Abstract
The scope of population density effects was investigated in steady-state continuous cultures of Escherichia coli in the absence of complications caused by transient environmental conditions and growth rates. Four distinct bacterial properties reflecting major regulatory and physiological circuits were analyzed. The metabolome profile of bacteria growing at high density contained major differences from low-density cultures. The 10-fold-elevated level of trehalose at higher densities pointed to the increased role of the RpoS sigma factor, which controls trehalose synthesis genes as well as the general stress response. There was an eightfold difference in RpoS levels between bacteria grown at 108 and at 109 cells/ml. In contrast, the cellular content of the DNA binding protein H-NS, controlling many genes in concert with RpoS, was decreased by high density. Since H-NS and RpoS also influence porin gene expression, the influence of population density on the intricate regulation of outer membrane composition was also investigated. High culture densities were found to strongly repress ompF porin transcription, with a sharp threshold at a density of 4.4 × 108 cells/ml, while increasing the proportion of OmpC in the outer membrane. The density-dependent regulation of ompF was maintained in rpoS or hns mutants and so was independent of these regulators. The consistently dramatic changes indicate that actively growing, high-density cultures are at least as differentiated from low-density cultures as are exponential- from stationary-phase bacteria.
As originally described for luminescent organisms, population density is an important factor in bacterial physiology (7, 23). Nevertheless, a distinct problem in studying population density-dependent regulation is that it is difficult to unravel the complex effects operating in commonly used batch cultures. There is no simple way with batch cultures to uncouple population density- and growth-phase effects, which affect expression of dozens of genes approaching stationary phase (11). Likewise, it is unclear whether the extracellular molecules found in spent media of Escherichia coli (3, 31, 36, 42) result from growth phase or population density effects. The purpose of this study was to identify the extent of regulation in E. coli affected purely by population density, as is possible in continuous cultures growing at the same growth rate but with predetermined, steady-state bacterial densities. As shown below, at least four global properties of E. coli are strongly influenced by population density in the absence of growth phase effects.
The macromolecular composition and metabolic activity of microbes are known to vary considerably with changes in growth conditions (24, 38). The metabolome, or global pattern of all metabolites, is a function of environmental conditions in steady-state cultures (39, 40). Population density may well affect the metabolome, if indeed the extracellular metabolites in spent media result from shifts in intracellular metabolism. In this study, we investigated the influence of steady-state cell density on metabolic pools in glucose-limited chemostats and report on extensive changes in the pool size of metabolites, including stress protectants.
A particularly important regulatory network suspected of being influenced by population density is the RpoS-dependent general stress response (12). RpoS controls large numbers of genes and influences many structural, resistance, and virulence properties of E. coli (16, 20). Cell density is generally included among possible factors affecting RpoS levels (12). Spent E. coli culture medium indeed affects RpoS regulation (33), but this and other (14) evidence for RpoS regulation by population density is indirect. We present experimental data relevant to the control of the dozens of genes influenced by this alternate sigma factor, as revealed by the extent of changes in RpoS levels at exponential growth rates but with high bacterial densities.
Another cellular component involved in global gene regulation is the histone-like H-NS protein. H-NS is an abundant, neutral, and heat-stable DNA binding protein, and the level of H-NS influences many components of a bacterium (41). Not only is H-NS involved in modulating the general stress response (12), but it also influences diverse cellular functions like flagellar synthesis (34), ribosomal promoters (1), virulence genes (8), and many components of the proteome (18). The cellular content of H-NS is subject to growth phase regulation (2), but the influence of population density has not been tested. Indeed, as for many cellular components, it is unclear whether the regulation of H-NS supposedly by growth phase is actually due to changes in population density in batch cultures. This point can be addressed in steady-state continuous cultures, as reported below.
The fourth global property investigated here is the interface of bacteria to the outside world. In gram-negative bacteria, outer membrane permeability is tightly regulated and governed by the balance of porin proteins (28). Aside from the EnvZ-OmpR two-component regulator, many global regulators including RpoS and H-NS influence transcription of the porin genes ompF and ompC (28). The sensitivity of porin regulation to environmental inputs made it worthwhile to study the influence of population density on outer membrane permeability. We demonstrate that high population densities indeed strongly affect porin regulation and hence the permeability of the outer membrane.
MATERIALS AND METHODS
Bacterial strains.
All bacterial strains used in this study are listed in Table 1. P1 transduction (21) with P1 cml clr100 lysates grown on ZK1171 and CU211 was used to introduce rpoS::Tn10 and hns::neo into MH513 and MC4100 to create strains BW3301, BW3305, BW3323, and BW3324 (Table 1).
TABLE 1.
