi: 10.1093/genetics/142.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simandan T, Sun J, Dix T A. Oxidation of DNA bases, deoxyribonucleosides and homopolymers by peroxyl radicals. Biochem J. 1998;335:233–240. doi: 10.1042/bj3350233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simon G, Méjean V, Jourlin C, Chippaux M, Pascal M-C. The torR gene of Escherichia coli encodes a response regulator protein involved in the expression of the trimethylamine-N-oxide reductase genes. J Bacteriol. 1994;176:5601–5606. doi: 10.1128/jb.176.18.5601-5606.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taddei F, Hayakawa H, Bouton M, Cirinesi A-M, Matic I, Sekiguchi M, Radman M. Counteraction by MutT protein of transcriptional errors caused by oxidative damage. Science. 1997;278:128–130. doi: 10.1126/science.278.5335.128. [DOI] [PubMed] [Google Scholar]
- 59.Venables W A, Guest J R. Transduction of nitrate reductase loci of Escherichia coli by phages P-1 and lambda. Mol Gen Genet. 1968;103:127–140. doi: 10.1007/BF00427140. [DOI] [PubMed] [Google Scholar]
- 60.Vogel H J, Bonner D M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 61.von Borstel R C. Measuring spontaneous mutation rates in yeast. Methods Cell Biol. 1978;20:1–24. doi: 10.1016/s0091-679x(08)62005-1. [DOI] [PubMed] [Google Scholar]
- 62.Weiner J H, MacIsaac D P, Bishop R E, Bilous P T. Purification and properties of Escherichia coli dimethyl sulfoxide reductase, an iron-sulfur molybdoenzyme with broad substrate specificity. J Bacteriol. 1988;170:1505–1510. doi: 10.1128/jb.170.4.1505-1510.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Woolfolk C A, Downard J S. Distribution of xanthine oxidase and xanthine dehydrogenase specificity types among bacteria. J Bacteriol. 1977;130:1175–1191. doi: 10.1128/jb.130.3.1175-1191.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamamoto I, Hinakura M, Seki S, Seki Y, Kondo H. Reduction of N-oxide and S-oxide compounds by Escherichia coli. J Gen Appl Microbiol. 1989;35:253–259. [Google Scholar]