Abstract
The retina is one of the highest oxygen-consuming tissues because visual transduction and light signaling processes require large amounts of ATP. Thus, because of the high energy demand, oxygen-rich environment, and tissue transparency, the eye is susceptible to excess production of reactive oxygen species (ROS) resulting in oxidative stress. Oxidative stress in the eye is associated with the development and progression of ocular diseases including cataracts, glaucoma, age-related macular degeneration, and diabetic retinopathy. ROS can modify and damage cellular proteins, but can also be involved in redox signaling. In particular, the thiol groups of cysteines can undergo reversible or irreversible oxidative post-translational modifications (PTMs). Identifying the redox-sensitive cysteines on a proteome-wide scale provides insight into those proteins that act as redox sensors or become irreversibly damaged upon exposure to oxidative stress. In this study, we profiled the redox proteome of the Drosophila eye under prolonged, high intensity blue light exposure and age using iodoacetamide isobaric label sixplex reagents (iodo-TMT) to identify changes in cysteine availability. Although redox metabolite analysis of the major antioxidant, glutathione, revealed similar ratios of its oxidized and reduced form in aged or light-stressed eyes, we observed different changes in the redox proteome under these conditions. Both conditions resulted in significant oxidation of proteins involved in phototransduction and photoreceptor maintenance but affected distinct targets and cysteine residues. Moreover, redox changes induced by blue light exposure were accompanied by a large reduction in light sensitivity that did not arise from a reduction in the photopigment level, suggesting that the redox-sensitive cysteines we identified in the phototransduction machinery might contribute to light adaptation. Our data provide a comprehensive description of the redox proteome of Drosophila eye tissue under light stress and aging and suggest how redox signaling might contribute to light adaptation in response to acute light stress.
Keywords: Drosophila, Redox proteomics, Cysteine oxidative modifications, Redox signaling eye, Aging, Light stress, Phototransduction, Ocular disease
1. Introduction
Oxidative stress increases in the aging eye, and strongly affects the development and progression of age-associated ocular disease [[1], [2], [3], [4]]. The retina experiences considerable oxidative stress because of its energy-intensive, oxygen-rich environment [1,2,5]. Moreover, while absorption of light is required for vision, light exposure also increases levels of oxidative stress in the eye [6]. Oxidative stress is driven by an imbalance between reactive oxygen species (ROS) and antioxidants, which can result in oxidative damage to proteins, lipids, and nucleic acids [7]. The specific type, level, and location of ROS is critical for its function in the cell; although high ROS levels lead to cell death, low concentrations of specific ROS such as hydrogen peroxide (H2O2) can act as signaling intermediates [7,8]. Thus, low levels of ROS during development are protective and enhance stress resistance during aging [9], but high levels of ROS are associated with damage and cell death [7,[10], [11], [12], [13]]. In addition, since proteins can be oxidized at specific residues, these oxidative modifications can either allow a protein to function as a redox sensor and/or impair its activity [7,[10], [11], [12], [13]]. For example, ROS signaling induces protective gene expression programs by releasing and activating the transcription factor Nrf2 [14], or by inhibiting activity of epigenetic factors such as the histone methyltransferase Set1/MLL via intramolecular disulfide bond formation [15].
Cysteine residues have multiple oxidation states that can be classified into two categories—reversible and irreversible [4]. Reversible modifications, which are often observed during redox signaling, include S-nitrosylation, S-glutathionylation, sulfenic acid, and inter- and intra-molecular disulfide bonds. In contrast, irreversible cysteine modifications such as sulfinic and sulfonic acids may alter or impair the activity of the protein, depending on the position of the cysteine residue [7]. In addition, recent work has shown that cysteines can form a covalent crosslink with lysine residues via a nitrogen–oxygen–sulfur (NOS) bridge [16,17]. In all of these cases, identifying the position of the cysteine residues that become modified can provide insight into how a given cell type responds to, or is damaged by, increasing ROS levels. The emergence of increased sensitivity mass-spectrometry instruments to characterize redox induced PTMs at not only the protein level, but at the specific amino acid residue, has enhanced researchers’ abilities to investigate the redox proteome [8].
The fruit fly, Drosophila melanogaster, is a widely used model system for studying the impact of oxidative stress on development and disease [10,11]. Drosophila provides advantages for proteomic studies because of the relatively small size of its proteome relative to humans due to less redundancy [12]. The Drosophila eye has been used extensively as a model to identify genes involved in development and patterning of the retina, and to characterize phototransduction [[18], [19], [20], [21]]. In flies, the photoreceptor neurons are organized into repeating units called ommatidia within the compound eye. The phototransduction machinery is housed within a specialized light-sensing signaling compartment, the rhabdomere, composed of ∼40,000 densely packed microvilli [9]. In Drosophila, phototransduction is mediated via a bistable photopigment rhodopsin (Rh1 in outer photoreceptors), which is a G-protein-coupled receptor (GPCR) [22]. Upon light absorption, the Rh1 photopigment is converted into the active pigment state, meta-rhodopsin [9,21,23]. This in turn activates the heterotrimeric Gq-protein where the Gqα protein subunit associates with the phospholipase Cβ (PLCβ) known as no receptor potential A (norpA gene) [9,21,23]. NorpA then hydrolyzes a minor membrane phosphatidylinositol 4,5-bisphosphate (PIP2) producing diacylglycerol (DAG), inositol tris phosphate (IP3), and a proton, leading in a still unclear mechanism to the opening of the light sensitive transient receptor potential (TRP) and TRP-like (TRPL) channels [9,21,23]. The major signaling proteins, TRP, PLCβ, and eye-specific protein kinase C (encoded by the inactivation no afterpotential C, inaC gene) are attached to a scaffolding protein, encoded by the inactivation no afterpotential D (inaD) gene. INAD in its reduced form binds the TRP channel [[24], [25], [26], [27]]. Visual transduction processes are known to be associated with light-dependent changes in redox potential [23,24]. For example, the scaffolding protein, INAD, can be oxidized forming a disulfide bond at cysteine residues 606 and 645, which releases INAD from the TRP channels and is associated with phototransduction termination [24]. In addition, redox glutathione ratios change in the Drosophila eye under normal diurnal light cycles relative to dark [28], suggesting that redox signaling may have roles in phototransduction [24]. However, increases in ROS levels caused by acute blue light stress are damaging, inducing retinal degeneration [[28], [29], [30]].
Prolonged exposure to blue light is an acute stress model in which light activates the GPCR Rh1, leading to a cascade of events that results in excessive calcium influx through the TRP channels [31,32]. Absorption of blue light converts the Rh1 rhodopsin (R) state (peak absorption ∼485 nm) into the physiologically active meta-rhodopsin (M) state (peak absorption ∼580 nm). During long wavelength illumination (green, orange, or white lights) the photopigment is continuously photoconverted between the R and M states, resulting in most photopigment molecules residing in the R state at light off [9,21,23]. However, if flies are exposed to intense, prolonged blue light in the absence of long wavelength light, there is a net photopigment conversion from R to M inducing prolonged activation of the phototransduction cascade at its maximal capacity called the prolonged depolarizing afterpotential (PDA) [33]. This prolonged activation leads to calcium excitotoxicity, increased oxidative stress, and retinal degeneration [29]. Specifically, blue light exposure increases levels of H2O2 and the lipid peroxidation indicator malondialdehyde in Drosophila eyes [29]. Prolonged blue light exposure also leads to excessive endocytosis of phosphorylated M attached to phosphorylated Arrestin 2 (Arr2), which also contributes to retinal degeneration [[34], [35], [36], [37], [38]]. In the acute light stress model, the retinal degeneration induced by exposing flies to strong blue light can be suppressed either by inactivating phototransduction or by decreasing levels of H2O2 and lipid peroxides [29]. Aging is also associated with increased oxidative stress and is a major risk factor for several ocular diseases [6,39,40]. In Drosophila, aging correlates with decreased visual function, altered gene expression, and retinal degeneration [41,42]. Identifying the proteins in the eye that are sensitive to redox changes under acute light stress or aging could provide insight into both the protective redox signaling mechanisms employed by cells to withstand high ROS levels, and into the events that lead to retinal degeneration during aging or under acute light stress.
Several strategies have been developed to identify changes in cysteine oxidation state on a proteome-wide scale [[43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53]] and some of these techniques have been applied to Drosophila tissues [[54], [55], [56], [57], [58], [59]]. Most recently, two approaches utilizing iodoacetamide (IAA) to label cysteine residues have been developed. The first approach uses stable isotope labeling with IAA (SICyLIA) [53], while the second method employs iodoacetyl tandem mass tags (iodo-TMT) that allow for sample multiplexing [60]. In SICyLIA, free thiol groups are labeled with heavy or light IAA, thereby enabling the identification of “nonreactive” cysteines that are potentially oxidized under experimental conditions. This labeling is then followed by a reduction step and a second labeling using N-Ethylmaleimide (NEM), enabling the identification of cysteines that were in a reversible oxidation state such as a disulfide bond. In contrast, iodo-TMT reagents are independent isobaric tags that possess a mass-reporter used to differentiate between biological samples during mass-spectrometry analysis [43,60]. These independent tags bind to free thiol groups followed by a reduction and second labeling step with IAA. The presence of an iodo-TMT tag in a sample indicates an increase in cysteine availability or reduction, whereas the absence of a tag indicates a decrease in cysteine availability corresponding to potential oxidation. A limitation to this method is that we cannot differentiate between reversible or irreversible modifications unless an IAA labeled cysteine is present on the same peptide as an iodo-TMT tagged cysteine. In contrast to SICyLIA, the iodo-TMT approach has an optional enrichment step for TMT-tagged peptides. This is especially important for redox profiling of cysteine availability because these residues are lowly abundant in the proteome, accounting for only 2.2% or less of amino acid residues in higher eukaryotes [45,[61], [62], [63]]. In this study, we evaluated iodo-TMT and SICyLIA approaches in cultured Drosophila cells and found that iodo-TMT was more cost-effective and efficient than SICyLIA, identifying more than twice as many oxidized or reduced peptides in a single mass spectrometry run. We then utilized iodo-TMT reagents to characterize the redox proteome of eyes exposed to acute light stress or during aging, revealing distinct and conserved changes in cysteine oxidation under both conditions. We identified distinct changes in cysteine availability in components of the phototransduction machinery upon exposure to acute light stress relative to aging, suggesting that these conditions have distinct effects on redox signaling that could differentially impact phototransduction.
2. Results
2.1. Iodo-TMT is an effective approach for redox proteomic profiling in cultured Drosophila cells
Several proteomic approaches have been developed to examine cysteine availability, which can indicate its oxidation status, but many of these approaches require specialized reagents that are not commercially available [45,63]. We sought to characterize the redox proteome of Drosophila tissues with approaches that could be performed using readily available commercial reagents, leading us to focus on SICyLIA and iodo-TMT [53,64]. To identify which of these approaches would work most effectively with Drosophila samples, we compared these techniques in cultured Schneider 2 (S2) cells. To do this, we exposed S2 cells to 15 min of treatment with 20 mM H2O2. We used a relatively high concentration of H2O2 based on previous studies [53,65] with the rationale that short-term high exposure to H2O2 would result in substantial changes in cysteine availability, allowing us to readily evaluate the two approaches in cultured cells. Under these conditions, we observed significant oxidative stress as determined by targeted metabolite analysis of redox ratios of nicotinamide adenine dinucleotide (NADH:NAD+), nicotinamide adenine dinucleotide phosphate (NADPH:NADP+), and glutathione (GSH:GSSG), with little cell death (Fig. S1).