Bacterial strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| BW2951 | MC4100 lamB::λplacMu55 Φ(lamB-lacZ) | 25 |
| BW3301 | MH513 rpoS::Tn10 | This study |
| BW3305 | MH513 hns::neo | This study |
| BW3323 | MC4100 rpoS::Tn10 | This study |
| BW3324 | MC4100 hns::neo | This study |
| CU121 | CSH26Δ(pro-lac) ara thi hns::neo | C. Ueguchi (43) |
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 deoC1 relA1 thiA ptsF25 flbB5301 | 32 |
| MH513 | MC4100 araD+ Φ(ompF′-lacZ+)16-13 | 10 |
| ZK1171 | W3110 ΔlacU169 tna-2 rpoS::Tn10 | 14 |
β-Galactosidase activity and catalase assay.
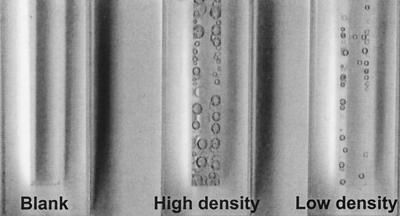
The β-galactosidase activity of lacZ fusions was assayed by the method of Miller (21). To assay catalase, bacteria from lower (2.15 × 108 cells/ml)- and higher (1.01 × 109 cells/ml)-density chemostats was subjected to H2O2 treatment to test the release of gaseous O2 produced by H2O2 hydrolysis (44). The higher-density culture was diluted with minimal medium A (MMA) to the same density as the lower-density culture; 1.6 ml of each culture was put into cuvettes, and 1 drop of 6% H2O2 was added. The sealed cuvettes were scanned with a Hewlett-Packard ScanJet 4C with the cuvettes in a horizontal position.
Growth medium and culture conditions.
The basal salts medium used in chemostats was MMA (21). Glucose-limited feed medium consisted of 0.005 to 0.12% (wt/vol) glucose and 1 g of ammonium sulfate per liter. Batch cultures used Luria broth and MMA containing 4 g of glucose and 1 g of ammonium sulfate per liter. Batch-cultured bacteria were grown at 37°C with constant shaking, and chemostat cultures were grown as previously described (5).
Protein extraction methods for outer membrane fractions and Western blotting analysis of RpoS and H-NS.
The outer membrane protein fraction of wild-type strain MC4100 was prepared as previously described (19). For Western blots, two methods were used. First, after sedimentation of outer membrane protein as described previously (19), the soluble proteins in the supernatant were treated by addition of saturated ammonium sulfate to a final concentration of 50% (4), incubated 30 min at 4°C, and centrifuged another 1 h at 35,000 × g at 4°C to sediment intracellular proteins. Second, samples for other blots were generated by boiling harvested bacteria in sodium dodecyl sulfate (SDS) sample buffer (19) directly, as specified in individual figure legends. Protein concentration was measured by the commercial bicinchoninic acid reagent (Pierce, Rockford, Ill.) and diluted with sample buffer to 20 μg/ml to equalize loadings for comparative analyses. Proteins were separated by electrophoresis in 15% acrylamide gels in the presence of 6 M urea (19).
Protein electrophoretic transfer and immunodetection of RpoS and H-NS.
For Western blotting analysis, the above acrylamide gels were directly subjected to electrophoretic transfer onto a sheet of cellulose nitrate membrane, using an Electrophor II transfer apparatus (Pharmacia, Uppsala, Sweden). The transfer buffer contained 48 mM Tris, 26 mM glycine, 0.0375% (wt/vol) SDS and 20% (vol/vol) methanol, adjusted pH to 9.3. The transfer was run for 2 h at 32 mA at room temperature. Following electrophoretic transfer, the membrane was blocked in TBSTM (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.05% Tween 20, 5% skimmed milk powder) overnight at 4°C. The membrane was then reacted with 5 μl of polyclonal antiserum against ςS (17) or H-NS (6) in 10 ml of TBSTM for 1.5 h. This was followed by three washes of the membrane with 20 ml of TBSTM for 10 min each. After washing, the membrane was soaked with 10 μl of goat anti-rabbit immunoglobulin G-alkaline phosphatase conjugate (Sigma, St. Louis, Mo.) in 10 ml of TBST (TBSTM with no milk powder) for 1.5 h, following by washing with TBST. The protein then was detected with the color reagent 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium (Sigma), and bands of RpoS and HNS were identified by comparison with control strains BW3323 and BW3324 harboring rpoS and hns mutations, respectively.
Metabolome analysis by TLC.
The methods for 14C-labeling of chemostat cultures, extraction, thin-layer chromatography (TLC) separation, and detection of 14C-metabolites were described previously (39). The published method was used for cultures of the bacterial strain BW2951 supplied with 0.02, 0.04, and 0.06% glucose, and the total 14C-glucose input was maintained at 300 μCi of [U-14C]glucose for each culture despite the different glucose concentrations. Constant pumping of label into the culture was maintained for 15 min at a dilution rate of 0.3 h−1 before harvesting.
RESULTS
Methodology.