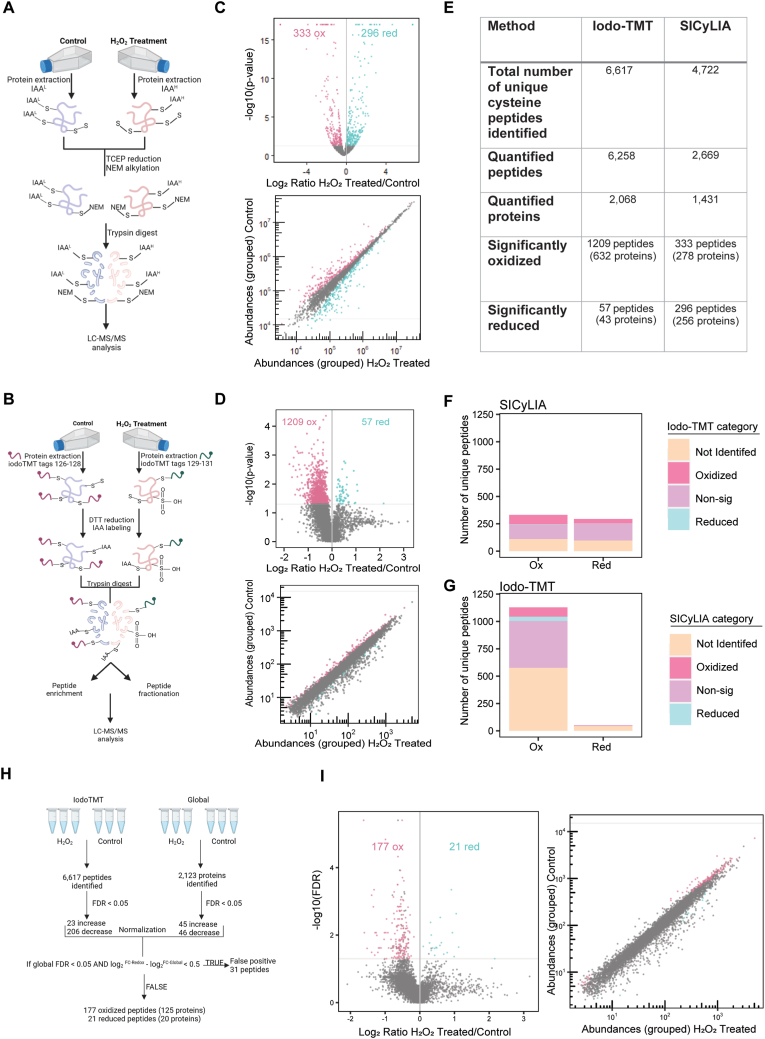
We then directly compared the redox proteome of S2 cells exposed to 15 min of H2O2 treatment relative to untreated controls using either SICyLIA, as described in Ref. [53], or using an iodo-TMT approach developed by our group (see methods). We examined two biological replicates for treated and control samples using SICyLIA, with each treated and control pair being run on an independent LC/MS/MS analysis (Fig. 1A). In contrast, because the iodo-TMT-sixplex reagents have independent isobaric tags that possess a mass-reporter, we could analyze three biological replicates each for treated and control within a single LC/MS/MS analysis (Fig. 1B). We also enriched the iodo-TMT-labeled peptides using anti-TMT resin and analyzed both the enriched and unenriched fractions to obtain the widest possible coverage of iodo-TMT-labeled peptides. We then compared the total number of cysteine peptides identified using either technique and found that SICyLIA identified 4722 unique total cysteine-containing peptides, of which 2669 were quantified, corresponding to 1431 proteins (Fig. 1E) (Table S1). In contrast, in the S2 cell-based experiment, our newly developed iodo-TMT approach identified 6617 unique total cysteine-containing peptides, of which 6258 were quantified, corresponding to 2068 proteins (Fig. 1E) (Table S2). We next identified significant changes in availability of the quantified cysteine-containing peptides (p < 0.05) using each approach; in both methods, decreased ratios (log2 Fold Change; log2FC < 0) represent potential cysteine oxidation while increased ratios (log2FC > 0) correspond to reduction. When comparing both approaches, we used unadjusted p-values because SICyLIA is performed with two biological replicates per treatment group whereas iodo-TMT has three biological replicates. However, for further analysis focusing solely on the results obtained using the iodo-TMT approach we obtained corrected FDRs via the PolySTest, which is a robust statistical test developed for proteomic data [66]. Using SICyLIA, we identified 333 oxidized and 296 reduced cysteine-containing peptides corresponding to 278 and 256 proteins, respectively (Fig. 1C and E). We identified more than twice as many significantly changed cysteine-containing peptides using the iodo-TMT approach: 1209 oxidized and 57 reduced peptides, corresponding to 632 and 43 proteins, respectively (Fig. 1D and E). Moreover, when we examined the abundance of the potentially oxidized and reduced peptides identified using each approach, we found that SICyLIA was biased towards moderately abundant peptides (Fig. 1C). In contrast, the iodo-TMT approach showed enhanced ability to identify greater fold changes across a variety of peptide abundances, potentially due to the enrichment of iodo-TMT labeled peptides prior to analysis (Fig. 1D).
Fig. 1.
Quantitative analysis of cysteine availability in Drosophila S2 cells comparing Iodo-TMT to SICyLIA.(A) Schematic of SICyLIA labeling method in S2 cells treated with 20 mM H2O2 versus control for 15 min followed by LC-MS/MS analysis. (B) Schematic of iodo-TMT labeling method in S2 cells treated with 20 mM H2O2 versus control for 15 min followed by LC-MS/MS analysis. (C) Volcano plot displaying log2 ratio of redox peptides identified in H2O2 Treated/Control versus the -log10(p-value) identified using SICyLIA method. Scatterplot displaying grouped abundances of redox peptides identified in H2O2 treated versus control. (D) Volcano plot and scatterplot as described in C using iodo-TMT method. Peptides with significant changes are highlighted in pink (oxidation) or blue (reduction). p-value <0.05. (E) Table outlining peptide identification and quantification for iodo-TMT and SICyLIA methods. (F) Bar plot displaying significantly oxidized (left) or reduced (right) peptides identified using SICyLIA that were either not-identified, oxidized, non-significant, or reduced in iodo-TMT method. (G) Bar plot as described in F using iodo-TMT compared to SICyLIA. (H) Overview of Iodo-TMT method normalization for S2 cells. Global protein abundance was used to account for changes in total protein levels pre- and post-treatment. (I) Normalized volcano and scatterplot displaying log2 ratio of redox peptides identified in H2O2 Treated/Control versus the -log10(p-value) using iodo-TMT method. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Next, we compared the peptides from each approach to see if we identified commonly oxidized or reduced cysteine-containing peptides using both approaches. Surprisingly, there was relatively little overlap between the two methods with only 86 oxidized peptides and one reduced peptide being identified by both SICyLIA and iodo-TMT (Fig. 1F and G). Since the cells were treated with the same concentration of H2O2, we attribute these discrepancies to differences in the two redox proteomic techniques. Notably, most of these differences were due to peptide identification because many of the peptides identified and quantified using iodo-TMT were not identified using SICyLIA. Moreover, because we had three biological replicates for the iodo-TMT analysis, we were able to identify high confidence oxidized and reduced events using FDR with this approach. Based on this preliminary data, we concluded that SICyLIA and iodo-TMT provide complementary approaches to characterizing the redox proteome of Drosophila, but that the enhanced peptide identification and multiplexing afforded by iodo-TMT increased our ability to identify oxidation and reduction of peptides across a wide range of abundance levels with high confidence. In addition, the iodo-TMT approach was more cost effective in our laboratory, despite the high initial cost of the reagents, because we were able to process samples in-house and multiplex samples within a single LC-MS/MS run.
Because significant changes in the availability of quantified cysteine-containing peptides between H2O2-treated S2 cells and the untreated control could be caused by changes in the abundance of those proteins, we next analyzed the global abundance of proteins in each sample used for the iodo-TMT labeling. Global protein abundance analysis is also available using SICyLIA; however, because of the superior peptide identification observed with the iodo-TMT method we only proceeded with global analysis for the iodo-TMT samples. To do this, we reserved a fraction of each sample prior to iodo-TMT labeling, and then labeled these using traditional tandem mass tags as performed in Ref. [67]. Because these TMT tags label free amines, which are present on the N-termini of most digested peptides rather than only the cysteine-specific peptides, we then used these to determine the relative abundance of each protein in H2O2-treated S2 cells versus control (n = 3). To account for oxidation of other residues such as Trp or His, which could occur under H2O2 treatment, we included these modifications in the proteomic search parameters. Using this approach, we quantified 2123 proteins of which 45 showed significantly increased abundance in H2O2-treated S2 cells while 46 showed decreased abundance (FDR <0.05) (Fig. 1H) (Table S3). We considered that any significant changes in cysteine availability that were accompanied by a significant change in protein abundance in the same direction (i.e., decreased cysteine availability and decreased protein abundance) and to the same extent (difference in log2FC < 0.5) were false positives. Using this approach, we excluded 31 significant peptides from the iodo-TMT analysis as false positives. Therefore, after normalization for changes in protein abundance, we identified 177 and 21 peptides that showed significant decreases or increases, respectively, corresponding to 125 and 20 proteins (Fig. 1H and I) (Table S4). We interpret these changes in cysteine availability as reflecting potential oxidation or reduction of the cysteine residues in these proteins upon exposure to H2O2 in S2 cells. Thus, our data indicate that under the strong H2O2 treatment conditions used for S2 cells, the majority of cysteine modifications represent oxidation events rather than reduction. Supporting this, we identified several proteins that are known to be oxidized reversibly upon exposure to ROS such as catalase (Cat), heat shock protein 83 (Hsp83), superoxide dismutase 1 (Sod1), and superoxide dismutase 2 (Mn) (Sod2) [40,[68], [69], [70], [71]] (Table S4). Based on these data, we proceeded forward with Drosophila tissue studies using the iodo-TMT approach.
2.2. Prolonged blue light exposure leads to oxidation of proteins involved in translational initiation, rhodopsin homeostasis, and phototransduction in the eye
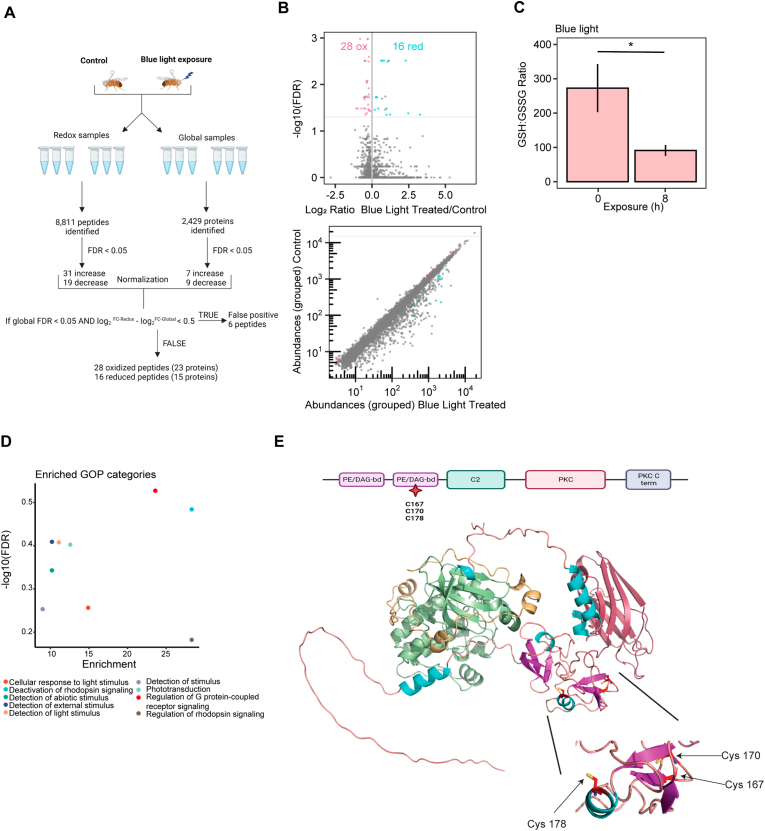
The retinal degeneration induced by blue light can be suppressed by genetic or pharmacological amelioration of oxidative stress [29]. However, it is unknown how acute blue light stress impacts the redox proteome of the eye, and whether these changes enhance or protect against retinal degeneration. Here, we sought to identify proteins with altered cysteine availability, indicative of oxidative modifications, upon blue light exposure in the eye. To do this, we exposed white-eyed w1118 flies to 8 h of blue light, which significantly decreases redox GSH:GSSG ratios relative to untreated controls (Fig. 2C) [30]. Retinal degeneration occurs progressively after the 8 h blue light exposure, and our previous studies generally assessed retinal degeneration 7–10 days post-exposure using confocal and/or electron microscopy [29,72]. Here to identify changes in the redox proteome that preceded retinal degeneration, we collected samples immediately after exposure to be processed using the same iodo-TMT approach that we developed for S2 cells. We identified 8811 unique total cysteine peptides in blue light versus control, of which 8421 could be quantified, corresponding to 2429 proteins (Fig. 2A) (Tables S5 and S6). We then identified significant changes in cysteine availability of 50 peptides (FDR <0.05), of which six were excluded as potential false positives after global normalization. Thus, after normalization, we identified 28 and 16 peptides with significant decreases or increases, respectively. This corresponds to a total of 23 and 15 potentially oxidized and reduced proteins under blue light exposure relative to controls (Fig. 2A &B) (Table S7).