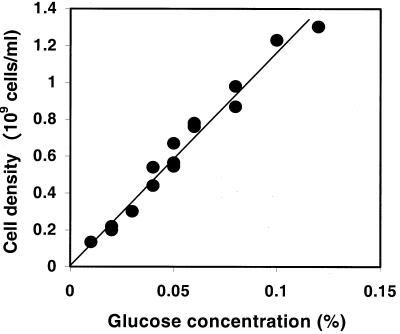
The chemostat culture approach was first used to clarify the relationship between cell density and luminescence, as discussed by Nealson (23). In this study, we used continuous cultures of E. coli to establish stable cell densities by fixed glucose concentrations in the feed medium. The feed glucose concentrations were set at 0.005 to 0.12% in the input medium and resulted in the steady-state bacterial densities shown in Fig. 1. The continuous cultures were allowed to equilibrate for 30 generations before sampling. These experiments were all conducted at a dilution rate of 0.3 h−1, corresponding to an exponential doubling time of approximately 2.5 h. This growth rate does not lead to appreciable expression of starvation or stress responses in glucose-limited continuous cultures (9). As expected from chemostat theory (27), the steady-state density was linearly related to input glucose concentration in the range used in these studies. The linearity also suggested that no other environmental limitation besides glucose limitation affected the growth in these cultures.
FIG. 1.
Bacterial density as a function of input glucose concentrations in glucose-limited chemostats. Chemostats inoculated with wild-type strain MC41000 were supplied with feed medium containing 0.005 to 0.12% glucose at a dilution rate of 0.3 h−1. Bacterial densities were obtained from measurements of optical density at 600 nm from individual chemostats and converted to cell numbers from viable plate counts of diluted cultures.
Metabolite pool changes affected by population density.
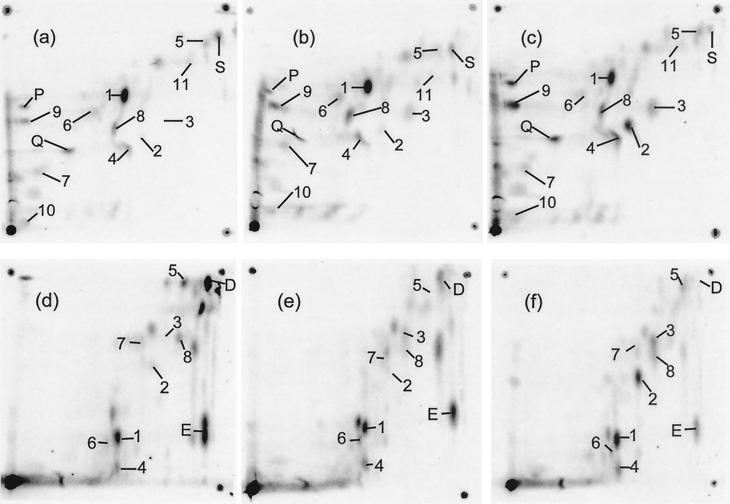
The metabolome patterns of E. coli growing at a dilution rate of 0.3 h−1 with three population densities (preset with input glucose concentrations of 0.02, 0.04, and 0.06%, corresponding to densities of 2.2, 4.5, and 6.5 × 108 bacteria/ml, respectively) are shown in Fig. 2 in two-dimensional separations with two solvent systems. The proportion of label in each of the major spots was quantitated as shown in Fig. 3. All identified metabolites (numbered 1 to 11) and some unidentified spots with obvious differences between densities were quantitated. However, a visually striking change in the unidentified spot labeled E was not quantitated because this spot shows considerable variation among extracts tested in triplicate (39).
FIG. 2.
Metabolome of E. coli growing on glucose at three different cell densities. Extracts were obtained from chemostat-grown strain BW2951 at a dilution rate of 0.3 h−1 with limiting glucose at 37°C. Glucose concentrations were 0.02% (wt/vol) (a and d), 0.04% (b and e), and 0.06% (c and f) (corresponding to densities of 2.2 × 108, 4.5 × 108, and 6.5 × 108 bacteria/ml, respectively). Isotopic labeling used the same amount of [U-14C]glucose (300 μCi) in each of the cultures. Extraction and separation techniques were exactly as described in reference 39, and the extracts were subjected to TLC in solvent system A specified previously (39) for panels a to c and solvent system B for panels d to f. Spots: 1, glutamate; 2, trehalose; 3, glucose; 4, UDP-glucose plus UDP-galactose; 5, adenosine; 6, aspartate; 7, lysine; 8, UDP-N-acetylglucosamine; 9, glutathione; 10, putrescine: 11, valine. The lettered spots were not identified but are mentioned in the text.
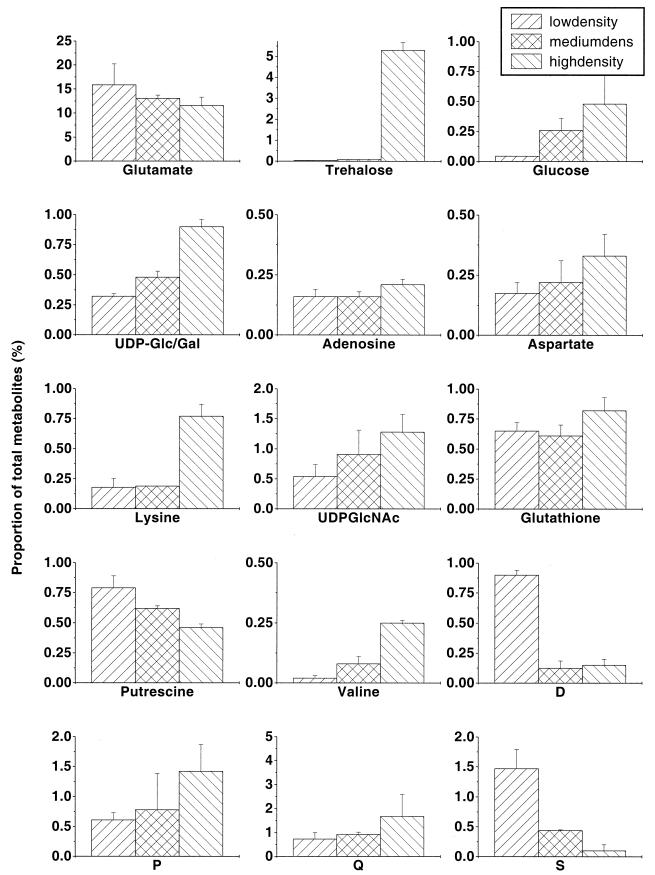
FIG. 3.
Changes in pool sizes as determined by metabolome analysis. Spots corresponding to the compounds represented in Fig. 2 were quantitated using ImageQuant software as described previously (39). Data are presented as the proportion of each metabolite in the total metabolite pool to minimize loading and extraction differences between the plates.
Many consistent changes in individual metabolites were detected between multiple extracts of these cultures. Most notably, the 3-fold increase in population density resulted in more than a 10-fold increase in trehalose pools. Smaller increases in response to higher population densities were seen with glucose, UDP-sugars, aspartate, and amino acids like valine and lysine. Glutamate was found to decline in the higher population density. In addition to these identified metabolites, some unidentified spots, labeled D and S, showed strongly decreased pool size at higher densities. Others, like P and Q, increased with high population densities. As seen below, the threshold for other major regulatory effects is also in the range of 2 × 108 to 6 × 108 bacteria/ml.
Aside from the extent of the changes, the most interesting result was that the trehalose level suggested that bacteria growing exponentially at a density of approximately 6 × 108 bacteria/ml were expressing the general stress response. The production of trehalose is controlled by the alternate sigma factor RpoS, which is required for expression of the otsA and -B genes (13). The possible linkage between RpoS and trehalose led us to investigate the extent of RpoS changes in response to population density.
Cell density influence on RpoS levels.
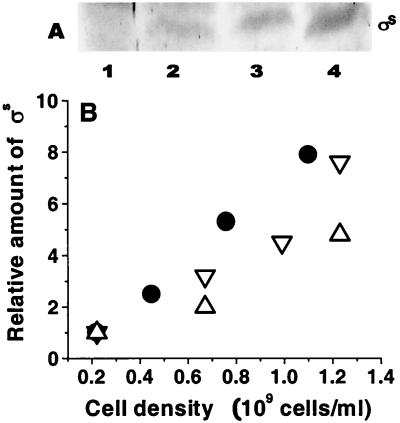
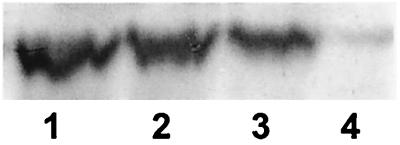
RpoS functions as a global regulator to influence stationary-phase gene expression and the general resistance properties of E. coli (12). RpoS protein was quantitated with polyclonal antibodies to ςS in bacteria growing at different densities in glucose-limited chemostats at a dilution rate of 0.3 h−1. The results of protein immunoblots (Fig. 4) show that the amount of RpoS protein was strongly affected by cell density. The level of RpoS protein increased about eightfold in the transition from 2 × 108 to 1.2 × 109 cells/ml. These results are consistent with the metabolome analysis that higher-density culture is itself sufficient to trigger the general response mediated by RpoS. As demonstrated in Fig. 5, an independent indication of enhanced stress resistance was the high catalase activity of higher-density bacteria, consistent with elevated katE expression controlled by RpoS (22).
FIG. 4.