Fig. 2.
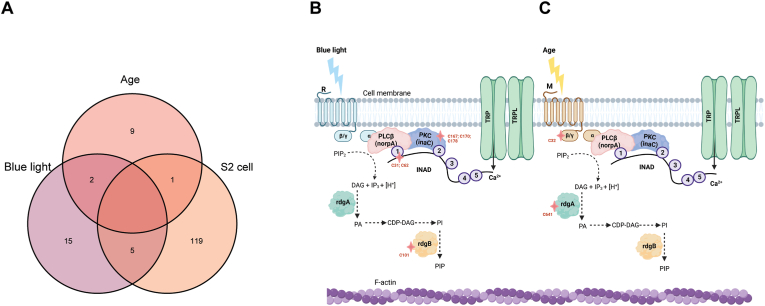
38 proteins show significant changes in redox cysteine availability upon blue light exposure of w1118flies.(A) Overview of Iodo-TMT method normalization for w1118 eyes. Global protein abundance was used to account for changes in total protein levels pre- and post-treatment (B) Normalized volcano and scatterplot as described in Fig. 1C. (C) Bar graph displaying redox GSH:GSSG ratios for blue light exposed w1118 flies relative to untreated control. Graph depicts ± standard deviation (S.D.) of three biological replicates. p-value (* < 0.05) Student's T-test, data reused from Ref. [30]. (D) Enriched GO process (GOP) categories for significantly oxidized proteins identified in the eye. (E) Domain map of inaC displaying protein kinase C-like, phorbol ester/diacylglycerol-binding domain (PE/DAG-bd) (purple) with oxidation modifications listed, C2 domain (green), protein kinase domain (pink), and protein kinase, C-terminal (blue) (Pfam). Alphafold prediction of Drosophila inaC with oxidized cysteine residues shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
We identified oxidized proteins involved in translation initiation and elongation, actin remodeling, transcription factors, and metabolism. For example, we identified eukaryotic translation initiation factor 3 subunit e (eIF3e) and eukaryotic translation elongation factor 5 (eEF5) as oxidized under blue light, both of which were also identified as oxidized following H2O2 exposure in S2 cells (Table S4). Additionally, we identified oxidation events on actin remodeling proteins such as twinstar (tsr) and failed axon connections (fax). Tsr is involved with several cytoskeleton processes such as F-actin turnover [73], and we observed potential oxidation at C64 which forms an intramolecular disulfide bond (Table 1). Interestingly, fax interacts with a tyrosine kinase known as Abelson (Abl) to form axon bundles [74]. Here, we observed oxidation of C332, which is on an alpha helix with the potential to disrupt the structure upon oxidation (Table 1). One of the most significantly oxidized proteins identified was adenosylhomocysteinase (Ahcy), which catabolizes the metabolite S-adenosylhomocysteine (SAH) into adenosine and homocysteine (Hcy) [[75], [76], [77], [78]]. Ahcy was oxidized at C195, which is a critical residue for its catalytic function [79] and may impair its enzymatic activity in vivo (Table 1). As in S2 cells, we identified thioredoxin reductase-1 (Trxr-1) as oxidized in blue light-treated eyes, consistent with its known reversible oxidation [80] (Table S7).
Table 1.
Oxidized proteins identified in the eye under blue light exposure.
| Protein | Cysteine Position | Function |
|---|---|---|
| Eukaryotic translation initiation factor 3 subunit e (Eif3E) | C11 | Translation initiation factor activity |
| Heat shock protein 83 (Hsp83) | C557 | Unfolded protein binding |
| Inactivation no afterpotential C (inaC) | C167; C170; C178 | Protein kinase C activity |
| Suppressor of Hairless (Su(H)) | C142; C146 | DNA-binding transcription factor activity |
| Retinal degeneration B (rdgB) | C101 | Phospholipid transporter activity |
| Twinstar (tsr) | C64 | Actin filament binding |
| Flapwing (flw) | C244 | Protein serine/threonine phosphatase activity |
| Glutamate dehydrogenase (Gdh) | C163 | Oxidoreductase activity |
| Thioredoxin reductase 1 (Trxr1) | C594; C595 | Oxidoreductase activity |
| Rudimentary-like (r-l) | C279 | Orotate phosphoribosyltransferase activity |
| Inactivation no afterpotential D (INAD) | C31; C62 | Signaling receptor complex adaptor activity |
| Helicase at 25E (Hel25E) | C195 | RNA helicase activity |
| Sine oculis (so) | C134 | Protein binding |
| Adenosylhomocysteinase (Ahcy) | C34; C195 | Catabolizes SAH |
| Failed axon connections (fax) | C332 | Axonogensis |
| Eukaryotic translation elongation factor 5 (eEF5) | C143 | Translation elongation factor activity |
| Regulatory particle non-ATPase 2 (Rpn2) | C105 | Enzyme regulator activity |
| Neither inactivation nor afterpotential B (ninaB) | C70 | Carotenoid dioxygenase activity |
| Barrier to autointegration factor (baf) | C81; C89 | DNA binding |
| Cytochrome P450 4d21 (Cyp4d21) | C79 | Oxidoreductase activity |
| GTPase activating protein and VPS9 domains 1 (Gapvd1) | C1293 | Guanyl-nucleotide exchange factor activity |
| Proteasome alpha1 subunit (Prosalpha1) | C78 | Peptidase activity |
Gene Ontology (GO) enrichment analysis revealed that the blue light-oxidized proteins were enriched for biological processes associated with the detection of and response to light stimulus or phototransduction (Fig. 2D). Many of these proteins are expressed predominantly in the eye including components of the phototransduction machinery, and proteins that maintain photoreceptor health. For example, multiple components of the phototransduction signaling cascade were oxidized under blue light including inaC, INAD, and retinal degeneration B (rdgB). The eye-specific protein kinase C, inaC, is necessary for the inactivation of light response [81], and it is known to interact with INAD and the TRP channels [82]. We did not observe oxidation at the active site of inaC; however, we identified oxidation at C167, C170, and C178 within the diacylglycerol-binding domain (Fig. 2E) (Table 1). The scaffolding protein INAD was oxidized at residues C31 and C62 in the PDZ1 domain, which interact with the eye-specific phospholipase C, norpA (Table 1) [83]. Lastly, rdgB was oxidized at residue C101 (Table 1), which is located on a beta sheet where an oxidation modification may disrupt its structure. The catalytic function of rdgB is to transfer phosphatidylinositol (PI) and phosphatidic acid (PA) between membranes [84], and rdgB mutants undergo rapid light-induced retinal degeneration [85].
2.3. Blue light suppresses the maximal ERG response amplitude, similar to light adaptation
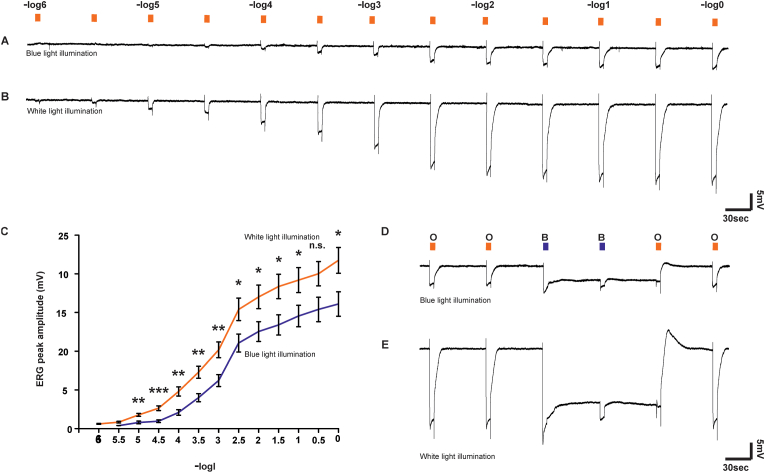
Because we identified several proteins involved in phototransduction that were oxidized upon exposure to prolonged blue light, we next asked if blue light treatment altered the ability of the eye to respond to light after the flies were removed from this acute light stress. Indeed, when we performed electroretinograms (ERGs) on flies exposed to prolonged blue light relative to white light, we found that the blue light-induced oxidation was accompanied by a large reduction in the sensitivity to light that did not arise from a reduction in the photopigment level (Fig. 3). Briefly, we exposed the flies to 8 h of blue illumination, using 8 h of white light exposure as a control, and then kept flies in the dark until tested. For each fly tested, intense orange light was applied for 5 min in order to convert maximal M molecules into the R state. Then, the fly was left in the dark for 20 min to ensure maximal dark adaptation. We tested the sensitivity to orange light by applying consecutive light pulses with an increasing light intensity, starting from dim lights (designated Intensity-Response Relationship). The peak amplitudes of the ERG responses (Fig. 3A and B) were plotted as a function of orange light intensity for the blue and white treated flies. Strikingly, prolonged blue light exposure shifted the Intensity-Response-Relationship to higher levels of light intensities and suppressed the maximal response amplitude reminiscent of light adaptation by continuous background light. This observation was unexpected given that the flies had been adapted to dark prior to performing the ERGs, and thus suggests that the prolonged blue light exposure alters the efficiency of phototransduction.
Fig. 3.
ERG measurements of the Intensity-Response Relationship and the PDA after blue and white light exposures.(A,B) ERG voltage responses to increasing intensity of orange (Schott 590 edge filter) light (in -log scale) of blue illuminated (A) and white illuminated (B) 5 days-old male flies. (C). The peak ERG amplitudes in response to the increasing light intensities were plotted as a function of relative light intensity. (D, E) PDA is induced by intense blue (B; Schott, BG 28 broad-band filter) light pulses and suppressed by orange (O; Schott OG 590 edge filter) light pulses applied to previously blue illuminated (D) (n = 11) and white illuminated (E) (n = 13) 5 day-old male flies. The paradigm of the assay included two intense orange light pulses followed by an intense blue light pulse, resulting in a prolonged depolarization of R1-6 photoreceptors (PDA, appears negative in the ERG) that continued in the dark long after the light was turned off. An additional intense blue light elicited small responses that originated from the central cells (R7-8) in which PDA was not induced, whereas the R1–6 cells were non-responsive (inactivated) due to maximal activation of the TRP/TRPL channels. The following orange light suppressed the PDA after light was turned off and allowed additional ERG responses to the orange light. Graphs depict ± standard error of the mean (SEM). p-value (* < 0.05, ** < 0.01, *** < 0.001) Student's T-test. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To test if the light adaptation phenotype resulting from prolonged blue light exposure was simply caused by a reduction in the levels of Rh1 photopigment, we used the PDA protocol. The PDA provides a convenient and reliable measure of the Rh1 photopigment level in vivo [33]. The paradigm of the assay included two intense orange light pulses followed by an intense blue light pulse, which converted ∼80% of the available Rh1 photopigment molecules from R to M pigment state in the flies (w1118), resulting in a PDA that continued in the dark long after the light was turned off. An additional intense blue light elicited small responses superimposed on the PDA that originated from the central cells (R7-8) in which PDA was not induced, whereas the R1-6 cells were non-responsive (inactivated) due to maximal activation of the TRP/TRPL channels. The following orange light, which converted virtually all M to R suppressed the PDA after light was turned off. The observation of maximal PDA after both white and blue prior 8 h lights indicated that the prolonged blue illumination did not reduce the photopigment level. Thus, we conclude that prolonged blue light exposure results in changes to the eye reminiscent of light adaptation, potentially via the cysteine oxidations in the phototransduction machinery observed in our redox proteomics.