RpoS protein level in bacteria at different culture densities. (A) Western blotting using anti-ςs antibodies was undertaken for extracts of bacteria grown at different cell densities in a glucose-limited chemostat at a dilution rate of 0.3 h−1. Tracks 1 to 4 contain MC4100 cultures at cell densities of 0.22 × 109, 0.45 × 109, 0.76 × 109, and 1.1 × 109 cells/ml, respectively. (B) Relative amount of RpoS protein as a function of cell density. Protein level was quantitated from densitometer scans using ImageQuant software in three separate blots, each containing samples from separate chemostat cultures of MC4100 growing at a dilution rate of 0.3 h−1 and with different steady-state densities. Samples were extracted by the ammonium sulfate precipitate method (●, ▿) or made using SDS extracts (▵). Quantitations were relative to the amount of RpoS found in bacteria grown with 0.02% glucose input in each series of experiments, which was set as 1.
FIG. 5.
Catalase activity of strain MC4100 at low (2.15 × 108 cells/ml) and high (1.01 × 109 cells/ml) densities. The bubbles represent the O2 produced from the hydrolysis of H2O2 catalyzed by the catalase activity of each culture.
H-NS changes in response to cell density.
Many genes controlled by RpoS, including rpoS itself, are negatively controlled by H-NS (12). Effects of cell density and growth rate on H-NS were not previously investigated. The H-NS levels in cultures grown at different cell densities but constant growth rate were compared in immunoblots. The results in Fig. 6 show that the H-NS content decreased as cell density increased. The range of the decrease in this and other experiments with H-NS protein was only about twofold but approximated the extent found with growth-phase differences (2). These results indicate that H-NS is reciprocally regulated with respect to RpoS and would lead to decreased repression of rpoS and other stress response genes at high population density.
FIG. 6.
H-NS protein level in bacteria at different culture densities. Western blotting was used to determine H-NS protein levels in strain MC4100 grown in glucose-limited chemostats at a dilution rate of 0.3 h−1. Tracks 1 to 3 represent cell densities of 0.22 × 109, 0.67 × 109, and 1.23 × 109 cells/ml. Track 4 shows the pattern in strain BW3324, the hns control. Samples were extracted by the SDS extraction method and are typical of samples on three separate blots from three different sets of samples.
Cell density influence on outer membrane composition.
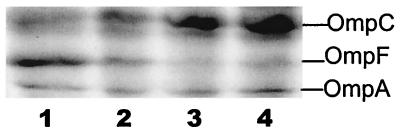
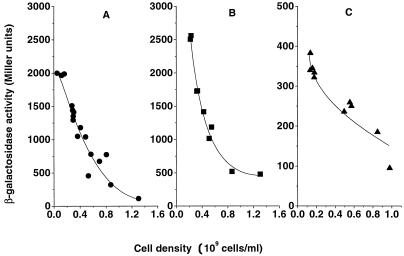
H-NS not only controls stress response genes but also affects porin gene transcription (37). RpoS was also reported to repress porin expression at the transcriptional level in some unspecified way (29). The alterations in H-NS and RpoS concentration reported above were therefore expected to influence porin expression. Porins determine outer membrane permeability and are subjected to intricate regulation (28). OmpF levels were highly induced by glucose limitation at a dilution rate of 0.3 h−1 (19). To investigate cell density influence on porin expression, porin protein content of wild-type bacteria at different culture densities was analyzed (Fig. 7). The β-galactosidase activity of a transcriptional ompF-lacZ fusion in E. coli MH513 was also investigated, as shown in Fig. 8. ompF expression, as shown in Fig. 7 and 8A, decreased with increasing cell density. The steep decrease occurred in the density range from 2 × 108 to 4.4 × 108 cell/ml but was consistently high in cultures growing at lower density (from 5 × 106 to about 2 × 108 cells/ml). Higher density, ranging from 4.4 × 108 to 1.2 × 109 cells/ml, resulted in a further slow decrease in ompF expression. The overall effect of population density on ompF transcription was greater than 10-fold. Consistent with the trend at the transcriptional level, the porin protein content in Fig. 7 showed that the OmpF protein amount decreased as the density increased. In contrast to OmpF, the production of the OmpC protein level increased in a reciprocal fashion.
FIG. 7.
Outer membrane protein composition at different cell densities. Strain MC4100 was grown in glucose-limited chemostats at cell densities of 0.22 × 109, 0.45 × 109, 0.76 × 109 and 1.1 × 109 cells/ml (tracks 1 to 4, respectively). The positions of the major proteins OmpF, OmpC, and OmpA are indicated. Two independent sets of samples gave results as presented.
FIG. 8.
Regulation of ompF transcription by cell density. Cultures were grown with a range of input glucose concentrations establishing the steady-state densities in the chemostat cultures. (A) ompF expression of strain MH513 as measured from the activity of the ompF-lacZ transcriptional fusion; (B) fusion activity in strain BW3323 mutated in rpoS; (C) fusion activity in strain BW3305 mutated in hns. All cultures were grown under glucose limitation at a dilution rate of 0.3 h−1.
Pratt and Silhavy (29) proposed that RpoS repressed ompF gene transcription, and so we checked whether the increase in RpoS was directly responsible for the decline in ompF expression in response to high density. An rpoS mutation was introduced into the ompF-lacZ strain, and the effect of population density was retested as shown in Fig. 8B. The level of ompF transcription was indeed higher in the rpoS mutant, increasing by 500 U at the dilution rate used in these experiments. However, there was only a minor effect of the rpoS mutation on the decrease in ompF expression at higher densities of 2 × 108 to 1.2 × 109 cells/ml. An hns mutation also affected porin transcription, decreasing expression about fivefold. This lowered expression in the hns mutant was also further depressed by higher population density, as shown in Fig. 8C. Hence, the population density effect on ompF was not mediated through RpoS or H-NS, and an independent pathway regulates ompF transcription at high population densities.
DISCUSSION
As reported above, bacterial cell density influences many aspects of metabolism and gene expression. The changes were independent of altered growth rate or other environmental perturbations in the cultures studied. Analysis of the metabolome pointed to the increased production of trehalose under conditions of high population density. Given that trehalose is a recognized stress protectant (35), and since the catalase activity of high-density cultures was also elevated (Fig. 5), these bacteria were well armed for general environmental challenges. The strikingly increased level of RpoS was consistent with the notion that high-density bacteria express the general stress response, even when growing exponentially in a continuous culture. It needs emphasizing that the level of glucose limitation and the growth rate of the cultures studied here was not by itself sufficient to trigger the RpoS-mediated stress response (26), but the possible synergistic effects of glucose limitation and high density need further investigation.
The global nature of RpoS and H-NS regulation and the considerable quantitative protein changes with different densities indicated that expression of large numbers of genes was affected by population density at the transcriptional level. The trehalose and catalase results provided indirect evidence consistent with changed gene expression. More directly, the ompF data provided striking evidence for order-of-magnitude changes in density-dependent transcriptional regulation. The finding that ompF density regulation was maintained in an rpoS mutant as well as an hns mutant reveals that the population density effect on porin regulation is mediated through yet another pathway independent of the two global regulators. The distinct targets and pathways and the sheer multiplicity of changes make it essential to analyze in more detail the regulators and signal transduction pathways in population density-stimulated regulation.
In the colon, E. coli may generally be in the high-density state, given the population size of E. coli in the intestine (30) and the possibility that other bacteria could also contribute density signals recognized by E. coli. The major transitions in RpoS, porin, metabolite, and H-NS levels in our study all occurred at approximately the same narrow range of bacterial densities, namely, between 2 × 108 and 4.4 × 108 cells/ml. This transition density was not particularly high and is readily achievable not only in the colon environment but also in the mid-exponential growth phase of experimental bacterial batch cultures. Indeed, we conclude that exponential batch cultures undergo regulatory and metabolic transitions based on population density-sensing mechanisms. There is still a widespread view that mid-exponential batch cultures are in a state of balanced growth from a physiological point of view (15), but our results suggest otherwise. It remains to be seen whether the threshold that we determined in continuous culture is also applicable to other media and growth rates generally used in physiological studies, such as those used in batch culture for production of the signal molecule(s) reported by Surette and Bassler (approximately 5 × 108 bacteria/ml) (36).
We have not addressed the question of which, if any, quorum-sensing mechanism or molecule is responsible for the extensive cellular adaptations. Much detailed work is required with respect to medium components such as the AI-2-like signal (36) in the individual regulatory changes and what point in transcriptional, translational, or protein stability regulation the signals attack. The potential role of other signals like homoserine lactones (14) and anhydro sugars (31) also needs assessment. The experimental system described in this report is ideally suited to future testing of the effects of extracellular signals in the absence of growth phase changes.
In conclusion, population density is itself an inducer of major changes in bacterial regulation, physiology, and metabolism. The dramatic shifts reported above were independent of altered growth rate or other environmental perturbations in the cultures studied. In this respect, the presence of smaller-channel porins, the increased synthesis of stress protectant, and the induction of the general stress response all point to the increased ability of E. coli to meet challenges, even in the absence of any external environmental threat. Presumably, high-density environments are associated with harsh situations in the lifestyle of E. coli.
ACKNOWLEDGMENTS
We thank Regine Hengge-Aronis and Erhard Bremer for gifts of antibodies and C. Ueguchi for bacterial mutants.
We thank the Australian Research Council for funding support.