2.4. Aging leads to oxidation of proteins involved in G-protein-coupled receptor signaling and regulation of rhodopsin in the eye
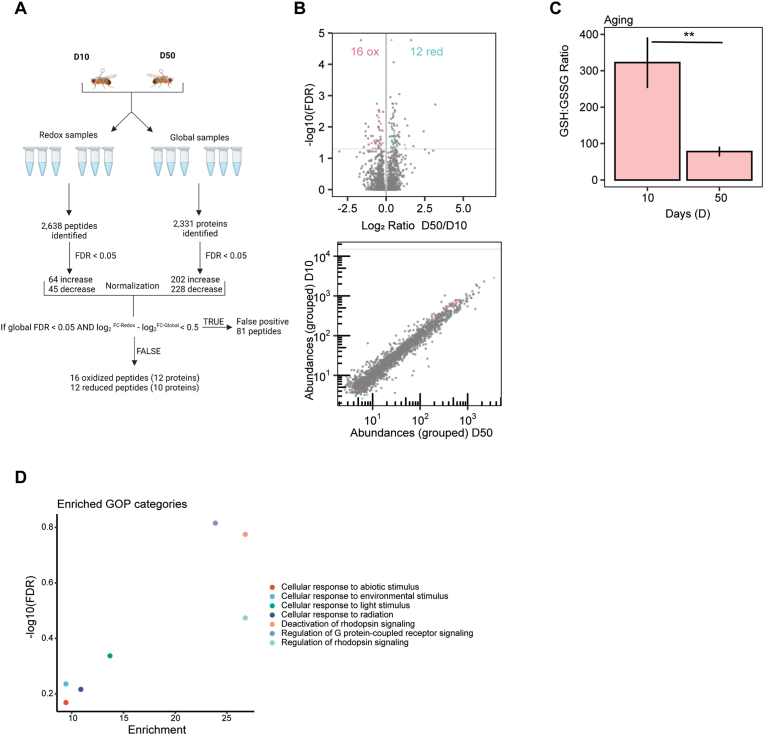
Because prolonged blue light exposure acts as an acute light stress on the Drosophila eye, we wondered how more chronic stress associated with aging in the eye [10,86] would impact the redox proteome. To do this, we compared OregonR (OrgR) flies at day 50 (D50) relative to day 10 (D10) in 12:12 light dark conditions. To assess relative oxidative stress, we measured redox glutathione ratios and observed a significant change in the ratios consistent with increased oxidative stress in old eyes versus young (Fig. 4C). Moreover, the decrease in redox glutathione ratios was similar in aging eyes relative to young eyes exposed to blue light (compare Fig. 2, Fig. 4). When we examined the redox proteome of aging eyes using the iodo-TMT approach, we identified 1847 quantifiable peptides out of 2638 unique peptides, corresponding to 2331 proteins (Fig. 4A) (Tables S9 and S10). Of these quantifiable peptides, 16 oxidized and 12 reduced peptides showed significant changes (FDR <0.05) with 81 peptides being excluded as false positives after global normalization (Fig. 4A and B). Thus, after normalization for changes in protein abundance, we identified 12 and 10 proteins with significant oxidation or reduction, respectively, in aged eyes relative to controls (Table S10). Interestingly, although aging and blue light exposure result in similar redox glutathione ratios, we observed fewer significantly oxidized proteins in the aging eye. We note that redox glutathione ratios only provide one measure of relative oxidative stress, and that lipid peroxidation was implicated in the retinal degeneration induced by blue light in young flies [29]. Moreover, we observed substantially more retinal degeneration in blue light-exposed young flies relative to old (D50) flies, consistent with the acute light stress induced by blue light exposure.
Fig. 4.
22 proteins show significant changes in redox cysteine availability observed in the aging eye of OregonR flies.(A) Overview of Iodo-TMT method normalization for aging eyes. Global protein abundance was used to account for changes in total protein levels pre- and post-treatment (B) Normalized volcano and scatterplot as described in Fig. 1C. (C) Bar graph displaying redox GSH:GSSG ratios for aging eyes relative to young eyes. Graph depicts ± S.D. of three biological replicates. p-value (** < 0.005) Student's T-test. (D) Enriched GO process (GOP) categories for oxidized proteins identified in the aging eye.
We next performed GO enrichment analysis on all significantly oxidized proteins and identified categories such as deactivation of and regulation of rhodopsin signaling or cellular response to light stimulus (Fig. 4D). Similar to the blue light redox dataset, we observed oxidation of phototransduction components including retinal degeneration A (rdgA) and G protein beta-subunit 76C (Gβ76C). RdgA is a DAG kinase, which converts DAG to PA [87]. RdgA mutants result in constitutive dark activity of the TRP and TRPL channels, most likely due to DAG accumulation, followed by a toxic increase in cellular Ca2+ leading to retinal degeneration [88]. RdgA was oxidized at C641, which has the potential to form disulfide bonds with two separate cysteine residues. The decrease in C641 availability we observe with age may indicate the formation of a disulfide with either of these available cysteine residues or could also represent an irreversible oxidation modification that may prevent the formation of these intramolecular disulfide bonds (Table 2). We also observed oxidation of Gβ76C, which is a subunit of the heterotrimeric G proteins that is responsible for the attachment of the Gqα to the plasma membrane and suppression of spontaneous activation of phototransduction [89]. In addition, several other eye-specific proteins were oxidized in old eyes including arrestin 1 (Arr1) and chaoptin (Chp). Arr1 is necessary for photoreceptor maintenance [90], and also participates indirectly in termination of the phototransduction cascade because it is essential for endocytosis of Rh1 [91]. Flies carrying mutations in arr1 undergo premature light-dependent retinal degeneration because of the overactivation of the phototransduction pathway [91]. We identified an oxidized C1252 within the catalytic domain of Chp, which is a glycosylphosphatidylinositol (GPI)-anchored glycoprotein necessary for photoreceptor cell morphogenesis [92,93]. These data suggest that the aging eye undergoes very different changes in the redox proteome relative to the acute blue light stress model, that based on the targets identified could contribute to an increased risk of retinal degeneration.
Table 2.
Oxidized proteins identified in the aging eye.
| Protein | Cysteine Position | Function |
|---|---|---|
| Ribosomal protein S21 (RpS21) | C56; C68 | Structural constituent of ribosome |
| Chaoptin (chp) | C1252 | Required for photoreceptor cell morphogenesis |
| Arrestin 1 (Arr1) | C182 | Regulates photoreceptor cell deactivation |
| Corkscrew (csw) | C326; C329 | Tyrosine phosphatase activity |
| G protein beta-subunit 76C (Gbeta76C) | C32 | Signaling receptor complex adaptor activity |
| Protein on ecdysone puffs (Pep) | C321; C324 | Single-stranded DNA/RNA binding |
| Retinal degeneration A (rdgA) | C641 | Diacylglycerol kinase activity |
| Cappuccino (capu) | C965 | Microtubule binding |
| Ribosomal protein S5a (RpS5a) | C90 | Structural constituent of ribosome/mRNA binding |
| CCHC-type zinc finger nucleic acid binding protein (CNBP) | C7; C70; C111 | Single-stranded DNA/RNA-binding |
| neither inactivation nor afterpotential B (ninaB) | C189; C303 | Carotenoid dioxygenase activity |
| Proteasome alpha1 subunit (Prosalpha1) | C78 | Peptidase activity |
Similar to S2 cells and blue light exposed flies, in the aging eye we identified oxidized proteins involved with translation such as ribosomal protein S21 (RpS21) and ribosomal protein S5a (RpS5a) (Table 2). RpS21 is not a core structural subunit of the ribosome, but it acts as a translation initiation factor [94]. RpS5a is a structural component of the ribosome with predicted function in assembly and translation [94,95]. Only two proteins were identified as being oxidized both in old eyes and under acute blue light stress: neither inactivation nor afterpotential B (ninaB) and proteasome alpha1 subunit (Prosα1). NinaB catalyzes the oxidative cleavage of carotenoids and is necessary for visual pigment production [96]. Although the modified cysteine residues in NinaB differ between blue light exposure and age, these cysteines are both located in areas that could disrupt the structure of the protein. Interestingly, Prosα1, a subunit of the proteasome complex, was oxidized at the same position under blue light exposure and age: C78, located inside the catalytic chamber on a beta sheet.
3. Discussion
Several studies have examined redox proteomic changes in Drosophila tissues [56,57] and other organisms [45,[48], [49], [50], [51], [52], [53],64], but to our knowledge, this is the first study to characterize the redox proteome of either Drosophila S2 cells or the fly eye. Tissue-specific studies for redox proteomics remain relatively rare and can provide valuable insight into those proteins and pathways that are susceptible to oxidative modifications in particular cell types or tissues. Here, we used iodo-TMT reagents to profile the cysteine redox proteome of Drosophila eyes and cultured cells exposed to different stress conditions. Previous Drosophila redox studies have utilized oxidative isotope-coded affinity tags (OxICAT) to profile the redox proteome; however, fewer than 500 proteins were identified due to limitations of this technique [56]. Menger et al., 2015 used OxICAT to measure cysteine residue redox states under aging and fasting conditions in heads and thoraces of flies [56]. They observed more significant changes in the redox state of the cell after 24 h of fasting compared to aging, specifically in proteins involved with translation, redox-sensing, and glycolysis [56]. More sensitive techniques such as SICyLIA and iodo-TMT multiplexing have been developed since OxICAT methods, and these newer techniques have proven to be more efficient for redox profiling. SICyLIA was designed to directly compare cysteine oxidation levels between two sample groups labeled with either light or heavy IAA and does not require enrichment steps [53]. However, in our hands, the lack of enrichment steps with SICyLIA resulted in biased proteome sensitivity leading to the identification of moderately abundant peptides at the expense of less abundant proteins. Based on these limitations of SICyLIA in our studies, we turned to the iodo-TMT multiplex reagents. These iodo-TMT reagents allow for increased statistical power and decreased mass spectrometry costs through multiplexing, resin enrichment for TMT-tagged peptides, additional controls for global protein abundances across treatment groups, and the ability to perform FDR corrections ensuring high confidence identification of oxidized and reduced proteins. Our data demonstrate that iodo-TMT is compatible with both cells and whole-tissue lysates using relatively small amounts of input material.
The redox proteome of mammalian cells and tissues has also been profiled using several different approaches, including an alternative method based on the iodo-TMT reagents used in our study. In this alternative approach, iodo-TMT reagents were used to identify reversibly oxidized cysteines in mouse hematopoietic stem and progenitor cells (HSPCs) comparing fetal and adult mice, and human embryonic stem cells (hESC) [64]. Reversibly oxidized cysteines such as disulfide bonds were identified by labeling free thiol groups with iodo-TMT tags, followed by a reduction step, and the addition of another iodo-TMT tag to label those thiols that has been reversibly modified [64]. In addition, a recent study characterized the proteome of human corneal endothelial cells (iHCEC) exposed to blue light using chemoproteomic probes, which are not currently commercially available [63]. When comparing this dataset to our in vivo blue light redox proteomics dataset, we observed oxidation of five proteins that have potential orthologs in Drosophila: heat shock protein 90 alpha family class B member 1 (HSP90AB1), cofilin 1 (CFL1), glutamate dehydrogenase 1 (GLUD1), DExD-box helicase 39B (DDX39B), and proteasome 20S subunit alpha 6 (PSMA6) (Table 3). The modified cysteine residues were not described for the iHCEC data, so could not be compared with our Drosophila redox proteomic data. Hsp83 (HSP90AB1) was also identified as oxidized in H2O2-treated S2 cells, suggesting that this protein might be a common target for oxidation across multiple cell types. We expect that the phototransduction proteins that were enriched in our analysis of blue light-treated eyes were not identified in the corneal cells because these proteins are not expressed in these cells.
Table 3.
Oxidized proteins identified in blue light treated Drosophile eyes and in blue light treated human iHCEC cells.
| Drosophila Protein | Human Protein Ortholog |
|---|---|
| Heat shock protein 83 (Hsp83) | Heat Shock Protein 90 Alpha Family Class B Member 1 (HSP90AB1) |
| Inactivation no afterpotential C (inaC) | Protein Kinase C Iota (PRKCI) |
| Twinstar (tsr) | Cofilin 1 (CFL1) |
| Glutamate dehydrogenase (Gdh) | Glutamate Dehydrogenase 1 (GLUD1) |
| Helicase at 25E (Hel25E) | DExD-Box Helicase 39B (DDX39B) |
| Proteasome alpha1 subunit (Prosα1) | Proteasome 20S Subunit Alpha 6 (PSMA6) |
Overlapping oxidized proteins identified in redox proteomic dataset of blue light-treated Drosophila and blue light-treated iHCEC cells [63]. Human orthologs of Drosophila proteins were obtained from DRSC integrative ortholog prediction tool (DIOPT).
When we compared the redox proteome induced by H2O2 treatment in cultured cells relative to eyes, either under acute light stress or during aging, we observed that very few proteins were oxidized under more than more than one condition and no proteins were oxidized in all three datasets (Fig. 5A). These data suggest that the specific changes in the redox proteome are heavily dependent on both the type of stress condition and on the tissue or cell type being examined. In eyes, we found that proteins involved in phototransduction were oxidized under both blue light and aging, although the specific proteins and cysteine residues differed under the two conditions (see next paragraph). In contrast, some broad categories of proteins were reproducibly oxidized in all three models including proteins involved in translation such as eIF and ribosomal subunits. Notably, these proteins were also identified as oxidized in mammalian HSPCs [64] and mouse embryonic kidney cells treated with H2O2 [53]. There were five overlapping oxidized proteins when we compared our two acute stress models, H2O2 treatment in cells and prolonged blue light exposure in eyes, including eIF subunits, Hsp83, regulatory particle non-ATPase 2 (Rpn2), a regulatory subunit of the 26S proteasome, and the metabolic enzyme Ahcy. Ahcy is an essential enzyme in methionine metabolism that breaks down S-adenosylhomocysteine (SAH), which can inhibit methyltransferase activity [[97], [98], [99], [100], [101], [102]]. Ahcy was oxidized at a conserved cysteine (C195) that was also identified in mouse kidney cells using the SICyLIA approach [53], and is essential for its activity [79], suggesting that Ahcy activity could be impacted negatively by redox signaling.
Fig. 5.
Phototransduction machinery is oxidized upon blue light exposure and during age in the eye.(A) Venn diagram of overlapping oxidized proteins identified from blue light exposed flies, aging flies, and S2 cells. (B, C) Model of phototransduction proteins with oxidized proteins identified under blue light exposure and age. Oxidation is indicated by a star symbol (pink). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
In the eye, we identified oxidation of key phototransduction components including inaC, INAD, and rdgB in blue light, and Arr1, rdgA, and Gβ76C in aging (Fig. 5B and C), suggesting that acute light stress and aging might differently affect phototransduction processes. Indeed, in vivo functional assays performed by measuring the sensitivity to light using the ERG response revealed a large (∼10 fold) reduction in the sensitivity to light after intense 8 h blue light relative to intense 8 h white light of equal energy. Strikingly, this reduction in sensitivity to light is reminiscent of light adaptation during background light [103], except that in the current study all measurements were carried out in a dark-adapted state. This reduction in sensitivity to light was not accompanied by a reduction in photopigment level, suggesting that there are longer-term changes in phototransduction efficiency caused by blue light. One of the blue light oxidized phototransduction kinases that regulates light response termination, inaC, is a candidate for regulating this alteration in phototransduction efficiency because inaC mutants display defects in light adaptation [81,104]. Moreover, inaC is inhibited by increased concentrations of Ca2+ [105], and preventing Ca2+ influx by trp mutations rescues blue light-induced retinal degeneration [29]. One possibility is that the cysteine oxidation in inaC shifts this kinase into an inactive conformation, similar to that resulting from high Ca2+ levels. However, identifying the mechanism that underlies the striking reduction in sensitivity to light caused by blue light exposure will require careful analysis of point mutants in these phototransduction proteins, and it is likely that more than one target could contribute to the phenotype we observed. Thus, one limitation to our study is that we have not directly shown which phototransduction proteins are responsible for the ERG phenotype. Other groups have performed ERGs in old flies and observed decreased transient amplitudes [106], differing from our observations under blue light, consistent with the idea that the different phototransduction proteins identified as oxidized under each condition result in different physiological outcomes. Since we only measured glutathione ratios in the eye under light stress and aging, which is a limitation of our study, it is possible that there could be changes in the levels and/or localization of specific ROS under these two different conditions. In addition, there are both overlapping and distinct gene expression changes in photoreceptors within the eye under blue light versus aging [30], suggesting that each of these conditions have some unique effects on the eye. For example, aging is associated with chronic inflammation and expression of proteins associated with the innate immune response [67] whereas blue light stress induces hyperactivation of the phototransduction pathway and perturbs calcium homeostasis in the eye [29]. Thus, further studies will be necessary to determine why there are such distinct changes in the redox proteome in light stress relative to aging, and to determine how these changes impact the physiology of the eye.
4. Materials and methods
4.1. S2 cell culture, H2O2 treatment, and viability assays
S2 cells were cultured at 1 × 106 cells/mL in 1X Schneider's Drosophila medium (Gibco, Cat #21720024), 1% Pen-Strep, and 10% FBS for 24 h prior to treatment with H2O2 (Sigma, Cat #7722-84-1). Cells were stained with Trypan blue to determine cell count and viability using a Thermo Countess II Cell Counter (Invitrogen, Cat #AMQAX1000).
4.2. Drosophila culture and blue light treatment
All experiments were conducted with male w1118 (blue light), or OregonR flies (aging). Flies were raised on cornmeal agar food (6.07 g agar type 2, 32 g sugar, 50 g yeast, 50 g cornmeal, 3.2 g methyl paraben for preservation in 1.3 L water) at 25 °C and 65–75% humidity under a 12:12 light:dark cycle. Flies were collected 3 days post eclosion and aged for 5 days with transferring to fresh food every 3 days. Flies were exposed to blue light using a custom designed light stimulator at 8000 lux (2 mW/cm2, λ = 465 nm) for 8 h [72].
4.3. GSH:GSSG, NAD:NADH, and NADP:NADPH targeted metabolomic assays
1 × 106 cells/mL S2 cells or 25 dissected eyes were analyzed per biological replicate for each targeted redox metabolite assay. GSH:GSSG ratios were analyzed as described previously [28]. For NAD:NADH and NADP:NADPH assays, S2 cells were washed in PBS and resuspended in methanol. We analyzed samples using an Agilent 1290 Infinity II liquid chromatography (LC) system coupled to an Agilent 6470 series QQQ mass spectrometer (MS/MS) (Agilent Technologies, Santa Clara, CA) with a Waters HSS T3 2.1 mm × 100 mm, 1.7 μm column used for LC separation (Water Corp, Milford, MA). Buffers included (A) water + 10 mM ammonium acetate (pH 6) and (B) acetonitrile with the following linear LC gradient: time 0 min, 0% B; time 5 min, 0% B; time 20 min, 40% B; time 21 min, 90% B; time 21.1 min, 0% B; time 25 min, 0% B at a flow rate of 0.3 mL/min. Data were acquired in positive electrospray ionization (ESI) mode. The jet stream ESI interface had a gas temperature of 325 °C, gas flow rate of 8 L/min, nebulizer pressure of 45 psi, sheath gas temperature of 250 °C, sheath gas flow rate of 7 L/min, capillary voltage of 4000 V in positive mode, and nozzle voltage of 1000 V. The ΔEMV voltage was 400 V. Agilent Masshunter Quantitative analysis software was used for data analysis (version 8.0). For targeted metabolite studies: significance analysis was performed using ANOVA and Tukey's post-hoc with p ≤ 0.05.
4.4. Drosophila S2 cell SICyLIA protein isolation and labeling
The following protocol was modified from a SICyLIA based approach as described in Ref. [53]. Cells were washed in PBS and lysed in 300 μL of 8 M urea/50 mM Tris-HCl pH 8.5. One replicate of the treated cells and one replicate of the untreated cells were labeled with IAA light (12C2H4INO) (Sigma, Cat #144-48-9), and the other corresponding replicates were labeled with IAA heavy (13C2D2H2INO) (Sigma). Samples were incubated for 1 h at room temperature (RT) protected from light. Protein was quantified with Qubit 4 fluorometer (Thermo Fisher Scientific™, Cat #Q33238). 150 μg of protein from light and heavy carbamidomethylated samples at each time point (0 light with 15 min heavy and 0 min heavy with 15 min light labeled) were mixed, reduced with TCEP and alkylated with N-methylmaleimide (NEM) at room temperature in the dark. Following overnight at 4 °C trichloroacetic acid precipitation (TCA added at 20% final v/v) and centrifugation for 30 min at 14,000×g, pellets were resuspended in 8 M Urea in 100 mM 100 mM Tris pH 8.5. Urea was diluted to <2 M with 50 mM Tris-HCl, pH 8.5 and proteins were digested overnight at room temperature with trypsin/LysC (ratio of 1:50, Promega, Cat #V5072). After quenching the digestion with 0.5% TFA, samples were fractionated with a Pierce high pH basic reversed-phase peptide fractionation kit (Thermo Fisher Scientific™, Cat #84858) using the protocol for label free peptide fractionation.
4.5. Nano-LC-MS/MS analysis of Drosophila cells using SICyLIA
Nano-LC-MS/MS analyses were performed on an EASY-nLC HPLC system (SCR: 014993, Thermo Fisher Scientific™) coupled to Orbitrap Lumos Tribrid™ mass spectrometer (Thermo Fisher Scientific™). 1/5 of each peptide fraction was loaded onto a reversed phase EasySpray™ C18 column (2 μm, 100 Å, 75 μm × 25 cm, Thermo Scientific™, Cat #ES902) at 400 nL/min. Peptides were eluted from 4 to 30% with mobile phase B (Mobile phases A: 0.1% FA, water; B: 0.1% FA, 80% Acetonitrile (Thermo Fisher Scientific™, Cat #LS122500)) over 160 min, 30–80% B over 10 min; and dropping from 80 to 10% B over the final 10 min. The mass spectrometer was operated in positive ion mode with a 3 s cycle time data-dependent acquisition method with advanced peak determination and Easy-IC (internal calibrant). Precursor scans (m/z 400–1600) were done with an orbitrap resolution of 120000, RF lens% 30, auto max inject time, standard AGC target, MS2 intensity threshold of 5e3, including charges of 2–7 for fragmentation with 60 s dynamic exclusion. MS2 scans were performed with a quadrupole isolation window of 1 m/z, 35% CID CE, Rapid ion trap scan rate, 20% normalized AGC target, dynamic maximum IT.