REFERENCES
- 1.Afflerbach H, Schroder O, Wagner R. Conformational changes of the upstream DNA mediated by H-NS and FIS regulate E. coli rmB P1 promoter activity. J Mol Biol. 1999;286:339–353. doi: 10.1006/jmbi.1998.2494. [DOI] [PubMed] [Google Scholar]
- 2.Azam T A, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baca-DeLancey R R, South M M T, Ding X D, Rather P N. Escherichia coli genes regulated by cell-to-cell signaling. Proc Natl Acad Sci USA. 1999;96:4610–4614. doi: 10.1073/pnas.96.8.4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouvier J, Gordia S, Kampmann G, Lange R, Hengge-Aronis R, Gutierrez C. Interplay between global regulators of Escherichia coli—effect of RpoS, Lrp and H-NS on transcription of the gene osmC. Mol Microbiol. 1998;28:971–980. doi: 10.1046/j.1365-2958.1998.00855.x. [DOI] [PubMed] [Google Scholar]
- 5.Death A, Notley L, Ferenci T. Derepression of LamB protein facilitates outer membrane permeation of carbohydrates into Escherichia coli under conditions of nutrient stress. J Bacteriol. 1993;175:1475–1483. doi: 10.1128/jb.175.5.1475-1483.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dersch P, Schmidt K, Bremer E. Synthesis of the Escherichia coli K-12 nucleoid-associated DNA-binding protein H-NS is subjected to growth-phase control and autoregulation. Mol Microbiol. 1993;8:875–889. doi: 10.1111/j.1365-2958.1993.tb01634.x. [DOI] [PubMed] [Google Scholar]
- 7.Dunny G M, Winans S C, editors. Cell-cell signaling in bacteria. Washington, D.C.: ASM Press; 1999. [Google Scholar]
- 8.Falconi M, Colonna B, Prosseda G, Micheli G, Gualerzi C O. Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS. EMBO J. 1998;17:7033–7043. doi: 10.1093/emboj/17.23.7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferenci T. Regulation by nutrient limitation. Curr Opin Microbiol. 1999;2:208–213. doi: 10.1016/S1369-5274(99)80036-8. [DOI] [PubMed] [Google Scholar]
- 10.Hall M N, Silhavy T J. The ompB locus and the regulation of the major outer membrane porin proteins of Escherichia coli K12. J Mol Biol. 1981;146:23–43. doi: 10.1016/0022-2836(81)90364-8. [DOI] [PubMed] [Google Scholar]
- 11.Hengge-Aronis R. Regulation of gene expression during entry into stationary phase. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- 12.Hengge-Aronis R. Interplay of global regulators and cell physiology in the general stress response of Escherichia coli. Curr Opin Microbiol. 1999;2:148–152. doi: 10.1016/S1369-5274(99)80026-5. [DOI] [PubMed] [Google Scholar]
- 13.Hengge-Aronis R, Klein W, Lange R, Rimmele M, Boos W. Trehalose synthesis genes are controlled by the putative sigma factor encoded by rpoS and are involved in stationary-phase thermotolerance in Escherichia coli. J Bacteriol. 1991;173:7918–7924. doi: 10.1128/jb.173.24.7918-7924.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huisman G W, Kolter R. Sensing starvation: a homoserine lactone-dependent signaling pathway in Escherichia coli. Science. 1994;265:537–539. doi: 10.1126/science.7545940. [DOI] [PubMed] [Google Scholar]
- 15.Ingraham J L, Maaloe O, Neidhardt F C. Growth of the bacterial cell. Sunderland, Mass: Sinauer Associates; 1983. [Google Scholar]
- 16.Kolter R, Siegele D A, Tormo A. The stationary phase of the bacterial life cycle. Annu Rev Microbiol. 1993;47:855–874. doi: 10.1146/annurev.mi.47.100193.004231. [DOI] [PubMed] [Google Scholar]
- 17.Lange R, Hengge-Aronis R. The cellular concentration of the sigma-S subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation and protein stability. Genes Dev. 1994;8:1600–1612. doi: 10.1101/gad.8.13.1600. [DOI] [PubMed] [Google Scholar]
- 18.Laurentwinter C, Ngo S, Danchin A, Bertin P. Role of Escherichia coli histone-like nucleoid-structuring protein in bacterial metabolism and stress response—identification of targets by two-dimensional electrophoresis. Eur J Biochem. 1997;244:767–773. doi: 10.1111/j.1432-1033.1997.00767.x. [DOI] [PubMed] [Google Scholar]
- 19.Liu X, Ferenci T. Regulation of porin-mediated outer membrane permeability by nutrient limitation in Escherichia coli. J Bacteriol. 1998;180:3917–3922. doi: 10.1128/jb.180.15.3917-3922.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loewen P C, Hu B, Strutinsky J, Sparling R. Regulation in the RpoS regulon of Escherichia coli. Can J Microbiol. 1998;44:707–717. doi: 10.1139/cjm-44-8-707. [DOI] [PubMed] [Google Scholar]
- 21.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 22.Mulvey M R, Switala J, Borys A, Loewen P C. Regulation of transcription of katE and katF in Escherichia coli. J Bacteriol. 1990;172:6713–6720. doi: 10.1128/jb.172.12.6713-6720.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nealson K H. Early observations defining quorum-dependent gene expression. In: Dunny G M, Winans S C, editors. Cell-cell signaling in bacteria. Washington, D.C.: ASM Press; 1999. pp. 277–289. [Google Scholar]