4.6. Drosophila iodo-TMT protein isolation, labeling, and enrichment
1 × 106 cells/mL or 250 eyes were analyzed per biological replicate (n = 3 control; n = 3 treatment) for cysteine availability analysis. Cells were harvested and resuspended in 1 mL cold PBS, and then spun at 20,000×g for 2 min at 4 °C. This was repeated twice to avoid media contamination. Cell pellets were lysed in 1 mL of HES (50 mM HEPES pH 8.0, 1 mM EDTA, 0.1% SDS) and ultrasonicated using a Covaris E220 (Covaris, Cat #500239). Eyes were manually dissected, directly added in HES buffer, and ultrasonicated as previously described. Lysates were collected centrifuged at 10,000×g for 10 min at 4 °C and quantified using Qubit 4 fluorometer (Thermo Fisher Scientific™, Cat #Q33238). All samples were set to 250 μg, and 50 μg was reserved from each sample to be used for global analysis. Each 200 μg replicate received an independent iodoTMT reagent (Thermo Fisher Scientific™, Cat #90066) that was solubilized in 10 μL LC/MS-grade methanol (Fisher Chemical, Cat #A452-4). The reaction proceeded for 1 h at 37 °C protected from light. All samples were reduced with 4 μL of 0.5 M DTT (Thermo Fisher Scientific™, Cat #A39255) for 15 min at 37 °C protected from light. After reduction, a second labeling with 55 mM IAA dissolved in (10 M Urea, 1 M Tris-HCl pH 8.5, and water) was added to each sample. Equal amounts of all samples were combined into one tube and precipitated using 6 vol of pre-chilled acetone overnight. The sample was then centrifuged at 10,000×g for 10 min at 4 °C, the acetone was decanted, and then centrifuged once more to remove remaining acetone. The acetone-precipitated pellet was dissolved with 50 mM ammonium bicarbonate buffer pH 8.0 and was digested with MS-grade trypsin protease (Thermo Fisher Scientific™, Cat #90057) overnight at 37 °C. After digestion, the sample was acidified by adding 4 μL of 10% trifluoroacetic acid (TFA) (Thermo Fisher Scientific™, Cat #28904). The sample was then flash frozen in liquid nitrogen and lyophilized. The sample was resuspended in 100 μL of 1X tris-buffered saline (TBS), and the anti-TMT resin (Thermo Fisher Scientific™, Cat #90076) was washed 3 times with 1 column volume 1 X TBS. 100 μg was reserved as an unfractionated sample for direct analysis of non-enriched peptides. The remaining sample was added to the resin and incubated for 2 h at RT with end-over-end rotation. The resin was then washed 3 times with one column volume of 1 X TBS, 3 times with one column volume of water, and then enriched peptides were eluted with 4 column volumes of TMT elution buffer (Thermo Fisher Scientific™, Cat #90104).
4.7. Global TMT protein isolation, labeling, and analysis
25–50 μg protein lysate (cells or eye tissue) was denatured by addition of 2x volume 8 M urea in 100 mM 100 mM Tris pH 8.5, reduced with 5 mM tris(2-carboxyethyl) phosphine hydrochloride (TCEP, Sigma-Aldrich, Cat #C4706) for 30 min at RT, and alkylated with 10 mM chloroacetamide (CAA, Sigma Aldrich, Cat #C0267) for 30 min at RT in the dark. Samples were diluted with 50 mM Tris HCl, pH 8.5 to a final urea concentration of 2 M and then digested overnight at 35 °C with 2 μg Trypsin/Lys-C (1:25 ratio, Mass Spectrometry grade, Promega Corporation, Cat #V5072). Digestions were acidified with trifluoracetic acid (TFA, 0.5% v/v) and desalted on Waters Sep-Pak cartridges, (Waters™, Cat #WAT054955) with a wash of 1 mL 0.1% TFA followed by elution in 70% acetonitrile containing 0.1% formic acid (FA). Peptides were dried by speed vacuum and resuspended in 24 μL of 50 mM triethylammonium bicarbonate pH 8.0 (TEAB). Peptide quantitation was performed using Pierce Colorimetric Peptide Assay kit (Thermo Fisher Scientific™, Cat #23275). For each global experiment, equivalent peptide amounts were then TMT labeled for 2 h at RT with 0.2 mg of Tandem Mass Tag (TMT, Thermo Fisher Scientific™, TMT™ Isobaric Label Reagent Set; Cat #90111, lot no. VH306777 (126, 127C and 128C for controls; 129C, 130C and 131 for experiment) for S2 cells and WC306775 (129C, 130C and 131 for control; 126, 127 N and 128 N for experiment) for eye tissue. Labelling reactions were quenched by adding 0.2% hydroxylamine (final v/v) to the reaction mixtures at RT for 15 min. Labeled peptides were then combined, mixed, and dried by speed vacuum. After drying, samples were fractionated with a Pierce high pH basic reversed-phase peptide fractionation kit (Thermo Fisher Scientific™, Cat #84858) as described above.
4.8. Nano-LC-MS/MS analysis of Drosophila cells and eye tissue using iodo-TMT reagents
Nano-LC-MS/MS analyses were performed on an EASY-nLC HPLC system (SCR: 014,993, Thermo Fisher Scientific™) coupled to Orbitrap Eclipse™ mass spectrometer (Thermo Fisher Scientific™) with a FAIMS pro interface. Peptides were separated on a 25 cm aurora column (IonOpticks, AUR2-25075C18A) at 400 nL/min with a gradient of 5–28% with mobile phase B (Mobile phases A: 0.1% FA, water; B: 0.1% FA, 80% Acetonitrile (Thermo Fisher Scientific™, Cat #LS122500)) over 160 min, 30–80% B over 10 min; and dropping from 80 to 10% B over the final 10 min. The mass spectrometer was operated in positive ion mode with 3 FAIMS CVs (−45, −55, −70) and 1.3 s cycle time per CV. Data-dependent acquisition method with advanced peak determination and Easy-IC (internal calibrant) were used. Precursor scans (m/z 400–1600) were done with an orbitrap resolution of 120,000, RF lens% 30, maximum inject time 105 ms, standard AGC target, MS2 intensity threshold of 2.5e4, precursor fit threshold of 70% and 0.7 m/z window, including charges of 2–6 for fragmentation with 60 s dynamic exclusion, single charge state per precursor dependent scan. MS2 scans were performed with a quadrupole isolation window of 0.7 m/z, 38% HCD CE, 50,000 resolution, 200% normalized AGC target, dynamic maximum IT fixed first mass of 100 m/z. The data were recorded using Thermo Fisher Scientific Eclipse Tune (v 3.3.2782.34) software (Thermo Fisher Scientific™).
4.9. Mass spectrometry data analysis
Resulting RAW files were analyzed in Proteome Discover™ 2.5 (Thermo Fisher Scientific™, RRID: SCR_014477) with a Drosophila melanogaster UniProt FASTA plus common contaminants (downloaded Feb 2021, 3629 total sequences). SEQUEST HT searches were conducted with a maximum number of 3 missed cleavages; precursor mass tolerance of 10 ppm; and a fragment mass tolerance of 0.02 Da. For iodoTMT samples, a max of 5 dynamic mods were allowed per peptide and dynamic modifications of oxidation (Met, His, Trp), iodoTMT6plex (Cys), carbamidomethyl (Cys); dynamic protein terminus modifications of acetylation (N-terminus), Met-loss or Met-loss plus acetylation (N-terminus). For global proteome samples, the same precursor/fragment/missed cleavage/max dynamic mod settings were used in addition to static modifications of TMT6plex on lysines (K) and carbamidomethylation on cysteine (C) residues; Dynamic modifications of oxidation on (M, H, W), TMT label on the N-termini of peptides, phosphorylation on (S,T,Y); dynamic Protein terminus modifications of acetylation (N-terminus), Met-loss or Met-loss plus acetylation (N-terminus). Percolator False Discovery Rate was set to a strict setting of 0.01 and a relaxed setting of 0.05. IMP-ptm-RS node was used for all modification site localization scores. In the Proteome Discover 2.5 consensus step, total peptide amount was used as the normalization mode. In this mode, the total sum of abundance values for each channel over all peptides is calculated, the channel with the highest total is used as a reference, and all other channels are then corrected by a constant factor so that at the end the total abundance is the same for all channels. Co-isolation thresholds of 50% and average reporter ion S/N cutoffs of 5 were used for quantification. Lot specific isotopic impurity levels were applied. Protein Abundance based protein ratio calculation with no imputation and ANOVA (individual proteins) was performed. The Reporter Ions Quantifier node was set to consider a peptide unique if it is included in only one protein group. Unique and razor peptides were used for protein quantification. All shared peptides were used for the quantification of the protein that has more identified peptides, but not for the other proteins they are contained in. Peptides are considered shared/not used for quantification if they reference proteins from different protein groups. The best master protein is the protein with the largest number of unique peptides, and if multiple proteins have the same identification score, equal number of peptides and equal number of PSMs, the protein with the longest sequence was designated the master protein. Strict protein grouping parsimony was applied so that all protein groups are excluded that are not strictly necessary to explain the identified peptides. Resulting normalized abundance values for each sample type (iodo-TMT labeled peptides or global proteins), abundance ratio and log2 (abundance ratio) values; and respective unadjusted p-values (t-test) from Proteome Discover™ were exported to Microsoft Excel. These normalized abundance values were used for subsequent statistical analysis, as outlined below and in the flow-charts shown in the figures. Six biological replicates were collected for iodo-TMT based redox proteomics and all targeted metabolomic studies. Two biological replicates were collected for SICyLIA-based redox proteomics. There were two experimental groups for S2 cells: H2O2-treated S2 cells and untreated/control S2 cells. For blue light exposure analysis there were two experimental groups for w1118 fly experiments: blue light exposed day 5 flies and non-exposed/control day 5 flies. For aging experiments, there were also two experimental groups: aged OregonR flies at D50 and young OregonR flies at D10. For all proteomic studies: oxidized proteins with an FDR <0.05 and log2FC < 0 with the difference between redox and global protein abundance log2FC > 0.5 were considered significant. Reduced proteins with FDR <0.05 and log2FC > 0 with the difference between redox and global protein abundance log2FC > 0.5 were considered significant. FDR corrections were calculated using a PolySTest [66].
4.10. Electroretinogram (ERG)
ERGs were recorded from immobilized male flies at the age of 5 days, as described previously [107]. Extracellular ERG light responses were measured at 20 °C with standard glass micropipettes filled with Ringer's solution (130 mM NaCl, 2 mM KCl, 5 mM MgCl2, 2 mM CaCl2, 10 mM Hepes, pH 7.0) introduced through the cornea into the extracellular matrix of the eye under dim red light (Schott 630 nm edge filter). Flies were grounded via a reference electrode that was placed on the thorax in a drop of electrode gel. Light from a xenon high-pressure lamp (PTI, LPS 220, operating at 50 W) passed via red (Schott 590 nm orange edge filter) or blue (Schott BG 28 broad band blue filter) filters was delivered to the compound eye via a light guide. The blue light, which is absorbed by fly Rh1, rhodopsin ensured strong activation of Rh1-rhodopsin. At the same time, the orange light ensures an efficient M to R photoconversion, but also activation of phototransduction during light. Accordingly, the use of orange light that is absorbed by the long wavelength range of R absorption spectrum prevented the induction of the prolonged depolarizing afterpotential (for a review see Ref. [33]), which constitutes a saturated response that extends in the dark for many minutes. The PDA was induced by intense blue (B; Schott, BG 28 broad-band filter) light pulses and suppressed by orange (O; Schott OG 590 edge filter) light pulses. Signals were amplified using a homemade amplifier. The maximal luminous intensity at the eye surface was about 3.5 logarithmic intensity units above the intensity for a half-maximal response. To measure the Intensity-Response Relationships the peak ERG amplitudes in response to increasing intensities of the orange light pulses were measured up to saturated amplitude.
Funding
This research was funded by the National Eye Institute of the NIH under Award Numbers R01EY024905 and R01EY033734 to V.M.W., the Bird Stair Research Fellowship (Biochemistry Department, Purdue University) to S.C.S., NIH training award 5T32GM125620 to S.C.S., and the Israel Science Foundation (ISF) to B.M. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Declaration of competing interest
None.
Acknowledgements
We thank the Weake lab for their comments on the manuscript. We thank Dr. Aruna Wijeratne and Dr. Hana Hall for their contributions to preliminary proteomic experimental design.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102723.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
Exposure to H2O2induces oxidative stress in Drosophila S2 cells.(A) Line graph displaying cell proliferation after H2O2 treatment. (B) Line graph displaying cell viability after H2O2 treatment. (C) Bar graphs displaying relative ratio of intracellular NADH:NAD+, NADPH:NADP+, and GSH:GSSG after treatment with 20 mM H2O2 for the indicated time relative to untreated S2 cells. All graphs depict mean standard deviation (S.D) of three biological replicates. p-value (* < 0.05, ** < 0.005, *** <0.001) Student's T-test.
Data availability
The mass spectrometry proteomic data for Drosophila melanogaster S2 cells and w1118 flies exposed to blue light have been deposited to the ProteomeXchange Consortium via the PRIDE [108] partner repository with the dataset identifier PXD034050 and 10.6019/PXD034050. Additionally, proteomic data for aging Drosophila melanogaster eyes have been deposited to the ProteomeXchange Consortium via the PRIDE [108] partner repository with the dataset identifier PXD036110 and 10.6019/ PXD036110.