- 24.Neidhardt F C, Umbarger H E. Chemical composition of Escherichia coli. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 13–16. [Google Scholar]
- 25.Notley L, Ferenci T. Differential expression of mal genes under cAMP and endogenous inducer control in nutrient stressed Escherichia coli. Mol Microbiol. 1995;160:121–129. doi: 10.1111/j.1365-2958.1995.tb02397.x. [DOI] [PubMed] [Google Scholar]
- 26.Notley L, Ferenci T. Induction of RpoS-dependent functions in glucose-limited continuous culture: what level of nutrient limitation induces the stationary phase of Escherichia coli? J Bacteriol. 1996;178:1465–1468. doi: 10.1128/jb.178.5.1465-1468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pirt S J. Principles of microbe and cell cultivation. Oxford, England: Blackwell Scientific Publications; 1975. [Google Scholar]
- 28.Pratt L A, Hsing W H, Gibson K E, Silhavy T J. From acids to OsmZ—multiple factors influence the synthesis of the OmpF and OmpC porins in Escherichia coli. Mol Microbiol. 1996;20:911–917. doi: 10.1111/j.1365-2958.1996.tb02532.x. [DOI] [PubMed] [Google Scholar]
- 29.Pratt L A, Silhavy T J. Crl stimulates RpoS activity during stationary phase. Mol Microbiol. 1998;29:1225–1236. doi: 10.1046/j.1365-2958.1998.01007.x. [DOI] [PubMed] [Google Scholar]
- 30.Savage D. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 31.Shiga Y, Kametani S, Kadokura T, Akanuma H. 1,5-Anhydroglucitol promotes glycogenolysis in Escherichia coli. J Biochem. 1999;125:166–172. doi: 10.1093/oxfordjournals.jbchem.a022254. [DOI] [PubMed] [Google Scholar]
- 32.Silhavy T J, Casadaban M J, Shuman H A, Beckwith J R. Conversion of beta-galactosidase to a membrane-bound state by gene fusion. Proc Natl Acad Sci USA. 1976;73:3423–3427. doi: 10.1073/pnas.73.10.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sitnikov D M, Schineller J B, Baldwin T O. Control of cell division in Escherichia coli—regulation of transcription of ftsQA involves both RpoS and SdiA-mediated autoinduction. Proc Natl Acad Sci USA. 1996;93:336–341. doi: 10.1073/pnas.93.1.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, Danchin A, Bertin P. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J Bacteriol. 1999;181:7500–7508. doi: 10.1128/jb.181.24.7500-7508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strom A R, Kaasen I. Trehalose metabolism in Escherichia coli: stress protection and stress regulation of gene expression. Mol Microbiol. 1993;8:205–210. doi: 10.1111/j.1365-2958.1993.tb01564.x. [DOI] [PubMed] [Google Scholar]
- 36.Surette M G, Bassler B L. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc Natl Acad Sci USA. 1998;95:7046–7050. doi: 10.1073/pnas.95.12.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki T, Ueguchi C, Mizuno T. H-NS regulates ompF expression through micF antisense RNA in Escherichia coli. J Bacteriol. 1996;178:3650–3653. doi: 10.1128/jb.178.12.3650-3653.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tempest D W, Neijssel O M. Physiological and energetic aspects of bacterial metabolite overproduction. FEMS Microbiol Lett. 1992;79:169–176. doi: 10.1111/j.1574-6968.1992.tb14036.x. [DOI] [PubMed] [Google Scholar]
- 39.Tweeddale H, Notley-McRobb L, Ferenci T. Effect of slow growth on metabolism of Escherichia coli, as revealed by global metabolite pool (“metabolome”) analysis. J Bacteriol. 1998;180:5109–5116. doi: 10.1128/jb.180.19.5109-5116.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tweeddale H, Notley-McRobb L, Ferenci T. Assessing the effect of reactive oxygen species on Escherichia coli using a metabolome approach. Redox Rep. 1999;4:237–241. doi: 10.1179/135100099101534954. [DOI] [PubMed] [Google Scholar]
- 41.Williams R M, Rimsky S. Molecular aspects of the E. coli nucleoid protein, H-NS—a central controller of gene-regulatory networks. FEMS Microbiol Lett. 1997;156:175–185. doi: 10.1111/j.1574-6968.1997.tb12724.x. [DOI] [PubMed] [Google Scholar]
- 42.Withers H L, Nordstrom K. Quorum-sensing acts at initiation of chromosomal replication in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:15694–15699. doi: 10.1073/pnas.95.26.15694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamada H, Yoshida T, Tanaka K, Sasakawa C, Mizuno T. Molecular analysis of the Escherichia coli hns gene encoding a DNA-binding protein, which preferentially recognizes curved DNA sequences. Mol Gen Genet. 1991;230:332–336. doi: 10.1007/BF00290685. [DOI] [PubMed] [Google Scholar]
- 44.Zambrano M M, Siegele D A, Almiron M, Tormo A, Kolter R. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science. 1993;259:1757–1760. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]