References
- 1.Rahal A., et al. Oxidative stress, prooxidants, and antioxidants: the interplay. BioMed Res. Int. 2014;2014 doi: 10.1155/2014/761264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrés Juan C., Manuel Pérez de la Lastra J., Plou F.J., Pérez-Lebeña E., Reinbothe S. Molecular sciences the chemistry of reactive oxygen species (ROS) revisited: outlining their role in biological macromolecules (DNA, lipids and proteins) and induced pathologies. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22094642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murray C.I., Van Eyk J.E. Chasing cysteine oxidative modifications: proteomic tools for characterizing cysteine redox status. Circ. Cardiovasc. Genet. 2012;5 doi: 10.1161/CIRCGENETICS.111.961425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poole L.B., Nelson K.J. Discovering mechanisms of signaling-mediated cysteine oxidation. Curr. Opin. Chem. Biol. 2008;12 doi: 10.1016/j.cbpa.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Domènech E.B., Marfany G. The relevance of oxidative stress in the pathogenesis and therapy of retinal dystrophies. Antioxidants. 2020;9 doi: 10.3390/antiox9040347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ozawa Y. Oxidative stress in the light-exposed retina and its implication in age-related macular degeneration. Redox Biol. 2020;37 doi: 10.1016/j.redox.2020.101779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forman H.J., Fukuto J.M., Torres M. Redox signaling: thiol chemistry defines which reactive oxygen and nitrogen species can act as second messengers. Am. J. Physiol. Cell Physiol. 2004;287 doi: 10.1152/ajpcell.00516.2003. [DOI] [PubMed] [Google Scholar]
- 8.McDonagh B. Detection of ROS induced proteomic signatures by mass spectrometry. Front. Physiol. 2017;8 doi: 10.3389/fphys.2017.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katz B., Minke B. Drosophila photoreceptors and signaling mechanisms. Front. Cell. Neurosci. 2009;3 doi: 10.3389/neuro.03.002.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lennicke C., Cochemé H.M. Redox signalling and ageing: insights from Drosophila. Biochem. Soc. Trans. 2020;48 doi: 10.1042/BST20190052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bilen J., Bonini N.M. Drosophila as a model for human neurodegenerative disease. Annu. Rev. Genet. 2005;39 doi: 10.1146/annurev.genet.39.110304.095804. [DOI] [PubMed] [Google Scholar]
- 12.Ugur B., Chen K., Bellen H.J. Drosophila tools and assays for the study of human diseases. DMM Dise. Models Mech. 2016;9 doi: 10.1242/dmm.023762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bateman A., et al. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49 doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sachdeva M.M., Cano M., Handa J.T. Nrf2 signaling is impaired in the aging RPE given an oxidative insult. Exp. Eye Res. 2014;119 doi: 10.1016/j.exer.2013.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bazopoulou D., et al. Developmental ROS individualizes organismal stress resistance and lifespan. Nature. 2019;576 doi: 10.1038/s41586-019-1814-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rabe von Pappenheim F., et al. Widespread occurrence of covalent lysine–cysteine redox switches in proteins. Nat. Chem. Biol. 2022;18 doi: 10.1038/s41589-021-00966-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wensien M., et al. A lysine–cysteine redox switch with an NOS bridge regulates enzyme function. Nature. 2021;593 doi: 10.1038/s41586-021-03513-3. [DOI] [PubMed] [Google Scholar]
- 18.Ready D.F., Hanson T.E., Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev. Biol. 1976;53 doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- 19.Tomlinson A. Cellular interactions in the developing Drosophila eye. Development. 1988;104 doi: 10.1242/dev.104.2.183. [DOI] [PubMed] [Google Scholar]
- 20.Wolff T., Ready D. Pattern formation in the Drosophila retina. Cold Spring Harb. Protoc. 1993;2:1277–1325. [Google Scholar]
- 21.Katz B., Minke B. The Drosophila light-activated TRP and TRPL channels - targets of the phosphoinositide signaling cascade. Prog. Retin. Eye Res. 2018;66 doi: 10.1016/j.preteyeres.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 22.Hardie R.C. vol. 1. Wiley Interdiscip Rev Membr Transp Signal; 2012. (Phototransduction Mechanisms in Drosophila Microvillar Photoreceptors). [Google Scholar]
- 23.Montell C. Drosophila visual transduction. Trends Neurosci. 2012;35 doi: 10.1016/j.tins.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu W., et al. The INAD scaffold is a dynamic, redox-regulated modulator of signaling in the Drosophila eye. Cell. 2011;145 doi: 10.1016/j.cell.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 25.Sheng M., Sala C. PDZ domains and the organization of supramolecular complexes. Annu. Rev. Neurosci. 2001;24 doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- 26.Tsunoda S., et al. A multivalent PDZ-domain protein assembles signalling complexes in a G- protein-coupled cascade. Nature. 1997;388 doi: 10.1038/40805. [DOI] [PubMed] [Google Scholar]
- 27.Huber A., et al. The transient receptor potential protein (Trp), a putative store-operated Ca2+ channel essential for phosphoinositide-mediated photoreception, forms a signaling complex with NorpA, InaC and InaD. EMBO J. 1996;15 [PMC free article] [PubMed] [Google Scholar]
- 28.Jauregui-Lozano J., et al. The Clock:Cycle complex is a major transcriptional regulator of Drosophila photoreceptors that protects the eye from retinal degeneration and oxidative stress. PLoS Genet. 2022;18 doi: 10.1371/journal.pgen.1010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X., et al. Cytochrome b5 protects photoreceptors from light stress-induced lipid peroxidation and retinal degeneration. NPJ Aging Mech Dis. 2017;3 doi: 10.1038/s41514-017-0019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Escobedo S.E., Stanhope S.C., Dong Z., Weake V.M. Aging and light stress result in overlapping and unique gene expression changes in photoreceptors. Genes. 2022;13:264. doi: 10.3390/genes13020264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hardie R.C., Minke B. The trp gene is essential for a light-activated Ca2+ channel in Drosophila photoreceptors. Neuron. 1992;8 doi: 10.1016/0896-6273(92)90086-s. [DOI] [PubMed] [Google Scholar]
- 32.Peretz A., et al. The light response of drosophila photoreceptors is accompanied by an increase in cellular calcium: effects of specific mutations. Neuron. 1994;12 doi: 10.1016/0896-6273(94)90442-1. [DOI] [PubMed] [Google Scholar]
- 33.Minke B. The history of the Prolonged Depolarizing Afterpotential (PDA) and its role in genetic dissection of Drosophila phototransduction. J. Neurogenet. 2012;26 doi: 10.3109/01677063.2012.666299. [DOI] [PubMed] [Google Scholar]
- 34.Lee S.J., Montell C. Suppression of constant-light-induced blindness but not retinal degeneration by inhibition of the rhodopsin degradation pathway. Curr. Biol. 2004;14 doi: 10.1016/j.cub.2004.11.054. [DOI] [PubMed] [Google Scholar]
- 35.Kiselev A., et al. A molecular pathway for light-dependent photoreceptor apoptosis in Drosophila. Neuron. 2000;28 doi: 10.1016/s0896-6273(00)00092-1. [DOI] [PubMed] [Google Scholar]
- 36.Alloway P.G., Howard L., Dolph P.J. The formation of stable rhodopsin-arrestin complexes induces apoptosis and photoreceptor cell degeneration. Neuron. 2000;28 doi: 10.1016/s0896-6273(00)00091-x. [DOI] [PubMed] [Google Scholar]
- 37.Satoh A.K., Ready D.F. Arrestin1 mediates light-dependent rhodopsin endocytosis and cell survival. Curr. Biol. 2005;15 doi: 10.1016/j.cub.2005.08.064. [DOI] [PubMed] [Google Scholar]
- 38.Wang T., Jiao Y., Montell C. Dissecting independent channel and scaffolding roles of the Drosophila transient receptor potential channel. JCB (J. Cell Biol.) 2005;171 doi: 10.1083/jcb.200508030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Handa J.T., Cano M., Wang L., Datta S., Liu T. Lipids, oxidized lipids, oxidation-specific epitopes, and Age-related Macular Degeneration. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2017;1862 doi: 10.1016/j.bbalip.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kasahara E., Lin L.R., Ho Y.S., Reddy V.N. SOD2 protects against oxidation-induced apoptosis in mouse retinal pigment epithelium: implications for age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2005;46 doi: 10.1167/iovs.05-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stegeman R., Hall H., Escobedo S.E., Chang H.C., Weake V.M. Proper splicing contributes to visual function in the aging Drosophila eye. Aging Cell. 2018;17 doi: 10.1111/acel.12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hall H., et al. Transcriptome profiling of aging Drosophila photoreceptors reveals gene expression trends that correlate with visual senescence. BMC Genom. 2017;18 doi: 10.1186/s12864-017-4304-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vajrychova M., et al. Quantification of cellular protein and redox imbalance using SILAC-iodoTMT methodology. Redox Biol. 2019;24 doi: 10.1016/j.redox.2019.101227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Topf U., et al. Quantitative proteomics identifies redox switches for global translation modulation by mitochondrially produced reactive oxygen species. Nat. Commun. 2018;9 doi: 10.1038/s41467-017-02694-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao H., et al. A quantitative tissue-specific landscape of protein redox regulation during aging. Cell. 2020;180 doi: 10.1016/j.cell.2020.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo J., et al. Resin-Assisted enrichment of thiols as a general strategy for proteomic profiling of cysteine-based reversible modifications. Nat. Protoc. 2014;9 doi: 10.1038/nprot.2013.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duan J., et al. Stochiometric quantification of the thiol redox proteome of macrophages reveals subcellular compartmentalization and susceptibility to oxidative perturbations. Redox Biol. 2020;36 doi: 10.1016/j.redox.2020.101649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weerapana E., et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468 doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shakir S., Vinh J., Chiappetta G. Quantitative analysis of the cysteine redoxome by iodoacetyl tandem mass tags. Anal. Bioanal. Chem. 2017;409 doi: 10.1007/s00216-017-0326-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Araki K., et al. Redox sensitivities of global cellular cysteine residues under reductive and oxidative stress. J. Proteome Res. 2016;15 doi: 10.1021/acs.jproteome.6b00087. [DOI] [PubMed] [Google Scholar]
- 51.Leichert L.I., et al. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc. Natl. Acad. Sci. U. S. A. 2008;105 doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.García-Santamarina S., et al. Monitoring in vivo reversible cysteine oxidation in proteins using ICAT and mass spectrometry. Nat. Protoc. 2014;9 doi: 10.1038/nprot.2014.065. [DOI] [PubMed] [Google Scholar]
- 53.Van Der Reest J., Lilla S., Zheng L., Zanivan S., Gottlieb E. Proteome-wide analysis of cysteine oxidation reveals metabolic sensitivity to redox stress. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-04003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Casas-Vila N., et al. The developmental proteome of Drosophila melanogaster. Genome Res. 2017;27 doi: 10.1101/gr.213694.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuznetsova K.G., et al. Brain proteome of Drosophila melanogaster is enriched with nuclear proteins. Biochemistry (Moscow) 2019;84 doi: 10.1134/S0006297919010097. [DOI] [PubMed] [Google Scholar]
- 56.Menger K.E., et al. Fasting, but not aging, dramatically alters the redox status of cysteine residues on proteins in Drosophila melanogaster. Cell Rep. 2015;11 doi: 10.1016/j.celrep.2015.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petrova B., et al. Dynamic redox balance directs the oocyte-to-embryo transition via developmentally controlled reactive cysteine changes. Proc. Natl. Acad. Sci. U. S. A. 2018;115 doi: 10.1073/pnas.1807918115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao Y., et al. Comparative proteomics analysis of dietary restriction in Drosophila. PLoS One. 2020;15 doi: 10.1371/journal.pone.0240596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fabre B., et al. Analysis of Drosophila melanogaster proteome dynamics during embryonic development by a combination of label-free proteomics approaches. Proteomics. 2016;16 doi: 10.1002/pmic.201500482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qu Z., et al. Proteomic quantification and site-mapping of S -nitrosylated proteins using isobaric iodoTMT reagents. J. Proteome Res. 2014;13 doi: 10.1021/pr401179v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Go Y.M., Chandler J.D., Jones D.P. The cysteine proteome. Free Radic. Biol. Med. 2015;84 doi: 10.1016/j.freeradbiomed.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miseta A., Csutora P. Relationship between the occurrence of cysteine in proteins and the complexity of organisms. Mol. Biol. Evol. 2000;17 doi: 10.1093/oxfordjournals.molbev.a026406. [DOI] [PubMed] [Google Scholar]
- 63.Toh K., et al. Chemoproteomic identification of blue-light-damaged proteins. J. Am. Chem. Soc. 2022 doi: 10.1021/jacs.2c07180. [DOI] [PubMed] [Google Scholar]
- 64.Pimkova K., et al. Quantitative analysis of redox proteome reveals oxidation-sensitive protein thiols acting in fundamental processes of developmental hematopoiesis. Redox Biol. 2022;53 doi: 10.1016/j.redox.2022.102343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fu L., et al. Systematic and quantitative assessment of hydrogen peroxide reactivity with cysteines across human proteomes. Mol. Cell. Proteomics. 2017;16 doi: 10.1074/mcp.RA117.000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwämmle V., Hagensen C.E., Rogowska-Wrzesinska A., Jensen O.N. PolySTest: robust statistical testing of proteomics data with missing values improves detection of biologically relevant features. Mol. Cell. Proteomics. 2020;19 doi: 10.1074/mcp.RA119.001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hall H., et al. Quantitative proteomic and metabolomic profiling reveals altered mitochondrial metabolism and folate biosynthesis pathways in the aging drosophila eye. Mol. Cell. Proteomics. 2021;20 doi: 10.1016/j.mcpro.2021.100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Montllor-Albalate C., et al. Sod1 integrates oxygen availability to redox regulate NADPH production and the thiol redoxome. Proc. Natl. Acad. Sci. U. S. A. 2022;119 doi: 10.1073/pnas.2023328119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baez S., Segura-Aguilar J., Widersten M., Johansson A.S., Mannervik B. Glutathione transferases catalyse the detoxication of oxidized metabolites (o-quinones) of catecholamines and may serve as an antioxidant system preventing degenerative cellular processes. Biochem. J. 1997;324 doi: 10.1042/bj3240025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nandi A., Yan L.J., Jana C.K., Das N. Role of catalase in oxidative stress- and age-associated degenerative diseases. Oxid. Med. Cell. Longev. 2019;2019 doi: 10.1155/2019/9613090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mueller S., Riedel H.D., Stremmel W. Direct evidence for catalase as the predominant H2O2-removing enzyme in human erythrocytes. Blood. 1997;90 [PubMed] [Google Scholar]
- 72.Chen X., Leon-Salas W.D., Zigon T., Ready D.F., Weake V.M. A programmable optical stimulator for the Drosophila eye. HardwareX. 2017;2 doi: 10.1016/j.ohx.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang L., et al. Regulation of cofilin phosphorylation and asymmetry in collective cell migration during morphogenesis. Development. 2011;138 doi: 10.1242/dev.046870. [DOI] [PubMed] [Google Scholar]
- 74.Hill K.K., Bedian V., Juang J.L., Hoffmann F.M. Genetic interactions between the Drosophila abelson (Abl) tyrosine kinase and failed axon connections (Fax), a novel protein in axon bundles. Genetics. 1995;141 doi: 10.1093/genetics/141.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Parkhitko A.A., et al. Tissue-specific down-regulation of S-adenosyl-homocysteine via suppression of dAhcyL1/dAhcyL2 extends health span and life span in Drosophila. Genes Dev. 2016;30 doi: 10.1101/gad.282277.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Turner M.A., et al. Structure determination of selenomethionyl S-adenosylhomocysteine hydrolase using data at a single wavelength. Nat. Struct. Biol. 1998;5 doi: 10.1038/nsb0598-369. [DOI] [PubMed] [Google Scholar]
- 77.Matthews R.P., et al. TNFα-dependent hepatic steatosis and liver degeneration caused by mutation of zebrafish s-adenosylhomocysteine hydrolase. Development. 2009;136 doi: 10.1242/dev.027565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vizán P., Di Croce L., Aranda S. Functional and pathological roles of AHCY. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.654344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yuan C.S., Ault-Riché D.B., Borchardt R.T. Chemical modification and site-directed mutagenesis of cysteine residues in human placental S-adenosylhomocysteine hydrolase. J. Biol. Chem. 1996;271 doi: 10.1074/jbc.271.45.28009. [DOI] [PubMed] [Google Scholar]
- 80.Wright D.E., Panaseiko N., O'Donoghue P. Acetylated thioredoxin reductase 1 resists oxidative inactivation. Front. Chem. 2021;9 doi: 10.3389/fchem.2021.747236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Smith D.P., et al. Photoreceptor deactivation and retinal degeneration mediated by a photoreceptor-specific protein kinase C. Science. 1991:254. doi: 10.1126/science.1962207. 1979. [DOI] [PubMed] [Google Scholar]
- 82.Huber A., Sander P., Bähner M., Paulsen R. The TRP Ca2+ channel assembled in a signaling complex by the PDZ domain protein INAD is phosphorylated through the interaction with protein kinase C (ePKC) FEBS Lett. 1998;425 doi: 10.1016/s0014-5793(98)00248-8. [DOI] [PubMed] [Google Scholar]
- 83.Minke B., Pak W.L. The light-activated TRP channel: the founding member of the TRP channel superfamily. J. Neurogenet. 2022;36:55–64. doi: 10.1080/01677063.2022.2121824. [DOI] [PubMed] [Google Scholar]
- 84.Garner K., et al. Phosphatidylinositol transfer protein, cytoplasmic 1 (PITPNC1) binds and transfers phosphatidic acid. J. Biol. Chem. 2012;287 doi: 10.1074/jbc.M112.375840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rubinstein C.T., Bar-Nachum S., Selinger Z., Minke B. Light-induced retinal degeneration in rdgB (retinal degeneration B) mutant of Drosophila: electrophysiological and morphological manifestations of degeneration. Vis. Neurosci. 1989;2 doi: 10.1017/s0952523800003473. [DOI] [PubMed] [Google Scholar]
- 86.Pinazo-Durán M.D., et al. Oxidative stress and its downstream signaling in aging eyes. Clin. Interv. Aging. 2014;9 doi: 10.2147/CIA.S52662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Masai I., Okazaki A., Hosoya T., Hotta Y. Drosophila retinal degeneration A gene encodes an eye-specific diacylglycerol kinase with cysteine-rich zinc-finger motifs and ankyrin repeats. Proc. Natl. Acad. Sci. U. S. A. 1993;90 doi: 10.1073/pnas.90.23.11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Raghu P., et al. Constitutive activity of the light-sensitive channels TRP and TRPL in the Drosophila diacylglycerol kinase mutant, rdgA. Neuron. 2000;26 doi: 10.1016/s0896-6273(00)81147-2. [DOI] [PubMed] [Google Scholar]
- 89.Dolph P.J., et al. An eye-specific Gβ subunit essential for termination of the phototransduction cascade. Nature. 1994;370 doi: 10.1038/370059a0. [DOI] [PubMed] [Google Scholar]
- 90.Lefkowitz R.J., Shenoy S.K. Transduction of receptor signals by β-arrestins. Science. 2005:308. doi: 10.1126/science.1109237. 1979. [DOI] [PubMed] [Google Scholar]
- 91.Dolph P.J., et al. Arrestin function in inactivation of G protein-coupled receptor rhodopsin in vivo. Science. 1993:260. doi: 10.1126/science.8316831. 1979. [DOI] [PubMed] [Google Scholar]
- 92.Krantz D.E., Zipursky S.L. Drosophila chaoptin, a member of the leucine-rich repeat family, is a photoreceptor cell-specific adhesion molecule. EMBO J. 1990;9 doi: 10.1002/j.1460-2075.1990.tb08325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gurudev N., Yuan M., Knust E. chaoptin, prominin, eyes shut and crumbs form a genetic network controlling the apical compartment of Drosophila photoreceptor cells. Biol Open. 2014;3 doi: 10.1242/bio.20147310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Marygold S.J., et al. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 2007;8 doi: 10.1186/gb-2007-8-10-r216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Alonso J., Santarén J.F. Characterization of the Drosophila melanogaster ribosomal proteome. J. Proteome Res. 2006;5 doi: 10.1021/pr0601483. [DOI] [PubMed] [Google Scholar]
- 96.Voolstra O., et al. NinaB is essential for Drosophila vision but induces retinal degeneration in opsin-deficient photoreceptors. J. Biol. Chem. 2010;285 doi: 10.1074/jbc.M109.056101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Parkhitko A.A., Jouandin P., Mohr S.E., Perrimon N. Methionine metabolism and methyltransferases in the regulation of aging and lifespan extension across species. Aging Cell. 2019;18 doi: 10.1111/acel.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu M., Barnes V.L., Pile L.A. Disruption of methionine metabolism in Drosophila melanogaster impacts histone methylation and results in loss of viability. G3: Genes, Genomes, Genetics. 2016;6 doi: 10.1534/g3.115.024273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Parkhitko A.A., et al. A genetic model of methionine restriction extends Drosophila health- and lifespan. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2110387118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dai Z., Mentch S.J., Gao X., Nichenametla S.N., Locasale J.W. Methionine metabolism influences genomic architecture and gene expression through H3K4me3 peak width. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-04426-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kitada M., Ogura Y., Monno I., Xu J., Koya D. Effect of methionine restriction on aging: its relationship to oxidative stress. Biomedicines. 2021;9 doi: 10.3390/biomedicines9020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Deguchi T., Barchas J. Inhibition of transmethylations of biogenic amines by S-adenosylhomocysteine. Enhancement of transmethylation by adenosylhomocysteinase. J. Biol. Chem. 1971;246 [PubMed] [Google Scholar]
- 103.B. Minks, “Light-induced Reduction in Excitation Efficiency in the Trp Mutant of Drosophila.”. [DOI] [PMC free article] [PubMed]
- 104.Hardie R.C., et al. Protein kinase C is required for light adaptation in Drosophila photoreceptors. Nature. 1993;363 doi: 10.1038/363634a0. [DOI] [PubMed] [Google Scholar]
- 105.Gu Y., Oberwinkler J., Postma M., Hardie R.C. Mechanisms of light adaptation in Drosophila photoreceptors. Curr. Biol. 2005;15 doi: 10.1016/j.cub.2005.05.058. [DOI] [PubMed] [Google Scholar]
- 106.Ueda A., et al. Two novel forms of ERG oscillation in Drosophila: age and activity dependence. J. Neurogenet. 2018;32 doi: 10.1080/01677063.2018.1461866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gutorov R., Katz B., Minke B. Electrophysiological methods for measuring photopigment levels in Drosophila photoreceptors. JoVE. 2022 doi: 10.3791/63514. [DOI] [PubMed] [Google Scholar]
- 108.Perez-Riverol Y., et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47 doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Exposure to H2O2induces oxidative stress in Drosophila S2 cells.(A) Line graph displaying cell proliferation after H2O2 treatment. (B) Line graph displaying cell viability after H2O2 treatment. (C) Bar graphs displaying relative ratio of intracellular NADH:NAD+, NADPH:NADP+, and GSH:GSSG after treatment with 20 mM H2O2 for the indicated time relative to untreated S2 cells. All graphs depict mean standard deviation (S.D) of three biological replicates. p-value (* < 0.05, ** < 0.005, *** <0.001) Student's T-test.
Data Availability Statement
The mass spectrometry proteomic data for Drosophila melanogaster S2 cells and w1118 flies exposed to blue light have been deposited to the ProteomeXchange Consortium via the PRIDE [108] partner repository with the dataset identifier PXD034050 and 10.6019/PXD034050. Additionally, proteomic data for aging Drosophila melanogaster eyes have been deposited to the ProteomeXchange Consortium via the PRIDE [108] partner repository with the dataset identifier PXD036110 and 10.6019/ PXD036110.